Summary
Systems biology offers cutting-edge tools for the study of complementary and alternative medicine (CAM). The advent of ‘omics’ techniques and the resulting avalanche of scientific data have introduced an unprecedented level of complexity and heterogeneous data to biomedical research, leading to the development of novel research approaches. Statistical averaging has its limitations and is unsuitable for the analysis of heterogeneity, as it masks diversity by homogenizing otherwise heterogeneous populations. Unfortunately, most researchers are unaware of alternative methods of analysis capable of accounting for individual variability. This paper describes a systems biology solution to data complexity through the application of parsimony phylogenetic analysis. Maximum parsimony (MP) provides a data-based modeling paradigm that will permit a priori stratification of the study cohort(s), better assessment of early diagnosis, prognosis, and treatment efficacy within each stratum, and a method that could be used to explore, identify and describe complex human patterning.
Keywords: Heterogeneity, Parsimony, Phylogenetics, Synapomorphies, Systems biology, Clinical trial design, Complementary and alternative medicine
Introduction
Systems biology offers sophisticated objective tools for investigating how complementary and alternative medicine (CAM) treatments could result in complex and individualized effects on the body [1–5]. Most scientific research aims to identify the hidden patterns that exist within a population or among populations by decoding data complexity. These patterns could be biological variations or behavioral patterns, depending on the hypothesis being probed. Statistical methods have dominantly been employed to support the existence or lack of such patterns, whether the subject of the study is a human population, a plant family, or a cell culture.
However, recent evidence suggests that data averaging has significant limitations when dealing with heterogeneity as it masks intrapopulation diversity and homogenizes otherwise heterogeneous subpopulations. Heterogeneity at all levels is a product of evolution [6]; it confers better fitness on individuals and thus positions populations to survive bottleneck events. Evolutionary processes produce heterogeneity at many levels from cellular (e.g., genes, chromosomes, genomes, epigenetics, and tissues) to behavioral patterns (e.g., dietary, exercise, health promotion patterns). Variations at these levels constitute the basis of natural selection [6, 7].
Recent whole-genome sequencing projects have shown the presence of millions of variations as single-nucleotide polymorphisms (SNPs), small insertions and deletions, and copy number variations (CNVs) [8]. However, the lack of proper analytical tools has reduced the significance of genetics studies and prevented meaningful interpretation of the data [9, 10].
The scientific community is addicted to statistical and phenetic approaches, and despite their inapplicability to certain high-throughput high-dimensional biological data, statistical parameters continue to be invoked even when their usefulness is doubtful [11]. The commonly cited reason for this is the perceived absence of an alternative; but as we will detail in this paper, such alternatives indeed exist, and they should be studied and employed. They are based on the fact that heterogeneity, whether in normal or disease conditions, is an evolution-based phenomenon that has to be dealt with by applying evolutionarily compatible methods.
Dealing with Data of Heterogeneous Populations
Although the paradigmatic and methodological argument is broadly applicable across domains and disciplines, we will present the case for the proposed approach using biological exemplars. Heterogeneity has implications for many aspects of research and clinical practice. It necessitates compensating for individual variations that produce significant differences in rates of treatments efficacy, effectiveness and side effects as well as responses to various therapeutic modalities, including whole systems of complementary and alternative medicine (WS-CAM) [12, 13]. For example, in a clinical trial where the study population encompasses individuals who are poor responders to a particular treatment, the treatment’s effects in good responders will be underestimated [12]. As in other fields, this is an issue that is particularly important in WS-CAM research, where personalized intervention packages and individualized trajectories of treatment response are the norm [14, 15].
