Abstract
We describe the characterization of an 80-kDa protein cross-reacting with a monoclonal antibody against the human La autoantigen. The 80-kDa protein is a variant of rabip4 with an N-terminal extension of 108 amino acids and is expressed in the same cells. For this reason, we named it rabip4′. rabip4′ is a peripheral membrane protein, which colocalized with internalized transferrin and EEA1 on early endosomes. Membrane association required the presence of the FYVE domain and was perturbed by the phosphatidylinositol 3-kinase inhibitor wortmannin. Expression of a dominant negative rabip4′ mutant reduced internalization and recycling of transferrin from early endosomes, suggesting that it may be functionally linked to rab4 and rab5. In agreement with this, we found that rabip4′ colocalized with the two GTPases on early endosomes and bound specifically and simultaneously to the GTP form of both rab4 and rab5. We conclude that rabip4′ may coordinate the activities of rab4 and rab5, regulating membrane dynamics in the early endosomal system.
INTRODUCTION
All eukaryotic cells internalize cell surface proteins and material from their environment by (receptor-mediated) endocytosis (Mellman, 1996; Mukherjee et al., 1997). The pathway is also used for the recycling of proteins of the secretory pathway (Lewis et al., 2000). Ligands are bound to receptors at the cell surface. The complex then enters clathrin-coated pits that pinch off the plasma membrane to form clathrin-coated vesicles. These rapidly lose their coat and subsequently fuse and deliver their cargo to early endosomes (EEs). The mildly acidic pH in the lumen of EEs causes dissociation of many ligand-receptor complexes and assists in sorting cargo molecules to different intracellular compartments. Ligands are often targeted to late endosomes/lysosomes for degradation, whereas most receptors are transported via recycling endosomes back to the cell surface for reutilization. Although the pathways of endocytosis are relatively well understood at the descriptive level, our knowledge of the molecular principles that regulate vesicular transport through EEs is still in its infancy.
The limiting membrane of EEs is enriched in phosphatidylinositol 3-phosphate [PI(3)P], whereas late endosomes and multivesicular bodies contain PI(3)P on the membrane of their internal vesicles (Fernandez-Borja et al., 1999; Gillooly et al., 2000). PI(3)P binds to peripheral membrane proteins containing a FYVE domain and may serve as docking site on EEs (Burd and Emr, 1998; Gaullier et al., 1998; Patki et al., 1998). The FYVE-finger consists of eight conserved cysteines at defined distances (C-X2-C-X12-C-X2-C-X4-C-X2-C-X16-C-X2-C) and folds into a so-called “cross-braced” topology in coordination with two Zn2+ ions (Schwabe and Klug, 1994; Stenmark et al., 1996). In addition, it contains a R(R/K)HHCRXCG motif encompassing the third and fourth cysteine (Stenmark and Aasland, 1999). Several FYVE-finger proteins, including EEA1 (Simonsen et al., 1998), rabenosyn-5 (Nielsen et al., 2000; de Renzis et al., 2002), and endofin (Seet and Hong, 2001) have been shown to be important for the orderly flow of membrane through EEs in mammalian cells.
Importantly, both EEA1 and rabenosyn-5 are effector molecules of the small GTPase rab5 that is associated with coated vesicles and EEs and required for homotypic EE fusion and heterotypic fusion of coated vesicles with EEs. rab5 is essential for the binding of EEA1 to EEs, and the combinatorial association of EEA1 with both PI(3)P and rab5 causes its highly restricted intracellular location to EEs (Gaullier et al., 1998; Simonsen et al., 1998). Because EEA1 functionally associates with the endosomal SNARE protein syntaxin 13 in an oligomeric protein complex (McBride et al., 1999), it is thought that rab5 in its GTP-bound form recruits cytoplasmic effector proteins and membrane-bound SNAREs to form a microdomain on EEs involved in tethering and fusion (Sönnichsen et al., 2000). rabenosyn-5 is also involved in biosynthetic transport of cathepsin D to lysosomes and linked to an unknown soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex through an interaction with the syntaxin binding protein hVps45p (Nielsen et al., 2000). Other FYVE-finger proteins are involved in signal transduction pathways at the cell surface and EEs. For instance, SARA is required for the recruitment of the effector molecules Smad2 and Smad3 to the activated transforming growth factor-β receptor on the cell surface and EEs (Tsukazaki et al., 1998; Panopoulou et al., 2002), and the localization of SARA to EEs is essential for nuclear translocation of Smad2 (Hayes et al., 2002). Smad2 also binds to the FYVE domain protein Hrs (Urbe et al., 2000) that together with SARA, stimulates activin receptor-mediated signaling (Miura et al., 2000). Because Hrs is a key component in sorting of ubiquitinated cell surface receptors in EEs (Raiborg et al., 2001, 2002; Bache et al., 2003), it is clear that FYVE proteins may serve as critical determinants in integrating membrane transport and signal transduction.
The La protein is an evolutionarily conserved RNA-binding phosphoprotein, originally identified as an autoantigen in patients suffering from the autoimmune diseases Sjögren's syndrome and systemic lupus erythematosus (Pruijn, 1994; Maraia and Intine, 2001). La has been implicated in RNA polymerase III transcription and internal initiation of translation of certain viral RNAs. Its best documented activity is the binding to and stabilization of newly synthesized RNA polymerase III transcripts, and La has been proposed to act as an RNA chaperone (Maraia and Intine, 2001). To further explore the function of La, we have used the monoclonal antibody (mAb) SW5, which recognizes a conformational epitope in the RNP domain of La (Pruijn et al., 1995). We here describe the characterization of a protein that cross-reacted with SW5. This protein, previously termed EEA2 (Fouraux et al., 2002a) and now renamed rabip4′, is a novel FYVE-finger domain protein that is functionally associated with EEs and may serve as a novel effector of rab5 and rab4.
MATERIALS AND METHODS
Antibodies
The monoclonal antibodies SW5 and SW3 against human La (Pruijn et al., 1995) and the rabbit antibodies raised against LAMP-1 (Harter and Mellman, 1992), GSTrab4 (10503) (Bottger et al., 1996), and a rab4 peptide (8091) (van der Sluijs et al., 1992a) have been described. The mouse monoclonal antibodies against the VSV-G and hemagglutinin (HA) epitope tags were purchased from Roche Diagnostics (Almere, The Netherlands), and against the FLAG from Sigma Chemie (Dordrecht, The Netherlands). The sheep antibody against TGN46 was from Serotec (Oxford, United Kingdom), and the goat antibody against rabaptin5 was from Santa Cruz Biotechnology (Santa Cruz, CA). The rabbit antibody against EEA1 and mouse mAb 4F11 against rab5 were generously provided by Silvie Urbe (Physiological Laboratory, Liverpool, United Kingdom) and Angela Wandinger-Ness (Department of Pathology, University of New Mexico, Albuquerque NM). rabip4′ antibodies were generated in rabbits by immunization with KLH-conjugated peptides corresponding to aa 93-107 and 302-316 (SN569, SN570), or with GSTrabip4′ (aa 201-400) (#289). Horseradish peroxidase-labeled secondary antibodies were from Dako Immunoglobulins (Glostrup, Denmark). Fluorescently labeled reagents were from Molecular Probes (Leiden, The Netherlands).
Cell Culture and Transfection
HEp-2 cells and HeLa cells were maintained in DMEM containing 10% heat-inactivated fetal calf serum, penicillin, and streptomycin. HeLa cells were transfected using FuGENE6 (Roche Diagnostics) according to the manufacturer's protocol. Expression from cytomegalovirus-driven constructs was induced with 5 mM sodium butyrate, and cells were used for experiments 18-24 h after transfection.
Subcellular Fractionation
S100 extracts were prepared from HeLa cells as described previously (Fouraux et al., 2002a). HEp-2 cells were washed with ice-cold phosphate-buffered saline, resuspended in 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and incubated on ice for 10 min. Cells were dounced using a type B pestle, and nuclei and unbroken cells were removed by centrifugation at 600 × g. The postnuclear supernatant was centrifuged at 10,000 × g to pellet mitochondria. Finally, the postmitochondrial supernatant was centrifuged at 100,000 × g to generate membrane and cytosol fractions. Pellets were solubilized in the same buffer supplemented with 1% SDS. Mitochondria and membrane fractions were resuspended in 20% of the volume of the cytosol and nuclear fractions.
Isolation and Cloning of rabip4′
Preparative immunoprecipitations from HeLa and Q-TOF mass spectrometry were done as described previously (Raymackers et al., 2000; Fouraux et al., 2002b). Peptide sequences were used to identify expressed sequence tags (ESTs), which enabled us to assemble a contig comprising ∼88% of full-length rabip4′. Screening of the human genome database with this contig led to the identification of a sequence that potentially corresponded to the N-terminal part of rabip4′. This sequence was corroborated by the database deposition of an expressed sequence tag (accession no. BE745186) that was identical to the sequence derived from the genomic library. The complete N-terminal part (aa 1-87) was cloned from genomic HeLa cell DNA by polymerase chain reaction (PCR) by using the Advantage-GC cDNA kit (BD Biosciences Clontech, Leusden, The Netherlands). The remaining part of the coding sequence (aa 87-708) was isolated by PCR from a human placental cDNA library and fused to the N-terminal part encoding aa 1-87 to generate a full-length rabip4′ cDNA (GenBank accession no. AF312367).
