Summary
Many lines of evidence suggest that memory in the mammalian brain is stored with distinct spatiotemporal patterns1,2. Despite recent progresses in identifying neuronal populations involved in memory coding3–5, the synapse-level mechanism is still poorly understood. Computational models and electrophysiological data have shown that functional clustering of synapses along dendritic branches leads to nonlinear summation of synaptic inputs and greatly expands the computing power of a neural network6–10. However, whether neighboring synapses are involved in encoding similar memory and how task-specific cortical networks develop during learning remain elusive. Using transcranial two-photon microscopy11, we followed apical dendrites of layer 5 (L5) pyramidal neurons in the motor cortex while mice practiced novel forelimb skills. Here we show that a third of new dendritic spines (postsynaptic structures of most excitatory synapses) formed during the acquisition phase of learning emerge in clusters, and the majority of such clusters are neighboring spine pairs. These clustered new spines are more likely to persist throughout prolonged learning sessions and even long after training stops, compared to non-clustered counterparts. Moreover, formation of new spine clusters requires repetition of the same motor task, and the emergence of succedent new spine(s) accompanies the strengthening of the first new spine in the cluster. We also show that under control conditions new spines appear to avoid existing stable spines, rather than being uniformly added along dendrites. However, succedent new spines in clusters overcome such a spatial constraint and form in close vicinity to neighboring stable spines. Our findings suggest that clustering of new synapses along dendrites is induced by repetitive activation of the cortical circuitry during learning, providing a structural basis for spatial coding of motor memory in the mammalian brain.
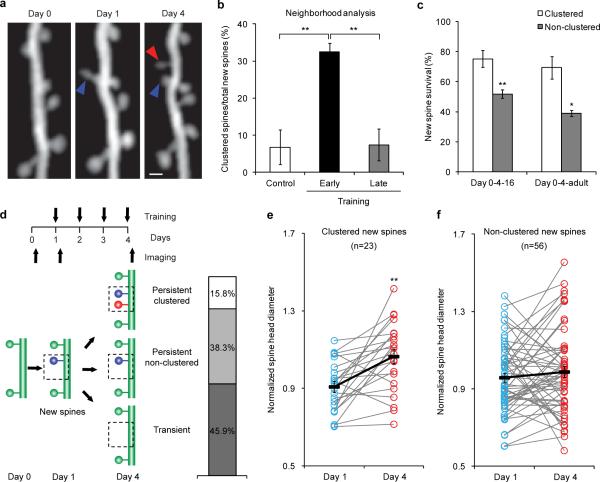
Spines are dendritic protrusions that contain all the essential components for postsynaptic signaling and are thus a good indicator of synaptic connectivity12,13. The clustered plasticity model suggests that neighboring spines tend to transmit similar information to the postsynaptic neuron6,7. To investigate the formation and functional significance of spine clusters during learning, we trained thy1-YFP-H mice14 with a seed-reaching task15 and followed the dynamics of spines on apical dendrites of L5 pyramidal neurons in the motor cortex contralateral to the trained limb during different learning phases. We found that 32.5±2.2% of new spines that formed during the acquisition phase of learning (early training, days 1–4) emerged in clusters, i.e., two or more neighboring new spines without interspersed existing spine(s) (Fig. 1a,b). The majority of such clusters (61 cases) comprised two contiguous new spines, and the other two clusters comprised three. In contrast, fewer new spine clusters emerged in untrained control mice over the same period (6.8±4.6%, P<0.01) or in trained mice during the consolidation phase of learning (late training, days 13–16, 7.4±4.3%, P<0.01, Fig. 1b). In addition to clustering of contiguous new spines, we observed a few cases in which two or more new spines formed in close vicinity to each other, but with up to three existing spines interspersed among them, as well as cases in which new filopodia clustered with new spines (Supplementary Fig. 1). We incorporated these cases in another set of analyses, in which a cluster was defined as a set of new spines/filopodia formed within 5 μm of each other, regardless of the presence or absence of existing spine(s) between them (Supplementary Notes). These analyses again revealed that a significantly higher percentage of new spines clustered during early training, compared to that in controls or during late training (P<0.01 for both cases, Supplementary Fig. 2). They also showed that filopodia only made a minor contribution to new protrusion clusters (Supplementary Fig. 3). More interestingly, among the new spines observed at the end of the acquisition phase (day 4), clustered new spines had a significantly higher survival rate than non-clustered ones (i.e., individual new spines flanked by two existing spines) by training day 16 (P<0.01), as well as 4 months after training stopped (P<0.05, Fig. 1c). Together, our results reveal that motor learning induces coordinated formation of clustered synapses, which presumably belong to the same neuronal circuit and persist over time to encode motor information.
