Abstract
Background and objective
The field of salivary diagnostics lacks an accepted and validated biomarker of alveolar bone remodeling. To address this we examined levels of salivary biomolecules specifically associated with biological aspects of bone remodeling in subjects with chronic periodontitis in a case-control study.
Methods
Levels of macrophage inflammatory protein (MIP)-1α, osteoprotegerin (OPG), C-telopeptide pyridinoline cross-links of type I collagen (ICTP), and β-C-terminal type I collagen telopeptide (β-CTX) in unstimulated whole saliva of 80 subjects (40 subjects with moderate to severe chronic periodontitis and 40 gender- and age-matched healthy control subjects) were measured using enzyme immunosorbent assays. Saliva was collected before clinical examination that included probing depth (PD), clinical attachment loss (CAL), and bleeding on probing (BOP).
Results
The mean level of MIP-1α in periodontitis subjects was 18-fold higher than in healthy subjects (p < 0.0001). Clinical periodontal indices significantly correlated with MIP-1α levels (p < 0.0001). MIP-1α, of the biomolecules examined, demonstrated the highest ability to discriminate between periodontal disease and health as determined by area under the curve (AUC = 0.94) and classification and regression tree analysis (sensitivity 94%, specificity 92.7%). OPG levels were elevated 1.6-fold (P = 0.055), whereas ICTP and β-CTX levels were below the level of detection in the majority of subjects.
Conclusion
These findings suggest that the chemokine MIP-1α may aid in identifying periodontitis. Future longitudinal studies are warranted to determine whether this biomarker can help to ascertain progression of bone loss in subjects with periodontal disease.
Keywords: periodontal health, periodontal disease; saliva; biomarkers; bone remodeling; macrophage inflammatory protein (MIP)-1α; osteoprotegerin (OPG); C-telopeptide pyridinoline cross-links of type I collagen (ICTP), β-C-terminal type I collagen telopeptide (β-CTX)
Periodontal disease results from interaction between oral bacteria and the host inflammatory response. This interaction triggers a cascade of inflammatory events, which in turn promote connective tissue destruction and alveolar bone remodeling.1-7 These unique biological events contain signatures of the microbial ecology, as well as downstream events involving inflammation, attachment loss and bone destruction. It is likely that identification of the dominant signatures for each of these biological phases could provide insight into biomarkers of periodontal disease in oral fluids. To this end, several investigators have identified salivary biomarkers of biological events associated with aspects of periodontal disease8-12 and reviews on this topic are available.13-15 However, lacking to date is a focused panel of biomarkers that encompass the early and later biological phases that would yield, at least theoretically, the specificity for the development of a real-time, accurate, biologically-based, periodontal disease diagnostic device.
Progress continues to be made towards the achievement of a salivary diagnostic device based on the knowledge that saliva is a rich pool of proteins and molecules that reflects aspects of oral health. In one recent example, we found that biomarkers of inflammation, connective tissue destruction and bone remodeling were elevated in concentrations in the saliva of patients with periodontal disease.11 In that study, we observed that salivary levels of IL-1β and MMP-8 were significantly associated with clinical parameters of periodontal disease, and levels of IL-1β and MMP-8 elevated above a threshold (i.e., 2 standard deviations above the mean of healthy controls), together demonstrated an odds ratio of 45 for periodontal disease. While these results demonstrated proof of principle that biomarkers from two distinct biological phases could aid in distinguishing periodontal disease from health, the identification of a biomarker associated with aspects of bone remodeling – a late biological event that could improve the accuracy of salivary diagnostics - remains elusive.
Bone resorption is mediated by osteoclasts that exhibit specific abilities to degrade organic and inorganic components of bone. Different mediators such as interleukin-1β, prostaglandin E2 (PGE2), tumor necrosis factor-alpha (TNF-α), macrophage inflammatory protein (MIP)-1α, interleukin-6 (IL-6), interleukin-11 (IL-11), and interleukin-17 (IL-17) act upstream as activators of osteoclastogenesis.16-22 Within the resorption lacunae, receptor activator of nuclear factor-kappa B ligand (RANKL) and osteoprotegerin (OPG) are important cytokines belonging to the TNF family that regulate differentiation of osteoclast progenitor cells into active osteoclasts or inhibiting the differentiation process, respectively.23, 24 As a result, type 1 collagen is degraded during bone destruction by proteolytic enzymes such as matrix metalloproteinases (MMPs) and cathepsin K which lead to release of cross-linked telopeptides into the circulation (serum, saliva, and urine) as stable fragments such as pyridinoline cross-linked carboxyterminal telopeptide of type 1 collagen (ICTP)25-27 and C-terminal type 1 collagen telopeptide (β-CTX).28-31 We and others have investigated salivary levels of several of these important molecules associated with cytokine signaling and alveolar bone resorption32-39 however no conclusive information has been yet reported on the best biomarker associated with alveolar bone remodeling in adults. Also, there is a lack of knowledge whether upstream pathways, midstream osteoclastogenic factors, or downstream degradation products are better salivary biomarkers of periodontal disease.