Rather than focusing on the commonalities of certain genes, metabolites, or proteins, profiling heterogeneity is better suited for dynamic systems [7]. Prior to 1966, natural populations were assumed to be more or less genetically uniform, even though Lewontin and Hubby [16] and Harris [17] demonstrated that polymorphisms are common throughout populations. Today, we recognize variation at several levels; a gene-nucleotide level of variation could manifest in mutations and genetic polymorphism, while a genome-chromosome level heterogeneity can be present as CNVs, loss of heterozygosity (LOH), and epigenetic heterogeneity such as DNA methylation, non-coding RNAs, or chromosomal folding [18]. Additionally, there are changes that take place independent of epigenetic alterations; these are influenced by environmental factors and are affected by nutrition, stress, exposure, and immune responses [18].
Analyzing Heterogeneous Data for Biological Significance
The recent clinical trials of targeted biomedical cancer treatments are examples of the current reductionist trend that has produced mostly disappointing results [19]. However, the failed targeted treatment approach has served the purpose of bringing the issue of heterogeneity to the forefront of scientific thought [18, 20].
More recently, by recognizing the ubiquity of heterogeneity in complex systems and the negative effects of ignoring it, statisticians and researchers are calling for the two-stage study, whereby in the first stage, the study group is stratified into well-defined but broad populations using traditional experimental methods, followed by the construction of subgroups in the second stage [12, 21]. Similarly, CAM researchers previously identified a need for two-stage diagnosis, with the conventional medicine disease entity group diagnosis followed by the individualized WS-CAM diagnosis [4]. Although the two-stage approach can be achieved fairly readily in a small non-complex situation with one to a few variables, it becomes difficult to conduct when the contributing variables are scaled up to the tens, hundreds, or thousands [22–26].
In a disease context, data heterogeneity can point out several phenomena, such as inter- and intra-specimen diversity in diseased specimens, a high rate of variability generation, and multiple pathways of disease development [18, 20]. Additionally, the disease process is further complicated by the multiphasic and dynamic nature of some pathologies, such as cancer and degenerative diseases, which pose the challenge of whether a multiphasic and dynamic process can be modeled by a bioinformatic paradigm.
Data interpretation requires analysis and synthesis compatible with the existing biological conceptual framework(s) and hypotheses. Thus, a biologically compatible analytical paradigm should incorporate four elements: the high-throughput data (e.g., genomics, metabolomics, proteomics), the disease phenotypes (e.g., hyperplasia, primary tumor, metastatic tumor), evolutionary theory, and bioinformatics (an analytical algorithm that processes the data). Parsimony phylogenetics offers an analytical algorithm that can bring these elements together to achieve novel multidimensional systems biology synthesis without the traditional overdependence on statistical methods.
Parsimony Phylogenetics for Analyzing and Modeling Heterogeneity
Phylogenetics, also termed cladistics, is an analytical paradigm based on the principles of evolution [27]. Its current codes known as phylogenetic systematics were laid down in the mid-1950s by the German systematist Willi Hennig [28]. Phylogenetics differs from other systems of classifications in that, rather than using overall similarity to classify objects, it utilizes shared derived similarity as evidence of relatedness. The practice has been applied in many fields such as botany, microbiology, and zoology to construct relationships among species, populations, and individuals in an evolutionary sense [27].
The goal of a phylogenetic analysis is to model the data to produce a hypothesis of relationships among the specimens under study that accurately reflects the biological processes that led to the diversity of specimens. Phylogenetics constructs the hypotheses of relationships by sorting data points into ancestral (normal/within the normal parameters) and derived (abnormal, above or below the selected baseline, or falling outside the normal range) categories, and then grouping together the specimens that share the same derived states [28, 29]. The process of sorting out data points into derived and ancestral states is termed polarity assessment, data polarization, or outgroup comparison. The derived states represent the aberrations or the new changes; for example, in a disease, the aberration can be an overexpression of a gene, up-regulation of a protein, or a mutation.
