Abstract
Enamelin is a secreted glycoprotein that is critical for dental enamel formation. Ameloblasts in enamelin (Enam) null mice develop atypical features that include the absence of a Tomes’ process, expanded endoplasmic reticulum, apparent loss of polarity, and pooling of extracellular matrix in all directions, including between ameloblasts and the stratum intermedium. We hypothesized that ameloblast pathological changes may be associated with increased cell apoptosis. Our objective was to assess apoptotic activity in maxillary first molars of wild-type, Enam+/−, and Enam−/− mice at postnatal days 5, 7, 9, 14, and 17. Mouse maxillae were characterized by light microscopy after terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL) or 5-bromo-2′-deoxyuridine (BrdU) staining. Following the initial deposition of dentin matrix, ameloblasts became highly dysplastic and no enamel crystal ribbons were deposited. Ameloblast apoptosis was observed in the Enam null mice starting in the secretory stage and with no apparent alteration in cell proliferation. We conclude that in the absence of enamelin and subsequent shutdown of enamel formation, ameloblasts undergo pathological changes early in the secretory stage that are evident as radically altered cell morphology, detachment from the tooth surface, apoptosis, and formation of ectopic calcifications both outside and inside the dystrophic enamel organ.
Keywords: amelogenesis imperfecta, enamel crystals, LacZ knockin, mineralization front, Tomes’ process
The three major proteins in the enamel matrix of developing teeth are amelogenin, enamelin, and ameloblastin (1). These proteins are expressed from related genes, all belonging to the secretory calcium-binding phosphoprotein (SCPP) family (2). Intact enamelin is a large, proline-rich, phosphorylated glycoprotein. Secreted human enamelin has 1,099 amino acids. Unlike amelogenin and ameloblastin, the enamelin primary RNA transcript does not undergo alternative splicing, so there is only a single isoform of enamelin. In pigs, secreted enamelin is a 186-kDa protein that localizes to the mineralization front along the secretory surface of ameloblasts (3). Matrix metalloproteinase 20 (MMP20) rapidly degrades it into many fragments, one of which is a stable 32-kDa domain that accumulates within the matrix (4, 5).
Expression of the enamelin gene (ENAM, 4q13) is largely restricted to developing teeth. The human enamelin expressed sequence tag (EST) profile (Hs667018) from normal tissues (which does not include developing teeth) lists only 10 enamelin cDNAs out of 3,360,307 characterized. No tissue has more than two enamelin ESTs, which is a pattern that is characteristic of trace expression. Defects in ENAM contribute to the aetiology of non-syndromic forms of amelogenesis imperfecta (AI), a collection of inherited diseases featuring enamel malformations as the only phenotype (6–8). ENAM mutations cause AI that generally follows an **autosomal-dominant pattern of inheritance (9), with variations in both penetrance and expressivity. When a single ENAM allele is defective, the phenotype can be non-penetrant (10), or be manifested as enamel pits (11), horizontal grooves (12), or generalized thin enamel (13). There is also a dose effect: when both ENAM alleles are defective the enamel is extremely thin or non-existent (11).
Enamelin gene and deduced amino acid sequences are available from species representing the main groups of tetrapods (14). There are 25 unchanging amino acid positions when the 36 known mammalian enamelin protein sequences are aligned with the few non-mammalian sequences that go back to amphibians (frog). Sites of enamelin post-translational modifications are highly conserved, including several potential and known phosphorylation and N-linked glycosylation sites, as well as six cysteines believed to form disulphide bridges (14). Evolutionary analyses have not only identified functionally critical positions in the enamelin protein, but they have also demonstrated that all of the major secretory-stage enamel proteins serve necessary functions only in tooth formation and specifically in dental enamel formation. Genes encoding enamel matrix proteins have degenerated into pseudogenes in multiple toothless or enamel-less species that descended from ancestors with teeth covered by enamel. The genes encoding amelogenin, ameloblastin, and enamelin have degenerated in birds (14, 15) and in toothless (baleen) whales (16). A functional enamelin gene is absent in enamel-less (Kogia) whales (16) and in four different orders of placental mammals with toothless and/or enamel-less taxa (17). Genetic studies in humans strongly corroborate the evolutionary findings. ENAM defects in 10 different kindreds with non-syndromic AI have been reported. One of the cases is caused by the substitution of leucine with a highly conserved phosphoserine (p.S216L) (9).