Therefore, the purpose of this study was to test the hypothesis that a specific salivary biomarker associated with bone remodeling could be identified that would distinguish healthy and periodontal disease subjects. Proteins associated with the upstream, midstream and downstream processes of osteoclastogenesis (i.e., MIP-1α, OPG, β-CTX, and ICTP) were selected for evaluation.
Material and Methods
Participants
Eighty patients were enrolled in this case-control, cross-sectional study. Participants were recruited from the general clinic population of the College of Dentistry as well as the surrounding counties by advertisement. Inclusion criteria included subjects older than 18 years of age who were in good general health (excluding the case definition) and had a minimum of 20 teeth. Participation in the control group (n=40) required bleeding on probing (BOP) in <10% of sites, probing depth (PD) ≥5 mm in <2% of sites, and clinical attachment loss (CAL) >2 mm in <1% of sites. Subjects of the test group (n=40) had the diagnosis of generalized moderate to severe chronic periodontitis based on the criteria defined by the American Academy of Periodontology.40, 41 Inclusion criteria of the test group were 5 sites in two quadrants with a minimum of two affected teeth in each quadrant. Each site had ≥5 mm PD, CAL ≥3 mm, and BOP with score ≥2 (0=one, 1=pinpoint, 2=interdental bleeding, 3=spontaneous/heavy bleeding). Exclusion criteria were a history of alcoholism; liver, kidney, or salivary gland dysfunction; infectious diseases; inflammatory bowel disease; rheumatoid arthritis; granulomatous diseases; diabetes or were undergoing or had undergone organ transplant or cancer therapy. Pregnancy or lactation, use of glucocorticoids, cyclooxygenase inhibitors, bisphosphonates, antibiotics or immunosuppressant medication within the last 6 months, need for antibiotics for infective endocarditis prophylaxis during dental procedures, symptoms of acute illness (i.e., fever, sore throat, body aches, and diarrhea), orthodontic appliances or presence of an oral mucosal inflammatory condition (e.g., aphthous, lichen planus, leukoplakia, and oral cancer) also were exclusion criteria. The study was performed at the University of Kentucky between August 2005 and August 2009 and was approved by the University Institutional Review Board. All subjects understood the study, provided written informed consent and received incentives (i.e., monetary compensation and a clinical examination) as part of the study protocol.
Clinical evaluation
Complete medical and dental histories were obtained from the patient's records and confirmed by interview. All subjects received comprehensive oral and periodontal examinations that included BOP, PD, CAL assessed as previously described.42 Briefly, PDs were measured at six locations per tooth (mesial-buccal, mid-buccal, distal-buccal, mesial lingual, mid-lingual, and distal-lingual) using a 15 UNC probe. After the measurement of PDs, all sites were observed for BOP. The degree of bleeding were estimated and recorded (0=no bleeding; 1=light bleeding; 2=moderate bleeding; and 3= heavy bleeding) for each probed site. Clinical attachment levels were measured at all six locations per tooth. Three periodontists, blinded to the participant group assignments, performed the clinical evaluations.
Saliva Collection
Unstimulated whole expectorated saliva (5 mL) was collected from each subject between 9 and 11 a.m. according to a modification in the method described by Navazesh.43 Subjects were asked to avoid oral hygiene measures (i.e. flossing, brushing, and mouth rinses), eating, drinking, or gum chewing for 1 hour before collection. Subjects rinsed their mouth with tap water, then expectorated whole saliva into sterile tubes containing a protease inhibitor solution (SIGMAFAST, Sigma, St. Louis, MO.) while seated in an upright position. Collected samples were placed immediately on ice and aliquoted prior to freezing at -80°C. Samples were thawed and analyzed within six months of collection. Samples from all participants were collected prior to the periodontal evaluation.
