Abstract
Background
Necrotizing enterocolitis (NEC) is a complex disease involving prematurity, enteral feeding, and bacterial effects. We propose that the underlying initial condition in its pathogenesis is reduced ability of the neonatal gut epithelial cells (NGECs) to clear oxidative stress (OS), and that when such a NGEC population is exposed to enteral feeding, the increased metabolic OS tips the population toward apoptosis, inflammation, bacterial activation, and eventual necrosis. The multi-factorial complexity of NEC requires characterization with computational modeling, and herein, we used an agent-based model (ABM) to instantiate and examine our unifying hypothesis of the pathogenesis of NEC.
Methods
An ABM of the neonatal gut was created with NGEC computational agents incorporating rules for pathways for OS, p53, tight junctions, Toll-like receptor (TLR)-4, nitric oxide, and nuclear factor-kappa beta (NF-κB). The modeled bacteria activated TLR-4 on contact with NGECs. Simulations included parameter sweeps of OS response, response to feeding, addition of bacteria, and alterations in gut mucus production.
Results
The ABM reproduced baseline cellular respiration and clearance of OS. Reduction in OS clearance consistent with clinical NEC led to senescence, apoptosis, or inflammation, with disruption of tight junctions, but rarely to NGEC necrosis. An additional “hit” of bacteria activating TLR-4 potentiated a shift to NGEC necrosis across the entire population. The mucus layer was modeled to limit bacterial–NGEC interactions and reduce this effect, but concomitant apoptosis in the goblet cell population reduced the efficacy of the mucus layer and limited its protective effect in simulated experiments. This finding suggests a means by which increased apoptosis at the cellular population level can lead to a transition to the necrosis outcome.
Conclusions
Our ABM incorporates known components of NEC and demonstrates that impaired OS management can lead to apoptosis and inflammation of NGECs, rendering the system susceptible to an additional insult involving regionalized mucus barrier failure and TLR-4 activation, which potentiates the necrosis outcome. This type of integrative dynamic knowledge representation can be a useful adjunct to help guide and contextualize research.
Necrotizing enterocolitis (NEC) is the leading gastrointestinal cause of death in neonatal infants. It is diagnosed in between one and three per 1,000 live births, with a mortality rate of 15%–25% [1]. Prematurity, initiation of enteral feeding, and a putative bacterial component are considered necessary for the development of NEC, with contributing factors including prenatal maternal or fetal insults and formula vs. maternal milk feedings [2]. Although animal models are used extensively to study NEC [3] and have provided valuable insight into its pathophysiologic mechanisms, the often-extreme degree of experimental manipulation required to generate the NEC phenotype generally represents a substantial departure from the clinical conditions associated with the disease. The relative infrequency of NEC suggests that even in neonatal infants with identified risk factors, there are robust systems in place that limit the occurrence of the disease.
The generation of NEC likely consists of a multi-factorial, cascading systems failure acting on a susceptible population. We suggest that the study of NEC requires an integrative conceptual framework to bring together experimentally identified pathophysiological mechanisms and to characterize the crucial combinations, sequences, and patterns of failure that lead to the clinical phenotype. To establish the baseline properties of this framework, we propose a minimally sufficient unifying hypothesis based on the premise that immature neonatal gut epithelial cells (NGECs) have a reduced capacity to clear reactive oxygen species (ROS) and to manage oxidative stress (OS). Impaired OS management is accentuated by the increased metabolic demands brought on by enteral feeding, leading to a population shift from apoptotic to inflammatory pathways. Alterations in the gut microenvironment, with subsequent bacterial virulence activation, can lead to exacerbations of mucosal inflammation and the eventual necrosis characteristic of the disease.
This unifying hypothesis is intended to provide the minimal set of baseline pathophysiologic factors on which additional putative mechanisms can be placed. Such an integrative analysis requires accounting for the spatio-temporal biocomplexity of NEC, and although techniques from systems biology can facilitate the integration, visualization, and manipulation of mechanistic knowledge and improve translational efforts [4–6], there is a clear need to expand beyond the level of individual cells to characterize the behavior of cell populations [7,8]. Pioneering work has been done in the application of mathematical modeling to NEC with this translational goal in mind, specifically in identifying the conditions in which the addition of probiotics might be beneficial or counter-intuitively detrimental [9]. This type of computational investigation allows the exploration of a wide range of conditions to describe, and potentially to explain, seemingly unanticipated and paradoxical behaviors. This work was performed using an ordinary differential equation model; herein, we present an alternative approach that utilizes agent-based modeling, a technique that offers specific capabilities for modeling spatially diverse, dynamic, multi-factorial systems such as NEC.
Dynamic Knowledge Representation Using Agent-Based Modeling
Agent-based modeling is an object-oriented, rule-based, discrete-event computational technique that employs a modular, scalable architecture to simulate biological systems [6,10,11]. Agent-based models (ABMs) are composed of virtual environments populated with objects (agents) that execute behaviors according to programmed rules that govern interactions with the local environment and with other agents. Individual agent behaviors can differ according to local conditions, and, in aggregate, produce population-level dynamics that represent the dynamics of the system as a whole. Agent-based modeling has been used to represent dynamically complex biological processes such as inflammation [4,12–17], cancer [18–21], infectious diseases [22–26], and wound healing [27,28]. In many biomedical applications, agents are used to represent individual cells in a system, with multiple classes of agents (cell types) sharing rules extrapolated from mechanistic knowledge obtained from in vitro experiments. The cell-as-agent is an intuitive level of resolution for biomedical agent-based modeling, as much basic research describes mechanistic processes that define cellular behavior, such as signal transduction, gene regulation, protein synthesis, and compound secretion. The advantages of cell-as-agent representation match a similar appreciation of the importance of spatial heterogeneity and population effects in ecology [29–31], immunology [24,25,32–34], and epidemiology [35,36]. By capturing the transition from individual agent behavior to the behavior of populations, ABMs are able to recapitulate non-intuitive, “emergent” behavioral patterns in dynamic systems, such as phase transitions in physical systems [37]; flocking/schooling behavior in birds [38], fish [39], and other ecological systems [40]; and quorum sensing in bacteria [41,42].
Capturing this type of aggregate behavior is crucial in the investigation of biomedical systems, because there are several hierarchies of organization between the level of the mechanism targeted for putative control (often a gene/molecule) and the clinical relevance/implications of that intervention (whole organism), each representing a potential epistemological boundary where inferred consequences at a higher level of organization cannot be assumed from mechanisms identified at a lower level. There is a need to establish a modeling relation between the model (be it an experimental biological system or simulation) and the biological referent, where the “modeling relation” is defined as the mapping of the generative processes and generated outputs between the model and its referent [10,43]. This is precisely the challenge in the translational interpretation of basic science research into NEC. A primary goal of agent-based modeling is to explore the dynamic consequences of a particular mechanistic hypothesis by extending the contextual range in which those mechanisms are manifest; i.e., at a higher level of biological organization. Dynamic knowledge representation aids in formalizing the modeling relation and fills in the gaps between the context in which mechanisms are identified (i.e., pathway information identified through in vitro experiments) and the multiple scales/contexts traversed to get to the clinical/organism level. One of the most important transitions occurs in the extrapolation of single-cell behavior into cellular population behavior. Therefore, we have chosen the cell-as-agent resolution as a means of bridging the intra-cellular molecular knowledge derived from basic experimental investigations to the population-level, space-incorporating, tissue- and organ-level context necessary to represent the system-level behavioral dynamics present in the clinical manifestations of NEC.
Materials and Methods
Overview of modeling process: Sequential and modular dynamic knowledge representation
Prior to a detailed discussion of the implementation of the NEC ABM, we relate the sequence and rationale for the model development process, as it is integral to the conclusions arrived at in this paper.
The NEC ABM was created using Netlogo 4.1.1 (available at http://bionetgen.org/SCAI-wiki/index.php/Main_Page; Supplementary Table 1, available at www.liebertonline.com/sur, contains a text description of the rules incorporated into the ABM). The agent classes were NGECs, Goblet Cells (GCs), and Bacteria. Table 1 lists these classes with brief descriptions of the cell type represented and a list of cellular functions incorporated into the agents' rule sets.
Table 1.