Expression Constructs
Constructs encoding N-terminally or C-terminally VSV-G-tagged rabip4′ and truncation mutants, N-RUN (aa 1-290), N-RUN-CC1 (aa 1-378), NRUN-CC12 (aa 1-514), LZ1-CC1 (aa 205-378), LZ1-CC12 (aa 205-514), LZ1-CC13 (205-636), LZ1-CC13-FYVE (205-708), N-RUN-CC13 (aa 1-636), and CC13-FYVE (aa 291-708) rabip4′ were described previously (Fouraux et al., 2002a). CC13-FYVE was also ligated into pGEX for recombinant protein production. rab4, rab5, and rab11 plasmids were described previously (Roberts et al., 2001; Deneka et al., 2003). rab7-pRP269 was generously provided by Jacques Neefjes (Dutch Cancer Institute, Amsterdam, The Netherlands), and rab21pGEX and rab22b-pGEX by Jack Fransen (Department of Cell Biology, Nijmegen University, Nijmegen, The Netherlands). cDNAs encoding rab15, rab17, and rab27 were provided by Lisa Elferink (Department of Biological Sciences, Wayne State University, Detroit, MI), Walter Hunziker (Institute of Biochemistry, University of Lausanne, Lausanne, Switzerland), and Bill Gahl (National Institutes of Health, Bethesda, MD), respectively, and were cloned in pGEX plasmids. FLAG-tagged RUFY1-pCMV and a plasmid containing the isolated FYVE domain of RUFY1 domain with an HA epitope tag, HA-RUFY1-FYVE-pCMV, were from Yun Qiu (Department of Pharmacology, University of Minnesota, Minneapolis, MN). Because the FYVE domain of RUFY1 is identical to that of rabip4′, we named this construct HA-rabip4′-FYVE-pCMV. The FENS-1-FYVE-pEGFP construct was provided by Phil Hawkins (Inositide Laboratory, The Babraham Institute, Cambridge, United Kingdom).
Binding Assays with35S-Labeled rabip4′ Constructs
GSTrab fusion proteins were expressed in Escherichia coli BL21(DE3), immobilized on glutathione (GSH)-Sepharose beads and charged with guanosine 5′-O-(3-thio)triphosphate (GTPγS) or GDP as described previously (Christoforidis et al., 1999).35S-Labeled rabip4′ constructs were produced by in vitro transcription-translation and incubated overnight at 4°C with the beads. Beads were washed three times at 4°C with 50 mM HEPES, pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol, 0.05% Tween 20, 1 mM GTPγS or GDP (Nielsen et al., 2000). Bound material was eluted with 25 mM reduced glutathione, resolved by SDS-PAGE on 10% SDS-PAA gels, and analyzed by phosphorimaging on a STORM 860. For mapping the binding domains on rabip4′, equal amounts of radiolabeled in vitro translated rabip4′ truncation mutants were used as input.
Binding Assays with Ternary Complexes
GST, GSTrabip4′ (CC13-FYVE), GSTrab4, and GSTrab5 were expressed in E. coli BL21 and immobilized on GSH-Sepharose beads. GSTrab4 and GSTrab5 were loaded with GTPγS as described previously (Christoforidis et al., 1999). Fusion proteins were cleaved with thrombin (Sigma, Dordrecht, The Netherlands) for 2 h at room temperature, unless indicated otherwise. Thrombin was then inactivated by 1 mM benzamidine, and released proteins were incubated twice with 250 μl of GSH beads for 1 h at 4°C to remove GST and noncleaved fusion protein. Twenty microliters of beads containing ∼200 μg of GSTrab5-GTPγS or GST were incubated for 2 h at 4°C with 100 μg of rabip4′ in buffer A containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), and 10 μM GTPγS in presence of 10 mg/ml bovine serum albumin (BSA). After washes with buffer A, beads were further incubated for 2 h at 4°C with 500 μg of rab4-GTPγS in buffer A containing 10 mg/ml BSA. Eluted proteins were separated by SDS-PAGE and analyzed by Western blotting. For competition experiments, 20 μl of beads containing ∼50 μg of GSTrabip4′ or GST was incubated for 2 h at 4°C with constant amounts of rab5-GTPγS in buffer A containing 10 mg/ml BSA, washed as described above, and incubated for another 2 h at 4°C with increasing amounts of rab4-GTPγS in buffer A supplemented with 10 mg/ml BSA. Beads were washed twice with 500 μl of buffer A; once with 500 μl of buffer B containing 20 mM HEPES, 250 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 10 μM GTPγS, pH 7.5; and once with 500 μl of buffer C containing 20 mM HEPES, 250 mM NaCl, and 1 mM DTT, pH 7.5. Bound proteins were eluted with buffer D containing 20 mM HEPES, 1.5 M NaCl, 20 mM EDTA, 1 mM DTT, and 5 mM GDP, pH 7.5. Eluted proteins were separated by SDS-PAGE and analyzed by Western blotting.
Internalization and Recycling of Alexa-labeled Transferrin (Tf)
HeLa cells grown on 10 mm round coverslips were incubated for 30 min in serum-free DMEM. For steady-state internalization experiments, cells were washed once with uptake medium (DMEM, 20 mM HEPES, pH 7.4, 0.5% BSA) and incubated for 30 min with 25 μg/ml Alexa594-Tf at 37°C. The cells were next fixed with 3% paraformaldehyde for 15 min at 37°C, followed by 30 min of 3% paraformaldehyde at room temperature. For pulse chase experiments, cells were washed with uptake medium and incubated with 25 μg/ml Alexa594-Tf for 30 min at 16°C. After one wash with DMEM, 20 mM HEPES, pH 7.4, 0.5% BSA, the cells were reincubated at 37°C with prewarmed medium (DMEM, 20 mM HEPES, pH 7.4, 0.5% BSA, 50 μM Desferal) for different periods of time and then fixed with 2.5% paraformaldehyde. Fixed cells were subsequently subjected to immunofluorescence microscopy as described below.
Miscellaneous Methods
SDS-PAGE, Western blotting, and confocal immunofluorescence microscopy was done as described previously (Sprong et al., 2001). The predicted protein sequence of rabip4′ was analyzed using COILS and SMART software (Lupas, 1996; Schultz et al., 1998). Gel filtration of a HeLa S100 extract was done on a Superdex 200 HR 10/30 column (Pharmacia, Roosendaal, The Netherlands), calibrated with thyroglobulin (670 kDa), bovine gammaglobulin (158 kDa), and chicken ovalbumin (44 kDa). Chromatography was performed at a flow rate of 0.5 ml/min in 25 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM dithioerythritol, 1 mM phenylmethylsulfonyl fluoride, and fractions were analyzed by Western blotting. Fusion proteins were expressed at 30°C in E. coli BL21(DE3) and retrieved on GSH beads. In vitro transcription translation reactions were done in the presence of [35S]methionine by using the TNT T7 Quick Coupled Reticulocyte Lysate system (Promega, Leiden, The Netherlands) as described previously (Bottger et al., 1996).
RESULTS
Identification and Cloning of rabip4′
To identify proteins interacting with La, we immunoprecipitated La from HeLa S100 extracts with the SW5 antibody. One of the coprecipitated proteins had an apparent molecular weight of ∼80 kDa (Fouraux et al., 2002a). We renamed this protein rabip4′ for reasons documented below. To identify the protein, we excised the 80-kDa band, incubated it with trypsin, and subjected the digest to tandem Q-TOF mass spectrometry. As shown in Figure 1A, this yielded several peptide sequences, none of which, however, allowed us to identify the protein.
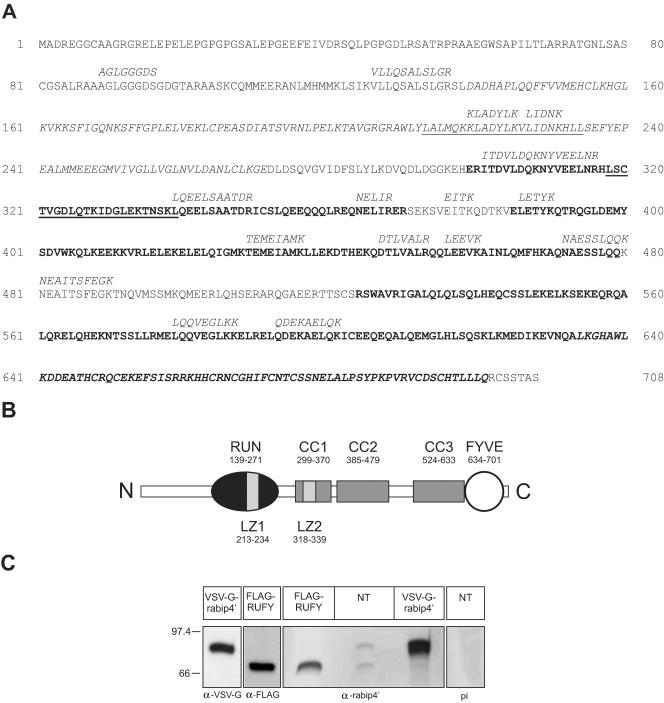
Figure 1.
(A) Sequence and domain structure of rabip4′. Deduced amino acid sequence of rabip4′. Peptide sequences obtained by mass spectrometry are depicted in italics above the amino acid sequence. Leucine zipper motifs are underlined, the RUN motif is in italics, predicted coiled-coil domains are in bold, and the FYVE-finger domain is in bold italics. (B) Schematic structure of rabip4′. Coiled-coil domains are indicated with CC1, CC2, and CC3, the FYVE-finger domain with FYVE, leucine zippers with LZ1 and LZ2, and the RUN motif by RUN. (C) A Western blot of lysate from nontransfected HeLa cells (NT) was probed with an antibody against rabip4′(aa201-400) that detects 2 bands at 69 kDa and 80 kDa that were not seen by the preimmune (pi) serum. As positive controls, we used lysates of HeLa cells that were transfected with either VSV-G-rabip4′ or FLAG-RUFY1 and detected with epitope tag antibodies.
