Abstract
CCR5 is the primary coreceptor for HIV entry. Early after infection, the HIV viral population diversifies rapidly into a quasispecies. It is not known whether the initial efficiency of the viral quasispecies to utilize CCR5 is associated with HIV disease progression or if it changes in an infected individual over time. The CCR5 and CXCR4 utilization efficiencies (R5-UE and X4-UE) of the HIV quasispecies were examined using a pseudovirus, single-round infection assay for samples obtained from known seroconverters from Rakai district, Uganda (n=88). Initial and longitudinal R5-UE values were examined to assess the association of R5-UE with HIV disease progression using multivariate Cox proportional hazard models. Longitudinal samples were analyzed for 35 seroconverters who had samples available from multiple time points. There was no association between initial or longitudinal changes in R5-UE and the hazard of HIV disease progression (p=0.225 and p=0.942, respectively). In addition, R5-UE increased significantly over time after HIV seroconversion (p<0.001), regardless of HIV subtype or the emergence of CXCR4-tropic virus. These data demonstrate that the R5-UE of the viral quasispecies early in HIV infection is not associated with disease progression, and that R5-UE levels increase in HIV-infected individuals over time.
Introduction
Human immunodeficiency virus type 1 (HIV) infects target cells through a multistage entry process that begins with binding of the viral envelope protein, gp120, to its primary cellular receptor, CD4. This triggers a conformational change in the gp120 protein that exposes the binding site for a cellular coreceptor, primarily CCR5 (R5) or CXCR4 (X4).1,2 Binding of HIV to the coreceptor induces further structural changes in gp120, which allows for unfolding of the viral gp41 protein, insertion of the viral fusion peptide into the cell membrane, and entry of the viral capsid into the target cell. CCR5-using (R5-tropic) viruses are found in almost all recently infected individuals, and are believed to be the predominant viral phenotype that is transmitted sexually.3 R5-tropic viruses isolated later in infection have been found to induce more robust CD4 T cell apoptosis than R5-tropic viruses found early in infection, which may be related to the pathogenesis of R5-tropic strains.4,5 R5-tropic viruses isolated later in disease often have an increased ability to utilize CCR5 to infect cells. The efficiency of CCR5 utilization can be assessed using a single-cycle replication assay that measures the replication of pseudoviruses containing the HIV envelope on CCR5-bearing cells (CCR5-utilization efficiency, R5-UE).6 Viruses isolated later in infection also have increased fusion kinetics compared to viruses isolated early in infection, but it is not known when in the course of disease progression these differences in R5-UE arise or if they vary by HIV subtype.6 Conversely, it has also been demonstrated that some HIV isolates that use both CCR5 and CXCR4 for cell entry (dual-tropic viruses) arise later in disease, and have decreased R5-UE depending on the composition of the V3-loop.7 CXCR4-using (X4-tropic) viruses are found during the late stages of HIV disease in approximately 50% of individuals infected with HIV subtypes B and D, but are found less frequently in individuals infected with subtypes A and C.2,8–10 The biological mechanisms responsible for differences in the emergence of X4-tropic viruses in different HIV subtypes are not fully understood. The emergence of X4-tropic HIV is associated with an increased rate of disease progression, which is likely due to an enhanced ability of X4-tropic strains to infect naive T cells.8,11–14 Given the strong association between the emergence of X4-tropic virus and disease progression, as well as the role that R5-UE plays in apoptosis, it is plausible that higher baseline R5-UE may itself affect disease progression.
A genetic bottleneck, which is influenced by many factors, occurs during sexual transmission of HIV, and leads to a relatively homogeneous viral population early in the disease.1–17 This population rapidly evolves into a diverse viral quasispecies with upregulation of the antiviral immune response.15 The majority of studies on viral coreceptor use and HIV disease progression have examined individual viral isolates.4,7,14,18,19 However, it is not known whether these isolates are representative of the viral quasispecies in infected individuals. With the recent introduction of antiretroviral entry inhibitors that target interactions between gp120 and CCR5, it is important to understand the natural evolution of R5-UE within the viral quasispecies and the association of R5-UE levels with HIV disease progression.