Figure 1. Acquisition of a novel motor skill induces formation of spine clusters.
a, Repeated imaging of the same dendritic branch during motor learning reveals that a second new spine that formed between days 1 and 4 (red arrowhead) is located next to a stabilized new spine that had formed on day 1 (blue arrowhead). Scale bar, 1 μm. b, A higher percentage of new spines formed in clusters over 4 days during early training (n=18 mice), compared to control (n=7) and late training (n=4). c, Clustered new spines observed on training day 4 have a higher survival rate than non-clustered counterparts by the end of the 16-day training (n=6), as well as 4 months after training stops (n=4). d, New spines formed on training day 1 are classified according to their fate and neighboring spine formation. e, Spine head sizes of persistent clustered new spines increase between training days 1 and 4. f, Spine head sizes of persistent non-clustered new spines show no change between training days 1 and 4. Spine head size is quantified by the normalized spine head diameter, defined as the ratio of the spine head diameter to the adjacent dendritic shaft diameter. *P<0.05, **P<0.01. Error bars, s.e.m.
Perfection of a motor skill requires repeated practice, usually through multiple training sessions. We therefore sought to find out whether clustered new spines observed on training day 4 were formed within the same training session or across different sessions. We imaged the mice three times (on the day prior to training, and following 1 and 4 days of training), and found that among new spine clusters observed on training day 4, only 2.4% were composed of spines that formed together between training days 0 and 1. On the other hand, 43.9% of clusters were composed of spines formed between days 1 and 4, and the remaining 53.7% of clusters consisted of one spine formed between days 0 and 1 (the first new spine) and another spine formed between days 1 and 4 (the second new spine). Thus the majority of new spine clusters emerged through recurrent training sessions. To determine how the formation of the second new spine in a cluster correlates with functional changes of the first new spine, we categorized first new spines into three groups based on their survival and neighboring spine addition: transient new spines (formed on training day 1 but lost by day 4), persistent clustered new spines (formed on training day 1, survived until day 4, with an adjacent new spine formed between days 1 and 4), and persistent non-clustered new spines (formed on training day 1, survived until day 4, with no adjacent new spine formation) (Fig. 1d). As spine head size closely correlates with synaptic strength, we followed head sizes of first new spines over time. On training day 1, we found that head sizes of both transient and persistent new spines were significantly smaller than those of existing stable spines along the same dendrite (P<0.001 for both cases, Supplementary Fig. 4). By training day 4, head sizes of persistent clustered new spines increased significantly (P<0.01, Fig. 1e, Supplementary Fig. 5a), whereas head sizes of persistent non-clustered new spines remained comparable to day 1 (P>0.2, Fig. 1f, Supplementary Fig. 5b). Since spine head size is a good proxy for synaptic strength, these data suggest that formation of the second new spines accompanies synaptic potentiation at first new spines during motor learning. It is consistent with previous reports that long-term potentiation at a single spine can facilitate formation and potentiation of neighboring spines16,17.