Biomarker analysis
Concentrations of β-CTP, ICTP, OPG, and MIP-1α in saliva were determined in duplicate for each subject using enzyme immunosorbent assays (EIA) kits, according to the manufacturer's directions, by technologists in the Clinical Laboratory Improvements Amendments (CLIA)-certified General Clinical Research Core laboratory at the University of Kentucky Medical Center. β-CTX Urine Crosslaps ELISA and ICTP serum Crosslaps ELISA (Nordic Bioscience Diagnostics A/S, Herlev, Denmark), OPG (Osteoprotegerin EIA kit, ALPCO Diagnostics, Salem, NH), and MIP-1α kit (Millipore/Milliplex® map kit, Billerica, MA) were used to evaluate the analytes. Standards were included on all runs, and all results were reported within the linearity of the assays.
Statistical analysis
Demographic variables and smoking status were compared between groups using Fisher's exact test. Comparison of analytes between test and control groups was performed by box-plots. Mean periodontal indices, age, and concentration of salivary biomarkers were compared between test and control groups using two analysis of covariance to adjust for differences in patient demographics. Relationships between analytes and periodontal indices were determined using Spearman's correlation coefficient. Analyte levels associated with clinical parameter of periodontitis as well as those that discriminated periodontitis from health were determined using logistic regression and Classification and Regression Tree (CART) analysis. In the latter analysis, measurements below the detection limit were set at one half the detection limit. All analyses were performed using the PC SAS 9.1 (SAS Institute Inc., Cary, NC, USA) with statistical significance determined at the 0.05 level.
Results
Eighty adults (40 with chronic periodontitis and 40 healthy controls) ranging in age from 21 to 60 years old were evaluated (Table 1). Participants were age- and gender-matched. Test subjects were predominantly non-Caucasian whereas the controls were predominantly Caucasian (p < 0.001). Smokers were only in the test group (p < 0.001). Each group had a similar mean number of teeth. As expected, all periodontal indices were significantly higher in the test group than in the control group (p < 0.0001).
Table 1.
Comparison of demographics and clinical characteristics between study groups.
| Health (n=40) | Periodontal Disease (n=40) | P Value | |
|---|---|---|---|
| Age (years; mean +/- SD) | 35.8 +/- 9.0 | 36.8 +/- 9.0 | |
| Female (%) | 50.0 | 50.0 | |
| White (%) | 90.0 | 35.0 | <0.0001* |
| Hispanic (%) | 2.5 | 17.5 | <0.0001* |
| African American | 0.0 | 10 | <0.0001* |
| Asian (%) | 2.5 | 10 | <0.0001* |
| Other | 5.0 | 27.5 | <0.0001* |
| Current tobacco use (%) | 0.0 | 30.0 | <0.0001* |
| # Teeth | 27.6 +/- 1.5 | 27.4 +/- 1.8 | |
| Periodontal indices (% sites; mean +/- SD) | |||
| BOP Sites | 3.2 +/- 6.2 | 64.5 +/- 24.6 | <0.0001 |
| PD Sites ≥4 mm | 1.6 +/- 2.6 | 25.8 +/- 13.6 | <0.0001 |
| PD Sites ≥5 mm | 0.3 +/- 0.5 | 13.3 +/- 9.4 | <0.0001 |
| CAL ≥2 mm | 0.1 +/- 0.2 | 19.3 +/- 13.3 | <0.0001 |
Demographical data determined by Exact Fisher's test.
Periodontal indices analyzed by ANCOVA
Salivary Analyte Levels
Comparison of salivary levels of OPG and MIP-1α are shown in Figure 1. Levels of β-CTX were at or below the level of detection (i.e., 0.80 μg/L) in all samples. Levels of ICTP were detectable (i.e., > 0.020 ng/mL) in only six control subjects and one periodontal disease subject. These datasets were too small to detect differences between the groups. OPG was detectable in all samples. The mean level of OPG was 1.6-fold higher in the periodontal disease group (p = 0.0553). In contrast, levels of MIP-1α were detected in all test subjects but only two controls. The mean level of MIP-1α was 18-fold higher in the periodontal disease group than the controls (p < 0.0001).
Figure 1.
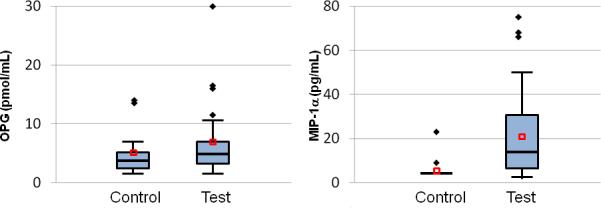
Boxplots for salivary levels of MIP-1α and OPG. Mean values of MIP-1α were significantly different between the two groups (p < 0.0001); comparisons between groups of the other analytes were not. Outliers shown as diamonds, means shown as red squares.