Agent Types, General Behaviors, and Functions Included in the Necrotizing Enterocolitis Agent-Based Model
| Agent | Description | Functions |
|---|---|---|
| NGECs | Agents that perform basic metabolic functions; secrete inflammatory mediators; regulate cell death pathways | • Cellular respiration: Nutrient consumption that leads to generation of ROS • ROS clearance: Decreases total ROS within each agent • Tight junction formation: Prevents interaction of bacteria with cells • Apoptosis: Programmed cell death without spillover of cell contents • Inflammation: Activation leads to production of mediators (TNF-α and NO·), which are both secreted. • Necrosis: Cell death by excessive inflammatory signaling with spillover of cell contents (i.e., DAMPs) |
| Goblet cells | Agents with ability to create protective mucus barrier for NGECs | • Same cellular and metabolic processes as NGECs • Secretion of mucus, which prevents interaction between NGECs and bacteria |
| Bacteria | Agents with ability to activate inflammatory pathways in NGECs through interaction with TLR-4 | • Generation of PAMPs, which activate NF-κB in NGECs via TLR-4 • Interaction with NGECs is inhibited by tight junctions and mucus |
| Reactive oxygen species | Produced by cells as part of metabolism secondary to consumption of nutrients | • Low amounts cause generation of p53 • Large amounts cause generation of free NF-κB |
| p53 | Generated by cells depending on amount of ROS; initial effector of apoptosis | • Generation of cytochrome c, which activates caspases to effect apoptosis |
| Nuclear factor-κB | Principal pro-inflammatory signaling molecule for cells | • Leads to generation of TNF-α and NO • Presence initiates production of I-κB, its inhibitor |
| Nitric oxide | Product of NF-κB inflammatory pathway | • Inhibits production of tight junction proteins • Secreted and absorbed by cells • Incorporated into total ROS |
| Tumor necrosis factor-α | Product of NF-κB inflammatory pathway | • Secreted and absorbed by cells • Activates NF-κB • Activates RIP kinase |
| RIP kinase | TNF-α receptor-mediated inflammatory pathway | • Leads to necrotic cell death • Spillover of NO and TNF-α to surrounding cells • Release of DAMPs |
| TLR-4 | Receptor that recognizes DAMPs and PAMPs | • Activates NF-κB |
| PAMPs | Secreted products of bacteria | • Activates NF-κB |
| Mucus | Secreted product of goblet cells | • Prevents interaction between PAMPs and TLR-4 |
DAMP=damage-associated molecular pattern; I-κB=nuclear factor kappa-beta inhibitor; NF=nuclear factor; NF-κB=nuclear factor kappa-beta; NO·=nitric oxide; NGEC=neonatal gut epithelial cells; PAMP=pathogen-associated molecular pattern; RIP=receptor-interacting protein; ROS=reactive oxygen species; TLR=Toll-like receptor; TNF-α=tumor necrosis factor-alpha.
Modeling NGECs as computational agents
The NGECs were modeled to perform nutrient metabolism, senescence, apoptosis, inflammation, and necrosis. The agent rules representing molecular processes such as receptor activation, signal transduction, metabolism, and transcription factor effects were expressed using a detailed qualitative approach [44,45] that consists of relatively detailed component representation (i.e., enzymes, molecular species, and genes) with qualitative representation of biochemical kinetics using a logic-based, algebraic rule construction. As a result, molecular interaction rules are expressed as conditional statements of the form:
If Ligand A is present, then bind to and activate Receptor B;
If Receptor B is activated, then increase Signal Transduction Enzyme C by 1; etc.
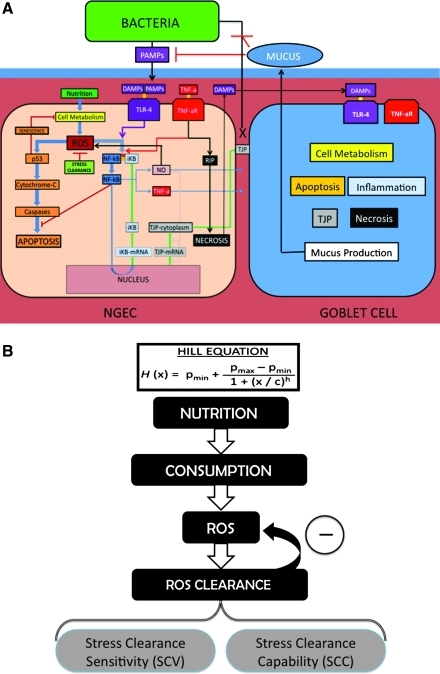
The exception to this type of rule construction is the use of Hill functions to represent feedback control loops for metabolic stress homeostasis (see “Modeling ROS Management” below). See Figure 1A for an overview of the pathways incorporated and represented as rules in the NGEC agents; Table 1 lists the agent classes, their descriptions, and their functions; Supplement 1 includes detailed descriptions of the specific rules that were programmed into the NEC ABM.
FIG. 1.
Overview of agent classes and functions in necrotizing enterocolitis (NEC) agent-based model (ABM) and schematic for Hill equations. (A) Interactions between states of the three cell types represented in the NEC ABM: Neonatal gut epithelial cells (NGECs), goblet cells, and abstracted bacteria. Intracellular pathways of NGECs are displayed in detail and color-coded according to function: Cellular metabolism (yellow), reactive oxygen species (ROS) generation and management (green), senescence and apoptosis (orange), nuclear factor kappa-beta (NF)-κB (blue), tight junction (TJ) metabolism (gray), Toll-like receptor (TLR)-4 (violet), tumor necrosis factor (TNF)-α (red), nitric oxide (pink), and necrosis (black). Goblet cells include all these pathways (represented abstractly as block labels) plus the ability to produce mucus. Abstracted bacteria have movement rules, release pathogen-associated molecular patterns (PAMPs), and can adhere to NGECs with impaired TJs if there is no mucus intervening. Detailed descriptions of the agent rules and interactions can be found in Supplement 1. (B) How NGEC metabolism and oxidative stress (OS) management are modeled with Hill equations. Each NGEC incorporates the following pathway for ROS management, with a unique Hill equation used to represent each part of the pathway. The replenishment rate for nutrients (nutrient inflow) in the system was adjusted on scale by a variable in its Hill equation. Hill equations for ROS and ROS clearance interact to produce a final, overall metabolic ROS value. The interactions between the Hill equations represent the control architecture of management of baseline metabolic OS. For details on the specific Hill equations, see Supplement 1. Abbreviations: DAMPs=damage-associated molecular patterns; IκB=NF-κB inhibitor; RIP=receptor-interacting protein kinase.
Modeling ROS management
Our central hypothesis requires representing how each NGEC produces and manages the ROS generation from cellular metabolism. The generation of ROS by cellular respiration has been well established [46]. We used a series of Hill function equations to model the relations among nutrients, cellular consumption, ROS generation, and ROS clearance (Fig. 1B; Supplement 1). Hill equations often are used to model the dose–response relations between a receptor and ligand mathematically, producing a sigmoid (S-shaped) curve. Hill equations were used in the NGECs to: (1) Approximate the kinetics of the cell's graduated generation of ROS secondary to metabolism; and (2) represent the response of the cell's intrinsic oxidative stress clearance machinery to remove the ROS produced. We limited the coverage of the Hill equations to the management of metabolism-induced stress; we recognize that there is a cross-over between this type of OS and that brought on by exogenous ROS, but for the purposes of separating internal cellular homeostasis and the NGEC response to external stimuli, we deal with these two types of OS with different modeling modules for different cellular pathways, which are merged downstream to determine the cellular fate (see Fig. 1A and the section on “Modeling the Effects of Stress”).
Variables were created to represent nutrients, cellular consumption, and metabolism-induced ROS generation. Nutrient availability was modeled using a patch variable, and spatial heterogeneity of the bioavailability of nutrients was simulated by a random distribution of the amounts of nutrients at the start of the experiment. Enteral feeding was simulated by having each patch regenerate nutrients at a set global rate based on the amount of the feeding (alterable for each experiment). Rates of consumption and ROS generation were calculated by Hill equations for each individual NGEC. Individual NGECs decreased their rate of consumption depending on the overall amount of p53 present within the cell, as discussed in detail below (“Modeling the Effects of Excess Stress”). Variables determining cellular ROS management were modeled on the basis of two concepts: stress clearance sensitivity (the ability to detect ROS) and stress clearance capability (the ability to neutralize ROS)(Fig. 1B). Both variables manipulated separate parts of a single Hill equation for stress clearance, but for these simulations, the choice was made to focus on alterations in stress clearance capability under the rationale that immaturity might be manifest as a decrease in enzyme capability rather than increased responsiveness to stimuli.
We hypothesized that NEC is manifested only within susceptible populations; i.e., those with reduced ability to clear OS across the population, such that some critical percentage of the NGECs undergo the transition to the inflammatory and necrosis-generating phenotype. A random-normal distribution of stress clearance capability, with a set upper bound for each population called the stress clearance capability maximum (SCCmax), was used to assign a stress clearance capability to individual NGECs, thereby simulating a heterogeneous population within the NEC ABM. The upper-bound stress clearance capability value was decreased incrementally for each experiment, thus simulating different NGEC populations. Stress clearance capability values were determined relative to an absolute value (AbSCCmax), arbitrarily set at 0.500 with no units and representing fully mature OS management. The variable for stress clearance sensitivity could be manipulated prior to the start of each experiment, but in our experiments, this value was kept constant for all NGECs for all experimental runs because our hypothesis focused on immaturity of the OS management capability, not sensing, although we recognize that the latter hypothesis may be a legitimate one.
Modeling the effects of stress: Role of p53 and pathway to apoptosis (Fig. 1A: Pathway Orange)
An NGEC state variable was used to represent the excess or “overflow” amount of metabolism-generated OS remaining after the NGEC clearance function (i.e., the amount of metabolism-derived ROS that could not be cleared by the baseline management system). We recognize that metabolism-derived ROS is not the only OS seen by the cells: Inflammatory conditions are associated with extracellular sources, such as ROS, nitric oxide (NO·), and secondary metabolites produced by inflammatory cells. For modeling purposes, we made distinctions between OS generated as a result of metabolism, externally generated ROS (produced by inflammatory cells and not included in the current ABM), and NO·.