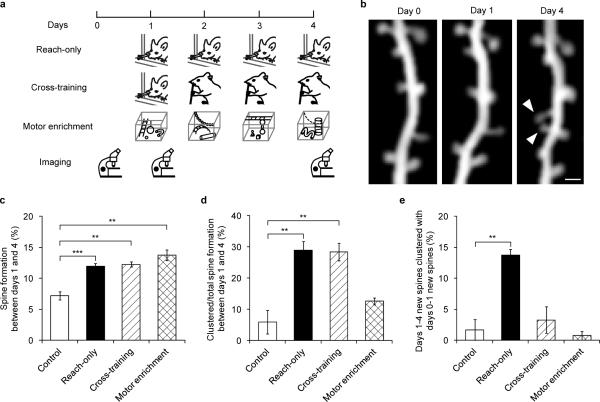
Different sets of synapses have been shown to be involved in different motor tasks15. We therefore trained the same mice sequentially with two motor skills (cross-training) to determine if spines induced by different motor tasks cluster. Cross-training started with the reaching task on day 1 and then switched to the capellini-handling task, which also requires forelimb coordination, on days 2 to 4 (Fig. 2a, Supplementary Table 1). We found that 12.3±0.4% new spines formed during the capellini-handling task between days 1 and 4, among which 28.4±2.8% occurred in clusters (Fig. 2b–d). Both the spine formation rate and the percentage of clustered new spines were comparable to those in mice continuously trained with the reaching task (reach-only) (P>0.5 in both cases), and were significantly higher than those in control mice over the same period of time (P<0.01 in both cases, Fig. 2c,d). Thus, the capellini-handling task itself can induce clustered spine formation. However, only 3.3±2.1% of capellini-handling-induced new spines clustered with reaching-induced new spines in cross-training. This contrasts with the outcome of reach-only training (13.8±1.0%, P<0.01, Fig. 2e), suggesting that new spines induced by different tasks have a low incidence of clustering with each other. To further characterize the task-specific nature of clustered spine formation, we housed animals in a motor enriched environment with daily change of motor challenges (Fig. 2a, see Methods). Motor enrichment also robustly enhanced spinogenesis: 13.7±0.8% new spines formed between days 1 and 4, comparable to the percentages under reach-only and cross-training conditions (P>0.1 for both cases). However, only 12.6±1.1% of these new spines appeared in clusters, a percentage comparable to controls (P>0.2, Fig. 2d) but significantly lower than that under reach-only or cross-training conditions (P<0.01 for both cases). Together these data indicate that, while novelty in learning stimulates spinogenesis, repetitive activation of the same cortical circuit is crucial in clustered spine formation (Supplementary Fig. 6).
Figure 2. Clustered new spines form over multiple training sessions of the same, but not different, motor tasks.
a, Timelines of reach-only, cross-training and motor enrichment experiments. b, Repeated imaging of the same dendritic branch revealed that two neighboring new spines (arrowhead) formed between days 1 and 4 during cross-training. Scale bar, 1 μm. c, Higher percentages of new spines formed between days 1 and 4 in reach-only, cross-training and motor enrichment, compared to controls. d, Higher percentages of new spines formed in clusters between days 1 and 4 in reach-only and cross-training, compared to controls. e, A higher percentage of new spines that formed between days 1 and 4 clustered with new spines that had formed between days 0 and 1 in the reach-only condition, compared to controls. Number of mice examined: 6 control, 9 reach-only, 5 cross-training and 6 motor enrichment. **P<0.01, ***P<0.001. Error bars, s.e.m.
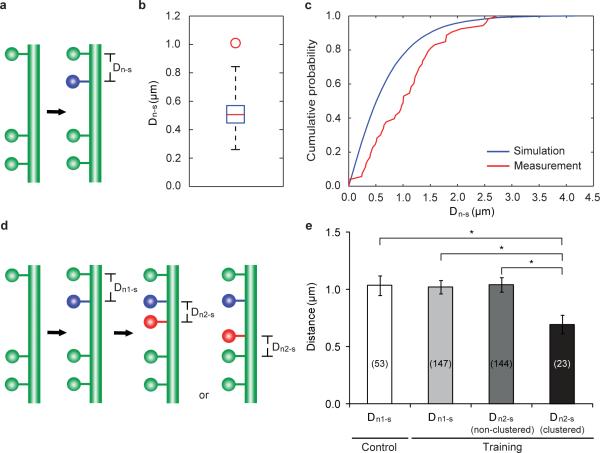
The phenomenon of learning-induced, coordinated spinogenesis led us to further investigate the spatial distribution of new spines. We first examined the distance between a new spine (n) and its nearest existing spine (s) (Dn-s, Fig. 3a) in control mice. We then simulated Dn-s distribution under the null hypothesis that new spines form uniformly and independently along the dendrite (see Methods). Compared to simulation results, the median of measured Dn-ss was significantly larger (Fig. 3b), and the cumulative probability distribution of measured Dn-ss was shifted towards longer distances (Fig. 3c). These results suggest that new spines are not randomly dispersed along dendritic segments, and their apparent avoidance of existing stable spines under control condition is consistent with the idea that neighboring spines share and compete for local resources18–21.