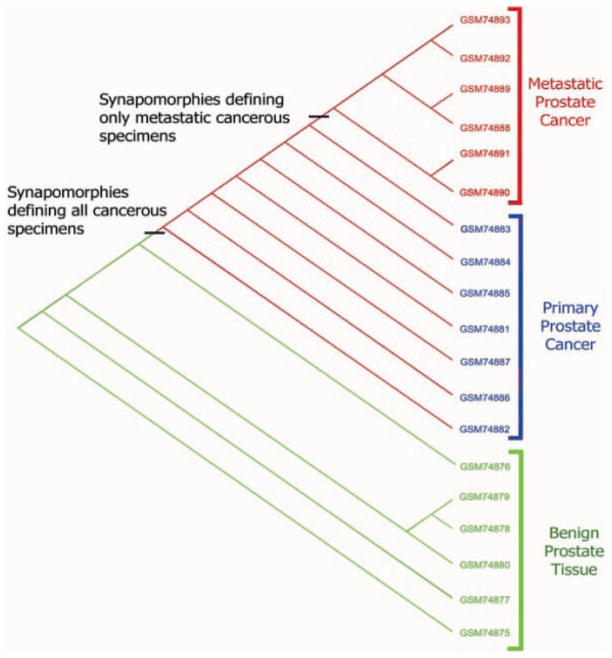
In phylogenetic terminology, a shared derived state is termed synapomorphy (a potential biomarker); because sharing a synapomorphy is indicative of a relationship, a group of specimens that share one or more synapomorphies is called a clade. Phylogenetics presents its hypotheses in a graphical tree format called the cladogram (fig. 1), which is a map of clades (groupings) and their supporting synapomorphies.
Fig. 1.
The most parsimonious cladogram of the gene expression microarray of dataset GDS1439 (www.ncbi.nlm.nih.gov/sites/entrez?db=gds). The study group contains specimens composed of 6 benign specimens, as well as 7 primary and 6 metastatic prostate carcinoma specimens [33]. The primary and metastatic specimens grouped separately from each other into two groups (clades); while the metastatic occupied the top of the cladogram, the primary was nestled in between the metastatic and benign clades.
There are a few methods to constructing phylogenetic cladograms (trees); among these are parsimony, maximum likelihood, and neighbor joining. They differ in their algorithmic functions and the type of data they handle. These methods have been compared, and parsimony has turned out to be the most suitable for the purposes of dealing with heterogeneous high-throughput data of various diseases. Parsimony, also known as Occam’s razor or the ‘principle of simplicity’, is generally defined as selecting the simplest hypothesis among competing ones. In phylogenetic analysis, it is the hypothesis that requires the least number of steps to construct, i.e., the shortest tree/cladogram, which is usually called the most parsimonious tree. A parsimonious approach produces a multidimensional analytical tool that is data based, not specimen based, which accounts for and integrates disease heterogeneity, nature of biological data, and principles of evolution [30, 31]. Yet, it is important that any analytical method has high predictive power; it must be able to differentiate between groups of people (e.g., those that are healthy from those with disease, or those who respond to treatment from those who do not), present the changes that distinguish between the two groups, show the transitional specimens that fall in between the two states [27], and stratify populations [32].
We provide an example to illustrate the application of a parsimony analysis of gene expression microarray data. We selected dataset GDS1439 from the National Center of Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov/sites/entrez?db=gds), which contains 6 benign specimens, 7 primary, and 6 metastatic prostate carcinoma specimens [33]. When dealing with large datasets that contain thousands of variables, especially datasets obtained from high-throughput microarrays or mass spectrometry, there are two steps in carrying out parsimony analysis; first, polarity assessment of data points through outgroup comparison into either derived (abnormal in case of disease phenotypes) or ancestral (normal) must be carried out. Polarity assessment transforms the continuous quantitative data points of gene expressions into discrete entities of zeros (0s) and ones (1s), where zero indicates that the value is ancestral (normal) and one indicates that the value is different and therefore assumed to be derived in an evolutionary sense. So, the new data matrix of polarized bivalent values has only zeros and ones, and it is this matrix that will be processed in a parsimony algorithm. The second step is the processing of the polarized values through a maximum parsimony algorithm to classify the specimens into a cladogram. The first step of the analysis was carrying out polarity assessment of the gene expression values that sorted the expression values into derived (abnormal) and ancestral (normal) by comparing the values of the cancerous specimens against the range of the benign specimens for every gene in the dataset. The new matrix was processed with the computer program MIX (the parsimony program of the PHYLIP package) using Wagner’s parsimony method [34], which produced only one most parsimonious tree/cladogram (fig. 1).
