Abstract
Photoreceptor nuclei in the Drosophila eye undergo developmentally regulated migrations. Nuclear migration is known to require the perinuclear protein Klarsicht, but the function of Klarsicht has been obscure. Here, we show that Klarsicht is required for connecting the microtubule organizing center (MTOC) to the nucleus. In addition, in a genetic screen for klarsicht-interacting genes, we identified Lam Dm0, which encodes nuclear lamin. We find that, like Klarsicht, lamin is required for photoreceptor nuclear migration and for nuclear attachment to the MTOC. Moreover, perinuclear localization of Klarsicht requires lamin. We propose that nuclear migration requires linkage of the MTOC to the nucleus through an interaction between microtubules, Klarsicht, and lamin. The Klarsicht/lamin interaction provides a framework for understanding the mechanistic basis of human laminopathies.
INTRODUCTION
The position of the cell nucleus is critical to many developmental processes. For example, in Drosophila, both the A/P and D/V axes of symmetry are defined by the position of the oocyte nucleus. First, signaling by the nucleus at the oocyte posterior defines the A/P axis. Subsequently, the oocyte nucleus migrates anteriorly to a random side of the oocyte and signals there to define the D/V axis of symmetry (van Eeden and St. Johnston, 1999). Later in Drosophila development, migration of syncitial nuclei to the cell cortex is a defining event in embryogenesis, which results in syncitial blastoderm formation, a phenomenon preceding cellularization (Foe et al., 1993). In Caenorhabditis elegans, nuclear migrations in P-cells, which give rise to neurons and the vulva, are important for cell viability (Malone et al., 1999).
Nuclear migration in the developing Drosophila eye is critical for shaping each individual cell and thus for normal morphology of the entire compound eye (Fischer-Vize and Mosley, 1994). The Drosophila compound eye develops within the larval eye imaginal disc, an epithelial monolayer (Figure 1; Wolff and Ready, 1993). Within the eye disc, the morphogenetic furrow marks the initiation of eye assembly. Rows of identical facets, or ommatidia, assemble posterior to the furrow, starting with the eight photoreceptors (R-cells), followed by the lens-secreting cone cells, and finally the pigment cells. Anterior to the furrow, cells are undifferentiated and their nuclei are positioned randomly within the monolayer. The nuclei dive basally within the furrow and posterior to the furrow, migrate apically as they are recruited into ommatidia (Tomlinson and Ready, 1987).
Figure 1.
Nuclear migration in the Drosophila eye disc. A longitudinal section of a wild-type larval eye disc is diagrammed with anterior right (A) and posterior left (P). Most of the cell volume surrounds the nucleus. Nuclei of undifferentiated cells are randomly positioned in the monolayer anterior to the morphogenetic furrow (mf), which is moving anteriorly, in the direction of the arrow. In the furrow, the nuclei are basal, and posterior to the furrow, nuclei rise as cells are recruited into ommatidia. Ommatidial clusters are progressively more mature from anterior to posterior. R-cells are gray, and cone cells are white. (Adapted from Tomlinson and Ready, 1986.)
Two Drosophila genes, klarsicht (previously known as marbles) and Glued, are essential for the apical migration of nuclei in differentiating R-cells (Fischer-Vize and Mosley, 1994; Fan and Ready, 1997). Glued encodes the large subunit of dynactin, a protein complex that regulates the minus-end-directed microtubule motor dynein (Holzbauer et al., 1991). The requirement for dynactin suggests that R-cell nuclear migration is a dynein- and microtubule-dependent process. Consistent with this idea, two other Drosophila genes, Bicaudal-D and D-Lis1, both of which may regulate dynein (reviewed in Reiner, 2000; Wynshaw-Boris and Gambello, 2001; Vallee et al., 2001), are implicated in R-cell nuclear migration (Swan et al., 1999), although their mutant phenotypes are weak compared with klarsicht and Glued (C.R. and J.A.F., unpublished observations). DLis-1, a WD40 repeat protein, is the homolog of the human disease gene Lissencephaly-1 (Reiner et al., 1993). Lissencephaly, or smooth brain, is a disorder resulting from defects in neuronal migrations essential for normal human brain development (reviewed in Morris et al., 1998; Morris, 2000; Wynshaw-Borris and Gambello, 2001). Neuronal migration requires nuclear migration, and the involvement of DLis-1 in Drosophila R-cell nuclear migration suggests that the two processes may be in part analogous.
klarsicht (klar) encodes a large protein, unique except for its small N-terminal KASH (Klarsicht, Anc-1, Syne-1 homology) domain, which localizes proteins to the nuclear membrane (Mosley-Bishop et al., 1999; Apel et al., 2000; Zhang et al., 2001; Starr and Han, 2002; Zhen et al., 2002; J.A.F., C.C., S.A., C.R. et al., unpublished results). The KASH-domain-containing protein Anc-1 and its vertebrate homolog, Syne-1 (also known as Myne-1, Nesprin, and NUANCE) are dystrophin-related proteins that anchor the nucleus to the actin cytoskeleton (Apel et al., 2000; Mislow et al., 2001, 2002; Zhang et al., 2001; Starr and Han, 2002; Zhen et al., 2002; Starr and Han, 2003). In addition to its role in nuclear migration in the eye, klar is required for the developmentally regulated migrations of lipid droplets during embryogenesis. In this role, it has been proposed that Klar regulates dynein and also the plus-end-directed motor kinesin (Welte et al., 1998; Gross et al., 2000).
Here, we use genetics and immunohistochemistry to investigate the role of Klar in R-cell nuclear migration. The results suggest that a connection between the MTOC and the nucleus is necessary for nuclear migration and that this connection is mediated by Klar and nuclear lamin. In addition to suggesting a specific role for Klar in nuclear migration, the results propose a general mechanistic explanation for the cytoplasmic effects of nuclear lamin, including human laminopathies.
MATERIALS AND METHODS
Drosophila Strains and Genetics
The following mutant lines were used in this work:
klarmBP and klarmBX3 are strong klar mutant alleles described in Fischer-Vize and Mosley (1994) and Mosley-Bishop et al. (1999).
Lam4643 (Spradling et al., 1999) was obtained from the Bloomington Stock Center.
LamP (Lenz-Bohme et al., 1997) was obtained from B. Schmitt.
Df(3L)emcE12 (61A-61D3), described in Lindsley and Zimm (1992) and Mosley-Bishop et al. (1999), was obtained from the Bloomington Drosophila Stock Center.
Df(2L)cl-h4 (25D6-25F3/4), described in Kotarski et al. (1983) and Lindsley and Zimm (1992), was obtained from the Umea Drosophila Stock Center.
Tw2-LamP, the Lam+ genomic rescue line described in Lenz-Bohme et al. (1997), was obtained from B. Schmitt.