Relationships between Salivary Analyte Levels and Parameters of Periodontal Disease
We next analyzed for relationships between levels of each analyte and clinical parameters of periodontitis. Here, MIP-1α demonstrated a strong positive correlation with Spearman correlation coefficients ranging from 0.75 to 0.8 for %BOP, %PD≥4 mm, %PD≥5 mm and %CAL (p < 0.0001). OPG also demonstrated a significant correlation with all four clinical parameters of periodontitis, with coefficients ranging from 0.22 to 0.36 (p ≤ 0.05). ICTP demonstrated a weak and negative correlation (P>0.05). Correlations with β-CTX were indeterminate due to too few samples having detectable levels.
To determine if any of the four analytes discriminated periodontal health from disease, logistic regression models were performed and analyzed. In the receiver operator characteristic analyses (Figure 2), only MIP-1α showed a significant association with periodontitis with an area under curve (AUC = 0.94; p < 0.0002). The CART analysis also agreed with the logistic model in that only MIP-1α was a significant discriminator of periodontitis. Here the CART determined the threshold for periodontitis was 1.12 pg/ml. This threshold displayed a sensitivity of 94.9%, specificity of 92.7%, positive predictive value of 94.9% and negative predictive value of 92.7% for periodontal disease. Additional pairing and combinations of the four biomarkers did not associate better with periodontitis.
Figure 2.
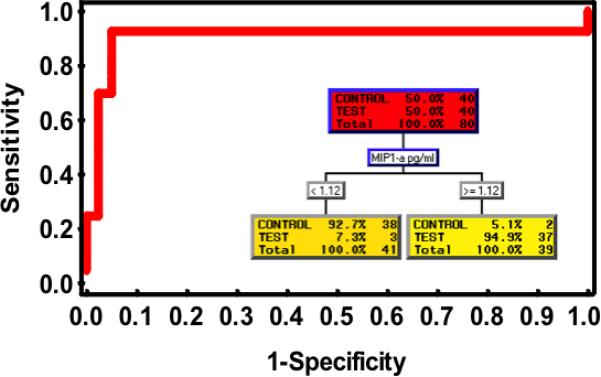
Receiver operator characteristics of MIP-1α for identification of periodontal disease. Inset shows CART analysis demonstrating MIP-1α is associated with periodontitis at the concentration ≥ 1.12 pg/mL.
Discussion
This cross-sectional, case-control study evaluated select bone remodeling biomarkers in whole unstimulated saliva from 40 adults who had chronic periodontitis and 40 healthy controls. The proteins evaluated are associated with important biological phases of bone remodeling. An upstream signaling protein (MIP-1α), a midstream anti-osteoclastic factor (OPG) and two downstream collagen type 1 degradation products (ICTP, β-CTX) were selected in an effort to address key aspects of alveolar bone remodeling that might be reflected in saliva. Our findings showed that MIP-1α performed well on several measures. Salivary MIP-1α levels were significantly elevated in subjects who had periodontal disease and demonstrated the strongest correlation with clinical parameters of periodontal disease. In the regression models, MIP-1α was the biomarker that best discriminated periodontal disease from health compared to OPG, ICTP and β-CTX.
Several studies have evaluated bone remodeling biomarkers in saliva in relation to periodontal disease, yet only a limited number have detected collagen degradation products in saliva. ICTP and β-CTX are two well recognized end products of the collagneolytic pathway. β-CTX is generated first from the organic matrix of type 1 collagen by lysosomal cathepsin K attack. Later in the resorption process matrix metalloproteinases (MMPs) breakdown crosslink peptides from the carboxy-terminal telopeptide areas of type I collagen into ICTP.44 Prior to the current study, neither β-CTX nor ICTP in saliva had been shown head-to-head to be a better discriminator of periodontal disease. Ng et al. reported in a cross-sectional study the inability to detect ICTP in whole, stimulated saliva in 110 untreated dental patients.35 In contrast, Kinney et al. reported the detection of ICTP in saliva and rising levels in patients with gingivitis and mild periodontitis over a 12 month observation period,32 and Gurlek et al. detected ICTP in the saliva of 67 otherwise healthy adults who had inflammatory periodontal disease.38 Our findings herein and in our previous study are similar with those of Ng et al. and in contrast to those of Kinney et al and Gurlek et al. Previously we detected levels of ICTP in only 4.8% of 74 subjects33 and here detected ICTP levels in only 8.8% of 80 dental patients, and only one patient in the test group. We believe that the use of different assay methods likely yielded higher sensitivity by Kinney et al and Gurlek et al., in as much as our detection limit was within the range reported by those authors.38 Of note, β-CTX in the current study was even more difficult to detect with all our samples having levels at or below the level of detection, and no previous salivary studies found in the published literature for comparison. Together these findings are consistent with the premise that ICTP is a more sensitive marker of lytic alveolar bone resorption in saliva than β-CTX45 and suggest that MMP-associated degradation products are predominant in saliva compared to cathepsin K-associated degradation products. Also, the overall context of the findings seem to indicate that downstream collagen degradation products are difficult to detect in the saliva of periodontitis patients when evaluation is performed in a cross-sectional manner, whereas their detection in longitudinal studies, where periods of active bone resorption are more likely to be demonstrated, could prove to be more useful in this area of diagnostic assessment.