The cladogram of figure 1 is a graphical summary that showed the groupings (clades), their synapomorphies, and a topology that reflects the relationships among the clades and the direction of change accumulation among the clades and their specimens. The analysis showed the primary and metastatic specimens grouped separately from each other in two groups; while the metastatic occupied the top of the cladogram, the primary was nested in between the metastatic and benign clades. Separating the metastatic cases from the primary ones on the basis of their gene expression is an excellent outcome that confers confidence that this approach has good validity. The primary and metastatic clades shared a list of 302 synapomorphies (uniquely shared derived expressions or potential biomarkers in a biomedical sense) that separate them from the benign clades. The metastatic specimens at the top end of the cladogram are separated from the primary specimens by a list of 577 synapomorphies that are shared by their respective specimens. The cladogram topology has directionality; the specimens with the highest number of derived states occupy the upper part of the cladogram. Therefore, one could interpret the tandem arrangement of the primary and metastatic groups as a sequential relationship, where the initiation of the cancer required 302 derived gene expressions, while the transformation to a metastatic phenotype required an additional 577 changes.
Implications of Parsimony Phylogenetics
As our example demonstrates, maximum parsimony has efficiently and accurately modeled the heterogeneous expression profiles of the diseased specimens, in this case, cancer with a rapid mutation rate. The analysis precisely classified the phenotypes (or genetic patterns) based on modeling of the disease genotypes (gene expressions); there was no mixing of the three phenotypes, and the gene expression data were perfectly congruent with their phenotypes.
The process of data polarization has the added advantage of reducing measurement variability. By transforming data points to distinct 1s and 0s, the comparison between specimens becomes qualitative rather than based on absolute quantitative values. Polarization of the data allows pooling of multiple experiments, and therefore facilitates intra- and inter-compatibility of the observed clades, types or classes. In this regard, the analysis is a systems biology approach that can pool data from related diseases to identify the common aberrations and differential features among them, e.g., several cancer types [30] or WS-CAM diagnostic subgroups [14, 15, 17, 35]. For example, Alraek and Baerheim [14] subgrouped their cystitis patients in three groups: (1) spleen yang/qi xu, (2) kidney yang/qi xu, and (3) liver qi stagnation; such subgrouping can more objectively be carried out by a phylogenetic analysis. Also, as Frei et al. [15] have shown, subgrouping of patients with attention deficit hyperactivity disorder (ADHD) before the commencement of a trial is important in order to avoid failure, since patients vary in their response to treatment and poor responders require alternative medication (see below on the use of phylogenetics for the stratification before clinical trial).
From a practical aspect, a parsimony approach can be translated into a clinical setting for diagnosis, prognosis, and post-treatment evaluation [27]. By constructing a comprehensive cladogram that incorporates many diseases (for example, a tree of cancer), the cladogram becomes an instrument for diagnosis. To diagnose a case, data can be entered into the comprehensive cladogram, thus placing the case on the cladogram. This approach might also facilitate more accurate prescriptive practices, like those used in homeopathy, in which the process of choosing an individualized remedy or therapeutic schema often requires categorizing each patient’s global homeopathic phenotype, i.e., remedy type, by kingdom (animal, plant, or mineral) and specific family [36].