BLAST searches yielded six human ESTs (accession nos. AI652188, AW978202, BE000558, AA324714, AW957168, and AA608559), each encoding a number of the peptides. Based on those, we assembled a contig comprising sequences encoding the identified peptides. A poly-A signal (AATAAA) was found near the 3′ end, but the 5′ end was not complete because the sequence encoding peptide AGLGGGDS (Figure 1A) was present in the contig upstream of the first in-frame methionine codon. Moreover, all ESTs of this region started with a NotI site, and NotI digestion had been performed to generate the clones encoding these ESTs. We screened the human genome database with this contig and obtained the complete 5′ end of the rabip4′ coding sequence (accession no. AC023559). This sequence has a high GC content (78%) in the 5′ region that may have precluded its synthesis by 5′ rapid amplification of cDNA ends analysis (our unpublished data). Based on these sequence data, we generated a full-length clone from a human placenta cDNA library and a genomic fragment isolated by PCR from HeLa cell DNA. The integrity of the 5′ region of the rabip4′ coding sequence was confirmed by a recent EST database entry (accession no. BE745186) comprising nucleotides 17-700. The full-length 2.63-kb rabip4′ clone contains an open reading frame of 2124 nucleotides, which encodes a polypeptide of 708 amino acids with a calculated molecular mass of 79.8 kDa (Figure 1A; GenBank accession no. AF312367). The 80-kDa protein is highly homologous to two recently identified proteins; the C-terminal 85% is virtually identical to human RUFY1 (Yang et al., 2002), and the same part shares 94% identity with mouse rabip4 (Cormont et al., 2001). In comparison with RUFY1 and rabip4, it contains a unique 108 aa N-terminal extension. Importantly, this region, which is most likely encoded by an alternative exon, contains peptide sequence AGLGGGDS that was identified by mass spectrometry of the 80-kDa band. For this reason, we named it rabip4′. Sequence analysis predicts that rabip4′ is a hydrophilic protein containing three regions that adopt a mainly α-helical conformation with heptad repeats characteristic of coiled-coil domains (CC1-CC3) (Lupas, 1996). Leucine zippers are present at amino acid positions 213-234 and 318-339, the second of which (LZ2) overlaps with the most N-terminal coiled-coil domain (CC1). rabip4′ contains a RUN domain in the N-terminal region (aa 139-271), thus encompassing LZ1, and a FYVE domain close to the C terminus (aa 634-701) (Figure 1B). The RUN domain is a recently identified protein sequence of unknown function, which occurs in several proteins that interact with rab and rap GTPases (Callebaut et al., 2001). To investigate whether rabip4′ and RUFY1 are both present in human cells, we generated an antibody (#289) against rabip4′(aa201-400), a region that is conserved between the two proteins. On a Western blot of nontransfected HeLa cells, the antibody detected two bands of the same intensity at 80 kDa and 69 kDa that correspond to the size of rabip4′ and RUFY1, respectively (Figure 1C). They also comigrated with individually overexpressed VSV-G-tagged rabip4′ and FLAG-tagged RUFY1, which served as positive controls. Thus endogenous rabip4′ and RUFY1 are present in HeLa cells at similar levels.
The FYVE Domain Is Essential for Membrane Association of rabip4′
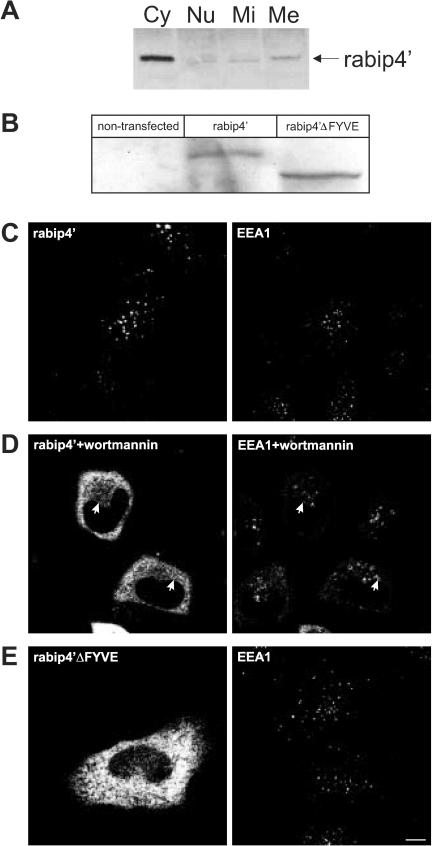
To assess the intracellular localization of rabip4′, we first fractionated HEp-2 cell lysates by differential centrifugation. The distribution of rabip4′ among nuclear, mitochondrial, microsomal, and cytosolic fractions was analyzed by Western blotting by using a peptide antibody against rabip4′ (SN569) (Fouraux et al., 2002a). As shown in Figure 2A, we found the majority of rabip4′ in the cytosolic fraction, whereas a significant amount was also present in the microsomal fraction. The nuclear and mitochondrial fractions contained little if any rabip4′. Because rabip4′ likely associates with membranes in a reversible manner, it may have dissociated in part from membranes during fractionation as was observed before for other proteins lacking a membrane anchor (Stenmark et al., 1995; Simonsen et al., 1998). To further explore membrane-association of rabip4′, we analyzed its distribution by confocal immunofluorescence microscopy. Because our antibodies did not allow reliable detection of endogenous rabip4′ in morphological assays, we transfected HeLa cells with rabip4′ constructs that were N-terminally tagged with the VSV-G epitope tag (Soldati and Perriard, 1991). To investigate whether the FYVE domain of rabip4′ is involved in its association with the cytoplasmic organelles, we transiently expressed the NRUN-CC13 rabip4′ mutant in which aa 636-708 were deleted. Western blots prepared from lysates of the transfectants showed that wild-type and mutant rabip4′ were expressed to the same level (Figure 2B).
Figure 2.
(A) The FYVE domain is required for membrane binding of rabip4′. HEp-2 cell lysates were fractionated as described in MATERIALS AND METHODS. Cytosolic (Cy), nuclear (Nu), mitochondrial (Mi), and microsomal (Me) fractions were analyzed by Western blotting by using a rabbit antibody against rabip4′. HeLa cells were transfected with VSV-G-tagged rabip4′ (B-D) or N-RUN-CC13 mutant (rabip4′ΔFYVE) lacking the FYVE domain (B and E). Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting with an antibody against the epitope tag. Both constructs are expressed at the same level (B). HeLa cells expressing VSVG-tagged rabip4′ (C) were incubated for 15 min at 37°C with 50 nM wortmannin (D). Cells were fixed 18-24 h after transfection, doubly labeled with mouse anti VSV-G and rabbit anti-EEA1, stained with Alexa488-labeled rabbit anti-mouse and Cy3-labeled goat anti-rabbit, and examined by fluorescence microscopy. Note extensive colocalization between rabip4′ and EEA1 in C. In wortmannin-treated cells, rabip4′ redistributed partially in the cytoplasm but also colocalized with EEA1 on enlarged endosomes (arrows in D). Double-label immunofluorescence of cells transfected with VSV-G-tagged N-RUN-CC13 and endogenous EEA1 shows that the construct lacking the FYVE domain is localized in the cytoplasm (E). Bar, 10 μm.
As shown in Figure 2C, rabip4′ was associated with discrete punctae scattered throughout the cytoplasm, which are reminiscent of endocytic organelles. The same localization was found with a construct in which the epitope tag was fused to the C-terminus (our unpublished data), documenting that the tag did not affect the distribution of rabip4′. In cells expressing high levels of rabip4′, part of the protein distributed into the cytoplasm, suggesting that membrane binding is saturable. Because the cytoplasmic structures containing VSV-G-tagged rabip4′ could be endocytic organelles, we performed double label immunofluorescence microscopy with EEA1 and found partial colocalization with this early endosome marker (Figure 2C). Expression of the rabip4′ truncation mutant lacking the FYVE domain revealed a cytoplasmic distribution (Figure 2E) suggesting that this domain is essential for localization to early endosomes. Preextraction of the cells with 0.05% saponin removed all rabip4′ from the cells (our unpublished data), confirming that the FYVE domain is required for binding of rabip4′ to membranes.
FYVE domains bind with moderate affinity to PI(3)P, a phosphorylated phosphatidylinositol derivative required for membrane association of EEA1(Lawe et al., 2002). We therefore next assessed whether PI(3)P might be important for membrane binding of rabip4′. HeLa cells transiently transfected with full-length rabip4′ were incubated for 15 min at 37°C with 50 nM wortmannin. The presence of the PI3-kinase inhibitor dramatically changed the distribution of rabip4′ from early endosomes to the cytoplasm (Figure 2D). Some rabip4′ remained associated with enlarged vacuolar structures that also labeled for EEA1, suggesting that another determinant in rabip4′ also contributes to its binding to early endosomes. The same effect of PI3-kinase inhibitors on EEA1 containing endosomes had been noted previously (van Dam and Stoorvogel, 2002) in HeLa cells. The results of this morphological assay thus show that rabip4′ is a peripheral membrane protein whose membrane association is dependent on a FYVE domain and perturbed by a PI3-kinase inhibitor. We attempted to confirm the effect of wortmannin in biochemical assays; however, whatever subcellular fractionation procedure and buffer we used, most of rabip4′ already distributed in the cytosol in the absence of wortmannin (our unpublished data).