Figure 3. The spatial distribution of new spines along dendrites.
a, A schematic illustrating the measurement of Dn-s. b, The median of measured Dn-ss (the red circle) is significantly larger than that of simulated Dn-ss (box plot of results from 1000 simulations, with whiskers representing the minimum and the maximum) in control mice. The simulation is based on the null hypothesis that new spines are added independently and uniformly along a linear dendrite. c, Cumulative probability distribution of measured Dn-s is shifted towards longer distances than the simulated Dn-s in control mice. d, A schematic illustrating the measurement of Dn2-s, clustered. The nearest spine to a clustered n2 could be either a persistent first new spine (n1) or a stable spine existing since day 0, depending on relative n2 location. e, Dn1-s in control mice is comparable to that of trained mice. In trained mice, Dn2-s, clustered is significantly smaller than Dn1-s, while Dn2-s, non-clustered is comparable to Dn1-s. The number of spines analyzed in each condition is indicated on each column. *P<0.05. Error bars, s.e.m.
To determine if motor learning alters the spatial distribution of new spines, we examined Dn-ss in mice trained with the reaching task. We found that the distance between a new spine formed on training day 1 (n1) and the nearest existing spine (Dn1-s) was comparable for trained and control mice (P>0.7). We classified new spines formed between training days 1 and 4 (n2) into two categories: clustered n2 (i.e., those that formed next to a stabilized first new spine, Fig. 3d) and non-clustered n2 (those that did not form next to a stabilized first new spine). We found that clustered n2s were significantly closer to their nearest stable spines (n1 or a stable spine existing since day 0, Fig. 3d) (Dn2-s, clustered) than were n1s (Dn1-s) (P<0.05). In contrast, the distance between a non-clustered n2 and its nearest stable spine (Dn2-s, non-clustered) was comparable to Dn1-s (P>0.9, Fig. 3e). In addition, when an n1 formed between two adjacent stable spines, the distance between the two stable spines (Ds-n1-s) was comparable for control and trained mice (P>0.7, Supplementary Fig. 7). However, the distance between a stabilized n1 and the adjacent stable spine, between which a clustered n2 formed (Dn1-n2-s), was significantly smaller than the distance between two adjacent stable spines, between which a non-clustered n2 formed (Ds-n2-s, P<0.01, Supplementary Fig. 7). These results suggest that learning-induced clustered new spines can overcome the spatial constraint of existing spines and be packed into tighter dendritic space.
Recent studies have shown that dendritic spines are dynamic in the living brain, and that rearrangement of cortical connections through de novo growth and loss of spines provides a structural substrate for experience-dependent plasticity22–25. Built upon these works, our study reveals a novel spatial rule of spinogenesis during motor learning. We found that learning-induced new spines tend to form in small clusters (mostly pairs). The correlation between the emergence of the second new spine and the strengthening of the first new spine also suggests their potential participation in the same neuronal circuit. These findings support the clustered plasticity model, which postulates that synapses located close together along the same dendritic branch are more likely to be allocated for the same information than synapses dispersed throughout the dendritic arbor7. Indeed, in the mouse auditory cortex although spines tuned for different frequencies are highly interspersed, 26% of neighboring spines exhibit similar effective frequencies, much more frequently than anticipated from random distribution (10%)26. Therefore, while neurons tend to maximize their overall connections27, clustered plasticity ensures strengthening of circuit-specific connections and enables spatial coding for task-related information.