OPG, an osteoblast-secreted decoy receptor that inhibits osteoclastogenesis, has been shown to be elevated in the saliva of patients with untreated chronic periodontitis,32, 46 correlate positively with probing depth and bleeding on probing11 and correlate with clinical attachment level.47 OPG also has been reported to be at lower levels in smokers than non-smokers who have chronic periodontitis47 and serves as a salivary biomarker when paired with MMP-8 and red-complex anaerobic periodontal pathogens.48 In patients with periodontal disease salivary OPG levels have diminished after scaling and root planing.32, 42 However, as a single biomarker OPG has shown a weaker correlation with clinical parameters of periodontal disease than IL-1β and MMP-8.11 Consistent with that finding, we observed that OPG was less of a discriminator for periodontal disease than MIP-1α, but better than ICTP and β-CTX. Together these findings suggest that OPG, a factor midstream in the bone remodeling process, is a more sensitive salivary biomarker of periodontal disease than factors further downstream in the process. Consistent with this, others have found salivary RANKL levels to decrease after periodontal therapy.49 However, we are cognizant that it is possible that different saliva management protocols could contribute to alternative interpretations of these data.
MIP-1α/CCL3 is a member of the cysteine-cysteine chemokine family which is secreted by macrophages, neutrophils, basophils, dendritic cells, lymphocytes and epithelial cells and mediates granulocyte migration and adhesion.50-52 It stimulates monocytes and/or osteoclast progenitor cells to become active osteoclasts in a RANK/RANKL and dose-dependent manner.53 MIP-1α has been detected at higher salivary levels (50-fold) in a longitudinal study of seven adolescents who had aggressive periodontitis compared with controls34, and is secreted by gingival fibroblasts and epithelial cells.51, 54 In our study, salivary concentrations of MIP-1α were 18-fold higher in periodontitis subjects than healthy subjects, (p < 0.0001) and periodontal indices demonstrated highly significant correlations with MIP-1α levels (p < 0.0001). In regression analyses, MIP-1α offered high specificity (94%) and sensitivity (92.5%) for distinguishing periodontal disease from health, with the CART analysis providing an optimal cut-point of MIP-1α at 1.12 pg/mL and high sensitivity and specificity. These findings suggest that the salivary level of MIP-1α could have clinical utility as a screening tool for moderate to severe periodontal disease. However, its utility for discriminating between intermediate levels of disease (gingivitis, mild periodontitis) and health is not yet known. Also, because our study is limited by several factors (i.e., only 40 subjects per group were studied, reproducibility of each subject was not analyzed longitudinally and we excluded patients with oral mucosal diseases and systemic inflammatory conditions), the overall specificity of these biomarkers for periodontal disease in the general population needs to be further investigated.
Conclusions
This study, which evaluated four biomolecules associated alveolar bone remodeling, found that salivary levels of MIP-1α can identify persons who have periodontal disease. Our data along with the findings from previous studies11, 32, 48, 55 suggest that the combined presence of elevated levels of a panel of salivary biomarkers representing the three biological phases (inflammatory, connective tissue destruction and bone remodeling) of periodontal disease may offer the sensitivity and specificity for screening for periodontal disease in non-dental settings as well as potentially providing an understanding of the dynamics of the periodontitis lesion. Validation of this premise could lead to the use of biofluid panels as adjuncts in the diagnostic assessment of periodontal disease in the near future.
Summary.
Salivary levels of MIP-1α distinguish periodontal disease from health.
Acknowledgements
The authors thank Donna Mischel, Vanessa Hodges-Reed, study coordinators, Jason Stevens, research analyst, and Malini Bharadwaj, data management specialist, of the Center for Oral Health Research of the University of Kentucky for clinical, laboratory and data management support. This study was supported by grants P20 RR020145 and M01-RR02602 from the National Institutes of Health, Bethesda, Maryland, and the University of Kentucky General Clinical Research Core and Graduate Periodontology fund. The authors report no conflicts of interest related to this study.