The first distinction was based on the need to separate metabolism-derived and inflammatory cell-generated ROS in order to represent explicitly the presumptive mechanism underlying our central hypothesis: That immature NGECs have reduced OS management capability, and, as a result, otherwise-manageable byproducts of cellular respiration will place these immature NGECs at risk for additional insults (i.e., increased system fragility). Although the effects of exogenous ROS may be important in the propagation of NEC, this process presupposes an initiating factor that starts the inflammatory cycle. The focus of our investigation was identification of those initiating factors and an attempt to establish a causative chain of events that meshed with the known subsequent contributory factors. As a result, exogenous sources of ROS, either from inflammatory cells or secreted by epithelial cells, were not included in the ABM.
Alternatively, because the NO effect on tight junction metabolism was incorporated into the ABM (see “Modeling Tight Junction Protein Metabolism” below), OS effects resulting from NO· and its secondary metabolites (abstractly represented together with a variable called “NO”) were included in the NGECs. The total amount of OS sensed by the NGEC was represented by the sum of internally generated metabolic stress (from the Hill equations) plus a scale-corrected value of NO· present in the cell. The OS level affects the role of the protein p53, which is generated by cells in the presence of damaged DNA resulting from oxidative stress [47,48]. This p53 is a crucial control point for senescence [49] to allow DNA repair; in these cells, metabolism is substantially decreased, allowing the ROS to be cleared and the cell eventually to return to a normal state. The p53 also is a direct effector of apoptosis through stimulating the release of mitochondrial cytochrome c and recruitment of caspases [50]. The exact mechanism by which cells choose between senescence and apoptosis is unknown: For purposes of the NEC ABM, we represented three categories of “stress,” each tied to a different means of handling p53:
Low Stress, where metabolism and consumption were governed by the baseline Hill equations; in this state, the NGEC p53 amounts would decrease over time.
Mid-Range Stress, where p53 was produced and, as it rose, shifted the metabolism-governing Hill function to reduce consumption (i.e., produced senescence). This allowed metabolism-derived OS to decrease until the amount of stress dropped into the Low Stress range, at which point, p53 decreased.
High Stress, reached when p53 continued to be produced until the concentration crossed a set threshold leading to the generation of variables for cytochrome c and caspase proteins that subsequently activated apoptotic mechanisms. This level of stress also activated inflammatory pathways (see below in “Modeling the Effects of Excess Stress.”)
These two thresholds for p53 activity (with the resulting three categories of behavior) were set during calibration of the model and are consistent with the different roles of p53 in different stress states.
In our model, a cell undergoing apoptosis would continue to exist in the ABM environment but would no longer interact with other agents in the system or carry out metabolic processes; this arrangement mimics the non-inflammation-generating characteristics of apoptosis. We abstracted out cellular reproduction and as a result made the modeling decision that apoptotic cells would be replaced through cellular division and migration. Each NGEC also had a minute random chance of undergoing apoptosis.
Pathways to inflammation and necrosis
The cellular pathways incorporated in the current NEC ABM up to this point did not include inflammatory pathways to necrosis. In fact, the p53 control mechanism operating within the Mid-Range Stress is extremely robust in terms of preventing apoptosis secondary to metabolism; this is consistent with the general recognition that metabolism-generated OS usually is not relevant pathophysiologically. Furthermore, because apoptosis is recognized as a non-inflammatory cell fate, the next modeling step involved adding pathways for inflammation and necrosis to the ABM in a fashion consistent with published knowledge. The first step was to simulate the initiation of inflammation arising from a failure point in the p53 OS response system where excess OS activated inflammatory pathways leading to nuclear factor kappa-Beta (NF-κB). Additionally, exogenous inflammatory triggering was included through a representative pathogen-associated molecular pattern (PAMP)/damage-associated molecular pattern (DAMP) signaling pathway, Toll-like receptor 4 (TLR-4). We hypothesized that both of these components are necessary in the cascading systems failure that leads to NEC.
Modeling effects of excess stress: NF-κB and tumor necrosis factor-α (TNF-α)(Fig. 1A: Pathways Blue and Red)
The analysis of the effect of excess OS overriding the p53 control system focused on the role of NF-κB. The behavior of NF-κB was modeled to reflect its dis-inhibition activation mechanism, well described in the literature [51]. At baseline non-activation, a variable for NF-κB was bound to a variable for iκB, its inhibitor. Activation of NF-κB resulted from its separation from iκB and the release of a free NF-κB variable. In a negative feedback control loop, free NF-κB resulted in the generation of variables for iκB mRNA and, consequently, iκB protein. Any free iκB would immediately bind to and decrease free NF-κB. Both iκB mRNA and protein were produced at the baseline state until all free NF-κB was bound. In the literature, NF-κB has been documented to be activated by the administration of exogenous free radicals [52,53]; we modeled this effect by having NO positively feed back on the total OS level of the NGECs. When the oxidative stress level rose into the High Stress category, the NF-κB pathways were activated. The presence of free NF-κB in the cell was made to increase the production of variables for TNF-α [54] and NO [55–57]. The TNF-α and NO were secreted to surrounding patches. Also, NF-κB directly decreased the variable for cytochrome c, thus inhibiting apoptosis [58,59].
The modeled effects of TNF-α were derived from the literature also [60]. After secretion, TNF-α activates the NF-κB pathway by interacting with a variable for TNF-α receptor (TNF-αR), which is expressed constitutively on all NGECs. By interacting with TNF-αR, TNF-α increases a variable representing receptor-interacting protein (RIP) kinase, an effector of necrosis [61]. As opposed to apoptotic cells, NGECs undergoing necrosis would diffuse their variables for TNF-α and NO· into surrounding patches; this reflects the pro-inflammatory nature of necrosis vs. the non-inflammation-inducing nature of apoptosis.
Modeling the influences of microbial pathogens and cellular damage: TLR-4 (Fig. 1A: Pathway Violet)
A variable for TLR-4 was created based on a review of the literature [62,63]. Variables for PAMPs and DAMPs could interact with TLR-4. We recognize that there are other pathways for responding to PAMPs/DAMPs; however, the contribution of TLR-4 has been studied extensively in NEC, and for abstraction purposes, we focused the representation of the PAMP/DAMP response within the current ABM to the TLR-4 pathway. The PAMPs were modeled as resulting from bacteria (see section below on “Modeling Bacteria as Computational Agents”), whereas DAMPs were generated by necrotic NGECs (represented by an abstract variable called “damage-signal”). Downstream signal transduction through the TLR-4 pathway was highly abstracted and resulted in activation of NF-κB. Following this point, NF-κB activity followed the same pathway as that resulting from TNF-α activation. The importance of this pathway is that it provided an additional and alternative route for the activation of NF-κB.
Modeling tight junction protein metabolism (Fig. 1A: Pathway Gray)
All NGECs maintain tight junctions, which prevent direct interaction with bacteria. Rules for tight junction formation and metabolism were derived from the literature [64]. Although the actual cellular tight junction complex is composed of several families of proteins, we abstracted it into a single, unified Tight Junction protein variable, which was generated from variables for tight junction protein mRNA, translated into a cytoplasmic tight junction protein, then localized to the cell membrane as the final, functional complex. This abstracted model was sufficient to interact with variables for other cellular products, such as NO·, which degraded tight junction mRNA and cytoplasmic proteins [65].
Modeling GC dynamics
An agent sub-class was created for GCs; they were represented as a subgroup of NEGC agents. In addition to incorporating all the stress management and response pathways present in the NGECS, they included rules governing the production of mucus [66–68]. Goblet cells increased patch variables for mucus, which was diffused to surrounding patches. Sufficient amounts of mucus prevent interaction between bacteria and NGECs. However, GCs could be subjected to the same risk of apoptosis as the rest of the NGECs, extending our unifying hypothesis of impaired OS management to GC populations. The importance of this model component is that it provides a pro-necrosis pathway (for the entire population) that arises from apoptosis, which normally is considered an inflammation-damping trajectory. As GCs were depleted through OS-induced apoptosis, there was a reduction of the mucus layer and a greater likelihood of bacteria–NGEC contacts. It should be noted that this latter effect manifested only at the mixed cellular population level and therefore constituted the type of emergent property that challenges the translation of cellular mechanism to tissue and organ function.
Modeling bacteria as computational agents
An agent class was created for generic “Bacteria.” The representation of bacteria in this ABM was extremely abstract: They were able to move independently and to adhere to NGECs in the absence of adequate mucus and intact tight junctions. On attachment, bacteria would increase a variable for PAMPs, which interacted with a variable for TLR-4.
Executing the model: Simulation time scale
A variable for time was created based on each “tick” of the model, which is one execution cycle of the ABM's code. One tick represents 5 min of simulated time. Twelve ticks were made to correspond to 1 h, which were subsequently measured in days.