P{w+, UAS-nod-lacZ}B3.3, a third chromosome insertion described in Giniger et al. (1993) and Clark et al. (1997), was obtained from I. Clark.
P{w+, elav-Gal4}H19 was hopped to chromosome 3. The original line was obtained from B. Zhang.
P{w+, glrs-klar} on X is described in Mosley-Bishop et al. (1999).
P{ry+, neoFRT}40A (Xu and Rubin, 1993) was obtained from the Bloomington Stock Center (Abbreviated FRT40A).
P{w+, GMR-hid}G1, l(2)CL-L1, P{ry+, neoFRT}40A/CyO; P{w+, ey-Gal4.H}SS5, P{w+ UAS-FLP1.D}JD2 (Stowers and Schwarz, 1999) was obtained from the Bloomington Drosophila Stock Center. (Abbreviated GMR-hid, CL2, FRT40A/CyO; EGUF.)
P{w+, UASp-6Xmyc-klar}NP5, a third chromosome insertion, was newly generated here. An AscI fragment containing 6Xmyc-klar (from pGLRS-6Xmyc-klar1; Mosley-Bishop et al., 1999) was ligated into a derivative of pUASp (Rorth, 1998) in which the NotI site was changed to AscI.
P element transformation of w1118 flies was as described (Fischer-Vize et al., 1992a). A CyO,GFP balancer (Flybase, 2003) was used to identify Lam homozygous mutant larvae, and a TM6B, Tb (Lindsley and Zimm, 1992) balancer was used to identify klar homozygous mutant larvae. Common marker strains used are described in Lindsley and Zimm (1992). All experimental flies were grown at 25°C.
Lam4643 homozygous eyes were generated in Lam4643/+ heterozygotes using the GMR-hid technique (Stowers and Schwarz, 1999). First, two stocks were generated: Lam4643 FRT40A/CyO,GFP and GMR-hid, CL, FRT40A/CyO, GFP; EGUF. Flies from the two stocks were intercrossed, and among the progeny, Lam4643 FRT40A/GMR-hid CL FRT40A; EGUF/+ larvae were detected by the absence of GFP expression. The eye discs that of these larvae are homozygous for Lam4643.
Modifier Screen
Males (pr; st) were mutagenized with EMS (Lewis and Bacher, 1968) or X-rays (4000 rads) and mated with w; P{w+; glrs-klar} females. The F1 males (14,500 with EMS-mutagenized autosomes and 35,000 with X-ray-mutagenized autosomes) were screened for enhanced roughness of the external eyes. The EMS screen yielded two egk1 alleles (Ari3 and Ari7), and the x-ray screen yielded seven egk1 alleles: A25, K2, 83, and four stronger alleles that were lethal in trans to Df(2L)cl-h4. (The lethal egk1 alleles have been lost.)
Genetic Mapping of egk1 Alleles
The egk1 alleles Ari3 and Ari7 were mapped meiotically between dp and b using the multiply marked chromosome aldp b pr Bl c px sp and by scoring the mutant eye phenotype of the egk1 homozygotes. Females that were al dp b pr Bl c px sp/egk1 were crossed with aldp b pr c px sp males, and ∼20 male progeny of each single recombinant class were collected. To determine which of the recombinant chromosomes contained egk1, each male was individually crossed to egk1/CyO females, and progeny were examined for the egk1 homozygous eye phenotype. Ari3 and Ari7 were subsequently tested for complementation with a variety of deficiency chromosomes spanning the region between dp and b, and Df(2L)cl-h4 (25D6-25F3/4) failed to complement egk1.
Molecular Analysis of Lam Alleles
Lam alleles were amplified by PCR using total genomic DNA prepared from a single fly homozygous or hemizygous (in trans to Df(2L)cl-h4) for each of the five viable or semiviable egk1 alleles. Genomic DNA was prepared as described in Chen and Fischer (2000). Six primer pairs (sequences available upon request) and standard PCR conditions were used. The DNA sequence of the PCR products were sequenced directly using automated fluorometric sequencing. Sequences were analyzed with MacVector (Accelrys, San Diego, CA) software.
Phenotypic Analysis of Eyes
Scanning electron micrographs (Huang et al., 1995) and plastic sections of adult eyes (Fischer-Vize and Mosley, 1994) were prepared as described previously. Light microscope analysis of eye discs immunostained with anti-Elav was performed exactly as described in Fischer-Vize and Mosley (1994). Light microscope images were produced with a Zeiss Axioplan microscope and a Zeiss AxioCam (Thornwood, NY), and processed with Adobe Photoshop software (Adobe, San Jose, CA). For confocal microscopy, immunostaining of eye discs was performed with PEMS fixation and PBST washes as described (Fischer-Vize et al., 1992a, 1992b). Eye discs were mounted in VectaShield (Vector Laboratories, Burlingame, CA). The primary antibodies used were as follows: mouse anti-Lam at 1:100 (mAbADL84; Stuurman et al., 1995), obtained from P. Fisher; mouse anti-Futsch at 1:5000 (mAb22C10; Fujita et al., 1982), obtained from the Developmental Studies Hybridoma Bank (DSHB, Iowa); rabbit anti-Myc at 1:500 (Santa Cruz Biochemicals, Santa Cruz, CA; sc-789 [c-myc/A-14]); rat anti-Elav at 90:100 (7E8A10, DSHB); mouse anti-βgal at 1:10 (40-1a, DSHB); rabbit anti-γ-tubulin (Rb 1015) at 1:40 (Tavosanis et al., 1997), obtained from C. Gonzalez. Secondary antibodies were from Molecular Probes (Eugene, OR; Alexa) and Jackson ImmunoResearch (West Grove, PA; Cy) and were used at 1:600. When double-labeling with rat and mouse primary antibodies, preadsorbed Jackson secondary antibodies were used. Green:Alexa488-goat anti-rabbit, Alexa488-goat anti-rat. Blue: Cy5-goat anti-rat, Alexa633-goat anti-rat. Red: Cy3-goat anti-mouse, Alexa568-goat anti-rabbit, or Alexa568-phalloidin.
Images were produced with a Leica TCS SP2 confocal microscope (Deerfield, IL) and processed with Adobe Photoshop software.
RESULTS
Klar Is Perinuclear and also Is Associated with Apical Microtubules
It was shown previously using light microscopy that Klar is associated with the nuclear membrane (Mosley-Bishop et al., 1999). Here, we investigate the subcellular localization of Klar in greater detail and at higher resolution using confocal microscopy. To visualize Klar protein, an epitope-tagged form of Klar, 6Xmyc-Klar, was expressed in R-cells by using a UAS-6Xmyc-klar transgene and an elav-Gal4 driver (elav>6Xmyc-klar). (The 6Xmyc-Klar protein is functional; Mosley-Bishop et al., 1999.) Otherwise wild-type eye discs expressing elav>6Xmyc-klar were labeled with anti-Myc and also with anti-Elav (Robinow and White, 1991), which marks R-cell nuclei after they have risen apically. As observed before, Klar is associated with the nuclear membrane (Figure 2D). In addition, dots of Klar are seen to extend from the nuclei toward the apical cell surface (Figure 2, A-C). The apical dots resemble the apical expression pattern of Futsch, also known as 22C10 (Figure 2, I-L), a neural-specific microtubule-associated protein (Hummel et al., 2000).