Previous electron microscopy studies have revealed that neighboring spines can form synapses with the same axon28–30 (see Supplementary Fig. 8a,c,e). Positioning multiple synapses between a pair of neurons in close proximity allows non-linear summation of synaptic strength, and potentially increases the dynamic range of synaptic transmission well beyond what can be achieved by random positioning of the same number of synapses. Alternatively, clustered new spines may synapse with distinct (but presumably functionally-related) pre-synaptic partners (Supplementary Fig. 8b,d). In this case, they could potentially integrate inputs from different neurons non-linearly and increase the circuit's computational power. Distinguishing between these two scenarios would likely require circuit reconstruction by electron microscopy following in vivo imaging to reveal the identities of pre-synaptic partners of newly-formed spines.
Profiling spine formation during novel experiences, our data revealed a critical role of repetitive activation of the same neuronal circuit. The fact that the second new spine in a cluster can overcome the spatial constraint imposed by existing spines suggests that repetitive activation of a neuronal circuit may modify or reallocate local “resources” for spinogenesis. Such resources may be permissive or instructive molecular cues at the pre- or post-synaptic site, or the availability of suitable partners (e.g., axonal boutons). Understanding the nature and regulation of such resources may hold the key to elucidating the cellular mechanisms of clustered spine formation. It will be conducive to the development of tools to label and manipulate specific synaptic populations, and ultimately to the dissection of the causal relationship between synaptic dynamics and learning.
METHODS SUMMARY
YFP-H line mice14 expressing yellow fluorescent protein in a small subset of cortical neurons were used in all the experiments. Mice of both sexes were trained with different motor skill tasks or housed in a motor enriched environment, starting at one month of age (see Methods). The procedure for transcranial two-photon imaging and quantification of spine dynamics have been described previously11,15. ImageJ was used to measure spine head size, as well as inter-spine distances. Simulation was performed with custom-written codes in Matlab (MathWorks, Natick, MA) and statistical analyses were performed using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA) (see Methods). All data were presented as mean ± standard error of mean (s.e.m.). P-values were calculated using the Mann-Whitney U test for independent samples, and the Wilcoxon signed-rank test for paired samples.
METHODS
Experimental animals
Thy1-YFP-H line mice were obtained from the Jackson Laboratory. Mice were group-housed and bred in the UCSC animal facility, with all experiments performed in accordance with approved animal protocols.
Motor skill training and motor enrichment
Both the mouse single-seed reaching task and capellini-handling task protocols have been previously described15. “Motor enriched” mice were reared in groups of 8–12 in large cages (90 cm×25 cm×15 cm) containing various toys, such as ropes, ladders, chains, hanging mesh/bars and etc., all of which require substantial motor coordination. The nature of toys was changed on a daily basis. Control mice were housed in standard mouse cages, with up to 5 mice per cage.
Surgical procedure for in vivo transcranial imaging
The procedure for transcranial two-photon imaging has been described previously11,31. Trained mice were imaged immediately following each training session.