Materials and Methods
Study population
Serum samples were obtained from adults with known dates of HIV seroconversion who were enrolled in the Rakai Community Cohort Study (RCCS) in Uganda. RCCS is an open cohort of all consenting persons aged 15 to 49 years, residing in 50 communities in the rural Rakai district, southwestern Uganda. The RCCS has previously been described in detail.20,21 Briefly, 10,000–14,000 participants are surveyed annually with a sociodemographic and behavioral interview and each participant is asked to provide a venous blood sample for HIV testing. Eighty-eight individuals with known times of HIV seroconversion and disease progression outcomes were evaluated to assess the associations between initial R5-UE and HIV disease progression (Table 1). The time of HIV infection was estimated as the mid-point between the last HIV-negative and first HIV-positive serologic test.20 Disease progression was estimated as the time between HIV seroconversion and death or diagnosis of AIDS (CD4 T cell count <250/μl).21 Eight study subjects did not reach an endpoint and were classified as long-term nonprogressors.20 Data from those individuals were right-censored at the time of last study visit. In addition, 35 individuals who had multiple samples collected at different time points after documented HIV seroconversion were assessed to examine longitudinal changes in R5-UE and CXCR4 utilization efficiency (R4-UE) of their viral quasispecies. The effect of longitudinal R5-UE measurements on disease progression was also estimated. These individuals were selected based on sample availability. All participants provided written informed consent and the study was approved by the Institutional Review Boards (IRBs) in Uganda (the Uganda Virus Research Institute's Science and Ethics Committee and the Uganda National Council for Science and Technology) and by the IRBs of collaborating institutions in the United States (the Walter Reed Army Institute of Research, Columbia University, and Johns Hopkins University).
Table 1.
Demographics of Study Subjects Included in the Analyses of HIV Disease Progression and Changes in Coreceptor Utilization Efficiency over Time
| Disease progression (n=88) | Longitudinal (n=35) | |
|---|---|---|
| Female gender | 48.9% | 45.7% |
| Age at seroconversion (years) | 31.6 (26.3–37.1) | 31.2 (25.4–36.6) |
| Viral load set point (log10 HIV RNA copies/ml) | 5.14 (4.73–5.57) | 5.14 (4.82–5.55) |
| Subtype A (%) | 25.0% | 20.0% |
| Subtype D (%) | 54.5% | 60.0% |
| Intersubtype recombinant (%) | 20.5% | 20.0% |
Median values and interquartile ranges are shown for age and viral load set point. Viral load set point was defined as the viral load measured in the first sample collected after HIV seroconversion.
Viral characteristics
HIV serostatus was determined using two ELISA assays (Organon Teknika, Charlotte NC and Cambridge Biotech Recombigen, Worcester, MA) and HIV seroconversion was confirmed by Western blot (Bio-Merieux-Vitek, St. Louis, MO). HIV viral load was measured using the standard Roche AMPLICOR assay version 1.5 (Roche Diagnostics Corporation, Indianapolis, IN). HIV viral load set point was defined as the viral load measured at the earliest annual sample obtained after HIV seroconversion. HIV subtype was determined using a multiple region hybridization assay or by direct sequence analysis of p24 and gp41 gene fragments.22 Viral isolates were determined to be infected with either subtype A or D if both regions segregated with those subtypes.23 If the subtype assignments were different for the p24 and gp41 regions the sample was classified as being infected with a recombinant strain. Individuals who had at least one sample that had X4-tropic strains detected were classified as having X4 emergence.
CCR5 and CXCR4 coreceptor utilization efficiency
The ability of the viral quasispecies to utilize CCR5 and CXCR4 to infect cells was determined by measuring luciferase activity [relative light units (RLU)] using the Trofile assay, as described previously (Monogram Biosciences, South San Francisco, CA).24 In this assay, HIV viral RNA is extracted from serum samples and the gp160 envelope region is reverse transcribed and amplified using the polymerase chain reaction (PCR). The resulting PCR products, which represent the individual's viral envelope quasispecies, are then cloned into expression vectors. The number of distinct viral strains captured differs between samples; however, previous work has demonstrated that this approach captures a representative population of the viral quasispecies.24 The plasmids containing the amplified envelope genes are then cotransfected into HEK 293 cells, along with a second plasmid that encodes a pseudovirus in which the HIV envelope gene has been replaced with a luciferase reporter gene.