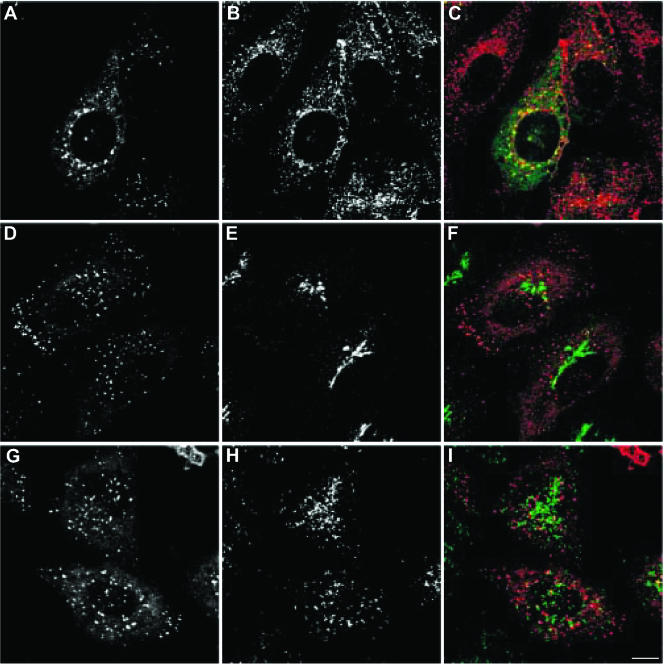
rabip4′ Is Associated with Early Endosomes
Because EEA1 distribution is restricted to early endosomes (Sönnichsen et al., 2000) and because some of the structures contained only EEA1, whereas others were positive for rabip4′ (Figure 2C), we also compared the localization of rabip4′ to other compartments of the transferrin receptor (TfR) pathway. For this purpose, Alexa594-Tf was internalized for 30 min in HeLa cells expressing VSV-G-tagged rabip4′. Most if not all of the endosomes that were decorated with the anti-VSV-G-tag antibody (Figure 3A) contained Alexa594-Tf, whereas a portion of Alexa594-Tf endosomes (Figure 3B) did not contain rabip4′ as evidenced by the merged images (Figure 3C). FYVE domain proteins do not exclusively localize to early endocytic compartments; for instance, PIKfyve is on late endocytic endosomes (Ikonomov et al., 2001), whereas Fgd1 and DFCP1 are on the Golgi complex (Estrada et al., 2001; Ridley et al., 2001). We therefore investigated whether rabip4′ was present in the trans-Golgi network, in double label experiments with antibodies against rabip4′ (Figure 3D) and the trans-Golgi network marker TGN46 (Figure 3E). As shown in the merged image (Figure 3F), rabip4′ and TGN46 did not colocalize. Double labeling experiments of rabip4′ (Figure 3G) and LAMP-1 (Figure 3H), a protein associated with late endosomes/lysosomes, showed very little overlap (Figure 3I). Together, these data indicate that the membranous structures to which rabip4′ binds are EEs.
Figure 3.
rabip4′ is associated with early endosomes. HeLa cells were transiently transfected with VSV-G-tagged rabip4′. To label the TfR pathway, cells were incubated for 30 min at 37°C with 25 μg/ml Alexa594-Tf (A-C). The cells were fixed and subjected to confocal immunofluorescence microscopy. rabip4′ was labeled with a mAb against the VSV-G tag and counterstained with Alexa488-labeled goat anti-mouse IgG (A and C) or Cy3-labeled goat anti-mouse IgG (D, F, G, and I). TGN46 detection was with a sheep antibody and Alexa488-goat anti-sheep (E and F), whereas LAMP-1 was detected with a rabbit antibody against its cytoplasmic tail and stained with Alexa488-goat anti-rabbit IgG (H and I). Merged images are shown in C, F, and I. Bar, 10 μm.
rabip4′ Regulates Transport through Early Endosomes
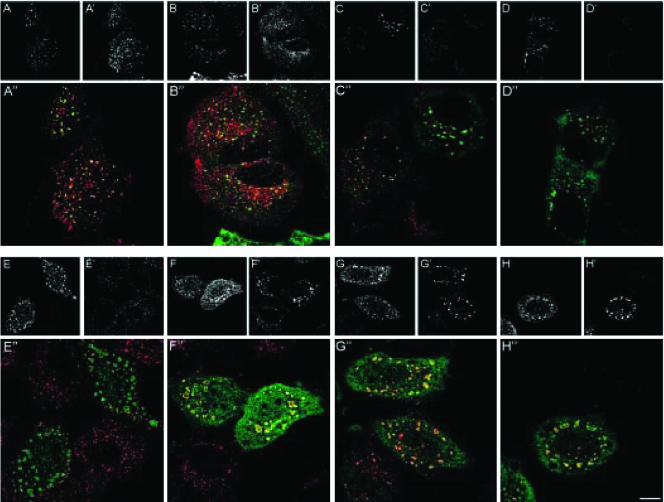
The extensive colocalization between rabip4′ and Tf in EEs and the absence of rabip4′ on the plasma membrane, suggested that it might function in a distal transport step of the TfR pathway. We therefore examined the effect of rabip4′ on transit of endocytosed Tf through EEs. HeLa cells were transiently transfected with wild-type rabip4′, or the CC13-FYVE rabip4′ mutant lacking the N-terminal aa 1-290, including the RUN domain (Figure 1B). Alexa594-Tf was internalized for 30 min at 16°C to accumulate the probe in EEs (Lim et al., 2001). Cells were then washed and chased at 37°C in the presence of Desferal to prevent reinternalization of recycled tracer. As shown in Figure 4E″, CC13-FYVE rabip4′ caused extensive swelling of EEs and inhibited entry of Alexa594-Tf in EEs at 16°C compared with cells expressing wild-type rabip4′ (Figure 4A″) and the nontransfected cells in Figure 4E″. During the first 10 min of chase, the tracer reached the swollen EEs in cells transfected with CC13-FYVE rabip4′, but exit of Alexa594-Tf from these structures was completely blocked during a 40-min chase (Figure 4F″-H″). In contrast, in nontransfected cells or wild-type rabip4′ transfectants, most if not all Alexa594-Tf was chased out of the cells after 40 min. Thus, CC13-FYVE rabip4′ seemed to act as an inhibitory mutant delaying entry into and inhibiting recycling from EEs.
Figure 4.
rabip4′ regulates Tf transport through early endosomes. HeLa cells were transfected with VSV-G-tagged rabip4′ (A-D) or the VSV-G-tagged CC13-FYVE rabip4′ mutant (E-H). Cells were labeled with mouse anti-VSV-G tag followed by Alexa488-goat anti-mouse IgG (A-H). Alexa594-Tf was taken up for 30 min at 16°C, washed, and chased at 37°C for 0 min (A′ and E′), 10 min (B′ and F′), 20 min (C′ and G′), and 40 min (D′ and H′) in the presence of 50 mM Desferal. The corresponding merged images are shown in A″-H″. Bar, 10 μm.
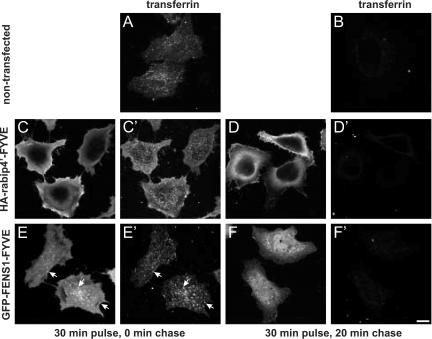
We next investigated whether the ectopically expressed rabip4′ FYVE domain affected endosome morphology and Tf recycling. To this aim, we transfected HeLa cells with the isolated FYVE domain (HA-rabip4′-FYVE) and assessed Tf recycling from endosomes. Nontransfected cells, and cells expressing the FYVE domain of the endosomal protein FENS1 (eGFP-FENS1-FYVE) served as controls. In contrast to FENS1-FYVE that was associated with early endosomes (Figure 5), rabip4′-FYVE localized to the cytoplasm and plasma membrane as was reported previously (Ridley et al., 2001; Yang et al., 2002). Neither construct altered the size of early endosomes or the kinetics of Tf recycling compared with nontransfected control cells (Figure 5). Using the same pulse-chase protocol as in Figure 4, we found that within 20 min of chase at 37°C, the entire pool of internalized tracer was cleared from the three cell lines. Thus, whereas some isolated FYVE domains contain sufficient information to be targeted to early endosomes as in case of EEA1 (Stenmark et al., 1996) and FENS-1 (Ridley et al., 2001), others, for instance, those of Hrs (Raiborg et al., 2001) and rabip4′ require additional sequence determinants. A common feature of the mentioned FYVE domains is that their overexpression does not affect the size of early endosomes, nor membrane recycling.