Data quantification
All analyses of spine dynamics were done using ImageJ software, blinded with regard to experimental conditions. Quantification criteria of dendritic spines have been described previously15. All dendritic protrusions were tracked manually in 3D stacks to ensure the consistency of protrusion identification across imaging sessions, despite possible tissue movement or rotation. The number and location of dendritic protrusions (defined as protrusion length larger than 1/3 of dendritic shaft diameter) were identified in each view. Filopodia were identified as long thin structures with the ratio of head diameter to neck diameter being smaller than 1.2 and the ratio of length to neck diameter being larger than 3. The remaining protrusions were classified as spines. Formation and elimination of spines and filopodia were determined by comparing images collected at two different time points. Spines or filopodia were considered identical between the two images, if they were within 0.7 μm of their expected positions, based on their spatial relationship to adjacent landmarks and/or their positions relative to immediately adjacent spines. A stable spine is defined as a spine that was present in both images. A new spine is a spine that appeared in a subsequent image but was absent from the initial image. Percentages of formed and eliminated spines (or dendritic protrusions) were normalized to the number of spines (or dendritic protrusions) in the initial image. Spine diameter analyses have been previously described11. Because imaging and animal conditions varied over time, the ratio of the spine head diameter to the adjacent dendritic shaft diameter was used as the normalized spine head diameter. Measurement of spine head intensity, as described previously32, was also performed to confirm these spine size results. Briefly, we determined the signal intensity (defined as the sum intensity of all pixels composing the spine in the best focal plane) and subtracted the background intensity (defined as the sum intensity of a region composed of the same number of pixels as the spine but with no YFP-labeled structure). The difference was then divided by the mean intensity of the adjacent dendritic shaft (defined similarly as the difference between the mean signal intensity of the shaft and the mean background intensity) in order to correct for varying imaging conditions. The final value is termed “integrated spine brightness.” All distance measurements were done in ImageJ. In order to simulate spine formation, we first obtained the relative location of stable spines by measuring inter-spine distances along traced dendrites in 7 control animals, and concatenated dendritic segments from each animal into a single “synthetic dendrite.” We then used custom-written Matlab codes to simulate the addition of new spines. As we observed, two spines can extend from the same linear location along the dendritic segment and point towards different directions, given the cylindrical shape of dendrites. In our analysis and simulation, we made the simplifying approximation that the dendritic segment is one-dimensional rather than a tube. Therefore, zero inter-spine distance in our analysis represents two spines overlapping in linear position but actually located at different sites around the circumference of the dendritic segment. In each round of simulation, the same number of new spines as observed in experiments were generated independently and uniformly along synthetic dendrites. The distance between each new spine and its nearest stable spine (Dn-s) was calculated. The simulation was repeated 1,000 times and the resultant data were pooled together to compute the simulated sample median and the cumulative probability curve. All data were presented as mean ± standard error of mean (s.e.m.). P-values were calculated using the Mann-Whitney U test for independent samples, and the Wilcoxon signed-rank test for paired samples.
Supplementary Material
Acknowledgements
We gratefully thank David States, Drs. Denise Garcia, Lindsay Hinck, Theresa Jones, Sen Song, Wes Thompson and Gordon Wang for critical comments on this manuscript. We thank Andrew Perlik and Dr. Tonghui Xu for technical support. This work was supported by grants from the DANA Foundation and the National Institutes of Mental Health to Y.Z.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.F. and X.Y. did the in vivo imaging and made the figures. M.F. performed behavioral training and all spine analyses, made figures for repetitive imaging. J.L. and M.F. performed Matlab simulation and statistical analyses. J.L., M.F. and X.Y. participated in paper discussion. Y.Z. initiated and designed the project, and wrote the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to Y.Z. (yizuo@ucsc.edu)
References
- 1.Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J. Molecular and cellular approaches to memory allocation in neural circuits. Science. 2009;326:391–395. doi: 10.1126/science.1174519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aimone JB, Wiles J, Gage FH. Potential role for adult neurogenesis in the encoding of time in new memories. Nature Neurosci. 2006;9:723–727. doi: 10.1038/nn1707. [DOI] [PubMed] [Google Scholar]