Figure 2.
Klar localization in eye discs. Shown are confocal images of eye discs double-labeled to reveal R-cell nuclei (anti-Elav; blue) and either 6Xmyc-Klar (anti-myc; green) or Futsch (mAb22C10; red), a microtubule-associated protein. (A, E, and I) Z-sections; (B-D, F-H, and J-L) progressively more basal XY-sections, whose positions correspond to the arrows in A, E, and I, respectively. (A-D) Wild-type eye discs expressing 6Xmyc-Klar in R-cells (elav>6Xmyc-klar). Klar is localized to microtubules apical to the nuclei (A-C) and is also perinuclear (D). (E-H) Lam83/Df(3L)cl-h4 eye discs expressing 6Xmyc-Klar. Klar is localized to apical microtubules (E and F), but is not present in perinuclear rings in the apical R-cell nuclei (G) or the basal ones (H). (I-L) Wild-type eye discs revealing Futsch protein localization on microtubules is shown. Futsch is apical to the R-cell nuclei (I-K) and extends around the nuclei (I and L), to the basal surface of the disc (I). Size bar in L is ∼10 μm and applies to all panels.
There is a variety of experimental evidence that the KASH domains of Anc-1 and Syne-1 localizes those proteins to the nuclear envelope (Mislow et al., 2001, 2002; Starr and Han, 2002; Zhang et al., 2001; Zhen et al., 2002). Similarly, we find that Klar is genuinely associated with the nuclear membrane, rather than appearing perinuclear only because it is associated with microtubules that extend around the nucleus. First, immunostaining with anti-Futsch reveals the microtubule cytoskeleton as it extends from the apical to basal cell surfaces, weaving around the nucleus (Figure 2I). Although Futsch is bound to microtubules, it does not appear perinuclear as does Klar (Figure 2L). Second, 6Xmyc-Klar colocalizes with the nuclear envelope protein lamin (Figure 3). Finally, the two aspects of Klar localization are separable: when an isolated Klar KASH domain is expressed, only nuclear membrane localization, not apical microtubule localization, is observed (J.A.F., C.C., S.A., C.R. et al., unpublished data). We conclude that 6Xmyc-Klar localizes to the apical microtubules and to the nuclear envelope in R-cells.
Figure 3.
Klar colocalizes with nuclear lamin. Shown are confocal images of a single developing ommatidium from otherwise wild-type eye discs that express 6Xmyc-Klar in R-cells (elav >6Xmyc-klar). The eye discs were double-labeled with anti-Myc and anti-Lam. Size bar, ∼2 μm.
The MTOC Detaches from R-cell Nuclei in klar Mutants
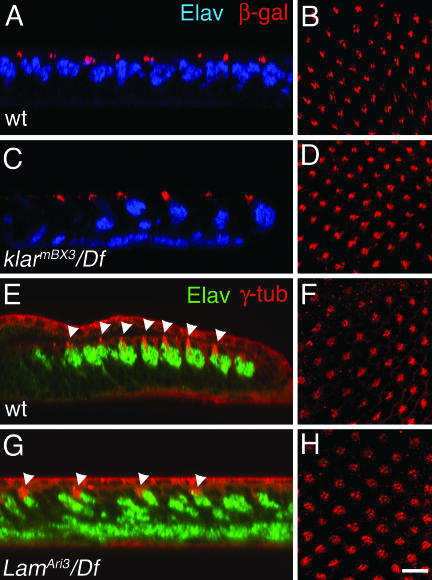
To probe the function of Klar in nuclear migration, we asked whether the cytoskeleton is organized differently in klar mutants than in wild-type eye discs. The MTOC was marked by expressing a Nod-βgal fusion protein, which accumulates at microtubule minus ends (Giniger et al., 1993; Clark et al., 1997). The MTOC is the point in the cell from which the microtubules grow: the slow-growing minus ends gather at the MTOC and the rapidly growing plus ends emanate from it. Nod-βgal was expressed using an elav-Gal4 driver and a UAS-nod-lacZ transgene. Otherwise wild-type and also klar mutant eye discs expressing elav>nod-lacZ were double-labeled with anti-Elav and anti-βgal. In wild-type, Nod-βgal is closely associated with the R-cell nuclei, and just apical to them (Figure 4A).
Figure 4.
Position of the MTOC and R-cell nuclei in eye discs. Confocal images of eye discs labeled to reveal R-cell nuclei (anti-Elav; blue or green) and the MTOC (anti-β-gal or anti-γ-tub; red) are shown. (A, C, E, and G) Z-sections; (B, D, F, and H) corresponding apical XY-sections, respectively. (A and B) Wild-type eye discs expressing Nod-β-gal in R-cells (elav>nod-lacZ). (C and D) klarBX3/Df(3L)emcE12 discs expressing Nod-β-gal in R-cells. (E and F) Wild-type eye discs. (G and H) LamAri3/Df(3L)cl-h4 eye discs. The arrowheads in E and G indicate the MTOC. (Anti-γ-tub has some background membrane staining.) Size bar in H is ∼10 μm and applies to all panels.
In klar mutants, most of the R-cell nuclei are basal, but all of the Nod-βgal is apical (Figure 4C). There are two possible interpretations of this result: 1) The klar mutant R-cells with basal nuclei (most of the cells) do not form an MTOC, or 2) the MTOC is separated from the abnormally basal nuclei of klar mutant R-cells. If most of the R-cells in klar mutant eye discs do not form an MTOC, then in klar discs we would expect to observe fewer apical dots of Nod-βgal in each ommatidial cluster than in wild-type. By contrast, we observe that the Nod-βgal dots in klar mutant discs appear wild-type in number and pattern (Figure 4, B and D). We conclude that in klar mutant R-cells the MTOC forms normally, but usually separates from the nucleus.