Simulation experiment outputs and conditions
All experiments were run for seven days of simulated time. To establish the metrics of model behavior, we defined five simulation outcomes. The first three were based on what cell fate type represented the majority of NGECs at the end of a simulation run (>50% or >312 cells): (1) Necrosis, when the majority of NGECs underwent necrosis; (2) Apoptosis, when the majority of NGECs underwent apoptosis; and (3) Healthy, when the majority of the NGEC population did not undergo either necrosis or apoptosis. There also were two classes of outcome interposed when no single outcome type was in the majority: (1) Mixed Necrosis–Apoptosis, when the number of necrotic and apoptotic NGECs was greater than the number of apoptotic and healthy NGECs; and (2) Mixed Apoptosis–Healthy, when the number of apoptotic and healthy NGECs was greater than the number of necrotic and apoptotic NGECs.
To map the behavior of the ABM to the clinical manifestation of NEC, we further defined a “survival outcome” as a non-Necrosis outcome. The rationale for this division is that, given the inflammation-damping effects of apoptosis, we did not consider apoptotic NGEC populations either as a proxy for the NEC phenotype or necessarily to be a pathway to the bowel necrosis seen in clinical NEC. We abstracted out NGEC replication and regeneration and therefore considered apoptotic NGECs as being replaced without negative consequences within the timeframe of the simulated experiments (seven days). Furthermore, although we defined the Necrosis outcome as representing the clinical manifestation of NEC, the presence of the Mixed Necrosis–Apoptosis outcome also represented a potentially interesting subset of behaviors by the NEC ABM. These simulations represented a group where “patches” of necrotic NGECs were present, but these areas did not propagate throughout the system. This group may therefore represent “NEC scares,” with corresponding implications for additional investigation. However, for the current set of experiments, the focus was on establishing the level and patterns of outcome present in the NEC ABM.
Successive parameter sweeps to indentify clinically relevant model configurations
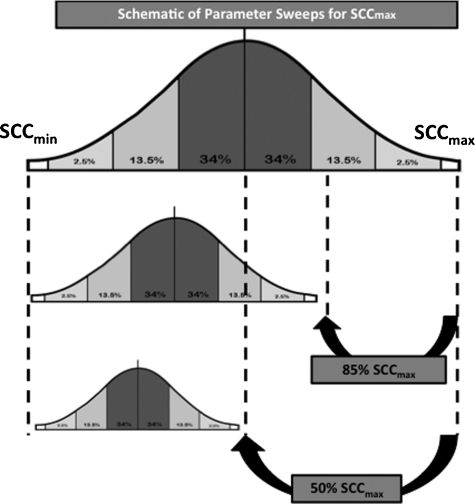
Initial experimental simulations consisted of parameter sweeps of the maximum Stress Clearance Capability (SCCmax) with maximum feeding and without bacteria or goblet cells. Maximum feeding in the absence of bacteria or goblet cells was used to establish a baseline behavior space of the NGEC populations and to determine tipping points of the NGEC OS management system in terms of health, apoptosis, or necrosis. The SCCmax was decreased incrementally across the entire range of values, thus placing the NGEC population distribution of Stress Clearance Capacity in progressively narrower ranges (see Fig. 2 for a schematic depiction of this process). The overall rates of the five outcome classes were recorded across the SCCmax range (from 0–0.500, representing 0%–100% AbSCCmax) to identify transition points between outcomes and thus establish an SCCmax value or range of values that could correspond to the clinical manifestations of an at-risk-for-NEC population. The defined properties of the at-risk-for-NEC population were: (1) The population could not always be healthy (i.e., there had to be a possibility that necrosis would arise); and (2) the population should not generate necrosis spontaneously with any significant frequency (i.e., clinical NEC is a relatively rare occurrence).
FIG. 2.
Schematic description of parameter sweeps performed for maximum stress clearance capability (SCCmax). Population of neonatal gut epithelial cells (NGECs) in the necrotizing enterocolitis (NEC) agent-based model (ABM) has a normal distribution of values for SCC for each cell; this allows intercellular heterogeneity within the population. This normal distribution has an upper bound, represented by the SCCmax. Absolute maximum SCCmax value in the NEC ABM was 0.500 (arbitrarily set and unit-less). When performing parameter sweep, the SCCmax was decreased sequentially, with the normal distribution of SCC values falling into progressively narrower ranges, resulting in NGEC population with progressively impaired oxidative stress (OS) management capability. This figure presents examples of distributions of NGEC SCC at SCCmax of 100% (representing fully mature OS management capability), 85%, and 50%.
A range of SCCmax values that met these criteria was then used as the basis for the addition of other ABM components, specifically bacteria (activating TLR-4) and GCs (producing a mucus barrier). Additional parameter sweeps were performed within the limited SCCmax range to narrow the at-risk-for-NEC population further. These additional model components were added as separate agent classes that provided “external” inputs to the NGECs, thereby altering the dynamic consequences of the NGEC's internal rules (i.e., the signaling and metabolic pathways seen in Fig. 1A) and resulted in alterations in the rates of apoptosis and necrosis for the NGEC population as a whole. For each simulation run, bacteria and GCs were added to the experiments after an NGEC ROS management equilibrium point was reached, defined as stabilization of tight junctions. The influences of bacteria and GCs were tested incrementally, bacteria first, then with GCs, to determine their effect on the rates of apoptosis and necrosis phenotype generation in the NGEC population.
Results
The rules and functions in the NEC ABM were implemented successfully, with the NGECs able to consume nutrients, generate ROS, and manage ROS on the basis of the Hill equations. An NGEC with 100% AbSCCmax (value 0.500) exhibited no generation of p53 or NF-κB activity regardless of the amount of nutrients available and consumed, consistent with the accepted concept that in mature cells under normal conditions, metabolism-generated OS do not represent a significant OS burden. As the parameter sweep progressed with lower values of SCCmax, the individual NGECs sequentially entered the functions associated with greater stress and correspondingly produced values for p53, NF-κB, NO·, and TNF-α.
Results of SCCmax parameter sweep with full feedings, no bacteria, and no GCs (−Bact/−GC)
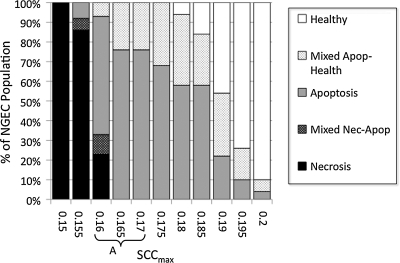
The initial parameter sweep of SCCmax was performed in the absence of bacteria but with full enteral feeding to establish the baseline behavior space of the NGEC population. The range of interest consisted of values of SCCmax between where the NEC ABM nearly always had a Healthy outcome (and thus representing “mature” OS management capability) and where the NEC ABM always underwent Necrosis (representing a non-survivable impairment of OS management capability). We identified this range as being bounded by values of 0.150, or 30% AbSCCmax, and 0.200, or 40% AbSCCmax, the percentages reflecting the proportional amount of reduction of the SCC distribution within each population. The NEC ABM outcomes within this range are shown in Figure 3. The relative trends of each of the five outcomes followed an expected pattern, where the number of more severe outcomes decreased as the values of SCCmax increased. Furthermore, with “survival” defined as non-Necrosis outcomes, there was a transition zone between SCCmax values of 0.160 (32% AbSCCmax) and 0.165 (33% AbSCCmax), where the Necrosis outcome disappeared. Given that this set of simulations did not include the bacterial component and the reasonable assumption that the addition of the bacterial component would increase the likelihood of severe outcomes, the range of interest was extended one interval to include the SCCmax 0.170 (34%) population to represent at-risk-for-NEC conditions. This range is denoted by Letter A in Figure 3 and was used for subsequent additions of bacteria and GCs. In order to provide a frame for comparison, a finer-grained parameter sweep with full feedings, no bacteria, and no GCs was run between SCCmax 0.160 (32%) and 0.170 (34%) in increments of 0.001. The result of this parameter sweep is shown in Table 2 and Figure 4 and demonstrates a transition point with respect to the presence of the Necrosis outcome between SCCmax 0.162 and 0.163.
FIG. 3.
Results of parameter sweep from absolute maximum stress clearance capacity (SCCmax) (AbSCCmax) from 30% to 40% with maximum feedings, no bacteria, and no goblet cells (−Bact/−GC). Shown are the results of experimental runs (n=100 each for seven days' simulated time) performed for SCCmax values between 0.150 and 0.200 at 0.005 increments representing 30% AbSCCmax and 40% AbSCCmax at 1% increments. Simulations in this range uniformly resulted in the Necrosis (Nec) outcome, whereas above this range, simulation runs essentially always produced the Healthy outcome. Trends of the five possible outcomes followed expected pattern, where more severe outcomes were present at lower levels of SCCmax (representing more impairment of SCC). There is a transition zone between SCCmax values of 0.160 (32% AbSCCmax) and 0.165 (33% AbSCCmax) in terms of “survival” (defined as non-Necrosis outcomes) where the Necrosis outcome disappears. Given the assumption that the addition of the bacteria would lead to more severe outcomes, SCCmax 0.170 (34% AbSCCmax) was included in the range of interest representing the at-risk-for-NEC conditions, denoted by A. Apop=apoptosis. NGEC=neonatal gut epithelial cells.
Table 2.