Enamelin knockout mice provide still more support for the tooth-specificity of enamelin function, as the phenotype of these mice is restricted exclusively to defects in the enamel layer (18–20). The genetically engineered Enam knockout/lacZ knockin mouse model has also provided strong support for ameloblast specificity of enamelin expression through X-gal histostaining for nuclear-localized β-galactosidase activity to mark tissues that normally express enamelin. To date, nuclear staining has been detected only in ameloblasts in developing teeth (18). Enam knockout mice also provide a means to investigate normal and pathological enamel formation.
Enamelin knockouts demonstrate that enamelin is not only required for the deposition of tooth enamel, but it is also necessary to maintain the ameloblast phenotype, as is the case for ameloblastin (21). Degeneration of the ameloblast layer and the subsequent appearance of cyst-like enamel organ-based structures suggested that enamelin and ameloblastin are cell-adhesion proteins. However, as no enamel layer forms in the Enam and ameloblastin (Ambn) knockout mice, ameloblasts could fail to adhere to the unnatural underlying surface even if their attachment apparatus was intact and did not normally contain enamelin or ameloblastin. RGD sequences are found in enamel proteins from some species, but they are not a conserved feature, and mouse enamelin and ameloblastin both lack these potential integrin-binding motifs. Recombinant ameloblastin shows some affinity for heparin (22) and fibronectin (23) in vitro, but these molecular partners have not been localized in ameloblasts or shown to be important for ameloblast adhesion in vivo.
The observations that enamelin is critical for dental enamel formation and for maintenance of the ameloblast phenotype emphasize the dynamic roles played by ameloblasts besides simply creating an extracellular space and environment conducive for mineral deposition. In this study we characterized ameloblasts in Enam heterozygous and null mice by documenting the extent of apoptosis in maxillary first molars starting in the secretory stage at day 5 and continuing until tooth eruption at day 17. We demonstrated that ameloblasts in restricted areas of developing teeth undergo apoptosis when enamelin is completely missing, and that reducing enamelin production by one-half, as occurs in heterozygous mice, may result in more apoptosis later in crown development, despite the fact that normal enamel eventually forms on these teeth.
Material and methods
Staining of apoptotic cells
The ApopTag Plus Peroxidase In Situ Apoptosis Kit (Chemicon International, Temecula, CA, USA) was used to detect apoptotic cells. This kit labels and detects DNA strand breaks by the indirect terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL) method. Wild-type, Enam+/−, and Enam−/− mice were collected at postnatal days 5, 7, 9, 14, and 17. The heads were removed, cut in half on the midsagittal plane and immersed overnight in ice-cold 4% paraformaldehyde in 0.08 M sodium cacodylate buffer (pH 7.3). The samples were washed three times (for 30–60 min each wash) in cacodylate buffer containing 7% sucrose, then demineralized in 4.13% EDTA (pH 7.3) for 3 wk (mice ≥ 5 d) or 6 wk (14- and 17-d-old mice). The samples were dehydrated using a graded ethanol series followed by xylene, embedded in paraffin, and sectioned at 5 µm. The sections were spread in a water bath (52°C), loaded on plus gold glass slides (Thermo Fisher Scientific, Waltham, MA, USA), and left to dry at room temperature overnight. Then, the sections were deparaffinized using three changes of xylene and rehydrated through a reverse ethanol series for 30 min. The sections were washed in PBS (135 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM Na2H2PO4; pH 7.3), incubated for 15 min with proteinase K (20 µg ml−1) at room temperature for 15 min, washed three times in distilled water, treated in 3% H2O2 for 5 min, washed twice in distilled water, incubated with equilibrium buffer, and then incubated with TdT at 37°C for 1 h. Control samples were run in PBS without TdT. Then, the sections were washed and incubated in stop/wash buffer for 10 min at room temperature followed by three washes in PBS. Anti-digoxigenin conjugate was applied to the sections for 30 min at room temperature, after which the sections were washed four times in PBS and developed for 3 min with peroxidase substrate [diaminobenzidine (DAB)]. The sections were then washed three times in distilled water, counterstained for 4 min with 0.5% methyl green, rinsed three times in distilled water, and immersed in three changes of 100% N-butanol. The sections were dehydrated in three changes of xylene, after which coverslips were attached using Fisher Scientific (Pittsburgh, PA, USA) mounting solution.