After a single round of transfection, the pseudoviruses are harvested and used to infect U87 cells that stably express CD4 and either CCR5 or CXCR4. Four reference control envelope samples are used to monitor assay performance and normalize luciferase activity of the test samples in every batch, including R5-, X4-, and dual-tropic envelope sequences. The amount of plasmid DNA (HIV genomic reporter vector and envelope expression vector) used in the assay was determined and transfection efficiencies were evaluated. Transfection efficiency is routinely monitored by determining the amount of luciferase produced following transfection of the virus producer cells. Infection is assessed by measuring luciferase activity in infected cells (following provirus formation) and is normalized for transfection efficiency. The Trofile assay is conducted using single transfections and infections. The amount of light produced (relative light units, RLU) on either CCR5- or CXCR4-bearing cells serves as a surrogate measure of the R5-UE and X4-UE of the viral quasispecies, respectively. R5-UE was measured for HIV from the first available sample (the sample collected closest to the time of HIV seroconversion, baseline), and the association between initial R5-UE and subsequent HIV disease progression was assessed.
Statistical analysis
Univariate and multivariate Cox proportional hazards models were used to estimate the hazard ratios (HR) of disease progression to death or AIDS associated with baseline R5-UE measurements (log10 transformed). Covariates included in the multivariate analysis were HIV viral load set point, age at time of HIV seroconversion, gender, X4 emergence, and HIV subtype. Initial R5-UE and set point viral load were compared using the Spearman correlation. The proportional hazards assumption was explored using cumulative Martingale residuals (SAS/STAT 9.22 User's Guide, SAS, Cary, NC).
Linear mixed effects regression models were used to model the longitudinal change of log10 transformed R5-UE or X4-UE. The intercept and slope parameters were modeled as random effects to allow individual variations from the average longitudinal trend. The assumption of a linear trend of R5-UE (or X4-UE) was examined by graphic inspection and by testing the statistical significance of the term for second order of time from seroconversion, both of which showed that the linear assumption was reasonable. Changes in R5-UE were also estimated by HIV subtype. Additionally, since initial R5-UE levels may influence the rate of R5-UE change over time, the baseline R5-UE was dichotomized as above and below the median baseline R5-UE, and the subsequent change in R5-UE was then assessed by low or high initial R5-UE level. To examine whether X4 emergence (as determined by the Trofile assay) was associated with R5-UE change, time-dependent measures of X4 emergence were included in the model to allow different slope estimates for R5-UE change.
The time-dependent Cox model was used to examine whether longitudinal R5-UE measurements over time are associated with disease progression. Specifically, the time-dependent covariate process of R5-UE was inferred using the repeated measurements of R5-UE for each patient, and the hazard at a time point was then modeled as a function of the most recent R5-UE measurement. All analyses were performed using SAS 9.2, using two-sided tests with α=0.05.
Results
To determine the relationship between the R5-UE of the viral quasispecies near the time of HIV seroconversion and disease progression, the baseline R5-UE results from 88 HIV seroconverters (Table 1) were used to estimate the hazard ratios (HR) of disease progression to death or AIDS (Table 2). In univariate analyses, the HR of disease progression associated with baseline R5-UE was not statistically significant [HR=1.15, 95% confidence interval (CI)=0.91–1.45; p=0.25] (Table 2). However, the HR of disease progression was significantly increased with older age at seroconversion (HR=1.04 per year, CI=1.01–1.07; p=0.006), log10 viral load set point (HR=1.51 per log increment, CI=1.07–2.19; p=0.019), and X4 emergence (HR=1.83, CI=1.01–3.32; p=0.045) (Table 2). Additionally, the HR for disease progression was significantly lower in individuals with subtype A infection than in individuals with subtype D infection (HR=0.53, CI=0.30–0.92; p=0.024) (Table 2). In multivariate analysis, age at seroconversion was the only factor that was significantly associated with disease progression. Since age was highly associated with viral load set point (Spearman correlation coefficient=0.86, p<0.05), it was subsequently excluded from the multivariate model, and only viral load set point was found to be significantly associated with disease progression (HR=1.49, CI=1.01-2.19; p=0.045). R5-UE and viral load were not significantly correlated (Spearman correlation coefficient=0.17, p=0.12).