Results of Parameter Sweeps of SCCmax between Values of 0.160 to 0.170 in Increments of 0.001 in Three Groups of Simulated Experimentsa
| |
No bacteria or goblet cells |
Bacteria, no goblet cells |
Both bacteria and goblet cells |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCCmax | Nec | Nec/apop | Apop | Apop/health | Health | Nec | Nec/apop | Apop | Apop/health | Health | Nec | Nec/Apop | Apop | Apop/health | Health |
| 0.160 | 23 | 10 | 60 | 7 | 0 | 92 | 4 | 0 | 4 | 0 | 30 | 26 | 0 | 44 | 0 |
| 0.161 | 6 | 15 | 75 | 4 | 0 | 74 | 19 | 0 | 7 | 0 | 14 | 17 | 7 | 62 | 0 |
| 0.162 | 7 | 9 | 79 | 5 | 0 | 63 | 23 | 0 | 14 | 0 | 4 | 11 | 1 | 86 | 0 |
| 0.163 | 0 | 8 | 77 | 15 | 0 | 45 | 29 | 0 | 26 | 0 | 1 | 6 | 6 | 89 | 0 |
| 0.164 | 0 | 1 | 77 | 22 | 0 | 40 | 26 | 0 | 34 | 0 | 0 | 2 | 5 | 97 | 0 |
| 0.165 | 0 | 0 | 76 | 24 | 0 | 17 | 23 | 0 | 60 | 0 | 0 | 0 | 8 | 92 | 0 |
| 0.166 | 0 | 0 | 82 | 18 | 0 | 9 | 21 | 0 | 70 | 0 | 0 | 0 | 3 | 97 | 0 |
| 0.167 | 0 | 0 | 77 | 23 | 0 | 5 | 12 | 0 | 83 | 0 | 0 | 1 | 11 | 88 | 0 |
| 0.168 | 0 | 0 | 74 | 26 | 0 | 2 | 2 | 0 | 96 | 0 | 0 | 0 | 8 | 92 | 0 |
| 0.169 | 0 | 0 | 76 | 24 | 0 | 0 | 3 | 0 | 97 | 0 | 0 | 0 | 5 | 95 | 0 |
| 0.170 | 0 | 0 | 71 | 29 | 0 | 1 | 3 | 0 | 96 | 0 | 0 | 0 | 9 | 91 | 0 |
FIG. 4.
Finer-grained parameter sweep of maximum stress clearance capability (SCCmax) between 0.160 and 0.170 with full feedings, no bacteria, and no goblet cells (−Bact/−GC). A parameter sweep to the range of SCCmax corresponding to Letter A in Figure 3 was performed between SCCmax 0.160 (32%) and 0.170 (34%) at increments of 0.001. This was done to form a frame of reference between the −Bact/−GC conditions reflecting solely the effects of maximum feedings with subsequent addition of bacteria and goblet cells (see Figs. 5–7). There is a transition point with respect to the Necrosis (Nec) outcome between SCCmax 0.162 and 0.163. Apop=apoptosis. NGEC=neonatal gut epithelial cells.
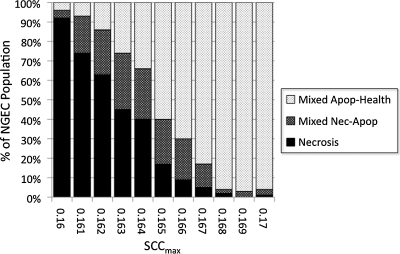
Results of SCCmax parameter sweep with full feedings, bacteria, and no GCs (+Bact/−GC)
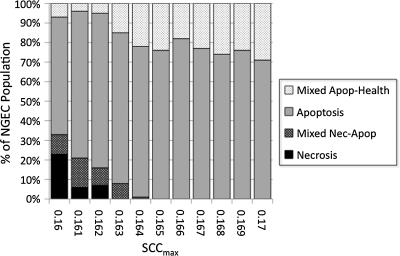
Bacterial agents were added to the NEC ABM, and a series of simulations was performed for SCCmax values between 0.160 (32% AbSCCmax) and 0.170 (34% AbSCCmax) at intervals of 0.001 (N=100 simulation runs). The results of these simulations is shown in Figure 5. There was the expected greater severity of outcome for each value of SCCmax. Notably, the absence of the Necrosis outcome was shifted from SCCmax 0.163 in the −Bact/−GC group to an SCCmax range of around 0.169 to 0.170 (sporadic) in the +Bact/−GC group. This shift demonstrated the inflammation- and necrosis-potentiating effect of activation of the TLR-4 pathway by bacteria and helped identify the potential range of SCCmax approximating the relative rarity of clinical NEC (values of 0.167–0.170 with a Necrosis outcome incidence of 0–6%; see Table 2). Furthermore, these experiments documented that the Apoptosis outcome disappeared in all simulation runs. Our interpretation is that NGECs tending toward apoptosis were forced into a binary decision by the activation of the PAMP/DAMP signaling pathways. This finding suggested that a substantial effect of bacteria in NEC could provide additional selection pressure, forcing those NGECs with sub-borderline OS management capability to progress to necrosis but, conversely, allowing NGECs just above this threshold to escape into recovery trajectories.
FIG. 5.
Parameter sweep of maximum stress clearance capability (SCCmax) between 0.160 and 0.170 with full feedings, bacteria, and no goblet cells (+Bact/−GC). Bacterial agents were added to the necrotizing enterocolitis agent-based model, and a series of simulations was performed for SCCmax values between 0.160 (32% AbSCCmax) and 0.170 (34% AbSCCmax) at intervals of 0.001 (n=100 simulation runs). There was an expected greater severity of outcome for each value of SCCmax at each value of SCCmax with the threshold for the absence of the Necrosis (Nec) outcome shifted from SCCmax 0.163 in the −Bact/−GC group to an SCCmax range of around 0.169 to 0.170. Furthermore, the Apoptosis (Apop) outcome disappeared in all simulation runs. Our interpretation is that bacterial activation of the pathogen-associated and damage-associated molecular pattern signaling pathways (represented in the NEC ABM by Toll-like receptor-4) provides a group-separating selection pressure on apoptotic neonatal gut endothelial cells (NGECs) to either progress to necrosis or “escape” toward recovery.
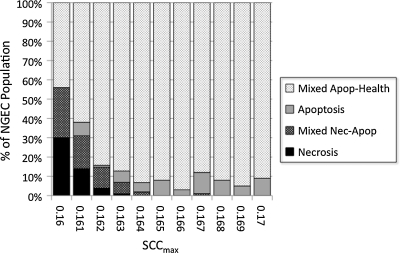
Results of SCCmax parameter sweeps with full feedings, bacteria, and GCs (+Bact/+GC)
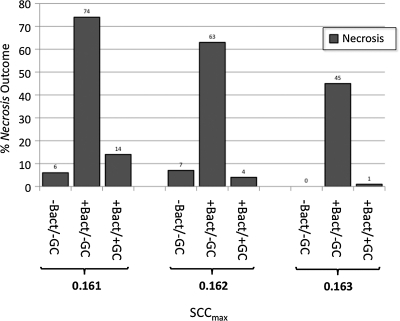
Goblet cells and a mucus layer were added to the NEC ABM, and a series of simulations was performed for SCCmax values between 0.160 (32% AbSCCmax) and 0.170 (34% AbSCCmax) at intervals of 0.001 (N=100 simulation runs). Mucus was implemented in the NEC ABM such that the presence of an intact layer of mucus blocked the interaction between bacteria and NGECs, preventing bacterial adherence and subsequent activation of the TLR-4 pathway by PAMP. Therefore, it was expected that there would be a leftward shift in the severity of outcome and perhaps a return of Apoptosis. This is seen to some degree in Figure 6, where there is a clear protective effect compared with the behaviors documented in the +Bact/−GC experiments (see Fig. 5), but not a complete return to the dynamics seen in the −Bact/−GC simulations (see Fig. 4). An incomplete protective effect of GCs and mucus is seen in Figure 7, a comparison of the relative behaviors of the three groups for a range of SCCmax (0.161–0.163) that produced Necrosis outcomes at a degree approximating the clinical rarity of NEC. Our interpretation of this finding is that apoptosis of GCs and subsequent depletion of their population led to reduced mucus production for the NEC ABM as a whole, with regional differences based on the location of apoptotic GCs. These areas depleted of mucus then became entry points for bacterial–NGEC interactions that tipped the NGECs toward inflammation and necrosis.
FIG. 6.
Parameter sweep of maximum stress clearance capability (SCCmax) between 0.160 and 0.170 with full feedings, bacteria, and goblet cells (+Bact/+GC). Goblet cells and mucus layer were added and simulations performed for SCCmax values between 0.160 (32% AbSCCmax) and 0.170 (34% AbSCCmax) at intervals of 0.001 (n=100 simulation runs). Mucus in the necrotizing enterocolitis agent-based model blocks interaction between bacteria and neonatal gut endothelial cells (NGECs), preventing bacterial adherence and subsequent activation of Toll-like receptor-4 pathway by pathogen-associated molecular patterns. There is an expected leftward shift in the severity of outcome (improved survival) compared with +Bact/−GC runs, seen in Figure 5, but not a complete return to the dynamics found in the −Bact/−GC simulations, seen in Figure 4. Apop=apoptosis; Nec=necrosis.