5-Bromo-2′-deoxyuridine staining
5-Bromo-2′-deoxyuridine (BrdU) is a thymidine analogue that is incorporated into proliferating cells during DNA replication. Following the administration of BrdU, recently divided cells are revealed by immunohistochemistry using a biotinylated monoclonal anti-BrdU antibiotics, which is detected using a streptavidin–biotin system that stains the nuclei dark brown. Cell proliferation was assayed during the secretory stage of amelogenesis in developing maxillary first molars at postnatal days 4 and 5 in wild-type (Enam+/+), enamelin heterozygous (Enam+/−), and enamelin homozygous (Enam−/−; null) mice (four mice in each group).
5-Bromo-2′-deoxyuridine labelling reagent (0.04 ml or 1 mg per 100 g body weight; Zymed Laboratories, Invitrogen Immunodetection, Carlsbad, CA, USA) was injected into the peritoneal space and the mice were killed 4 h later. Mouse heads were fixed and demineralized as described under ‘Staining of apoptotic cells’, sectioned at 5 µm thickness, and processed using the Zymed BrdU staining kit (Zymed Laboratories). Experiments were conducted on four samples at postnatal days 4 and 5 for each genotype with negative controls included throughout, and the sections were photographed using a Nitcon Eclipse TE300 inverted microscope.
Statistical analyses
Apoptosis (determined using the TUNEL method) in molar sections was assessed using Imagej software (http://rsb.info.nih.gov/ij/). The combined ameloblast + stratum intermedium layers on images of histological sections through the three buccal cusps of maxillary first molars were manually traced (outlined in yellow) and their areas determined. Image contrast was adjusted to highlight positive TUNEL staining, and its area within the boundaries of the ameloblast + stratum intermedium layers was determined. The areas of the ameloblast + stratum intermedium layers from heterozygous and homozygous (null) samples were divided by the area of the wild-type sample from the same day to normalize for variations in tooth size. This ratio was then multiplied by the area of positive TUNEL staining determined using Imagej to calculate the normalized area of apoptosis. The mean value of the normalized positive (TUNEL) staining