Parsimony phylogenetics could also be applied to WS-CAM and integrative therapies research. CAM clinicians have often claimed that a portion of the population responds positively to a particular therapy (responders) while others seem to have little to no change in outcomes (non-responders) [14, 15, 35, 37, 38]. Clinical trial limitations, often cited in the CAM literature, have fueled a call for new methodological strategies and sophisticated analyses. The successful analytical tool is the one that does not obviate or underplay heterogeneity-driven variability. Solutions such as genomic control, which adjusts association statistics for each marker by a uniform overall inflation factor, compensate only partially for heterogeneity [39, 40].
Parsimony phylogenetic methods can be used to differentiate among responder types by classifying into responder-type clades, based on shared synapomorphies or sets of intra-population characteristics. Thus, a wider set of study participants could be enrolled into CAM clinical trials, more closely aligning the trial population with those seen in clinical practice. CAM researchers could better evaluate treatment-effect variability and treatment-related risks, while predicting those persons who are most likely to benefit from a particular CAM therapy in a given solution [15, 41].
Others have suggested conducting multiple trials of treatment on each individual in an ‘n-of-one’-type design in order to minimize the data heterogeneity. Phylogenetic analysis would allow pooling of data across these types of studies to again identify responders and non-responders. Thus, information gained from clinical trials could be more informative and more easily extrapolated to the clinic [42].
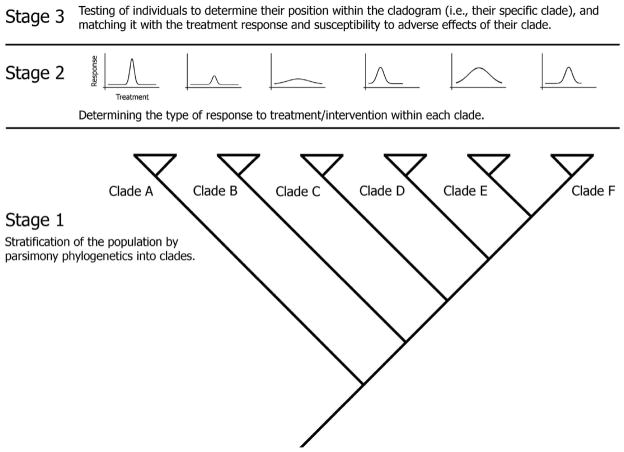
We propose a three-stage clinical trial model that starts with a stratification of the study sample based on phenotypic and genotypic characters. A pre-trial phase using a priori stratification by parsimony phylogenetics will delimit the subpopulations that share common biological traits (classes) (fig. 2). A small blood sample subjected to high-throughput analysis could provide the data needed for stratification. Because the stratification is done without a priori weighing of variables, parsimony may also reveal the variables that define the subpopulation partitioning. Essentially, in the first stage, the recruitment could include a wide spectrum of inclusion criteria in order to embrace a heterogeneous study population that would reflect the ‘real-world’ setting. Based on the identified clades that reveal relatedness among groups and subgroups, the second stage of the trial could be implemented knowing that we reached a level of homogeneity at baseline within each clade. Thus, each clade could then be randomized to either control or intervention, depending on the clinical trial type.
Fig. 2.
Diagram illustrating the three-stage clinical trial design. Stage 1: Stratification of the population based on their genotype into various clades, and randomization of each clade into control and intervention. Stage 2: Classification of individuals based on their response to treatment/intervention (conventional and/or CAM) and characterization of clades. Stage 3: Translation of findings to the clinic and application of personalized medicine; patients visiting their physician will be tested against the master clade, using a simple blood test (omics-type of data or equivalent), and based on their position within a clade an individualized treatment can be planned.