Identification of egk1 as a Modifier of the Overexpressed klar Phenotype
A transgene called glrs-klar overexpresses klar+ in the developing eye, resulting in defects in eye morphology (Mosley-Bishop et al., 1999; Figure 5A). To identify additional genes that function in nuclear migration in the Drosophila eye, we performed a mutagenesis screen for dominant enhancers of the glrs-klar rough eye phenotype (Figure 5B). Nine mutant alleles of a complementation group that we named egk1 (enhancer of glrs-klar) were isolated (Figure 5A). The nine egk1 alleles were divided into three groups based on the severity of their mutant phenotype: 1) four alleles are lethal as homozygotes or in trans to each other, 2) four alleles (Ari3, Ari7, K2, 83) are semiviable as homozygotes, and 3) one allele (A25) is homozygous viable. Initial observation of the egk1 mutants suggested that the egk1 gene has an essential role in eye development; adults homozygous for any of the semiviable or viable alleles have external eye defects (Figure 5A). Meiotic mapping localized egk1 between the markers dp and b on chromosome 2, and subsequent physical mapping localized egk1 to polytene position 25E3-6, the region uncovered by the deletion chromosome Df(2L)cl-h4. In trans to Df(2L)cl-h4, the lethal egk1 alleles are lethal and the semiviable or viable egk1 alleles are semiviable. Below we show that the weak egk1 alleles are loss-of-function mutants and that they display nuclear migration defects. Because egk1 loss-of-function mutants have a similar mutant eye phenotype to klar mutants and egk1 interacts genetically with klar, we conclude that the egk1 gene is likely to function in the klar pathway.
Figure 5.
Identification and characterization of egk1 mutants. (A) Scanning electron micrographs of eyes of the genotypes indicated are shown. gk is glrs-klar. Y is the Y chromosome. The egk1 allele shown is LamAri3. (B) The cross scheme used in the F1 mutagenesis screen for enhancers of the glrs-klar rough eye phenotype is shown. (C) The positions of the nonsense or frameshift mutations in each of the five homozygous viable Lam alleles isolated as egk1 mutations are shown. The allele names are at the top, and the number beneath each indicates the first amino acid affected. (The M residue of the start codon is 1.) NLS is the nuclear localization signal, and CaaX refers to the motif used to localize lamin to the cytoplasm at the inner nuclear membrane. (D) The precise nucleotide and predicted amino acid changes of the five mutant Lam alleles in C.
egk1 Is Lamin Dm0
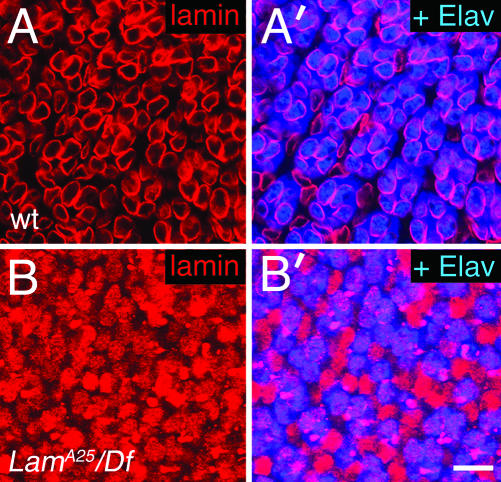
Among the ∼25 genes in 25E3-6 (Flybase, 2003), Lam Dm0 (Lam) was chosen as a candidate for egk1. Lam encodes type B nuclear lamin, an intermediate filament protein that is a major component of the inner nuclear envelope (Lenz-Bohme et al., 1997; Stuurman et al., 1998; Gruenbaum et al., 2000). To determine if egk1 is Lam, we tested several of the egk1 alleles for complementation with two previously identified homozygous lethal Lam mutants, Lam4643 and LamP. Neither Lam mutant complements any of the egk1 alleles tested. In addition, we determined the DNA sequences of the Lam genes in flies homozygous for each of the semiviable or viable egk1 alleles. In each case, a nonsense or frameshift mutation was found within the Lam coding region (Figure 5, C and D). An antibody to Lam (mAbADL84) recognizes no protein in immunostained eye discs carrying any one of the semiviable alleles in trans to Df(2L)cl-h4. This result is consistent with the DNA sequence analysis of the four semiviable Lam alleles, which predicts that severely truncated Lam proteins are the most likely gene products. Even if these truncated proteins are produced and stable, they need not contain the epitope recognized by mAbADL84. The weakest allele, A25, has a frameshift that results in the deletion of the C-terminal CaaX box, which localizes lamin to the inner nuclear membrane (Holtz et al., 1989; Kitten and Nigg, 1991). Consistent with this observation, A25/Df(2L)cl-h4 eye discs immunostained with mAbADL84 reveal that Lam protein does not localize to the membrane, but instead is found throughout the nucleus (Figure 6). Finally, a P element containing Lam+ genomic DNA (Tw2-LamP) rescues the lethality and eye phenotypes of the egk1 alleles. We conclude that egk1 is Lam.
Figure 6.
Lamin localization in eye discs. Confocal images of eye discs double-labeled to reveal R-cell nuclei (anti-Elav; blue) and nuclear lamin (anti-Lam; red) are shown. (A and A′) Wild-type discs. (A) Lamin expression in apical nuclei; (A′) a merge of lamin and Elav. The apical nuclei in A′ that have lamin but no Elav are cone cell nuclei. The pink appearance of the R-cell nuclear lamin in A′ is due to colocalization of lamin and Elav within the nucleus. (B and B′) LamA25/Df(3L)cl-h4 discs. (B) Lamin expression; (B′) a merge of lamin and Elav. The pink is where lamin and Elav overlap. The purely red nuclei are of cone cells. The plane in B and B′ is more basal than in A and A′, in order to detect Lam mutant R-cell nuclei that are not as apical as in wild-type discs. Size bar in B′ is ∼10 μm.