area for two samples at each of days 5, 7, 9, 14, and 17 and its SD was calculated. The paired t-test was performed, and bar graphs prepared, using statistica for Windows version 10 (Statsoft, Tulsa, OK, USA). Statistical significance was determined when P < 0.05. Contrast-enhanced images used for quantification, as well as the negative controls, are shown in Fig. S1.
Results
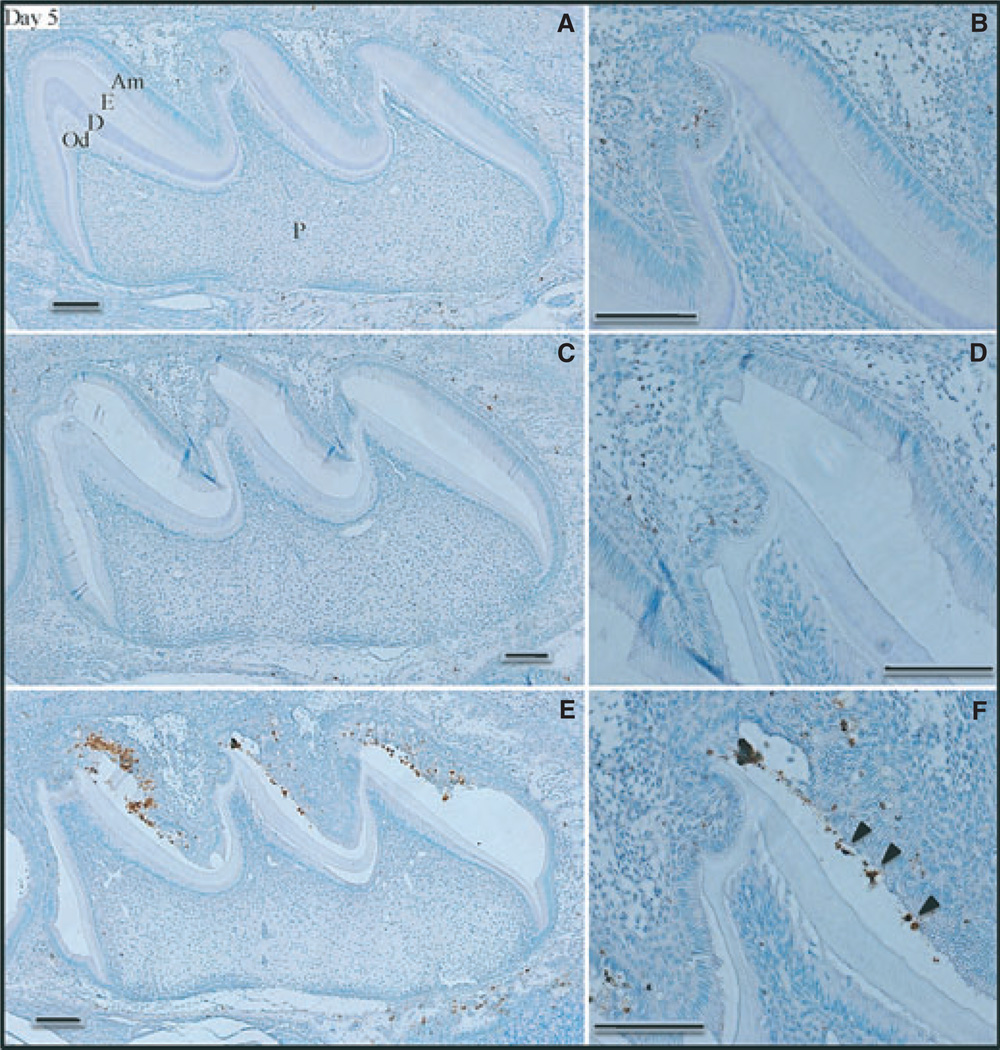
We examined the ameloblast layers of developing maxillary first molars in wild-type, Enam heterozygous (+/−), and Enam null (−/−) mice at postnatal days 5, 7, 9, 14, and 17. At postnatal day 5, ameloblasts in the maxillary first molars were all in the secretory stage. At this early stage, only trace levels of positive TUNEL staining were observed in the ameloblast layers of wild-type (Fig. 1A,B) and Enam heterozygous (Fig. 1C,D) maxillary first molars, and the staining was restricted to the enamel-free area (24) of the central cusp. Apoptosis was previously reported in the enamel-free area of rat molars (25). In contrast, the Enam null mouse molars showed extensive positive TUNEL staining (Fig. 1E,F) along the mesial slopes of all cusps. We observed that the enamel space of Enam null mice undergoes two types of pathological expansion. The first is irregular, mineralized, and associated with ameloblast apoptosis. The second is smooth, apparently unmineralized, cyst-like (20), often filled with organic matrix, and is not associated with apoptosis. The TUNEL staining on day 5 showed greatest intensity nearest the cusp tips, in areas of aberrant mineral deposition. Some of the stained nuclei appeared to be embedded in the mineral layer.
Fig. 1.
Apoptosis [determined using terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL)] in day-5 mouse maxillary first molars from wild-type (A, B), enamelin (Enam) heterozygous (C, D), and Enam null (E, F) mice. Panels on the right (B, D, F) detail the apoptotic staining in the ameloblast layer of the central cusp tip. Arrowheads in panel F indicate apoptotic nuclei apparently embedded in the mineral layer. Am, ameloblasts; De, dentin; En, enamel; Od, odontoblasts; P, pulp. Bars = 100 µm. Ameloblasts are in the secretory stage of amelogenesis.