Sources of Support: This study was supported by grants from the National Institute of Health P20 RR020145 and M01-RR02602, the University of Kentucky General Clinical Research Core and Graduate Periodontology fund.
Footnotes
Conflict of interest: The authors report no conflicts of interest related to this study.
References
- 1.Offenbacher S. Periodontal diseases: pathogenesis. Ann Periodontol. 1996;1:821–878. doi: 10.1902/annals.1996.1.1.821. [DOI] [PubMed] [Google Scholar]
- 2.Ranney RR. Immunologic mechanisms of pathogenesis in periodontal diseases: an assessment. J Periodontal Res. 1991;26:243–254. doi: 10.1111/j.1600-0765.1991.tb01650.x. [DOI] [PubMed] [Google Scholar]
- 3.Page RC. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodontal Res. 1991;26:230–242. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 4.Seymour GJ. Importance of the host response in the periodontium. J Clin Periodontol. 1991;18:421–426. doi: 10.1111/j.1600-051x.1991.tb02310.x. [DOI] [PubMed] [Google Scholar]
- 5.Genco RJ. Host responses in periodontal diseases: current concepts. J Periodontol. 1992;63:338–355. doi: 10.1902/jop.1992.63.4s.338. [DOI] [PubMed] [Google Scholar]
- 6.Dennison DK, Van Dyke TE. The acute inflammatory response and the role of phagocytic cells in periodontal health and disease. Periodontol 2000. 1997;14:54–78. doi: 10.1111/j.1600-0757.1997.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 7.Gemmell E, Yamazaki K, Seymour GJ. Destructive periodontitis lesions are determined by the nature of the lymphocytic response. Crit Rev Oral Biol Med. 2002;13:17–34. doi: 10.1177/154411130201300104. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman E, Lamster IB. Analysis of saliva for periodontal diagnosis--a review. J Clin Periodontol. 2000;27:453–465. doi: 10.1034/j.1600-051x.2000.027007453.x. [DOI] [PubMed] [Google Scholar]
- 9.Lamster IB, Kaufman E, Grbic JT, Winston LJ, Singer RE. Beta-glucuronidase activity in saliva: relationship to clinical periodontal parameters. J Periodontol. 2003;74:353–359. doi: 10.1902/jop.2003.74.3.353. [DOI] [PubMed] [Google Scholar]
- 10.Christodoulides N, Mohanty S, Miller CS, et al. Application of microchip assay system for the measurement of C-reactive protein in human saliva. Lab Chip. 2005;5:261–269. doi: 10.1039/b414194f. [DOI] [PubMed] [Google Scholar]
- 11.Miller CS, King CP, Jr., Langub MC, Kryscio RJ, Thomas MV. Salivary biomarkers of existing periodontal disease: a cross-sectional study. J Am Dent Assoc. 2006;137:322–329. doi: 10.14219/jada.archive.2006.0181. [DOI] [PubMed] [Google Scholar]
- 12.Herr AE, Hatch AV, Throckmorton DJ, et al. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc Natl Acad Sci U S A. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller CS, Foley JD, Bailey AL, et al. Current developments in salivary diagnostics. Biomarkers in Medicine. 2010;4:1–18. doi: 10.2217/bmm.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannobile WV, Beikler T, Kinney JS, Ramseier CA, Morelli T, Wong DT. Saliva as a diagnostic tool for periodontal disease: current state and future directions. Periodontol 2000. 2009;50:52–64. doi: 10.1111/j.1600-0757.2008.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buduneli N, Kinane DF. Host-derived diagnostic markers related to soft tissue destruction and bone degradation in periodontitis. J Clin Periodontol. 2011;38(Suppl 11):85–105. doi: 10.1111/j.1600-051X.2010.01670.x. [DOI] [PubMed] [Google Scholar]
- 16.Pfeilschifter J, Chenu C, Bird A, Mundy GR, Roodman GD. Interleukin-1 and tumor necrosis factor stimulate the formation of human osteoclastlike cells in vitro. J Bone Miner Res. 1989;4:113–118. doi: 10.1002/jbmr.5650040116. [DOI] [PubMed] [Google Scholar]
- 17.Bertolini DR, Nedwin GE, Bringman TS, Smith DD, Mundy GR. Stimulation of bone resorption and inhibition of bone formation in vitro by human tumour necrosis factors. Nature. 1986;319:516–518. doi: 10.1038/319516a0. [DOI] [PubMed] [Google Scholar]