Table 2.
Hazard Ratio Analysis of Demographic and Virological Parameters with HIV Disease Progression
| Univariate HR (95% CI) | p-value | Multivariate HR (95% CI) | p-value | |
|---|---|---|---|---|
| Age at seroconversion | 1.04 (1.01–1.07) | 0.006 | Excluded | — |
| Female gender | 1.08 (0.69–1.67) | 0.746 | — | — |
| Viral load set point | 1.51 (1.07–2.12) | 0.019 | 1.49 (1.01–2.19) | 0.045 |
| Baseline CCR5-UE | 1.15 (0.91–1.45) | 0.249 | — | — |
| X4 emergence | 1.83 (1.01–3.32) | 0.045 | 1.69 (0.88–3.23) | 0.112 |
| Subtype A vs. D | 0.53 (0.30–0.92) | 0.024 | 0.66 (0.36–1.19) | 0.164 |
| Subtype A vs. R | 0.56 (0.28–1.10) | 0.091 | — | — |
95% confidence intervals (CI) are shown in parentheses. HR, hazard ratio. CCR5-UE: utilization efficiency of viral replication on CCR5-bearing cells in the Trofile assay. X4 emergence: detection of CXCR4-using viruses. Viral load set point was defined as the viral load measured in the first sample collected after HIV seroconversion. Statistically significant associations are shown in bold and were determined at p<0.05. Age was excluded since it has a disproportionate effect on disease progression.
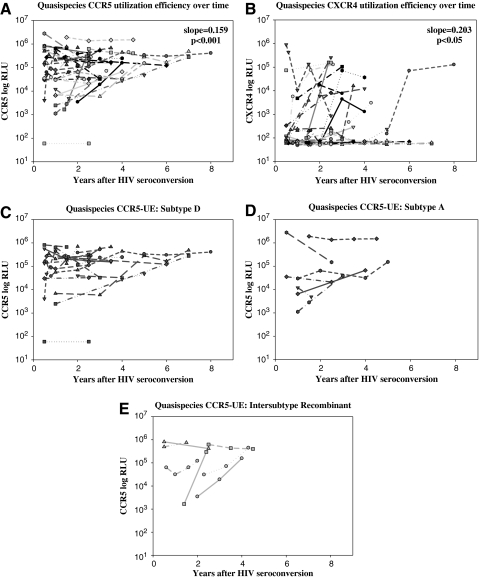
HIV quasispecies obtained from 35 of the 88 seroconverters who had samples at multiple time points available were tested with the Trofile assay to measure longitudinal changes in R5-UE and X4-UE (Table 1 and Fig. 1).24 The R5-UE of the viral quasispecies increased significantly with time after HIV seroconversion, at a rate of 0.159 log10 RLU per year (CI=0.081–0.236; p<0.001, Fig. 1A). Moreover, using the time-dependent Cox model, it was found that longitudinal measurements of R5-UE overtime were not associated with the hazard of disease progression (HR=1.02, CI 0.68–1.76, p=0.942).
FIG. 1.
Longitudinal measurements of CCR5 and CXCR4 utilization efficiency [R5-UE (A) and X4-UE (B)] are shown as relative light units (RLU) for all samples tested after seroconversion. Data from each individual are indicated by a unique symbol. A linear fixed effects model was used to examine changes in R5-UE and X4-UE over time and differences in R5-UE and X4-UE in individuals infected with different HIV subtypes; slopes and p-values are shown. No differences were observed in R5-UE or X4-UE among individuals infected with subtype A, subtype D, or intersubtype recombinant HIV (C–E, p=0.49).