Photoreceptor Nuclear Migration Fails in Lam Loss-of-Function Mutants
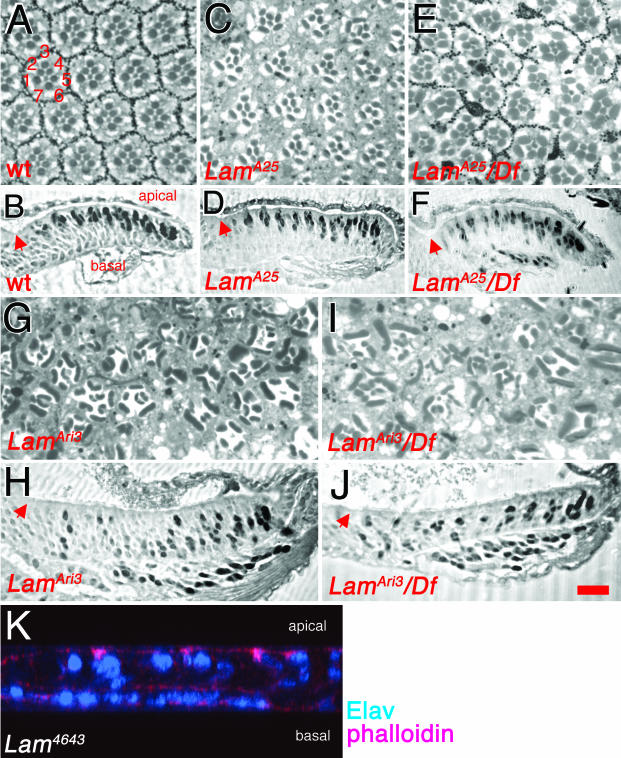
To determine if the eye morphology flaws in Lam mutants are due to nuclear migration defects, anti-Elav was used to label R-cell nuclei in Lam eye discs. All of the semiviable and viable Lam mutants (Figure 5, C and D) were analyzed; LamA25, LamAri3, and LamAri7 homozygotes were assayed, and all five weak Lam alleles were analyzed in trans to Df(2L)cl-h4. With the exception of the weakest allele, LamA25, all homozygotes and hemizygotes showed similar phenotypes; as in klar mutants, R-cell nuclei are present throughout the apical/basal axis of the eye disc, and most of them are basal (Figure 4G and Figure 7, H and J). In LamA25 homozygous discs the R-cell nuclear positions are indistinguishable from wild type (Figure 7, B and D). By contrast, LamA25/Df(2L)cl-h4 discs show nuclear migration defects, but they are less severe than those of the other alleles analyzed (Figure 7F). The difference in severity of the nuclear migration defects in the different alleles is mirrored in their adult eye morphology. Adult retinas with R-cell nuclear migration defects, like those of klar mutants, typically have misshapen rhabdomeres (Fischer-Vize and Mosley, 1994; Mosley-Bishop et al., 1999). Rhabdomeres are light-gathering organelles that project from each photoreceptor cell throughout the apical/basal plane of the eye disc. When R-cell nuclei fail to migrate apically, the cell shapes are aberrant, resulting in oddly shaped or missing rhabdomeres in tangential retinal sections. The retinas of LamA25 homozygotes are nearly wild type (Figure 7, A and C), LamA25 hemizygotes are defective (Figure 7E), and the eyes of LamAri3 homozygotes or hemizygotes have more severe defects (Figure 7, G and H).
Figure 7.
Eye phenotypes of Lam mutants. Light micrographs of apical tangential sections through adult compound eyes are shown in A, C, E, G, and I, and of sections through third instar larval eye discs immunostained with anti-Elav to label R-cell nuclei in B, D, F, H, J, and K. (A and B) Wild-type eyes and discs. The numbers in A refer to the seven R-cells in each ommatidium visible in apical sections. The R-cell nuclei are apical in B. (C and D) LamA25 homozygotes. In C, some ommatidia are defective. The disc in D is indistinguishable from wild-type. (E and F) LamA25/Df(2L)cl-h4 hemizygotes are shown. Severe eye morphology (E) defects and nuclear migration (F) defects are observed. (G and H) LamAri3 homozygotes. The eye morphology (G) and nuclear migration (H) defects are more severe than in LamA25 hemizygotes. (I and J) LamAri3/Df(2L)cl-h4 hemizygotes. The adult eye and disc defects are similar to those of LamAri3 homozygotes. The red arrows in B, D, F, H, and J indicate the morphogenetic furrow. (K) A confocal image of a Lam4643 homozygous eye disc generated by mitotic recombination is shown, labeled with anti-Elav to mark R-cell nuclei, and phalloidin to mark apical and basal cell membranes. Size bar in J is ∼20 μm in all panels except B, D, and F, where it is ∼25 μm.
Several results described above indicate that the five weak Lam mutant alleles are partial loss-of-function mutants, as opposed to gain-of-function mutants: 1) Both lethal and viable Lam alleles were isolated as enhancers of glrs-klar. This result indicates that all classes of Lam alleles have a similar (detrimental) effect on nuclear migration. 2) LamA25 homozygotes have a weaker phenotype than LamA25/Df(2L)cl-h4 hemizygotes. 3) All phenotypes of the weak and strong Lam mutants (lethality and eye phenotypes) are complemented completely by one copy of the transgene Tw2-LamP, which contains a Lam+ gene (see above). Nevertheless, to be certain that the nuclear migration defects represent loss-of-function phenotypes, we observed eye discs homozygous for Lam4346, a strong lethal, loss-of-function mutation caused by P element insertion (Guillemin et al., 2001). We find that Lam4346 eye discs have nuclear migration defects similar to those of LamAri3 homozygotes (Figure 7, H and K) or LamAri3/Df(2L)cl-h4 (compare Figure 7K with Figure 4G). We conclude that like klar+, the Lam+ gene is normally required for R-cell nuclear migration.
Nuclei Disconnect from the MTOC in Lam Mutants
The position of the MTOC was monitored in wild-type and Lam mutant eye discs with antibodies to γ-tubulin, a constituent of a protein complex that binds the MTOC (Joshi, 1993). In wild-type discs, we observe dots of γ-tubulin just apical to the R-cell nuclei. Moreover, the γ-tubulin dots are present only in differentiating cells, whose nuclei are normally apical (Figure 4E). Undifferentiated cells that have not yet been recruited into ommatidial clusters surround the developing facets and their nuclei are basal (Figure 1). No γ-tubulin is observed associated with the basal nuclei or at the apical surface of the undifferentiated cells (Figure 4F); all of the γ-tubulin dots are within the developing clusters (Figure 4F; see also Swan et al.; 1999.) This result suggests that the cytoskeleton becomes organized and an MTOC forms in differentiating cells as they are recruited into the ommatidia.
As in klar mutants, in Lam mutant discs the MTOCs of all of the R-cells are apical, even though most of the R-cell nuclei are basal (Figure 4, G and H). We conclude that like Klar, lamin is required for nuclear migration and to link the MTOC to the nucleus.
Lamin Is Required for Perinuclear Localization of Klar
To determine if lamin and Klar function together, the localization of each protein was monitored in the mutant background of the other. In Lam mutant eye discs that express elav>6Xmyc-klar, Klar localization on microtubules apical to the nucleus appears normal (Figure 2, A-C and E-G). Perinuclear Klar, however, is largely absent in Lam mutants (Figure 2, C, D, G, and H). In contrast, lamin localization appears normal in klar mutant eye discs (our unpublished results). We conclude that localization of Klar to the nuclear envelope requires nuclear lamin.