- 18.Chiang CY, Kyritsis G, Graves DT, Amar S. Interleukin-1 and tumor necrosis factor activities partially account for calvarial bone resorption induced by local injection of lipopolysaccharide. Infect Immun. 1999;67:4231–4236. doi: 10.1128/iai.67.8.4231-4236.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Khansari A, Shapira L, Graves DT, Amar S. Contribution of interleukin-11 and prostaglandin(s) in lipopolysaccharide-induced bone resorption in vivo. Infect Immun. 2002;70:3915–3922. doi: 10.1128/IAI.70.7.3915-3922.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi SJ, Cruz JC, Craig F, et al. Macrophage inflammatory protein 1-alpha is a potential osteoclast stimulatory factor in multiple myeloma. Blood. 2000;96:671–675. [PubMed] [Google Scholar]
- 21.Kotake S, Udagawa N, Takahashi N, et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 1999;103:1345–1352. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishimi Y, Miyaura C, Jin CH, et al. IL-6 is produced by osteoblasts and induces bone resorption. J Immunol. 1990;145:3297–3303. [PubMed] [Google Scholar]
- 23.Lerner UH. Inflammation-induced bone remodeling in periodontal disease and the influence of post-menopausal osteoporosis. J Dent Res. 2006;85:596–607. doi: 10.1177/154405910608500704. [DOI] [PubMed] [Google Scholar]
- 24.Roodman GD. Regulation of osteoclast differentiation. Ann N Y Acad Sci. 2006;1068:100–109. doi: 10.1196/annals.1346.013. [DOI] [PubMed] [Google Scholar]
- 25.Mogi M, Otogoto J. Expression of cathepsin-K in gingival crevicular fluid of patients with periodontitis. Arch Oral Biol. 2007;52:894–898. doi: 10.1016/j.archoralbio.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Giannobile WV, Al-Shammari KF, Sarment DP. Matrix molecules and growth factors as indicators of periodontal disease activity. Periodontol 2000. 2003;31:125–134. doi: 10.1034/j.1600-0757.2003.03108.x. [DOI] [PubMed] [Google Scholar]
- 27.Risteli L, Risteli J. Biochemical markers of bone metabolism. Ann Med. 1993;25:385–393. doi: 10.3109/07853899309147301. [DOI] [PubMed] [Google Scholar]
- 28.Delmas PD. Bone marker nomenclature. Bone. 2001;28:575–576. doi: 10.1016/s8756-3282(01)00480-x. [DOI] [PubMed] [Google Scholar]
- 29.Bonde M, Qvist P, Fledelius C, Riis BJ, Christiansen C. Immunoassay for quantifying type I collagen degradation products in urine evaluated. Clin Chem. 1994;40:2022–2025. [PubMed] [Google Scholar]
- 30.Garnero P, Ferreras M, Karsdal MA, et al. The type I collagen fragments ICTP and CTX reveal distinct enzymatic pathways of bone collagen degradation. J Bone Miner Res. 2003;18:859–867. doi: 10.1359/jbmr.2003.18.5.859. [DOI] [PubMed] [Google Scholar]
- 31.Garnero P. Biomarkers for osteoporosis management: utility in diagnosis, fracture risk prediction and therapy monitoring. Mol Diagn Ther. 2008;12:157–170. doi: 10.1007/BF03256280. [DOI] [PubMed] [Google Scholar]
- 32.Kinney JS, Morelli T, Braun T, et al. Saliva/pathogen biomarker signatures and periodontal disease progression. J Dent Res. 2011;90:752–758. doi: 10.1177/0022034511399908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frodge BD, Ebersole JL, Kryscio RJ, Thomas MV, Miller CS. Bone remodeling biomarkers of periodontal disease in saliva. J Periodontol. 2008;79:1913–1919. doi: 10.1902/jop.2008.080070. [DOI] [PubMed] [Google Scholar]
- 34.Fine DH, Markowitz K, Furgang D, et al. Macrophage inflammatory protein-1alpha: a salivary biomarker of bone loss in a longitudinal cohort study of children at risk for aggressive periodontal disease? J Periodontol. 2009;80:106–113. doi: 10.1902/jop.2009.080296. [DOI] [PubMed] [Google Scholar]
- 35.Ng PY, Donley M, Hausmann E, Hutson AD, Rossomando EF, Scannapieco FA. Candidate salivary biomarkers associated with alveolar bone loss: cross-sectional and in vitro studies. FEMS Immunol Med Microbiol. 2007;49:252–260. doi: 10.1111/j.1574-695X.2006.00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scannapieco FA, Ng P, Hovey K, Hausmann E, Hutson A, Wactawski-Wende J. Salivary biomarkers associated with alveolar bone loss. Ann N Y Acad Sci. 2007;1098:496–497. doi: 10.1196/annals.1384.