Figure 5.
rabip4′ FYVE domain does not affect endosome morphology and Tf recycling. Pulse-chase experiments in control HeLa cells (A and B) or HeLa cells transfected with HA-rabip4′-FYVE (C-D′) or GFP-FENS1-FYVE (E-F′). Cells expressing HA-rabip4′-FYVE were labeled with mouse anti-HA tag followed by Alexa488-goat anti-mouse IgG (C and D), whereas expression of GFP-FENS1-FYVE was visualized by the GFP signal (E and F). Alexa594-Tf was internalized for 30 min at 16°C, washed, and chased at 37°C for 0 min (A, C′, and E′), and 20 min (B, D′, and F′) in the presence of 50 mM Desferal. Arrows denote colocalization of Alexa594-Tf and GFP-FENS1-FYVE. Bar,10 μm.
rabip4′ Directly Interacts with the GTP-bound Forms of rab4 and rab5
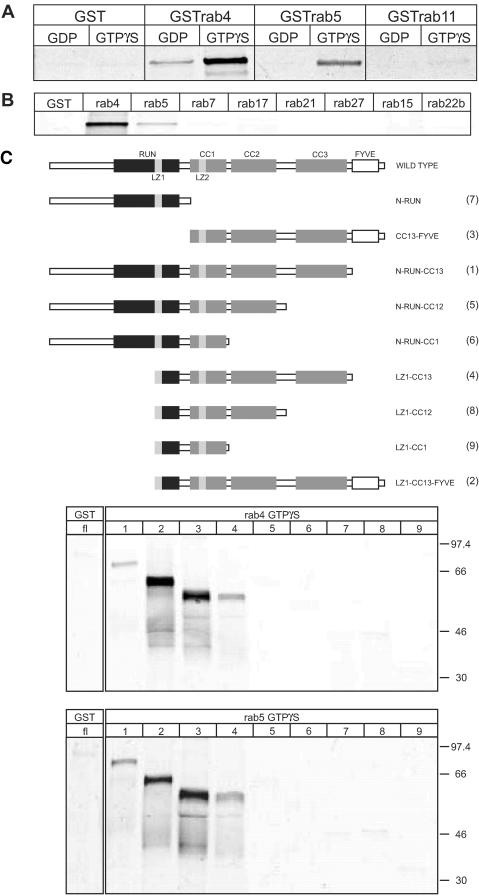
The phenotypic effects of the CC13-FYVE mutant on TfR kinetics suggested a cooperative role of rabip4′ and the monomeric GTPases rab5, rab4, and rab11. rab5 is known to control vesicular transport to EEs (Bucci et al., 1992), whereas rab4 and rab11 regulate recycling from this compartment (van der Sluijs et al., 1992b; Ullrich et al., 1996). We therefore investigated whether rabip4′ directly interacted with these rab proteins by using a GST pull-down assay (Nielsen et al., 2000). Bacterially expressed GSTrab4, GSTrab5, and GSTrab11 were bound to GSH beads. The proteins were then charged with either GTPγS or GDP and incubated with 35S-labeled VSV-G-tagged rabip4′, produced in an in vitro transcription-translation reaction. As shown in Figure 6A, rabip4′ directly and preferentially bound to the active form (>20 times the GDP form) of rab4 and rab5, whereas rab5 seemed to bind 2-3 times less rabip4′ than rab4. The interaction was specific for rab4 and rab5 because rabip4′ did not bind to rab11. To further determine the specificity of the interaction, we tested a number of rab proteins that localize to endocytic compartments (rab7, rab15, rab17, and rab22b) the Golgi complex (rab21) or other organelles (rab27). None of these, however, bound to rabip4′ (Figure 6B). Given that rab4 and rab5 bound to rabip4′, we next investigated whether their binding sites map to the same or distinct domains on rabip4′. To this aim, we generated a series of VSV-G-tagged rabip4′ deletion mutants (Figure 6C) and tested their ability to bind to guanine nucleotide-loaded GSTrab5 or GSTrab4 in the GST pull-down assay. As shown in Figure 6C, deletion of CC3 abolished binding to both rab4 and rab5. Truncations that retained the ability to bind rab5 or rab4 did so in a GTP-dependent manner and none bound to GDP-charged rab4 and rab5 (our unpublished data).
Figure 6.
rab4 and rab5 bind directly to rabip4′. (A) GST, GSTrab4, GSTrab5, and GSTrab11 were loaded with GTPγS or GDP and incubated overnight with35S-labeled VSV-G-tagged rabip4′. Beads were washed three times, and proteins were eluted with 25 mM reduced glutathione. Eluates were resolved on 10% SDS-PAA gels and analyzed by phosphorimaging. (B) GST, GSTrab4, GSTrab5, GSTrab7, GSTrab15, GSTrab17, GSTrab21, GSTrab22b, and GSTrab27 were loaded with GTPγS and incubated with 35S-labeled VSV-G-tagged rabip4′ as described in A. rabip4′ was eluted with glutathione and analyzed by SDS-PAGE and phosphorimaging. (C) Schematic representation of rabip4′ truncation mutants. See legend to Figure 1B for domain nomenclature. GSTrab4 and GSTrab5 were loaded with GTPγS. Binding and analysis of 35S-labeled VSV-G-tagged rabip4′ (fl) and truncations N-RUN-CC13 (1), LZ1-CC13-FYVE (2), CC13-FYVE (3), LZ1-CC13 (4), NRUN-CC12 (5), N-RUN-CC1 (6), N-RUN (7), LZ1-CC12 (8), and LZ1-CC1 (9) was done as described in MATERIALS AND METHODS.
rab4 and rab5 Bind Simultaneously to rabip4′
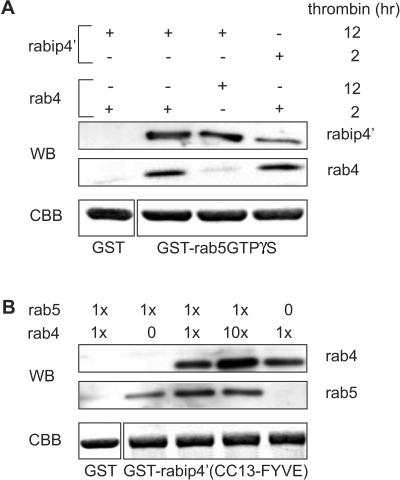
These results showed that amino acids comprising CC3 are required for binding to rab4 and rab5. We then determined whether rab4 and rab5 bound simultaneously or competitively to rabip4′. To this aim, we immobilized GSTrab5 on GSH beads, charged it with GTPγS, and added rabip4′ (from which the GST moiety had been removed by thrombin) for 2 h at 4°C. The beads were then washed and further incubated at 4°C with rab4-GTPγS (isolated from GSTrab4-GTPγS that had been incubated for 2 h with thrombin). After 2 h, beads were washed, and bound proteins were eluted with EDTA and high salt. This buffer dissociates rab effector complexes because EDTA chelates away Mg2+ ions that are required for binding of guanine nucleotide (Christoforidis et al., 1999). As shown in Figure 7A, rab4 (and rabip4′) were bound on beads that contained GSTrab5, but not on control beads that contained GST. If GSTrab4GTPγS was treated for 12 h with thrombin and then used in the binding assay, it did not bind to the GSTrab5-rabip4′ complex. The extended incubation with thrombin inactivated rab4 (our unpublished data), and prevented binding of rab4 to rabip4′, suggesting that formation of a ternary complex was specific and required the active forms of the two rab proteins. To confirm these results, we performed the assay in a complementary format in which GSTrabip4′ was immobilized on beads that were subsequently incubated with a saturating amount of rab5GTPγS and up to 10-fold more rab4GTPγS. The total amount of rab proteins in the assay was between 2 and 10 times more than rabip4′. At the end of the incubation, we analyzed the rab proteins bound to the beads by Western blotting. As shown in Figure 7B, a 10-fold molar excess of rab4 did not compete off rab5. These results are consistent with the data in Figure 7A, thus rab4 and rab5 can bind independently to rabip4′.
Figure 7.
rab4 and rab5 bind simultaneously to rabip4′. (A) GSTrab5 (loaded with GTPγS) and GST were immobilized on GSH beads and incubated with rabip4′ (CC13-FYVE). After washing, a second incubation was performed with rab4-GTPγS at a molar ratio rab5:rab4 of 5. Bound proteins were eluted, resolved by SDS-PAGE, and analyzed by Western blotting by using antibodies against rabip4′ (SN570) and rab4 (8091). Then, 0.5% of GSTrab5 and GST was loaded on a SDS-PAA gel and stained with Coomassie Brilliant Blue (CBB) to control for input in the assays. rab4 bound to the GSTrab5 beads in the presence rabip4′, but not to GST beads when these were incubated with rabip4′. Extending the thrombin cleavage time of GSTrab4 from 2 to 12 h inactivates GTPγS-loaded rab4 and reduced rab4-binding to background level. (B) GSTrabip4′(CC13-FYVE) and GST were immobilized on GSH beads and first incubated with rab5-GTPγS. After washing, increasing amounts of rab4-GTPγS (x stands for 50 μg) were added and beads were further incubated. Bound proteins were eluted, resolved by SDS-PAGE and analyzed by Western blotting with antibodies against rab4 (8091) and rab5 (4F11). CBB staining shows that similar amounts of GSTrabip4′ (CC13-FYVE) and GST have been used in each reaction (1% of the input). Note that addition of a 10-fold molar excess of rab4 does not compete out bound rab5.
rabip4′ Colocalizes with rab4 and rab5
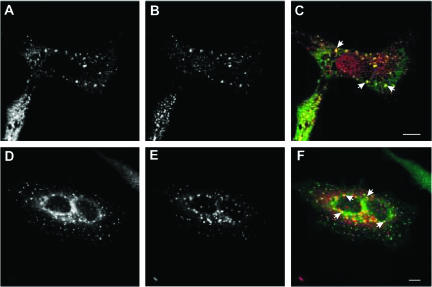
If rabip4′ binds to rab4 and rab5 in vivo, it should colocalize at least in part with rab4 and rab5. To investigate this, we cotransfected cells with VSV-G-rabip4′ and rab4 or GFP-rab5 and analyzed their distributions by confocal fluorescence microscopy. As shown in Figure 8, we found a nearly complete colocalization of rabip4′ (Figure 8A) and rab4 (Figure 8B) on EEs that were enlarged with respect to those seen in Figure 3. rabip4′ (Figure 8D) also colocalized with GFP-rab5 (Figure 8E) as shown in the merged image in Figure 8F. rab5, in contrast to rab4, however, less extensively colocalized with rabip4′, as might be expected from the partial colocalization of EEA1 and rabip4′ (Figure 3) and the reported codistribution of rab5 and EEA1 (Sönnichsen et al., 2000). This suggested that rabip4′ might bind both rab5 and rab4 in vivo, which is supported by the ability of rabip4′ and the two GTPases to cause morphological alterations to EEs, that are similar to those seen after overexpression of rab4 and rab5 and effectors (Vitale et al., 1998; Nagelkerken et al., 2000; de Renzis et al., 2002). Thus, rabip4′ not only colocalized with rab4 and rab5 but also directly interacted with the active forms of both proteins.