FIG. 7.
Comparison of relative number of Necrosis (Nec) outcomes with addition of bacteria (Bact), and then goblet cells (GCs)/mucus for range of stress clearance capability (SCCmax) 0.161 to 0.163). This range of SCCmax values produces Necrosis outcomes to a degree approximating clinical rarity of necrotizing enterocolitis. Although protective effect of mucus is pronounced (compare +Bact/−GC with +Bact/+GC), it is not complete (−Bact/−GC vs. +Bact/+GC). This latter finding suggests that apoptosis (Apop) of GCs and subsequent depletion of their population leads to regionally reduced mucus production. These areas depleted of mucus then become entry points for bacterial–neonatal gut epithelial cell (NGEC) interactions that tip NGECs toward inflammation and necrosis.
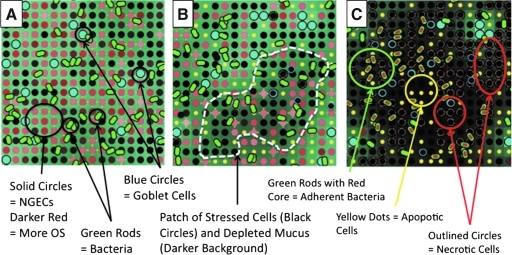
A representative series of NEC ABM screenshots can be seen in Figure 8, demonstrating: (1) Normal intact mucus in Low Stress conditions; (2) a High Stress circumstance with patchy areas of apoptotic NGECs and GCs and corresponding mucus depletion; and (3) the subsequent progression to necrosis, with adherent bacteria and areas of necrotic cells. Significantly, this mixed population dynamic provides a means by which apoptosis, normally considered an inflammation-dampening process, could contribute to the generation of an overall inflammation–necrosis behavioral pattern at the system level.
FIG. 8.
Representative screenshots of necrotizing enterocolitis agent-based model, demonstrating intact mucus, patchy apoptosis, and necrosis. The topology of the model is a square grid. Neonatal gut epithelial cell (NGEC) agents are represented as solid circles of various shades of red: Lighter shades denote Low Stress, whereas deepening red corresponds to greater levels of stress, leading to black solid circles. Bacterial agents are rod-shaped: Green if non-adherent, green outline with red cores if adherent and activated. Goblet cells are represented by larger light blue circles. Apoptotic cells are shown as small yellow circles. Necrotic cells are represented by outlined (in red for NGECs, blue for GCs) black circles. The amount of mucus is represented by green shading of the background: Lighter shades correspond to adequate amounts of mucus, whereas progressively darker areas reflect mucus depletion. (A) Viable goblet cells with an intact mucus layer under only mild to moderate stress. Bacteria are present but impeded from interacting with NGECs. (B) A more highly stressed system, with patchy areas of highly stressed cells with corresponding areas of mucus depletion (white outlined areas). Bacteria have not yet become adherent, and there are rare necrotic NGECs. (C) Progression to necrosis, with clusters of adherent bacteria and patches of apoptotic and necrotic cells. OS=oxidative stress.
Discussion
Despite decades of research, there still is no clear delineation of the pathogenesis of NEC. We have applied ABM to the body of literature regarding this disease to test our hypothesis that inability to manage OS adequately in NGEC populations is an initiating factor in the pathogenesis of NEC. Our ABM implemented cellular signaling pathways in NGECs concerning cellular respiration, OS management, apoptosis, and inflammation and integrated them with factors known to affect the pathogenesis of NEC—enteral feeding, bacterial effects, and mucus barrier integrity—to generate a plausible set of behaviors consistent with the clinical incidence and dynamics of NEC.
The difficulty in solving the puzzle of the pathogenesis of NEC is reflected in its broad clinical presentation, ranging from simple feeding intolerance and abdominal distention to pneumatosis intestinalis and full-thickness necrosis. There is an ill-defined relation between therapy and outcome. For instance, a “NEC scare,” treated by bowel rest and antibiotics, may, in one situation, be associated with improvement, whereas in another, it may be followed by full-blown NEC and patient death. Our current understanding of the pathophysiological trajectory of NEC is insufficient to allow the parsing of what works, what does not, and what might be incidental to the outcome. The literature concerning NEC is broad, covering detailed mechanisms of cell injury, inflammation, and immunity [69]; properties of the gut microbiome [70]; and the contributory effects of both commensal [71] and pathogenic [72] organisms. However, despite this extensive study and characterization, the transition from the presence of risk factors and a clinical suspicion of NEC to full-blown NEC remains a significant diagnostic challenge and an area of intensive investigation. This is partly because the paucity of real-time “pre-”surgical NEC data at the intestinal level related to local biochemical and microbiome information in human beings does not allow characterization across a spectrum of severity of NEC-like conditions. Much existent data of this type are from the rodent experiments discussed above, with the associated limitations of characterizing the “pre”-NEC state effectively. It is exactly for this reason that we assert that the type of computational modeling presented herein can provide a useful adjunct to current studies of NEC. In silico experiments integrate these contributory mechanisms in a form that allows a researcher to explore temporal states at a much higher degree of resolution and control than is feasible in the real world and permits the “movie” to be run backward to examine those conditions that led up to the development of the clinical phenotype.
Our model proposes a single unifying mechanism-based hypothesis, in which the immaturity of a single innate characteristic (OS clearance) of epithelial cells can affect an entire system, ultimately producing necrosis in a susceptible population. The ROS leading to OS have long been recognized as playing an important role in the pathogenesis of NEC [73,74], but the question remains about the initial source of excess ROS, which, once present, can initiate the inflammatory cycle. We posit that there is a functional manifestation of prematurity and immaturity in the NGECs that leads them to be susceptible to the cascading series of events that can cause NEC eventually. The NEC ABM may explain how generalized mucosal inflammation can lead to necrosis, given the right set of circumstances and perturbations. Outside severe, population-wide ROS management immaturity (<SCCmax 0.163 and 32.6% AbSCCmax), feeding alone cannot produce necrosis. This is consistent with the body of literature regarding the clinical manifestation of NEC and further emphasizes the importance of additional factors, such as inflammatory signaling, of which TLR-4 activation is an example [75], in exacerbating the progression of the disease. Furthermore, as a single causative organism has not been identified in the pathogenesis of NEC [76], we implemented generic bacterial agents with certain virulence properties; thus, it is not the type of organism that is important in the generation of NEC but rather what said organism is able to do when it interacts with NGECs. Our ABM also demonstrated, through the simulations including GC function and the mucus layer, a system-level consequence of higher apoptotic potential, a type of cellular behavior/fate that generally is considered to dampen the propagation of inflammation. These simulated behaviors are consistent with reports that there is some degree of GC impairment associated with prematurity [67,77,78] and suggests that the dynamics of GC population behavior and function represent an intriguing target for future research.
The limitations of the ABM are reflected primarily by the degree of abstraction present at its current state of development. For instance, only epithelial necrosis has been modeled thus far, whereas the clinical entity of NEC spans the full intestinal thickness of affected segments. In addition, the effect of innate and adaptive immune response mechanisms has yet to be incorporated into the ABM. Other notable factors not yet present in the ABM include the effect of probiotics in protecting NGECs, the effect of antibiotics in aiding the eradication of pathogens, different metabolic consequences resulting from different types of enteral feedings, the effect of maternal milk vs. formula on bacterial population characteristics, and ecological shifts and virulence dynamics associated with gut bacterial populations. These factors will be the subjects of future development, in conjunction with current basic research projects.
It is important to emphasize that the goal of our ABM is not to prove our hypothesis; rather, it is to demonstrate that our hypothesis is plausible and to provide a framework suggesting that additional research in the realm of ROS management may help elucidate the pathogenesis of NEC. The primary goal of this type of dynamic knowledge representation is to allow the generation and exploration of potentially plausible explanatory hypotheses, which can then be used to direct and guide future research, both in terms of attempting to find evidence of the underlying hypothesis (i.e., that premature NGECs actually do have decreased OS management capability) and being able to suggest new descriptive paradigms to characterize NEC. For instance, transcriptomic and proteomic analysis of stress management components can be performed to see if there is a difference between premature and mature enterocytes. This would allow the determination of the actual OS clearance capacity associated with prematurity. Additionally, immaturity might be characterized as causing greater fragility or impaired resistance to environmental alterations; this raises the question of what constitutes the mechanism by which maturity leads to greater robustness. One particular aspect of the current NEC ABM that might represent an area for further investigation is the Mixed Necrosis–Apoptosis outcome group: It is plausible that this set of simulation outcomes corresponds to pre-NEC states, manifest clinically as NEC scares. Characterization of the dynamics and properties of this simulation may provide insight into potential diagnostic targets for identifying those NEC scares that can be managed effectively with bowel rest and those that will mandate more aggressive interventions. The NEC ABM offers the possibility that alterations in feeding (specific metabolic profiles reducing the generation of oxidative metabolites), bacterial interdiction (through synthetic mucus augmentation), epithelial protection strategies (through growth factors such as epidermal growth factor), or suppression of DAMP/PAMP signaling (anti-TLR) might be simulated to evaluate their relative and situational efficacy. The determination of the right therapy in the right situation at the right time can be obtained comprehensively only through the use of computational models that provide high-resolution information on system states and trajectories. Finally, although we know that there are no specific microbial species causal for NEC, there might be certain properties associated with “NEC-genic” microbial species. Some possibilities include:
Metabolic capacity to generate high-stress compounds such as short-chain fatty acids, known to be involved in inflammatory processes associated with NEC [79–81];
Mucus-degrading capacity that would impair mucus barrier function and thereby increase bacterial–NGEC encounters;
Inflammatory signaling potential, such as is seen in Pseudomonas aeruginosa;
Low threshold of virulence activation.