Among the individuals of each clade there will probably be variable levels of responsiveness to the intervention, but most likely less variability than between clades. The cladogram will serve as a dynamic database for the implementation of the third stage, which corresponds to the translation of the clinical trial findings to the clinic. This means that, prior to using a new therapy or intervention, the patient will need to submit a blood sample to determine his/her clade. A clade membership determines the treatment options; what dosage he/she will need or how responsive to treatment the patient will be. Thus, the health care provider could make an informed decision when recommending a particular therapy or implementing a particular type of treatment. Potentially, this could lead to decreased treatment-related risk, improved outcomes, decreased costs, and treatment efficiency. Furthermore, in WS-CAM where the practitioner stratifies patients on categorical yet unifying classes (types of doshas, humors, temperaments, or imbalances in interrelationships among elements), this method could offer a modern verification of these concepts and potentially include them in the clinical design.
The advantages of this three-stage clinical trial design could be summarized in three significant points: (i) By employing parsimony analysis to carry out the pre-trial populational stratification into natural clades, the baseline heterogeneity per clade will significantly be reduced; (ii) individuals’ positions within a clade determine their treatment options; and (iii) by using the clades as a dynamic data base, the physician could prescribe the suitable treatment on the basis of the patient’s clade membership. Our proposed three-stage clinical trial design encompasses the practice of personalized medicine using a systems biology approach that addresses most of the currently debated issues of baseline heterogeneity, data heterogeneity, treatment-related risk, and translation of trials findings to the clinic.
Conclusions
Using parsimony phylogenetics as a means to account for heterogeneity from the subcellular to the whole-human behavioral level of function holds extraordinary promise in expanding clinical knowledge related to CAM therapies, and clinical treatment in general. Using a systems biology approach and putting data into an algorithm that can accurately model subtypes of people/phenomena in an evolutionary context offers a novel methodology to clinical design. This stands to deepen clinician understanding and confidence for matching interventions to those who are most likely to benefit.
Footnotes
Disclosure Statement
The authors declared no conflict of interest.
References
- 1.Ahn AC, Tewari M, Poon CS, Phillips RS. The limits of reductionism in medicine: Could systems biology offer an alternative? PLoS Med. 2006;3:e208. doi: 10.1371/journal.pmed.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 3.Wang M, Lamers RJ, Korthout HA, van Nessel-rooij JH, Witkamp RF, van der Heijden R, Voshol PJ, Havekes LM, Verpoorte R, van der Greef J. Metabolomics in the context of systems biology: Bridging traditional Chinese medicine and molecular pharmacology. Phytother Res. 2005;19:173–182. doi: 10.1002/ptr.1624. [DOI] [PubMed] [Google Scholar]
- 4.Vincent C, Furnham A. Complementary medicine: A research perspective. New York: John Wiley & Sons; 1997. [Google Scholar]
- 5.Zhou X, Liu B, Wu Z, Feng Y. Integrative mining of traditional Chinese medicine literature and medline for functional gene networks. Artif Intell Med. 2007;41:87–104. doi: 10.1016/j.artmed.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 6.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. Genetic and epigenetic heterogeneity in cancer: A genome-centric perspective. J Cell Physiol. 2009;220:538–547. doi: 10.1002/jcp.21799. [DOI] [PubMed] [Google Scholar]