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teles RP, Likhari V, Socransky SS, Haffajee AD. Salivary cytokine levels in subjects with chronic periodontitis and in periodontally healthy individuals: a cross-sectional study. J Periodontal Res. 2009;44:411–417. doi: 10.1111/j.1600-0765.2008.01119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurlek O, Lappin DF, Buduneli N. Effects of smoking on salivary C-telopeptide pyridinoline cross-links of type I collagen and osteocalcin levels. Arch Oral Biol. 2009;54:1099–1104. doi: 10.1016/j.archoralbio.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Ozcaka O, Nalbantsoy A, Buduneli N. Salivary osteocalcin levels are decreased in smoker chronic periodontitis patients. Oral Dis. 2011;17:200–205. doi: 10.1111/j.1601-0825.2010.01721.x. [DOI] [PubMed] [Google Scholar]
- 40.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 41.Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontol 2000. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- 42.Sexton WM, Lin Y, Kryscio RJ, Dawson DR, 3rd, Ebersole JL, Miller CS. Salivary biomarkers of periodontal disease in response to treatment. J Clin Periodontol. 2011;38:434–441. doi: 10.1111/j.1600-051X.2011.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navazesh M. Methods for collecting saliva. Ann N Y Acad Sci. 1993;694:72–77. doi: 10.1111/j.1749-6632.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- 44.Everts V, Delaisse JM, Korper W, Beertsen W. Cysteine proteinases and matrix metalloproteinases play distinct roles in the subosteoclastic resorption zone. J Bone Miner Res. 1998;13:1420–1430. doi: 10.1359/jbmr.1998.13.9.1420. [DOI] [PubMed] [Google Scholar]
- 45.Koizumi M, Matsumoto S, Takahashi S, Yamashita T, Ogata E. Bone metabolic markers in the evaluation of bone scan flare phenomenon in bone metastases of breast cancer. Clin Nucl Med. 1999;24:15–20. doi: 10.1097/00003072-199901000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Costa PP, Trevisan GL, Macedo GO, et al. Salivary interleukin-6, matrix metalloproteinase-8, and osteoprotegerin in patients with periodontitis and diabetes. J Periodontol. 2010;81:384–391. doi: 10.1902/jop.2009.090510. [DOI] [PubMed] [Google Scholar]
- 47.Buduneli N, Biyikoglu B, Sherrabeh S, Lappin DF. Saliva concentrations of RANKL and osteoprotegerin in smoker versus non-smoker chronic periodontitis patients. J Clin Periodontol. 2008;35:846–852. doi: 10.1111/j.1600-051X.2008.01310.x. [DOI] [PubMed] [Google Scholar]
- 48.Ramseier CA, Kinney JS, Herr AE, et al. Identification of pathogen and host-response markers correlated with periodontal disease. J Periodontol. 2009;80:436–446. doi: 10.1902/jop.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El-Sharkawy H, Aboelsaad N, Eliwa M, et al. Adjunctive treatment of chronic periodontitis with daily dietary supplementation with omega-3 Fatty acids and low-dose aspirin. J Periodontol. 2010;81:1635–1643. doi: 10.1902/jop.2010.090628. [DOI] [PubMed] [Google Scholar]
- 50.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 51.Ryu OH, Choi SJ, Linares AM, et al. Gingival epithelial cell expression of macrophage inflammatory protein-1alpha induced by interleukin-1beta and lipopolysaccharide. J Periodontol. 2007;78:1627–1634. doi: 10.1902/jop.2007.070066. [DOI] [PubMed] [Google Scholar]
- 52.Maurer M, von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36:1882–1886. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 53.Terpos E, Politou M, Viniou N, Rahemtulla A. Significance of macrophage inflammatory protein-1 alpha (MIP-1alpha) in multiple myeloma. Leuk Lymphoma. 2005;46:1699–1707. doi: 10.1080/10428190500175049. [DOI] [PubMed] [Google Scholar]
- 54.Morandini AC, Sipert CR, Gasparoto TH, et al. Differential production of macrophage inflammatory protein-1alpha, stromal-derived factor-1, and IL-6 by human cultured periodontal ligament and gingival fibroblasts challenged with lipopolysaccharide from P. gingivalis. J Periodontol. 2010;81:310–317. doi: 10.1902/jop.2009.090375. [DOI] [PubMed] [Google Scholar]
- 55.Christodoulides N, Floriano PN, Miller CS, et al. Lab-on-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Ann N Y Acad Sci. 2007;1098:411–428. doi: 10.1196/annals.1384.035. [DOI] [PubMed] [Google Scholar]