Emergence of X4-tropic strains occurred in 15 of the 35 (42.9%) longitudinal study subjects and X4-UE tended to increase over time (slope=0.203, CI=0.041–0.364; p<0.05) (Fig. 1B). Eleven of those 15 individuals were infected with HIV subtype D and four were infected with intersubtype recombinant HIV strains. X4-tropic viruses were not detected in samples from any individuals infected with subtype A (n=7). It should also be noted that the increase in R5-UE over time was similar in individuals with or without X4 emergence (slope=0.151, CI=0.064–0.238 and slope=0.173, CI=0.004–0.341, respectively; p=0.81).
There were no significant differences in the change in R5-UE over time (slope of R5-UE plotted against time) when stratified by HIV subtype (A, D or intersubtype recombinant, p=0.56) (Fig. 1C–E). The change in R5-UE over time was also evaluated by stratifying the baseline R5-UE (above vs. below the median baseline value of 4.95 log10 RLU). An increase in R5-UE was observed among individuals whose baseline R5-UE value was below the median (slope=0.117 log10 RLU/year, CI=–0.006–0.239), but this was of borderline statistical significance (p=0.06). There was no trend in R5-UE over time among individuals with baseline R5-RE levels above the median (slope=0.007 log10 RLU/year, CI=–0.062–0.076, p=0.84).
Discussion
These data suggest that the R5-UE of the viral quasispecies near the time of HIV seroconversion is not associated with HIV disease progression in this population. In contrast, the emergence of X4-tropic strains was associated with advanced HIV disease, as observed in other studies.4,11,12,14 There was also no association with change in R5-UE over time and disease progression. Due to the strong association of X4-tropism with increased disease progression, we hypothesized that higher initial R5-UE would have a similar effect and associate with increased disease progression. One possibility for the lack of association of R5-UE and disease progression is that the increase in R5-UE over time makes up for any initial effects that variation in R5-UE has on disease progression. Another possibility for the lack of association is the sample size of the study. However, disease progression was associated with set point viral load, HIV subtype D, age at seroconversion, and X4 emergence, which suggests that the population should be sufficient.
The increase in R5-UE of the quasispecies over time supports the finding that R5-UE is higher later in disease.6,25 The mean rate of increase in R5-UE was approximately one log10 over 6–7 years, although this varied depending on the R5-UE level early in infection. There was no association between HIV subtype and R5-UE change over time; however, the limited numbers of individuals infected with subtype A and intersubtype recombinant HIV strains in this study may have limited the power to detect differences.
The continued increase in R5-UE within the quasispecies, even after X4 emergence, supports the previous suggestion by Van Rij et al. that there are distinct evolutionary paths for R5 and X4-tropic viral strains in the same subject.25 This may be due to a compartmentalization of the distinct viral species within different susceptible cell populations based on their HIV coreceptor expression patterns.25 In addition, we speculate that the increase in R5-UE over time could be associated with decreased R5-containing cells in the body or a decrease in the amount of R5 expressed on target cells. Because we did not examine the tropism of individual HIV variants, it is not possible to determine whether the increase in R5-UE that we observed after X4 emergence reflected more efficient CCR5 utilization by R5-tropic variants within the viral quasispecies or an increase in R5-UE in dual-tropic viruses.7,19
Acknowledgments
The authors would like to thank the participants of the RCCS and the employees of the Rakai Health Science Program. Funding for this project was provided by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). Additional funding was provided by the HIV Prevention Trials Network (HPTN), sponsored by the NIAID, National Institutes of Child Health and Human Development (NICHD), National Institute on Drug Abuse, the National Institute of Mental Health, and the Office of AIDS Research, of the NIH, Department of Health and Human Services (U01-AI068613); US Army Medical Research and Material Command, Department of the Army (cooperative agreement DAMD17-98-2-8007); Henry M. Jackson Foundation (Grants 5D43TW00010 and 2D TW000010-19 from the Fogarty Foundation, NIH); NIAID (Grant R01 AI34826); NICHD (Grant 5P30HDS06826); Fogarty Foundation (Grant 5D43W00010); and the World Bank Uganda STI Project.