- 8.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, Zhang Z, Ranade SS, Dimalanta ET, Hyland FC, Sokolsky TD, Zhang L, Sheridan A, Fu H, Hendrickson CL, Li B, Kotler L, Stuart JR, Malek JA, Manning JM, Antipova AA, Perez DS, Moore MP, Hayashibara KC, Lyons MR, Beaudoin RE, Coleman BE, Laptewicz MW, Sannicandro AE, Rhodes MD, Gottimukkala RK, Yang S, Bafna V, Bashir A, MacBride A, Alkan C, Kidd JM, Eichler EE, Reese MG, De La Vega FM, Blanchard AP. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galvan A, Ioannidis JP, Dragani TA. Beyond genome-wide association studies: Genetic heterogeneity and individual predisposition to cancer. Trends Genet. 2010;26:132–141. doi: 10.1016/j.tig.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orozco LD, Cokus SJ, Ghazalpour A, Ingram-Drake L, Wang S, van Nas A, Che N, Araujo JA, Pellegrini M, Lusis AJ. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet. 2009;18:4118–4129. doi: 10.1093/hmg/ddp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziliak ST. The validus medicus and a new gold standard. Lancet. 2010;376:324–325. doi: 10.1016/s0140-6736(10)61174-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Zhao W, Shaffer ML, Icitovic N, Chase GA. Modelling clinical trials in heterogeneous samples. Stat Med. 2005;24:2765–2775. doi: 10.1002/sim.2144. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Icitovic N, Shaffer ML, Chase GA. The impact of population heterogeneity on risk estimation in genetic counseling. BMC Med Genet. 2004;5:18. doi: 10.1186/1471-2350-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alraek T, Baerheim A. The effect of prophylactic acupuncture treatment in women with recurrent cystitis: Kidney patients fare better. J Altern Complement Med. 2003;9:651–658. doi: 10.1089/107555303322524508. [DOI] [PubMed] [Google Scholar]
- 15.Frei H, Everts R, von Ammon K, Kaufmann F, Walther D, Schmitz SF, Collenberg M, Steinlin M, Lim C, Thurneysen A. Randomised controlled trials of homeopathy in hyperactive children: Treatment procedure leads to an unconventional study design. Experience with open-label homeopathic treatment preceding the Swiss ADHD placebo controlled, randomised, double-blind, cross-over trial. Homeopathy. 2007;96:35–41. doi: 10.1016/j.homp.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics. 1966;54:595–609. doi: 10.1093/genetics/54.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris H. Enzyme polymorphisms in man. Proc R Soc Lond B Biol Sci. 1966;164:298–310. doi: 10.1098/rspb.1966.0032. [DOI] [PubMed] [Google Scholar]
- 18.Heng HH, Liu G, Stevens JB, Bremer SW, Ye KJ, Ye CJ. Genetic and epigenetic heterogeneity in cancer: The ultimate challenge for drug therapy. Curr Drug Targets. 2010;11:1304–1316. doi: 10.2174/1389450111007011304. [DOI] [PubMed] [Google Scholar]
- 19.Roukos DH. Mea culpa with cancer-targeted therapy: New thinking and new agents design for novel, causal networks-based, personalized biomedicine. Expert Rev Mol Diagn. 2009;9:217–221. doi: 10.1586/erm.09.7. [DOI] [PubMed] [Google Scholar]
- 20.Heng HH, Stevens JB, Bremer SW, Ye KJ, Liu G, Ye CJ. The evolutionary mechanism of cancer. J Cell Biochem. 2010;109:1072–1084. doi: 10.1002/jcb.22497. [DOI] [PubMed] [Google Scholar]
- 21.Davidoff F. Heterogeneity is not always noise: Lessons from improvement. JAMA. 2009;302:2580–2586. doi: 10.1001/jama.2009.1845. [DOI] [PubMed] [Google Scholar]
- 22.Rizzo-Sierra CV. Ayurvedic genomics, constitutional psychology, and endocrinology: The missing connection. J Altern Complement Med. 2011;17:465–468. doi: 10.1089/acm.2010.0412. [DOI] [PubMed] [Google Scholar]
- 23.van der Greef J, van Wietmarschen H, Schroen J, Wang M, Hankemeier T, Xu G. Systems biology-based diagnostic principles as pillars of the bridge between Chinese and Western medicine. Planta Med. 2010;76:2036–2047. doi: 10.1055/s-0030-1250450. [DOI] [PubMed] [Google Scholar]