Enhancing the ability to engage in these speculations is exactly the justification for the use of ABMs for dynamic knowledge representation: Methods and tools that allow the in silico exploration and testing of putative hypothesis configurations and suggest future experiments that could query the key points of difference between proposed hypotheses. For a disease as broad as NEC, in which several normally co-existent systems are disrupted in the generation of the disease, new and wholly integrative approaches such as ABM are valuable adjuncts to established clinical and basic science research.
Supplementary Material
Acknowledgments and Author Disclosure Statement
GA acknowledges support from National Science Foundation grant 0830-370-V601, National Institutes of Health grant 2P50GM053789-13, and National Institute of Disability and Rehabilitation Research grant H133E070024.
GA is a consultant for Immunetrics, Inc.
References
- 1.Henry MC. Moss RL. Necrotizing enterocolitis. Annu Rev Med. 2009;60:111–124. doi: 10.1146/annurev.med.60.050207.092824. [DOI] [PubMed] [Google Scholar]
- 2.McGuire W. Anthony MY. Donor human milk versus formula for preventing necrotising enterocolitis in preterm infants: Systematic review. Arch Dis Child Fetal Neonatal Ed. 2003;88:F11–F14. doi: 10.1136/fn.88.1.F11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodhi C. Richardson W. Gribar S. Hackam DJ. The development of animal models for the study of necrotizing enterocolitis. Dis Model Mech. 2008;1:94–98. doi: 10.1242/dmm.000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An G. Concepts for developing a collaborative in silico model of the acute inflammatory response using agent-based modeling. J Crit Care. 2006;21:105–110. doi: 10.1016/j.jcrc.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.An G. Hunt CA. Clermont G, et al. Challenges and rewards on the road to translational systems biology in acute illness: Four case reports from interdisciplinary teams. J Crit Care. 2007;22:169–175. doi: 10.1016/j.jcrc.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer AL. Beauchemin CA. Perelson AS. Agent-based modeling of host–pathogen systems: The successes and challenges. Inform Sci. 2009;179:1379–1389. doi: 10.1016/j.ins.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An G. Faeder J. Vodovotz Y. Translational systems biology: Introduction of an engineering approach to the pathophysiology of the burn patient. J Burn Care Res. 2008;29:277–285. doi: 10.1097/BCR.0b013e31816677c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vodovotz Y. Csete M. Bartels J, et al. Translational systems biology of inflammation. PLoS Comp Biol. 2008;4:e1000014. doi: 10.1371/journal.pcbi.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arciero JC. Ermentrout GB. Upperman JS, et al. Using a mathematical model to analyze the role of probiotics and inflammation in necrotizing enterocolitis. PloS One. 2010;5:e10066. doi: 10.1371/journal.pone.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt CA. Ropella GE. Lam TN, et al. At the biological modeling and simulation frontier. Pharm Res. 2009;26:2369–2400. doi: 10.1007/s11095-009-9958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An G. Mi Q. Dutta-Moscato J. Vodovotz Y. Agent-based models in translational systems biology. Wiley Interdiscip Rev. 2009;1:159–171. doi: 10.1002/wsbm.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An G. Introduction of an agent-based multi-scale modular architecture for dynamic knowledge representation of acute inflammation. Theor Biol Med Model. 2008;5:11. doi: 10.1186/1742-4682-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey AM. Thorne BC. Peirce SM. Multi-cell agent-based simulation of the microvasculature to study the dynamics of circulating inflammatory cell trafficking. Ann Biomed Eng. 2007;35:916–936. doi: 10.1007/s10439-007-9266-1. [DOI] [PubMed] [Google Scholar]
- 14.Li NY. Verdolini K. Clermont G, et al. A patient-specific in silico model of inflammation and healing tested in acute vocal fold injury. PloS One. 2008;3:e2789. doi: 10.1371/journal.pone.0002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vodovotz Y. An G. Systems biology and inflammation. Methods Mol Biol. 2010;662:181–201. doi: 10.1007/978-1-60761-800-3_9. [DOI] [PubMed] [Google Scholar]
- 16.Vodovotz Y. Clermont G. Chow C. An G. Mathematical models of the acute inflammatory response. Curr Opin Crit Care. 2004;10:383–390. doi: 10.1097/01.ccx.0000139360.30327.69. [DOI] [PubMed] [Google Scholar]
- 17.Dong X. Foteinou PT. Calvano SE, et al. Agent-based modeling of endotoxin-induced acute inflammatory response in human blood leukocytes. PloS One. 2010;5:e9249. doi: 10.1371/journal.pone.0009249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH. Debnath J. Mostov K, et al. A computational approach to resolve cell level contributions to early glandular epithelial cancer progression. BMC Syst Biol. 2009;3:122. doi: 10.1186/1752-0509-3-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansury Y. Diggory M. Deisboeck TS. Evolutionary game theory in an agent-based brain tumor model: Exploring the “genotype–phenotype” link. J Theor Biol. 2006;238:146–156. doi: 10.1016/j.jtbi.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L. Strouthos CG. Wang Z. Deisboeck TS. Simulating brain tumor heterogeneity with a multiscale agent-based model: Linking molecular signatures, phenotypes and expansion rate. Math Comp Model. 2009;49:307–319. doi: 10.1016/j.mcm.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L. Wang Z. Sagotsky JA. Deisboeck TS. Multiscale agent-based cancer modeling. J Math Biol. 2009;58:545–559. doi: 10.1007/s00285-008-0211-1. [DOI] [PubMed] [Google Scholar]
- 22.Beauchemin C. Samuel J. Tuszynski J. A simple cellular automaton model for influenza A viral infections. J Theor Biol. 2005;232:223–234. doi: 10.1016/j.jtbi.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Chao DL. Davenport MP. Forrest S. Perelson AS. Stochastic stage-structured modeling of the adaptive immune system. Proc IEEE Comp Soc Bioinformat Conf. 2003;2:124–131. [PubMed] [Google Scholar]
- 24.Funk GA. Barbour AD. Hengartner H. Kalinke U. Mathematical model of a virus-neutralizing immunglobulin response. J Theor Biol. 1998;195:41–52. doi: 10.1006/jtbi.1998.0779. [DOI] [PubMed] [Google Scholar]
- 25.Funk GA. Jansen VA. Bonhoeffer S. Killingback T. Spatial models of virus-immune dynamics. J Theor Biol. 2005;233:221–236. doi: 10.1016/j.jtbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Peleg M. Penchina CM. Modeling microbial survival during exposure to a lethal agent with varying intensity. Crit Rev Food Sci Nutr. 2000;40:159–172. doi: 10.1080/10408690091189301. [DOI] [PubMed] [Google Scholar]
- 27.Walker DC. Hill G. Wood SM, et al. Agent-based computational modeling of wounded epithelial cell monolayers. IEEE Trans Nanobiosci. 2004;3:153–163. doi: 10.1109/tnb.2004.833680. [DOI] [PubMed] [Google Scholar]
- 28.Walker D. Wood S. Southgate J, et al. An integrated agent-mathematical model of the effect of intercellular signalling via the epidermal growth factor receptor on cell proliferation. J Theor Biol. 2006;242:774–789. doi: 10.1016/j.jtbi.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Young WR. Roberts AJ. Stuhne G. Reproductive pair correlations and the clustering of organisms. Nature. 2001;412:328–331. doi: 10.1038/35085561. [DOI] [PubMed] [Google Scholar]
- 30.Durrett R. Levin S. Spatial aspects of interspecific competition. Theor Pop Biol. 1998;53:30–43. doi: 10.1006/tpbi.1997.1338. [DOI] [PubMed] [Google Scholar]
- 31.Durrett R. Levin SA. Stochastic spatial models: A user's guide to ecological applications. Phil Transact R Soc London B Biol Sci. 1994;343:329–350. [Google Scholar]
- 32.Strain MC. Richman DD. Wong JK. Levine H. Spatiotemporal dynamics of HIV propagation. J Theor Biol. 2002;218:85–96. doi: 10.1006/jtbi.2002.3055. [DOI] [PubMed] [Google Scholar]
- 33.Castiglione F. Pappalardo F. Bernaschi M. Motta S. Optimization of HAART with genetic algorithms and agent-based models of HIV infection. Bioinformatics. 2007;23:3350–3355. doi: 10.1093/bioinformatics/btm408. [DOI] [PubMed] [Google Scholar]
- 34.Beauchemin C. Probing the effects of the well-mixed assumption on viral infection dynamics. J Theor Biol. 2006;242:464–477. doi: 10.1016/j.jtbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Lloyd AL. May RM. Spatial heterogeneity in epidemic models. J Theor Biol. 1996;179:1–11. doi: 10.1006/jtbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 36.Hagenaars TJ. Donnelly CA. Ferguson NM. Spatial heterogeneity and the persistence of infectious diseases. J Theor Biol. 2004;229:349–359. doi: 10.1016/j.jtbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Chopard B. Luthi P. Droz M. Reaction-diffusion cellular automata model for the formation of Leisegang patterns. Phys Rev Lett. 1994;72:1384–1387. doi: 10.1103/PhysRevLett.72.1384. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds CW. Flocks, herds, and schools: A distributed behavioral model. Comp Graph. 1987;21:25–34. [Google Scholar]