Figure 8.
rab4 and rab5 colocalize with rabip4′. HeLa cells expressing VSV-G-rabip4′ and rab4 (A-C), or VSV-G-rabip4′ and EGFP-rab5 (D-F) were labeled with a mAb against the VSV-G tag and counterstained with either Alexa488-goat anti-mouse IgG (A) or Alexa594-goat anti-mouse (D). rab4 was labeled with a rabbit antibody and stained with Cy3-goat anti-rabbit IgG (B), whereas rab5 was detected by EGFP fluorescence (E). Merged images are shown in C and F, and colocalization is indicated by arrows. Bar, 10 μm.
rabip4′ Is Associated with a Large Cytosolic Protein Complex
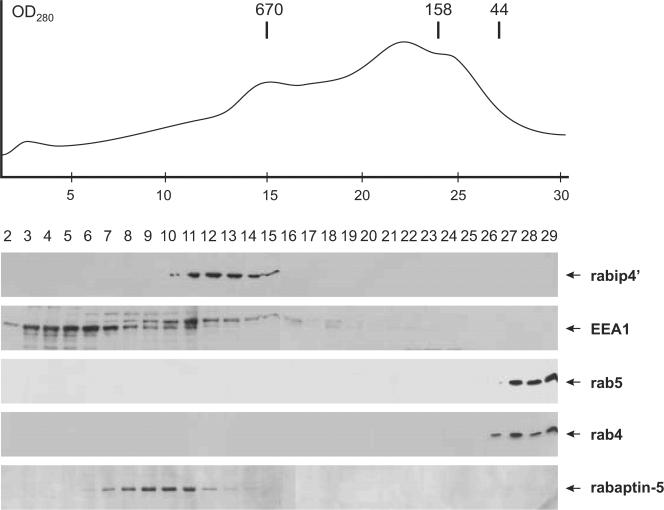
Recent evidence shows that rab proteins act through the local assembly of highly specific effector complexes that contain a myriad of proteins, of which some have been identified so far. For instance, at least 26 cytosolic proteins could be isolated on a rab5GTP affinity column, including rabenosyn-5, EEA1, and rabaptin5 (Christoforidis et al., 1999). Having found that rabip4′ directly binds to rab4 and rab5, we were interested to know whether endogenous rabip4′ was present as a monomer or within a protein complex in the cytoplasm. To this aim, we fractionated a HeLa S100 extract by gel filtration on a Superdex 200 HR column. The majority of rabip4′ eluted in fractions corresponding to a molecular mass >670 kDa (Figure 9). On overexposure of the Western blot, we detected trace amounts of rabip4′ in the fractions corresponding to its monomeric state (our unpublished data). The rabip4′ complex (or complexes) are devoid of nucleic acids because we did not find RNA or DNA in immunoprecipitations of [32P]orthophosphate-labeled cells by using SN569 antibody against-rabip4′ (our unpublished data). We also analyzed the column fractions for the presence of EEA1, the majority of which eluted in fractions 3-7, showing no overlap with rabip4′. rabaptin5, an effector of rab4 and rab5, eluted in fractions between the rabip4′ and EEA1 peaks, whereas rab5 and rab4 eluted from the column after the 44-kDa marker. These data demonstrate that the majority of cytosolic rabip4′ is associated with a relatively large protein complex that is distinct from complexes containing other rab5 effectors.
Figure 9.
Cytosolic rabip4′ is associated with a large protein complex. A HeLa S100 extract was fractionated by Superdex 200 HR 10/30 gel filtration chromatography as described in MATERIALS AND METHODS. The position of marker proteins is indicated above the elution profile. Column fractions were analyzed by Western blotting with antibodies against rabip4′, EEA1, rab5, rab4, and rabaptin5. The positions of the various proteins are indicated with arrows.
DISCUSSION
We here describe the identification of rabip4′, a novel 80-kDa protein colocalizing with endocytosed transferrin, rab4, and albeit to a lesser extent with EEA1, and rab5 on EEs. We isolated rabip4′ from HeLa cells with the high-affinity SW5 mAb against La. The molecular basis for the interaction between SW5 and rabip4′ likely involves charged amino acid residues that adopt a α-helical coil in the SW5 epitope present in both La and rabip4′ (Fouraux et al., 2002a). The primary structure of rabip4′ displays some homology to EEA1, particularly in the FYVE-finger domain. rabip4′, however, is distinct from EEA1. First, rabip4′ contains an N-terminal RUN domain and has a significantly lower coiled-coil content than EEA1. Second, both rab4 and rab5 bind to rabip4′, a property that is not shared by EEA1. These features suggest that rabip4′ is an effector of rab5 and rab4 and relies on the presence of a FYVE-domain to localize to EEs.
During the preparation of this manuscript, Yang et al. reported the identification of RUFY1, a human protein interacting with Etk that serves as a substrate of this tyrosine kinase (Yang et al., 2002), whereas Cormont et al. identified rabip4, a murine rab4 effector (Cormont et al., 2001; Mari et al., 2001). RUFY1 and rabip4 are nearly identical (95%) and possibly represent orthologs, although their tissue distribution does not seem to be identical. Sequence comparison of rabip4′ with RUFY1 and rabip4 revealed extensive homology except for the N-terminal 108 aa of rabip4′, which are not present in RUFY1 and rabip4. BLAST searches with this region of rabip4′ revealed that the mouse genome encodes a long variant of rabip4 with an N-terminal extension of 112 aa (accession no. BAC32815) that is highly homologous to the N terminus of rabip4′. There are two more variants in mouse with the N terminal extension (BAC34548 and BAC32811). However, these contain C-terminal truncations, lack the FYVE domain and encode shorter proteins than rabip4. Because there is only one gene encoding these polypeptides, this shows that at least two isoforms are expressed, a short one, RUFY1/rabip4, and a long version represented by rabip4′/BAC 32815. The results from a recent EST database search strongly suggest that the long and short isoforms result from transcription initiation at alternative promoters in the rabip4′/RUFY1 gene (our unpublished data). Because the N-terminal 108 aa of rabip4′ does not constitute known protein motifs, it is not immediately predictable in what respect this region might contribute to the functional properties of the protein. We do note, however, that this part of rabip4′ lies directly adjacent to the RUN domain, a conserved element found in many proteins that are known to functionally associate with GTPases of the rab and rap subfamilies (Callebaut et al., 2001). Importantly, rabip4′ binds to both rab4 and rab5 and is involved in transport steps that are regulated by rab5 and rab4, whereas rabip4 reportedly does not interact with rab5 (Cormont et al., 2001).
Analysis of the rabip4′ N-RUN-CC13 deletion mutant showed that the FYVE-finger domain plays an essential role in the binding of rabip4′ to early endosomal membranes. Low concentrations of wortmannin selectively inhibit PI3-kinase and resulted in the redistribution of rabip4′ from EEs to the cytoplasm, suggesting that PI(3)P is required for the early endosomal association of rabip4′. Currently, it is not known whether other proteins are involved in the association of rabip4′ with EEs, as is the case for rabenosyn-5 (Nielsen et al., 2000) and EEA1 (Simonsen et al., 1998). Endosome association of these rab5 effectors also involves binding to rab5-GTP, which in EEA1 interacts with a region comprising the FYVE domain and flanking amino acids (Lawe et al., 2000), and dimerization (Dumas et al., 2001). Aligning this region of EEA1 with rabip4′ revealed a homologous element immediately upstream (aa 562-647) of the FYVE-domain in rabip4′ with 31% identity and 42% similarity. Interestingly, this part of rabip4′ is 89% identical to the rab4 binding domain of rabip4 (Cormont et al., 2001) and contains the rab4 and rab5 binding domain in CC3 (Figure 6C). Because N-RUN-CC13 rabip4′ (Figure 2E) is localized in the cytoplasm, it is clear that binding of rabip4′ to rab4 and/or rab5 is not sufficient for the association with EEs. Because PI(3)P is not exclusively localized to EEs but also occurs in late endocytic compartments as well as in the nucleus (Gillooly et al., 2000), we inferred that rab4 and/or rab5 binding and the FYVE domain are minimally required for endosomal recruitment of rabip4′.