DISCUSSION
A Model for the Roles of Klar and Lam in R-cell Nuclear Migration
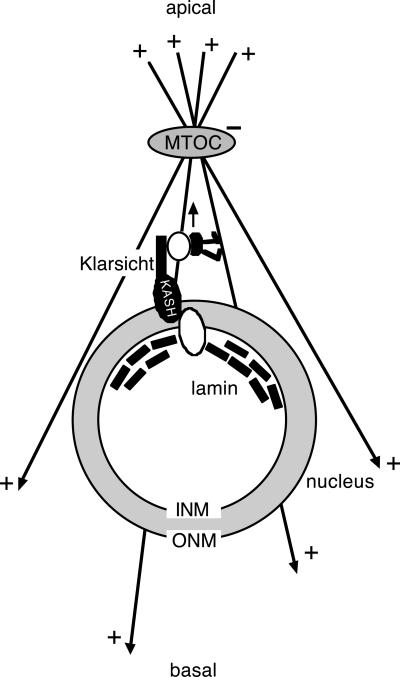
To understand the role of Klar in R-cell nuclear migration, we have investigated Klar subcellular localization and the position of the MTOC in klar mutant eye discs. In addition, we have used genetics to identify another protein, nuclear lamin, that functions in the same pathway with Klar. We find that Klar is perinuclear and also is associated with microtubules apical to the nucleus. In addition, we find that in klar and Lam mutant discs, MTOCs form normally in R-cells, but are often not associated with the nucleus as they are in wild-type eyes. Finally, we find that Lam+ is required for Klar localization to the nuclear membrane. These observations, taken together with previous results, suggest a model for the function of Klar in nuclear migration where Klar, held in the nuclear envelope by nuclear lamin, links the nucleus to the MTOC (Figure 8).
Figure 8.
Model for the roles of Klar and lamin in R-cell nuclear migration. A diagram showing how Klar links the nucleus to the MTOC is shown. INM and ONM are the inner and outer nuclear membranes, respectively. The two unfilled ovals indicate possible intermediate proteins that link the C-terminal KASH domain of Klar to lamin, and also link Klar to dynein. Dynein, in black, is walking in the direction of the arrow. (See text for details.)
The interaction between Klar and lamin may be indirect, but it is likely to be specific, rather than a generalized failure of nuclear envelope assembly in Lam mutants. Although we observe that most R-cell nuclei fail to migrate apically even in weak, viable Lam mutants, >90% of nuclear envelopes are intact even in stronger, lethal Lam mutants (Lenz-Bohme et al., 1997; Guillemin et al., 2001).
We propose (Figure 8) that one or more proteins may form a bridge between the KASH domain of Klar, present in the outer nuclear membrane, and nuclear lamin, in the inner nuclear envelope. The observation that in addition to its perinuclear localization, Klar is cytoplasmic (on apical microtubules) supports the idea that Klar is in the outer, as opposed to the inner, nuclear membrane. Similarly, C. elegans Anc-1 is present in the cytoplasm as well as the nuclear membrane, and a model has been proposed where the Anc-1 KASH domain is held in the outer nuclear membrane by an inner nuclear membrane protein, Unc-84 (Malone et al., 1999; Starr and Han, 2002). Although nuclear lamin has not been shown directly to be required for Anc-1 nuclear membrane localization, nuclear envelope localization of Unc-84 requires lamin (Lee et al., 2002). For Syne-1, the vertebrate homolog of Anc-1, experiments where the detergent digitonin was used to allow antibody access to the outer but not the inner nuclear membrane provide direct evidence that the KASH domain is in the outer nuclear membrane (Zhen et al., 2002). There is, however, some conflicting data (Zhang et al., 2001; Mislow et al., 2001, 2002).
We speculate that the N-terminal portion of Klar is linked to microtubules by dynein. At present, we cannot test for colocalization of Klar and dynein because there are no available reagents that allow detection of dynein or dynactin in the eye disc. Nevertheless, there is much evidence to support an essential role for dynein in R-cell nuclear migration and Klar function. As mentioned above, dynactin, a regulator of dynein, is essential for R-cell nuclear migration in the eye; mutants in the p150 dynactin subunit (Glued) have a phenotype similar to that of klar mutants in the eye disc (Fan and Ready, 1997). In addition, dynein linkage could explain why Klar is localized to microtubules only apical to the nucleus; Klar that escapes the hold of the nuclear envelope, still attached to dynein, would walk along microtubules to the MTOC. Finally, Klar has been implicated as a regulator of dynein in Drosophila embryos (Welte et al., 1998). In addition to its role in R-cell nuclear migration, Klar is required for developmentally regulated migration of lipid storage vesicles during embryogenesis. Lipid droplets at the center of the cellular blastoderm embryo normally migrate cortically during gastrulation. In embryos from klar mutant mothers, the lipid droplets fail to migrate. A variety of data support a model where dynein transports the lipid droplets along microtubules, whose minus ends are at the cell periphery. The results of biophysical experiments led to a model where Klar may attach the appropriate types of motor to lipid droplets, control the number of actively engaged motors on a droplet, or coordinate the activities of kinesins and dyneins bound simultaneously to the same droplet (Jackle and Reinhard, 1998; Welte et al., 1998; Gross et al., 2000). Notably, dynein is required for nuclear attachment to centrosomes (the MTOCs) during mitosis in Drosophila embryos (Robinson et al., 1999). Klar, however, is not essential for this process (Fischer-Vize and Mosley, 1994).
Migration versus Anchoring
The observation that the MTOC is normally apical to the R-cell nuclei, at the leading edge of nuclear movement, suggests that a force pulls on the MTOC from above. We speculate that the mechanism for this force could be analogous to the means by which the nucleus of budding yeast are pulled into the bud neck before cell division. One pathway for migration of the nucleus into the bud neck involves dynein, anchored at the cell cortex to which the nucleus is moving. Cortically tethered dynein “reels in” the nucleus by walking along microtubules whose plus ends are at the cortex, toward the MTOC, which is anchored to the nucleus (reviewed in: Morris et al., 1995; Beckwith et al., 1995; Bloom, 2000, 2001, Segal and Bloom, 2001; Morris, 2003). In support of this idea, microtubule plus-ends are present apically in R-cells (Mosley-Bishop et al., 1999), and as discussed above, dynactin is essential for R-cell nuclear migration (Fan and Ready, 1997).
Whether a force emanating from the apical membrane pulling on the MTOC would drive nuclear migration or serve as an anchor after the nucleus has migrated depends on where the MTOC initially forms. The γ-tubulin antibody detects MTOCs only apically in differentiating cells. Transiently basal MTOCs associated with nuclei that are about to rise could have escaped detection. However, if the MTOC does form apically, then the force that drives nuclear migration would come from below the nucleus, that is, dynein, linked to the nuclear membrane by Klar and lamin, walking on microtubules up toward the MTOC.