- 39.Railsback SF. Lamberson RH. Harvey BC. Duffy WE. Movement rules for individual-based models of stream fish. Ecol Model. 1999;123:73–89. [Google Scholar]
- 40.Grimm V. Revilla E. Berger U, et al. Pattern-oriented modeling of agent-based complex systems: Lessons from ecology. Science. 2005;310:987–991. doi: 10.1126/science.1116681. [DOI] [PubMed] [Google Scholar]
- 41.Netotea S. Bertani I. Steindler L, et al. A simple model for the early events of quorum sensing in Pseudomonas aeruginosa: Modeling bacterial swarming as the movement of an “activation zone.”. Biol Direct. 2009;4:6. doi: 10.1186/1745-6150-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goryachev AB. Toh DJ. Wee KB, et al. Transition to quorum sensing in an Agrobacterium population: A stochastic model. PLoS Comp Biol. 2005;1:e37. doi: 10.1371/journal.pcbi.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosen R. Life Itself. New York: Columbia University Park; 1991. [Google Scholar]
- 44.An GC. Faeder JR. Detailed qualitative dynamic knowledge representation using a BioNetGen model of TLR-4 signaling and preconditioning. Math Biosci. 2009;217:53–63. doi: 10.1016/j.mbs.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An G. A model of TLR4 signaling and tolerance using a qualitative, particle-event-based method: Introduction of spatially configured stochastic reaction chambers (SCSRC) Math Biosci. 2009;217:43–52. doi: 10.1016/j.mbs.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Finkel T. Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 47.Polyak K. Xia Y. Zweier JL, et al. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 48.Johnson TM. Yu ZX. Ferrans VJ, et al. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc Natl Acad Sci USA. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z. Trotman LC. Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuler M. Bossy-Wetzel E. Goldstein JC, et al. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- 51.Siebenlist U. Franzoso G. Brown K. Structure, regulation and function of NF-kappa B. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 52.Schreck R. Rieber P. Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt KN. Traenckner EB. Meier B. Baeuerle PA. Induction of oxidative stress by okadaic acid is required for activation of transcription factor NF-kappa B. J Biol Chem. 1995;270:27136–27142. doi: 10.1074/jbc.270.45.27136. [DOI] [PubMed] [Google Scholar]
- 54.Trede NS. Tsytsykova AV. Chatila T, et al. Transcriptional activation of the human TNF-alpha promoter by superantigen in human monocytic cells: Role of NF-kappa B. J Immunol. 1995;155:902–908. [PubMed] [Google Scholar]
- 55.Kone BC. Schwobel J. Turner P, et al. Role of NF-kappa B in the regulation of inducible nitric oxide synthase in an MTAL cell line. Am J Physiol. 1995;269:F718–F729. doi: 10.1152/ajprenal.1995.269.5.F718. [DOI] [PubMed] [Google Scholar]
- 56.Flodstrom M. Welsh N. Eizirik DL. Cytokines activate the nuclear factor kappa B (NF-kappa B) and induce nitric oxide production in human pancreatic islets. FEBS Lett. 1996;385:4–6. doi: 10.1016/0014-5793(96)00337-7. [DOI] [PubMed] [Google Scholar]
- 57.Nadler EP. Dickinson E. Knisely A, et al. Expression of inducible nitric oxide synthase and interleukin-12 in experimental necrotizing enterocolitis. J Surg Res. 2000;92:71–77. doi: 10.1006/jsre.2000.5877. [DOI] [PubMed] [Google Scholar]
- 58.Van Antwerp DJ. Martin SJ. Kafri T, et al. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 59.Wang CY. Mayo MW. Baldwin AS., Jr. TNF- and cancer therapy-induced apoptosis: Potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 60.Beg AA. Finco TS. Nantermet PV. Baldwin AS., Jr. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of I kappa B alpha: A mechanism for NF-kappa B activation. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan FK. Shisler J. Bixby JG, et al. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 62.Verstrepen L. Bekaert T. Chau TL, et al. TLR-4, IL-1R and TNF-R signaling to NF-kappaB: Variations on a common theme. Cell Mol Life Sci. 2008;65:2964–2978. doi: 10.1007/s00018-008-8064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caplan MS. Simon D. Jilling T. The role of PAF, TLR, and the inflammatory response in neonatal necrotizing enterocolitis. Semin Pediatr Surg. 2005;14:145–151. doi: 10.1053/j.sempedsurg.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 64.Schneeberger EE. Lynch RD. The tight junction: A multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 65.Han X. Fink MP. Delude RL. Proinflammatory cytokines cause NO*-dependent and -independent changes in expression and localization of tight junction proteins in intestinal epithelial cells. Shock. 2003;19:229–237. doi: 10.1097/00024382-200303000-00006. [DOI] [PubMed] [Google Scholar]
- 66.Einerhand AW. Renes IB. Makkink MK, et al. Role of mucins in inflammatory bowel disease: Important lessons from experimental models. Eur J Gastroenterol Hepatol. 2002;14:757–765. doi: 10.1097/00042737-200207000-00008. [DOI] [PubMed] [Google Scholar]
- 67.Ballance WA. Dahms BB. Shenker N. Kliegman RM. Pathology of neonatal necrotizing enterocolitis: A ten-year experience. J Pediatr. 1990;117:S6–S13. doi: 10.1016/s0022-3476(05)81124-2. [DOI] [PubMed] [Google Scholar]
- 68.Ryley HC. Rennie D. Bradley DM. The composition of a mucus glycoprotein from meconium of cystic fibrosis, healthy pre-term and full-term neonates. Clin Chim Acta. 1983;135:49–56. doi: 10.1016/0009-8981(83)90387-x. [DOI] [PubMed] [Google Scholar]
- 69.Sharma R. Tepas JJ., 3rd Microecology, intestinal epithelial barrier and necrotizing enterocolitis. Pediatr Surg Int. 2010;26:11–21. doi: 10.1007/s00383-009-2536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Magne F. Abely M. Boyer F, et al. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006;57:128–138. doi: 10.1111/j.1574-6941.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 71.Khailova L. Dvorak K. Arganbright KM, et al. Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2009;297:G940–G949. doi: 10.1152/ajpgi.00141.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y. Hoenig JD. Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hsueh W. Caplan MS. Qu XW, et al. Neonatal necrotizing enterocolitis: Clinical considerations and pathogenetic concepts. Pediatr Dev Pathol. 2003;6:6–23. doi: 10.1007/s10024-002-0602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clark DA. Fornabaio DM. McNeill H, et al. Contribution of oxygen-derived free radicals to experimental necrotizing enterocolitis. Am J Pathol. 1988;130:537–542. [PMC free article] [PubMed] [Google Scholar]
- 75.Leaphart CL. Cavallo J. Gribar SC, et al. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J Immunol. 2007;179:4808–4820. doi: 10.4049/jimmunol.179.7.4808. [DOI] [PubMed] [Google Scholar]
- 76.Morowitz MJ. Poroyko V. Caplan M, et al. Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics. 2010;125:777–785. doi: 10.1542/peds.2009-3149. [DOI] [PubMed] [Google Scholar]
- 77.Hackam DJ. Upperman JS. Grishin A. Ford HR. Disordered enterocyte signaling and intestinal barrier dysfunction in the pathogenesis of necrotizing enterocolitis. Semin Pediatr Surg. 2005;14:49–57. doi: 10.1053/j.sempedsurg.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 78.Clark JA. Doelle SM. Halpern MD, et al. Intestinal barrier failure during experimental necrotizing enterocolitis: Protective effect of EGF treatment. Am J Physiol Gastrointest Liver Physiol. 2006;291:G938–G949. doi: 10.1152/ajpgi.00090.2006. [DOI] [PubMed] [Google Scholar]
- 79.Lin J. Too much short chain fatty acids cause neonatal necrotizing enterocolitis. Med Hypoth. 2004;62:291–293. doi: 10.1016/S0306-9877(03)00333-5. [DOI] [PubMed] [Google Scholar]
- 80.Waligora-Dupriet AJ. Dugay A. Auzeil N, et al. Short-chain fatty acids and polyamines in the pathogenesis of necrotizing enterocolitis: Kinetics aspects in gnotobiotic quails. Anaerobe. 2009;15:138–144. doi: 10.1016/j.anaerobe.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 81.Sanderson IR. Short chain fatty acid regulation of signaling genes expressed by the intestinal epithelium. J Nutr. 2004;134:2450S–2454S. doi: 10.1093/jn/134.9.2450S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.