The in vitro binding studies showed that rab5 and rab4 in their GTP-bound form directly interact with rabip4′. The interaction with the two GTPases was specific because none of the other rab proteins that we tested bound to rabip4′, including rab11, which is also functionally associated with compartments of the TfR pathway (Ullrich et al., 1996; Wilcke et al., 2000). These interactions suggest that rabip4′ may coordinate rab4 and rab5 function in regulating transport through EEs. This idea is supported by several of our findings. First, rabip4′ is associated with EEs that contain rab4 and rab5. Second, the CC13-FYVE rabip4′ mutant acts as an inhibitor of Tf transport to or entry into EEs, a step that is typically controlled by rab5. Third, CC13-FYVE rabip4′ generated enlarged EEs, a phenotype also arising from the expression of rab5 and its effector proteins. Fourth, CC13-FYVE rabip4′ inhibits the exit or recycling of Tf from EEs, an event that is regulated by rab4.
What would be the purpose of a protein that binds two endosomal GTPases? Previous studies have shown that rab5, rab4, and rab11 containing endosomes are reached in this order by endocytosed Tf (Trischler et al., 1999). Triple label fluorescence microscopy of Zerial's group confirmed these biochemical data (Sönnichsen et al., 2000) and also showed that pairs of these sequentially acting rab proteins had distinct and partially overlapping distributions. It is thought that rab proteins may form domains in the lateral plane of a membrane by recruiting effectors and other interacting molecules to control a specific membrane transport event (McBride et al., 1999). Extrapolation of this hypothesis suggests that the morphological and biochemical heterogeneity of early endocytic organelles can be materialized in terms of domains that are formed by rab5, rab4, and rab11 (Sönnichsen et al., 2000). The spatial coordination of rab5-dependent entry and rab4-mediated exit of Tf can then simply be brought about by connecting the rab5 and rab4 domains via bivalent effector molecules that have the ability to bind both rab5 and rab4. Coupling of the domains might be strengthened by the cooperative activities of bivalent effectors such as rabenosyn-5, rabaptins, or rabip4′. In this respect, rabip4′ is more similar to Rab coupling protein, which binds to rab4 and rab11 in a short C-terminal domain (Lindsay et al., 2002), than to the rabaptins in which the binding sites are separated by a long interspacing sequence (Vitale et al., 1998).
An alternative model for rab5- and rab4-dependent binding in rabip4′ function that is consistent with our data is suggested by findings of others on RUFY1. This rabip4′ homolog binds to the tyrosine kinase Etk in a phosphorylation-dependent manner (Yang et al., 2002). Etk-mediated phosphorylation occurs on two tyrosine residues that are completely conserved between RUFY1, rabip4, rabip4′, and BAC32815. Interestingly, the RUFY1-Etk complex regulates epidermal growth factor-induced internalization of the epidermal growth factor receptor (Yang et al., 2002), which is a rab5-dependent event (Tall et al., 2001). Possibly members of the RUFY1, rabip4, rabip4′, and BAC32815 family might link cell surface signaling to rab4-dependent receptor recycling.
Cytosolic rabip4′ is associated with a large (>670-kDa) protein complex that does not contain EEA1 and rabaptin5, suggesting that it is distinct from the previously identified rab5 effector complex consisting of EEA1-rabaptin5-rabex5-syntaxin13 (McBride et al., 1999). Although we have not yet identified other components of the complex, the modular structure of rabip4′ affords association with multiple proteins via its leucine zippers and/or coiled-coil domains, which are established protein-protein interaction domains (Alber, 1992; Lupas, 1996). Cytosolic rab4 and rab5 did not cofractionate with the complex, possibly because they are present in the cytosol in the GDP-bound form (Stenmark et al., 1994; Gerez et al., 2000) that does not bind to effector proteins. Alternatively, the rabip4′ association with the GTPases might be transient and determined by cycles of GTP-hydrolysis and GDP exchange reactions that preclude their cofractionation. Similarly, the multimeric rab5 endosomal docking/fusion complex (McBride et al., 1999) did not contain rab5. Possibly, the cytosolic rabip4′ complex might serve as a precomplex that is activated upon binding to rab4/rab5 on EEs.
Acknowledgments
We thank our colleagues for the generous supply of reagents, and members of our laboratories for insightful comments. We also thank Dr. L. Meheus (Innogenetics N.V.) for mass spectrometry and Erik Vossenaar for help in the preparation of figures. This work was supported by grants from the Netherlands Organization for Scientific Research (NWO-CW to G.P., and NWOALW to P.v.d.S.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-05-0343. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0343.
References
- Alber, T. (1992). Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2, 205-210. [DOI] [PubMed] [Google Scholar]
- Bache, K.G., Brech, A., Mehlum, A., and Stenmark, H. (2003). Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162, 435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottger, G., Nagelkerken, B., and van der Sluijs, P. (1996). Rab4 and rab7 define distinct endocytic compartments. J. Biol. Chem. 271, 29191-29197. [DOI] [PubMed] [Google Scholar]
- Bucci, C., Parton, R., Mather, I., Stunnenberg, H., Simons, K., and Zerial, M. (1992). The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70, 715-728. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., and Emr, S.D. (1998). Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 2, 157-162. [DOI] [PubMed] [Google Scholar]
- Callebaut, I., de Gunzberg, J., Goud, B., and Mornon, J.P. (2001). RUN domains: a new family of domains involved in ras-like GTPase signaling. Trends Biochem. Sci. 26, 79-83. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., McBride, H., Burgoyne, R.D., and Zerial, M. (1999). The rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621-625. [DOI] [PubMed] [Google Scholar]
- Cormont, M., Mari, M., Galmiche, A., Hofman, P., and Le Marchand-Brustel, Y. (2001). A FYVE finger-containing protein, rabip4, is a rab4 effector protein involved in early endosomal traffic. Proc. Natl. Acad. Sci. USA 98, 1637-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renzis, S., Sönnichsen, B., and Zerial, M. (2002). Divalent rab effectors regulate the sub-compartmental organization and sorting function of early endosomes. Nat. Cell Biol. 4, 124-133. [DOI] [PubMed] [Google Scholar]
- Deneka, M., Neeft, M., Popa, I., van Oort, M., Sprong, H., Oorschot, V., Klumperman, J., Schu, P., and van der Sluijs, P. (2003). rabaptin-5α/rabaptin-4 serves as a linker between rab4 and γ1-adaptin in membrane recycling from endosomes. EMBO J. 22, 2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, J.J., Merithew, E., Sudharshan, E., Rajamani, D., Hayes, S., Lawe, D., Corvera, S., and Lambright, D.G. (2001). Multivalent endosome targeting by homodimeric EEA1. Mol. Cell 8, 947-958. [DOI] [PubMed] [Google Scholar]
- Estrada, L., Caron, E., and Gorski, J.L. (2001). Fgd1, the cdc42 guanine nucleotide exchange factor responsible for faciogeneital dysplasia, is localized to the subcortical actin cytoskeleton and Golgi membrane. Hum. Mol. Gen. 10, 485-495. [DOI] [PubMed] [Google Scholar]
- Fernandez-Borja, M., Wubbolts, R., Calafat, J., Janssen, H., Divecha, N., Dusseljee, S., and Neefjes, J. (1999). Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr. Biol. 9, 55-58. [DOI] [PubMed] [Google Scholar]
- Fouraux, M., van der Heijden, A., van Venrooij, W.J., and Pruijn, G.J.M. (2002a). Cross-reactivity of the anti-La mAb SW5 with early endosome antigen 2. Immunology 106, 336-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouraux, M.A., Bouvet, P., Verkaart, S., van Venrooij, W.J., and Pruijn, G.J.M. (2002b). Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J. Mol. Biol. 320, 475-488. [DOI] [PubMed] [Google Scholar]
- Gaullier, J.M., Simonsen, A., D'Arrigo, A., Bremnes, B., Stenmark, H., and Aasland, R. (1998). FYVE fingers bind PtdIns(3)P. Nature 394, 432-433. [DOI] [PubMed] [Google Scholar]
- Gerez, L., Mohrmann, K., van Raak, M., Jongeneelen, M., Zhou, X.Z., Lu, K.P., and van der Sluijs, P. (2000). Accumulation of rab4 GTP in the cytoplasm and association with the peptidyl-prolyl isomerase Pin1 during mitosis. Mol. Biol. Cell 11, 2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly, D.J., Morrow, I.C., Lindsay, M., Gould, R., Bryant, N.J., Gaullier, J.M., Parton, R.G., and Stenmark, H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cels. EMBO J. 19, 4577-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harter, C., and Mellman, I. (1992). Transport of the lysosomal membrane glycoprotein lgp120 (lgp-A) to lysosomes does not require appearance on the plasma membrane. J. Cell Biol. 117, 311-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, S., Chawla, A., and Corvera, S. (2002). TGFbeta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J. Cell Biol. 158, 1239-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov, O.C., Sbrissa, D., and Shisheva, A. (2001). Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 276, 26141-26147. [DOI] [PubMed] [Google Scholar]
- Lawe, D.C., Chawla, A., Merithew, E., Dumas, J., Carrington, W., Fogarty, K., Lifshitz, L., Tuft, R., Lambright, D., and Corvera, S. (2002). Sequential roles for phosphatidylinositol 3-phosphate and rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J. Biol. Chem. 277, 8611-8617. [DOI] [PubMed] [Google Scholar]