- 24.van Wietmarschen H, Yuan K, Lu C, Gao P, Wang J, Xiao C, Yan X, Wang M, Schroen J, Lu A, Xu G, van der Greef J. Systems biology guided by Chinese medicine reveals new markers for sub-typing rheumatoid arthritis patients. J Clin Rheumatol. 2009;15:330–337. doi: 10.1097/RHU.0b013e3181ba3926. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Sun H, Zhang A, Sun W, Wang P, Wang Z. Potential role of metabolomics apporoaches in the area of traditional Chinese medicine: As pillars of the bridge between Chinese and Western medicine. J Pharm Biomed Anal. 2011;55:859–868. doi: 10.1016/j.jpba.2011.01.042. [DOI] [PubMed] [Google Scholar]
- 26.Zhang A, Sun H, Wang Z, Sun W, Wang P, Wang X. Metabolomics: Towards understanding traditional Chinese medicine. Planta Med. 2010;76:2026–2035. doi: 10.1055/s-0030-1250542. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Asab M, Chaouchi M, Amri H. Evolutionary medicine: A meaningful connection between omics, disease, and treatment. Proteomics Clin Appl. 2008;2:122–134. doi: 10.1002/prca.200780047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennig W. Phylogenetic Systematics. Urbana: University of Illinois Press; 1966. [Google Scholar]
- 29.Abu-Asab M, Chaouchi M, Amri H. Phyloproteomics: What phylogenetic analysis reveals about serum proteomics. J Proteome Res. 2006;5:2236–2240. doi: 10.1021/pr0504485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abu-Asab MS, Chaouchi M, Amri H. Phylogenetic modeling of heterogeneous gene-expression microarray data from cancerous specimens. OMICS. 2008;12:183–199. doi: 10.1089/omi.2008.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abu-Asab M. Microarrays need phylogenetics. Sci STKE. http://stke.sciencemag.org/cgi/eletters/sigtrans;1/51/eg11.
- 32.Sridhar S, Lam F, Blelloch GE, Ravi R, Schwartz R. Direct maximum parsimony phylogeny reconstruction from genotype data. BMC Bioinformatics. 2007;8:472. doi: 10.1186/1471-2105-8-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Felsenstein J. Phylip: Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 35.Pelsser LM, Frankena K, Toorman J, Savelkoul HF, Dubois AE, Pereira RR, Haagen TA, Rommelse NN, Buitelaar JK. Effects of a restricted elimination diet on the behaviour of children with attention-deficit hyperactivity disorder (INCA study): A randomised controlled trial. Lancet. 2011;377:494–503. doi: 10.1016/S0140-6736(10)62227-1. [DOI] [PubMed] [Google Scholar]
- 36.Thompson EA, Geraghty J. The vital sensation of the minerals: Reducing uncertainty in homeopathic prescribing. Homeopathy. 2007;96:102–107. doi: 10.1016/j.homp.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Bell IR, Lewis DA, 2nd, Schwartz GE, Lewis SE, Caspi O, Scott A, Brooks AJ, Baldwin CM. Electroencephalographic cordance patterns distinguish exceptional clinical responders with fibromyalgia to individualized homeopathic medicines. J Altern Complement Med. 2004;10:285–299. doi: 10.1089/107555304323062275. [DOI] [PubMed] [Google Scholar]
- 38.Elder C, Aickin M, Bauer V, Cairns J, Vuckovic N. Randomized trial of a whole-system Ayurvedic protocol for type 2 diabetes. Altern Ther Health Med. 2006;12:24–30. [PubMed] [Google Scholar]
- 39.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.Setakis E, Stirnadel H, Balding DJ. Logistic regression protects against population structure in genetic association studies. Genome Res. 2006;16:290–296. doi: 10.1101/gr.4346306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyland ME, Lewith GT. Oscillatory effects in a homeopathic clinical trial: An explanation using complexity theory, and implications for clinical practice. Homeopathy. 2002;91:145–149. doi: 10.1054/homp.2002.0025. [DOI] [PubMed] [Google Scholar]
- 42.Kelley JM, Kaptchuk TJ. Group analysis versus individual response: The inferential limits of randomized controlled trials. Contemp Clin Trials. 2010;31:423–428. doi: 10.1016/j.cct.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]