Author Disclosure Statement
Wei Huang is an employee of Monogram Biosciences, the company that performed the Trofile assays.
References
- 1.Wyatt R. Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Nedellec R. Coetzer M. Shimizu N, et al. Virus entry via the alternative coreceptors CCR3 and FPRL1 differs by human immunodeficiency virus type 1 subtype. J Virol. 2009;83:8353–8363. doi: 10.1128/JVI.00780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean M. Carrington M. Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 4.Wade J. Sterjovski J. Gray L, et al. Enhanced CD4+ cellular apoptosis by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with progressive HIV-1 infection. Virology. 2009;396:246–255. doi: 10.1016/j.virol.2009.10.029. [DOI] [PubMed] [Google Scholar]
- 5.Yu X. McLane MF. Ratner L, et al. Killing of primary CD4+ T cells by non-syncytium-inducing macrophage-tropic human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1994;91:10237–10241. doi: 10.1073/pnas.91.21.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etemad B. Fellows A. Kwambana B, et al. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. J Virol. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W. Eshleman SH. Toma J, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: High prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81:7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorndal A. Deng H. Jansson M, et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjorndal A. Sonnerborg A. Tscherning C. Albert J. Fenyo EM. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retroviruses. 1999;15:647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- 10.Peeters M. Vincent R. Perret JL, et al. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: Syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 11.Blaak H. van't Wout AB. Brouwer M. Hooibrink B. Hovenkamp E. Schuitemaker H. In vivo HIV-1 infection of CD45RA(+)CD4(+) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(+) T cell decline. Proc Natl Acad Sci USA. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor RI. Sheridan KE. Ceradini D. Choe S. Landau NR. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platt EJ. Kozak SL. Kabat D. Critical role of enhanced CD4 affinity in laboratory adaptation of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000;16:871–882. doi: 10.1089/08892220050042819. [DOI] [PubMed] [Google Scholar]
- 14.Goetz MB. Leduc R. Kostman JR, et al. Relationship between HIV coreceptor tropism and disease progression in persons with untreated chronic HIV infection. J Acquir Immune Defic Syndr. 2009;50:259–266. doi: 10.1097/QAI.0b013e3181989a8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salazar-Gonzalez JF. Salazar MG. Keele BF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu T. Mo H. Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 17.Keele BF. Giorgi EE. Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollakis G. Abebe A. Kliphuis A, et al. Phenotypic and genotypic comparisons of CCR5- and CXCR4-tropic human immunodeficiency virus type 1 biological clones isolated from subtype C-infected individuals. J Virol. 2004;78:2841–2852. doi: 10.1128/JVI.78.6.2841-2852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coetzer M. Nedellec R. Salkowitz J, et al. Evolution of CCR5 use before and during coreceptor switching. J Virol. 2008;82:11758–11766. doi: 10.1128/JVI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laeyendecker O. Redd AD. Lutalo T, et al. Frequency of long-term nonprogressors in HIV-1 seroconverters from Rakai Uganda. J Acquir Immune Defic Syndr. 2009;52:316–319. doi: 10.1097/QAI.0b013e3181bc08f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutalo T. Gray RH. Wawer M, et al. Survival of HIV-infected treatment-naive individuals with documented dates of seroconversion in Rakai, Uganda. AIDS. 2007;21(Suppl 6):S15–19. doi: 10.1097/01.aids.0000299406.44775.de. [DOI] [PubMed] [Google Scholar]
- 22.Wawer MJ. Gray RH. Sewankambo NK, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 23.Collinson-Streng AN. Redd AD. Sewankambo NK, et al. Geographic HIV type 1 subtype distribution in Rakai district, Uganda. AIDS Res Hum Retroviruses. 2009;25:1045–1048. doi: 10.1089/aid.2009.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitcomb JM. Huang W. Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rij RP. Blaak H. Visser JA, et al. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J Clin Invest. 2000;106:1039–1052. doi: 10.1172/JCI7953. [DOI] [PMC free article] [PubMed] [Google Scholar]