- Lawe, D.C., Patki, V., Harrison, R.H., Lambright, D., and Corvera, S. (2000). The FYVE domain of early endosome associated antigen 1 is required for both phosphatidylinositol 3-phosphate and rab5 binding. J. Biol. Chem. 275, 3699-3705. [DOI] [PubMed] [Google Scholar]
- Lewis, M.J., Nichols, B.J., Baschong, C.P., Riezman, H., and Pelham, H.R.B. (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11, 23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S.N., Bonzelius, F., Low, S.H., Wille, H., Weimbs, T., and Herman, G.A. (2001). Identification of discrete classes of endosome-derived small vesicles as major cellular pool for recycling membrane proteins. Mol. Biol. Cell 12, 981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, A.J., Hendrick, A.G., Cantalupo, G., Matuglia, F.S., Goud, B., Bucci, C., and McCaffrey, M.W. (2002). Rab coupling protein (RCP), a novel rab4 and rab11 effector protein. J. Biol. Chem. 277, 12190-12199. [DOI] [PubMed] [Google Scholar]
- Lupas, A. (1996). Coiled coils: new structures and new functions. Trends Biochem. Sci. 21, 375-382. [PubMed] [Google Scholar]
- Maraia, R.J., and Intine, R.V.A. (2001). Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell Biol. 21, 367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari, M., Maci, E., Le Marchand-Brustel, Y., and Cormont, M. (2001). Role of the FYVE finger and the RUN domain for the subcellular localization of rabip4. J. Biol. Chem. 276, 42501-42508. [DOI] [PubMed] [Google Scholar]
- McBride, H.M., Rybin, V., Murphy, C., Giner, A., Teasdale, R., and Zerial, M. (1999). Oligomeric complexes link rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell 98, 377-386. [DOI] [PubMed] [Google Scholar]
- Mellman, I. (1996). Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575-626. [DOI] [PubMed] [Google Scholar]
- Miura, S., et al. (2000). Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol. Cell Biol. 20, 9346-9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., Ghosh, R.N., and Maxfield, F.R. (1997). Endocytosis Physiol. Rev. 77, 759-803. [DOI] [PubMed] [Google Scholar]
- Nagelkerken, B., van Anken, E., van Raak, M., Gerez, L., Mohrmann, K., van Uden, N., Holthuizen, J., Pelkmans, L., and van der Sluijs, P. (2000). Rabaptin4, a novel effector of rab4a, is recruited to perinuclear recycling vesicles. Biochem. J. 346, 593-601. [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E., Christoforidis, S., Utenwiler-Joseph, S., Miaczynska, M., Dewitte, F., Wilm, M., Hoflack, B., and Zerial, M. (2000). Rabenosyn-5, a novel rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151, 601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulou, E., Gillooly, D.J., Wrana, J.L., Zerial, M., Stenmark, H., Murphy, C., and Fotsis, T. (2002). Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 277, 18046-18052. [DOI] [PubMed] [Google Scholar]
- Patki, V., Lawe, D.C., Corvera, S., Virbasius, J.V., and Chawla, A. (1998). A functional PtdIns(3P)-binding motif. Nature 394, 433-434. [DOI] [PubMed] [Google Scholar]
- Pruijn, G.J., Thijssen, J.P., Smith, P.R., Williams, D.G., and van Venrooij, W.J. (1995). Anti-La monoclonal antibodies recognizing epitopes with the RNA-binding domain of the La protein show differential capacities to immunoprecipitate RNA-associated La protein. Eur. J. Biochem. 232, 611-619. [DOI] [PubMed] [Google Scholar]
- Pruijn, G.J.M. (1994). The La (SS-B) antigen. In: Manual of Biological Markers of Disease, ed. W.J. van Venrooij and R.N. Maini, Dordrecht: Kluwer Academic Publishers, B4.2/1-B4.2/14.
- Raiborg, C., Bache, K.G., Gillooly, D.J., Madshus, I.H., Stang, E., and Stenmark, H. (2002). Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4, 394-398. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Bache, K., Mehlum, A., Stang, E., and Stenmark, H. (2001). Hrs recruits clathrin to early endosomes. EMBO J. 20, 5008-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C., Bremnes, B., Mehlum, A., Gillooly, D., D'Arrigo, A., Stang, E., and Stenmark, H. (2001). FYVE and coiled coil domains determine the specific localization of Hrs to early endosomes. J. Cell Sci. 114, 2255-2263. [DOI] [PubMed] [Google Scholar]
- Raymackers, J., Daniels, A., de Brabandere, V., Missiaen, C., Dauwe, M., Verhaert, P., van Mechelen, E., and Meheus, L. (2000). Identification of two-dimensionally separated human cerebrospinal fluid proteins by N-terminal sequencing, matrix assisted laser desorption/ionization-mass spectrometry, nanoliquid chromatography electrospray ionization-time of flight-mass spectrometry, and tandem mass spectrometry. Electrophoresis 21, 2266-2283. [DOI] [PubMed] [Google Scholar]
- Ridley, S.H., et al. (2001). FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 114, 3991-4000. [DOI] [PubMed] [Google Scholar]
- Roberts, M., Barry, S., Woods, A., van der Sluijs, P., and Norman, J. (2001). Rab4-dependent recycling of αvβ3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 11, 1392-1402. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95, 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe, J.W., and Klug, A. (1994). Zinc mining for protein domains. Nat. Struct. Biol. 1, 345-349. [DOI] [PubMed] [Google Scholar]
- Seet, L.F., and Hong, W. (2001). Endofin, an endosomal FYVE domain protein. J. Biol. Chem. 276, 42445-42454. [DOI] [PubMed] [Google Scholar]
- Simonsen, A., Lippe, R., Christoforidis, S., Gaullier, J.M., Brech, A., Callaghan, J., Toh, B.H., Murphy, C., Zerial, M., and Stenmark, H. (1998). EEA1 links PI(3)K function to rab5 regulation of endosome fusion. Nature 394, 494-498. [DOI] [PubMed] [Google Scholar]
- Soldati, T., and Perriard, J.C. (1991). Intracompartmental sorting of essential myosin light chains: molecular dissection and in vivo monitoring by epitope tagging. Cell 66, 277-289. [DOI] [PubMed] [Google Scholar]
- Sönnichsen, B., de Renzis, S., Nielsen, E., Rietdorf, J., and Zerial, M. (2000). Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of rab4, rab5, and rab11. J. Cell Biol. 149, 901-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprong, H., Degroote, S., Claessens, T., van Drunen, J., Oorschot, V., Westerink, B.H.C., Hirabayashi, Y., Klumperman, J., van der Sluijs, P., and van Meer, G. (2001). Glycosphingolipids are required for sorting melanosomal proteins in the Golgi complex. J. Cell Biol. 155, 369-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark, H., and Aasland, R. (1999). FYVE finger proteins-effectors of an inositol lipid. J. Cell Sci. 112, 4175-4183. [DOI] [PubMed] [Google Scholar]
- Stenmark, H., Aasland, R., Toh, B.H., and D'Arrigo, A. (1996). Endosomal localization of the autoantigen EEA1 is mediated by a Zinc-binding FYVE finger. J. Biol. Chem. 271, 24048-24054. [DOI] [PubMed] [Google Scholar]
- Stenmark, H., Parton, R., Steele-Mortimer, O., Lutcke, A., Gruenberg, J., and Zerial, M. (1994). Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13, 1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark, H., Vitale, G., Ullrich, O., and Zerial, M. (1995). Rabaptin-5 is a direct effector of the small GTPase rab5 in endocytic membrane fusion. Cell 83, 423-432. [DOI] [PubMed] [Google Scholar]
- Tall, G.G., Barbieri, M.A., Stahl, P.D., and Horazdovsky, B.F. (2001). Ras-activated endocytosis is mediated by the rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1, 73-82. [DOI] [PubMed] [Google Scholar]
- Trischler, M., Stoorvogel, W., and Ullrich, O. (1999). Biochemical analysis of distinct rab5-and rab11 positive endosomes along the transferrin pathway. J. Cell Sci. 112, 4773-4783. [DOI] [PubMed] [Google Scholar]
- Tsukazaki, T., Chiang, T.A., Davison, A.F., Attisano, L., and Wrana, J.L. (1998). SARA, a FYVE domain protein that recruits Smad2 to the TGF-β receptor. Cell 95, 779-791. [DOI] [PubMed] [Google Scholar]
- Ullrich, O., Reinsch, O., Urbe, S., Zerial, M., and Parton, R. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbe, S., Mills, I., Stenmark, H., Kitamura, N., and Clague, M.J. (2000). Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor regulated tyrosine kinase substrate. Mol. Cell Biol. 20, 7685-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam, E., and Stoorvogel, W. (2002). Dynamin-dependent transferrin recycling by endosome-derived clathrin-coated vesicles. Mol. Biol. Cell 13, 169-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs, P., Hull, M., Huber, L.A., Male, P., Goud, B., and Mellman, I. (1992a). Reversible phosphorylation-dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J. 11, 4379-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs, P., Hull, M., Webster, P., Goud, B., and Mellman, I. (1992b). The small GTP binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70, 729-740. [DOI] [PubMed] [Google Scholar]
- Vitale, G., Rybin, V., Christoforidis, S., Thornqvist, P.O., McCaffrey, M., Stenmark, H., and Zerial, M. (1998). Distinct rab-binding domains mediate the interaction of rabaptin-5 with GTP-bound rab4 and rab5. EMBO J. 17, 1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcke, M., Johannes, L., Galli, T., Mayou, V., Goud, B., and Salamero, J. (2000). Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans Golgi network. J. Cell Biol. 151, 1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Kim, O., and Qiu, Y. (2002). Interaction between tyrosine kinase Etk and a RUN domain and FYVE domain containing protein RUFY1. J. Biol. Chem. 277, 30219-30226. [DOI] [PubMed] [Google Scholar]