The Klar/lamin Interaction, Cytoplasmic Phenomena, and Human Laminopathies
The model we propose whereby Klar forms a bridge between nuclear lamin in the inner nuclear membrane and cytoplasmic microtubules provides a general framework for explaining how nuclear lamin affects cytoplasmic events. Guillemin et al. (2002) showed that Drosophila Lam mutations result in D/V polarity defects in eggs, and tracheal branching defects in embryos. Moreover, a variety of human diseases are the result of mutations in the LMNA gene, which encodes lamin A (reviewed in Hutchison, 2002; Burke and Stewart, 2002; Goldman et al., 2002; Holaska et al., 2002). The Drosophila Lam Dm0 gene encodes type B lamin, whereas the Drosophila LamC gene encodes lamin C, which is most similar to human lamin A (Riemer et al., 1995). The A/C- and B-type lamins are similar proteins, with some different structural features, and some expression pattern differences (Gruenbaum et al., 2000). LMNA-associated human diseases affect the heart, skeletal muscles, and the nervous system (Emery-Dreifuss muscular dystrophy, limb-girdle muscular dystrophy, cardiomyopathy, and Charcot-Marie-Tooth disorder), and metabolism (Dunnigan-type lipodystrophy). The two main hypotheses as to how nuclear lamin defects can result in these disease phenotypes are that the mutations result in nuclear envelope fragility or result in changes in gene expression. An alternative hypothesis is that the inner nuclear envelope interacts with the cytoplasm through proteins like Klar or Anc-1/Syne-1, which connect the inner nuclear envelope to the microtubule, or actin cytoskeletons, respectively.
Acknowledgments
We thank everyone mentioned in MATERIALS AND METHODS for antibodies and flies, the UT Austin DNA sequencing facility, John Mendenhall for the SEMs, Paul Macdonald for the use of his confocal microscope, Gwen Gage for help with Figure 1, and Dan Starr for discussions. This work was supported by grants to J.A.F. from the Eye Institute of the National Institutes of Health (NIH; R01 EY13958) and the National Science Foundation (IBN-9809937), a Postdoctoral Fellowship to K.P. from the Eye Institute of the NIH (F32 EY06978), and a UT Austin Undergraduate Research Fellowship to A.B.M.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-06-0374. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0374.
References
- Apel, E.D., Lewis, R.M., Grady, R.M., and Sanes, J.R. (2000). Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275, 31986-31995. [DOI] [PubMed] [Google Scholar]
- Beckwith, S.M., Roghi, C.H., and Morris, N.R. (1995). The genetics of nuclear migration in fungi. Genet. Eng. 17, 165-180. [PubMed] [Google Scholar]
- Bloom, K. (2000). It's a kar9ochore to capture microtubules. Nat. Cell Biol. 6, E96-E98. [DOI] [PubMed] [Google Scholar]
- Bloom, K. (2001). Nuclear migration: cortical anchors for cytoplasmic dynein. Curr. Biol. 11, R326-R329. [DOI] [PubMed] [Google Scholar]
- Burke, B., and Stewart, C.L. (2002). Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell. Biol. 3, 575-585. [DOI] [PubMed] [Google Scholar]
- Chen, X., and Fischer, J.A. (2000). In vivo structure/function analysis of the Drosophila fat facets deubiquitinating enzyme gene. Genetics 156, 1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, I.E., Jan, L.Y., and Jan, Y.N. (1997). Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development 124, 461-470. [DOI] [PubMed] [Google Scholar]
- Fan, S.-S., and Ready, D.F. (1997). Glued participates in distinct microtubule-based activities in Drosophila eye development. Development 124, 1497-1507. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize, J.A., and Mosley, K. (1994). marbles mutants: uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development 120, 2609-2618. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize, J.A., Rubin, G.M., and Lehmann, R. (1992a). The fat facets gene is required for Drosophila eye and embryo development. Development 116, 985-1000. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize, J.A., Vize, P.D., and Rubin. G.M. (1992b). A unique mutation in the Enhancer of split gene complex affects the fates of the mystery cells in the developing Drosophila eye. Development 115, 89-101. [DOI] [PubMed] [Google Scholar]
- Flybase. (2003). The FlyBase database of the Drosophila genome projects and community literature. Available from http://flybase.bio.indiana.edu/ Nucleic Acids Res. 31, 172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe, V.E., Odell, G.M., and Edgar, B.A. (1993). Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In: The Development of Drosophila melanogaster, Vol. I, ed. M. Bate and A. Martinez-Arias, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 149-300. [Google Scholar]
- Fujita, S.C., Zipursky, S.L., Benzer, S., Ferrus, A., and Shotwell, S.L. (1982). Monoclonal antibodies against the Drosophila nervous system. Proc. Natl. Acad. Sci. USA 79, 7929-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger, E., Wells, W., Jan, L.Y., and Jan, Y.N. (1993). Tracing neurons with a kinesin-β-galactosidase fusion protein. Roux's Arch. Dev. Biol. 202, 112-122. [DOI] [PubMed] [Google Scholar]
- Goldman, R.D., Gruenbaum, Y., Moir, R.D., Shumaker, D.K., and Spann, T.P. (2002). Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16, 533-547. [DOI] [PubMed] [Google Scholar]
- Gross, S.P., Welte, M.A., Block, S.M., and Wieschaus, E.F. (2000). Dynein-mediated cargo transport in vivo: a switch controls travel distance. J. Cell Biol. 148, 945-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum, Y., Wilson, K.L., Harel, A., Goldberg, M., and Cohen, M. (2000). Nuclear lamins-structural proteins with fundamental functions. J. Struct. Biol. 129, 313-323. [DOI] [PubMed] [Google Scholar]
- Guillemin, K., Williams, T., and Krasnow, M.A. (2001). A nuclear lamin is required for cytoplasmic organization and egg polarity in Drosophila. Nat. Cell Biol. 3, 848-851. [DOI] [PubMed] [Google Scholar]
- Holaska, J.M., Wilson, K.L., and Mansharamani, M. (2002). The nuclear envelope, lamins and nuclear assembly. Curr. Opin. Cell Biol. 14, 357-364. [DOI] [PubMed] [Google Scholar]
- Holtz, D., Tanaka, R.A., Hartwig, J., and McKeon, F. (1989). The CaaX motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell 59, 969-977. [DOI] [PubMed] [Google Scholar]
- Holzbauer, E.L., Hammarback, J.A., Paschal, B.M., Kravit, N.G., Pfister, K.K., and Vallee, R.B. (1991). Homology of a 150K cytoplasmic dynein-associated polypeptide with the Drosophila gene Glued. Nature 351, 579-583. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Baker, R.T., and Fischer-Vize, J.A. (1995). Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science 270, 1828-1831. [DOI] [PubMed] [Google Scholar]
- Hummel, T., Krukkert, K., Roos, J., Davis, G., and Klambt, C. (2000). Drosophila Futsch/22C10 is a Map1B-like protein required for dendritic and axonal development. Neuron 26, 357-370. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.J. (2002). Lamins: building blocks or regulators of gene expression? Nat. Rev. Mol. Cell. Biol. 3, 848-858. [DOI] [PubMed] [Google Scholar]
- Jackle, H., and Reinhard, J. (1998). Vesicle transport: klarsicht clears up the matter. Curr. Biol. 8, R542-R544. [DOI] [PubMed] [Google Scholar]
- Joshi, H.C. (1993). Gamma-tubulin: the hub of cellular microtubule assemblies. Bioessays 15, 637-643. [DOI] [PubMed] [Google Scholar]
- Kitten, G.T., and Nigg, E.A. (1991). The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J. Cell Biol. 113, 13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotarski, M.A., Pickert, S., and MacIntyre, R.J. (1983). A cytogenetic analysis of the chromosomal region surrounding the alpha-glycerophosphate dehydrogenase locus of Drosophila melanogaster. Genetics 105, 371-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.K., Starr, D., Cohen, M., Liu, J., Han, M., Wilson, K.L., and Gruenbaum, Y. (2002). Lamin-dependent localization of Unc-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol. Biol. Cell 13, 892-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme, B., Wismar, J., Fuchs, S., Reifergerste, R., Buchner, E., Betz, H., and Schmitt, B. (1997). Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear port complexes, and accumulation of annulate lamellae. J. Cell Biol. 137, 1001-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, E.B., and Bacher, F. (1968). Method of feeding ethane methylsulfonate (EMS) to Drosophila males. Dros. Inf. Serv. 43, 193. [Google Scholar]
- Lindsley, D.L., and Zimm, G.G. (1992). The Genome of Drosophila melanogaster. San Diego: Academic Press.