- 3.Han JH, et al. Selective erasure of a fear memory. Science. 2009;323:1492–1496. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- 4.Komiyama T, et al. Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature. 2010;464:1182–1186. doi: 10.1038/nature08897. [DOI] [PubMed] [Google Scholar]
- 5.Kee N, Teixeira CM, Wang AH, Frankland PW. Preferential incorporation of adult-generated granule cells into spatial memory networks in the dentate gyrus. Nature Neurosci. 2007;10:355–362. doi: 10.1038/nn1847. [DOI] [PubMed] [Google Scholar]
- 6.Larkum ME, Nevian T. Synaptic clustering by dendritic signalling mechanisms. Curr. Opin. Neurobiol. 2008;18:321–331. doi: 10.1016/j.conb.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Govindarajan A, Kelleher RJ, Tonegawa S. A clustered plasticity model of long-term memory engrams. Nature Rev. Neurosci. 2006;7:575–583. doi: 10.1038/nrn1937. [DOI] [PubMed] [Google Scholar]
- 8.Poirazi P, Mel BW. Impact of active dendrites and structural plasticity on the memory capacity of neural tissue. Neuron. 2001;29:779–796. doi: 10.1016/s0896-6273(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 9.Sjostrom PJ, Hausser M. A cooperative switch determines the sign of synaptic plasticity in distal dendrites of neocortical pyramidal neurons. Neuron. 2006;51:227–238. doi: 10.1016/j.neuron.2006.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Losonczy A, Magee JC. Integrative properties of radial oblique dendrites in hippocampal CA1 pyramidal neurons. Neuron. 2006;50:291–307. doi: 10.1016/j.neuron.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Zuo Y, Lin A, Chang P, Gan WB. Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 2005;46:181–189. doi: 10.1016/j.neuron.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Segal M. Dendritic spines and long-term plasticity. Nature Rev. Neurosci. 2005;6:277–284. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- 13.Harms KJ, Dunaevsky A. Dendritic spine plasticity: looking beyond development. Brain Res. 2007;1184:65–71. doi: 10.1016/j.brainres.2006.02.094. [DOI] [PubMed] [Google Scholar]
- 14.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, et al. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450:1195–1200. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Roo M, Klauser P, Muller D. LTP promotes a selective long-term stabilization and clustering of dendritic spines. PLoS Biol. 2008;6:e219. doi: 10.1371/journal.pbio.0060219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey CD, Yasuda R, Zhong H, Svoboda K. The spread of Ras activity triggered by activation of a single dendritic spine. Science. 2008;321:136–140. doi: 10.1126/science.1159675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose J, Jin SX, Craig AM. Heterosynaptic molecular dynamics: locally induced propagating synaptic accumulation of CaM kinase II. Neuron. 2009;61:351–358. doi: 10.1016/j.neuron.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuriel S, et al. Local sharing as a predominant determinant of synaptic matrix molecular dynamics. PLoS Biol. 2006;4:e271. doi: 10.1371/journal.pbio.0040271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu X, Zuo Y. Spine plasticity in the motor cortex. Curr. Opin. Neurobiol. 2011;21:169–174. doi: 10.1016/j.conb.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JL, Nedivi E. Neuronal structural remodeling: is it all about access? Curr. Opin. Neurobiol. 2010;20:557–562. doi: 10.1016/j.conb.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu M, Zuo Y. Experience-dependent structural plasticity in the cortex. Trends Neurosci. 2011;34:177–187. doi: 10.1016/j.tins.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nature Rev. Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Leischner U, Rochefort NL, Nelken I, Konnerth A. Functional mapping of single spines in cortical neurons in vivo. Nature. 2011;475:501–505. doi: 10.1038/nature10193. [DOI] [PubMed] [Google Scholar]
- 27.Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knott GW, Holtmaat A, Wilbrecht L, Welker E, Svoboda K. Spine growth precedes synapse formation in the adult neocortex in vivo. Nature Neurosci. 2006;9:1117–1124. doi: 10.1038/nn1747. [DOI] [PubMed] [Google Scholar]
- 29.Fiala JC, Allwardt B, Harris KM. Dendritic spines do not split during hippocampal LTP or maturation. Nature Neurosci. 2002;5:297–298. doi: 10.1038/nn830. [DOI] [PubMed] [Google Scholar]
- 30.Toni N, Buchs PA, Nikonenko I, Bron CR, Muller D. LTP promotes formation of multiple spine synapses between a single axon terminal and a dendrite. Nature. 1999;402:421–425. doi: 10.1038/46574. [DOI] [PubMed] [Google Scholar]
- 31.Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- 32.Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hubener M. Experience leaves a lasting structural trace in cortical circuits. Nature. 2009;457:313–317. doi: 10.1038/nature07487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.