- Malone, C.J., Fixsen, W.D., Horvitz, H.R., and Han, M. (1999). Unc-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126, 3171-3181. [DOI] [PubMed] [Google Scholar]
- Mislow, J.M.K., Kim, M.S., Davis, D.B., and McNally, E.M. (2001). Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J. Cell Sci. 115, 61-70. [DOI] [PubMed] [Google Scholar]
- Mislow, J.M.K., Holaska, J.M., Kim, M.S., Lee, K.K., Segura-Totten, M., Wilson, K.L., and McNally, E.M. (2002). Nesprin-1a self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 525, 135-140. [DOI] [PubMed] [Google Scholar]
- Morris, N.R., Xiang, X., and Beckwith, S.M. (1995). Nuclear migration advances in fungi. Trends Cell Biol. 5, 278-282. [DOI] [PubMed] [Google Scholar]
- Morris, N.R., Efimov, V.P., and Xiang, X. (1998). Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 8, 467-470. [DOI] [PubMed] [Google Scholar]
- Morris, N.R. (2000). Nuclear migration: from fungi to the mammalian brain. J. Cell Biol. 148, 1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, N.R. (2003). Nuclear positioning: the means is at the ends. Curr. Opin. Cell Biol. 15, 54-59. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop, K.L., Li, Q., Patterson, K., and Fischer, J.A. (1999). Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cell of the Drosophila eye. Curr. Biol. 9, 1211-1220. [DOI] [PubMed] [Google Scholar]
- Reiner, O. (2000). LIS 1, Let's interact sometimes. (part 1). Neuron 28, 633-636. [DOI] [PubMed] [Google Scholar]
- Reiner, O., Carrozza, R., Shen, Y., Wehnert, M., Faustinella, F., Dobyns, W.B., Caskey, C.T., and Ledbetter, D.H. (1993). Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364, 717-721. [DOI] [PubMed] [Google Scholar]
- Riemer, D., Stuurman, N., Berrios, M., Hunter, C., Fisher, P.A., and Weber, K. (1995). Expression of Drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. J. Cell Sci. 108, 3189-3198. [DOI] [PubMed] [Google Scholar]
- Robinow, S., and White, K. (1991). Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, 443-461. [DOI] [PubMed] [Google Scholar]
- Robinson, J.T., Wojcik, E.J., Sanders, M.A., McGill, M., and Hays, T.S. (1999). Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146, 597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, P. (1998). Gal4 in the Drosophila female germline. Mech. Dev. 78, 113-118. [DOI] [PubMed] [Google Scholar]
- Segal, M., and Bloom, K. (2001). Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 11, 160-166. [DOI] [PubMed] [Google Scholar]
- Spradling, A.C., Stern, D., Beaton, A., Rhem, E.J., Laverty, T., Mozden, N., Misra, S., and Rubin, G.M. (1999). The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of the vital Drosophila genes. Genetics 153, 135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D., and Han, M. (2002). Role of Anc-1 in tethering nuclei to the actin cytoskeleton. Science 298, 406-409. [DOI] [PubMed] [Google Scholar]
- Starr, D., and Han, M. (2003). ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 116, 211-216. [DOI] [PubMed] [Google Scholar]
- Stowers, R.S., and Schwarz, T.L. (1999). A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152, 1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman, N., Maus, N., and Fisher, P.A. (1995). Interphase phosphorylation of the Drosophila nuclear lamin: site-mapping using a monoclonal antibody. J. Cell Sci. 108, 3137-3144. [DOI] [PubMed] [Google Scholar]
- Stuurman, N., Heins, S., and Aebi, U. (1998). Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122, 42-66. [DOI] [PubMed] [Google Scholar]
- Swan, A., Nguyen, T., and Suter, B. (1999). Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1, 444-449. [DOI] [PubMed] [Google Scholar]
- Tavosanis, G., Llamazares, S., Goulielmos, G., and Gonzalez, C. (1997). Essential role for gamma-tubulin in the acentriolar female meiotic spindle of Drosophila. EMBO J. 16, 1809-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, A., and Ready, D.F. (1986). sevenless: a cell-specific homeotic mutation of the Drosophila eye. Science 231, 400-402. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., and Ready, D.F. (1987). Neuronal differentiation in the Drosophila ommatidium. Dev. Biol. 120, 366-376. [DOI] [PubMed] [Google Scholar]
- Vallee, R.B., Tai, C., and Faulkner, N.E. (2001). LIS 1, cellular function of a disease-causing gene. Trends Cell Biol. 11, 155-160. [DOI] [PubMed] [Google Scholar]
- van Eeden, F., and St Johnston, D. (1999). The polarization of the anterior-posterior and dorsal-ventral axes during Drosophila oogenesis. Curr. Opin. Genet. Dev. 9, 396-404. [DOI] [PubMed] [Google Scholar]
- Welte, M.A., Gross, S.P., Postner, M., Block, S.M., and Wieschaus, E.F. (1998). Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell 92, 547-557. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and Ready, D.F. (1993). Pattern formation in the Drosophila retina. In: The Development of Drosophila melanogaster, Vol. II, ed. M. Bate and A. Martinez-Arias, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1277-1326. [Google Scholar]
- Wynshaw-Borris, A., and Gambello, M.J. (2001). LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 15, 639-651. [DOI] [PubMed] [Google Scholar]
- Xu, T., and Rubin, G.M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. et al. (2001). Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114, 4485-4498. [DOI] [PubMed] [Google Scholar]
- Zhen, Y.-Y., Libotte, T., Munck, M., Noegel, A.A., and Korenbaum, E. (2002). NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115, 3207-3222. [DOI] [PubMed] [Google Scholar]