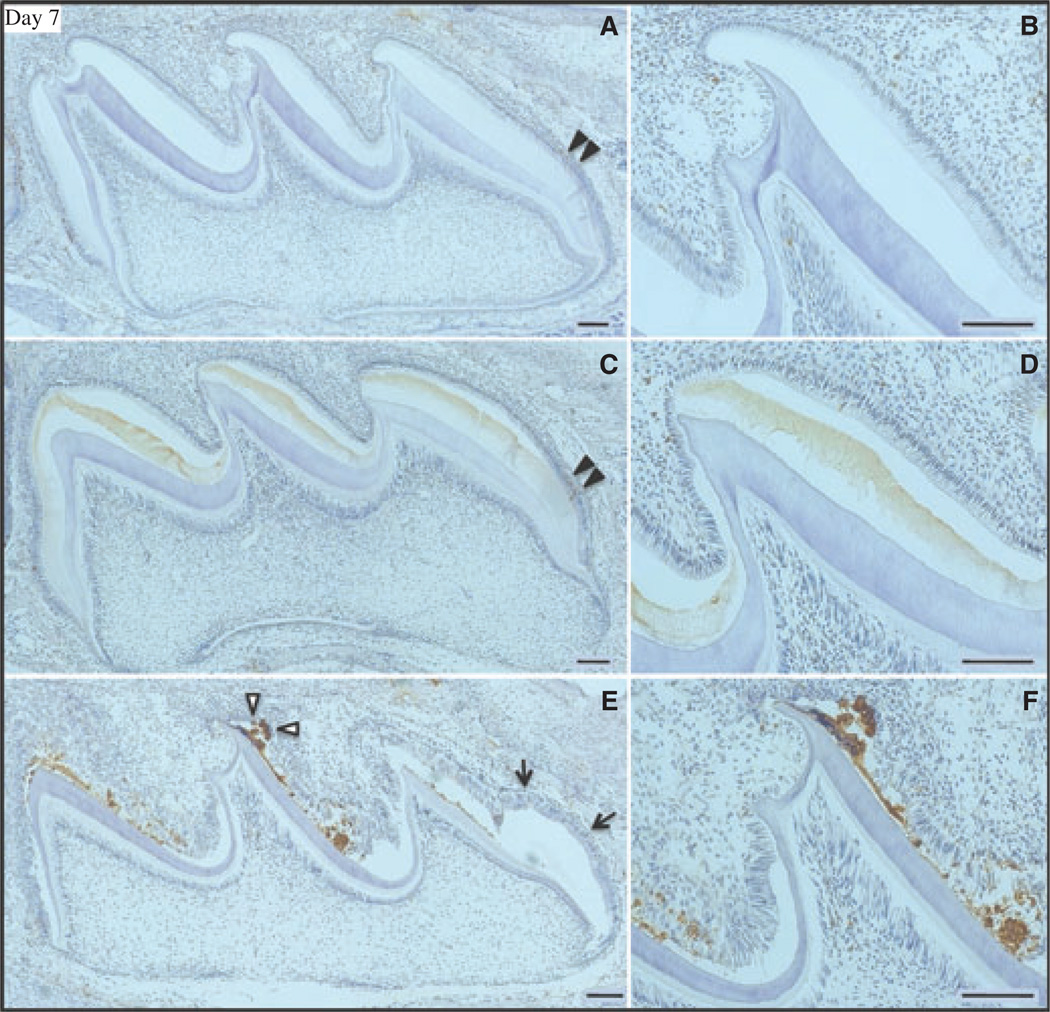
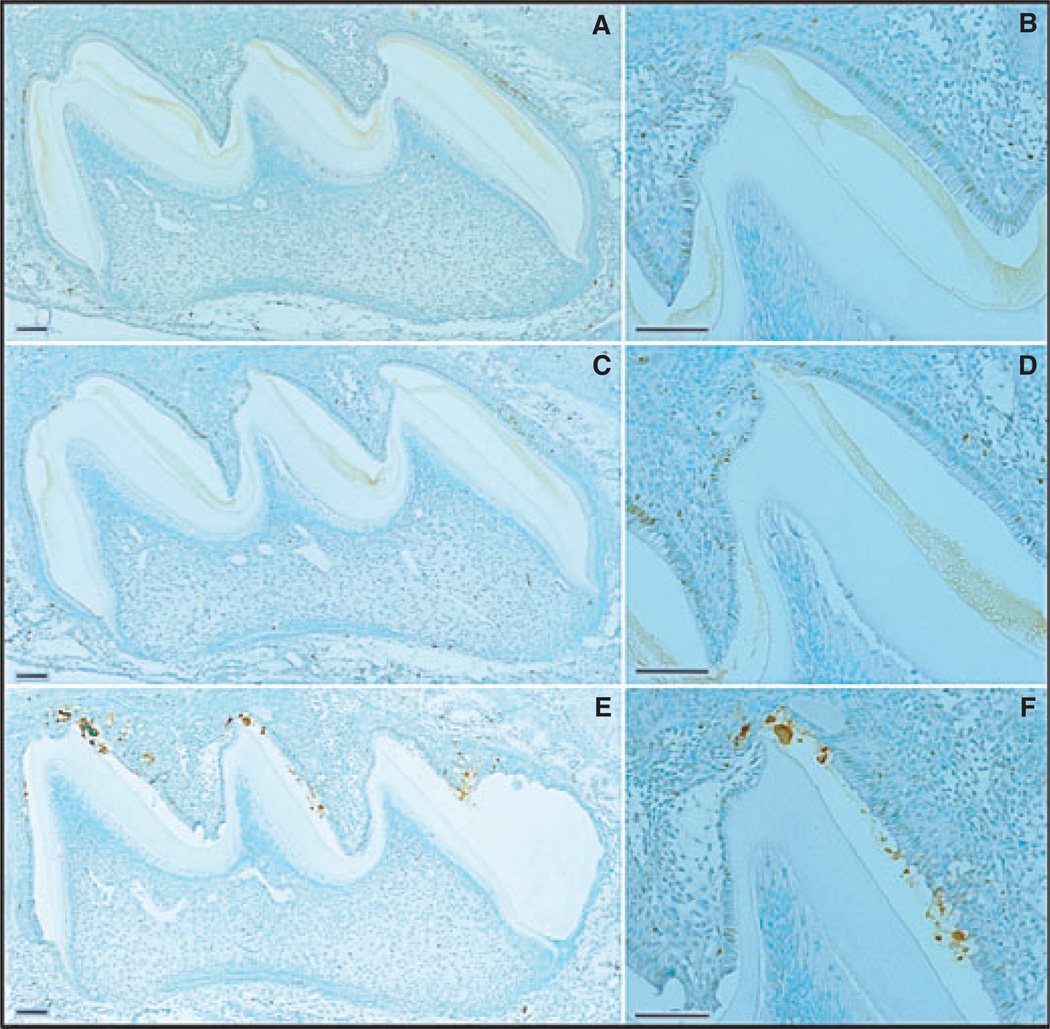
During postnatal day 7 the maxillary first molars are still mostly in the secretory stage, except in and near the enamel-free area. Only trace, sporadic TUNEL staining was observed in the ameloblast layers of day-7 wild-type and Enam heterozygous mice (Fig. 2A–D). As in day 5, the Enam null mouse maxillary first molars showed extensive positive TUNEL staining (Fig. 2E,F), especially along the mesial slopes of all cusps. The staining was again most intense in areas of mineral nodule formation. By day 9, ameloblasts in the maxillary first molars are in the early maturation stage, which is normally associated with an increase in normal ameloblast apoptosis during the transition from secretory to modulating ameloblasts (26, 27). Apoptosis (positive TUNEL staining) was observed in the day-9 wild-type and Enam heterozygous mice (Fig. 3A–D). The positive signal was evident in the nuclei of well-shaped columnar ameloblasts, with more staining appearing in the Enam heterozygous mice than in the wild-type mice. In the day-9 Enam null mouse molars, TUNEL staining was extensive and included nuclei of ameloblasts that had apparently become trapped within the mineral layer, which appeared to disrupt the integrity of the ameloblast sheet-like layer (Fig. 3E,F).
Fig. 2.
Apoptosis [determined using terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL)] in day-7 mouse maxillary first molars from wild-type (A, B), enamelin (Enam) heterozygous (C, D), and Enam null (E, F) mice. Arrowheads in panels A and C indicate trace apoptotic staining in the molars from wild-type and Enam heterozygous mice. Panels on the right (B, D, F) detail the ameloblast layer of the central cusp tip. In panel E the arrowheads mark a mineralized nodule associated with apoptotic nuclei. Light brown staining of the extracellular enamel matrix in the Enam heterozygote is background. Arrows indicate a cyst-like expansion not associated with apoptotic nuclei. Bars, panels on left = 200 µm; panels on right = 100 µm. Ameloblasts near the cusp tips are in the maturation stage; ameloblasts nearer the cervical loop are in the secretory stage.
Fig. 3.
Apoptosis [determined using terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL)] and assessment of day-9 mouse maxillary first molars from wild-type (A, B), enamelin (Enam) heterozygous (C, D), and Enam null (E, F) mice. Panels on the right (B, D, F) detail the apoptotic staining in the ameloblast layer of the central cusp tip. Bars = 100 µm. Ameloblasts are in the maturation stage of amelogenesis.
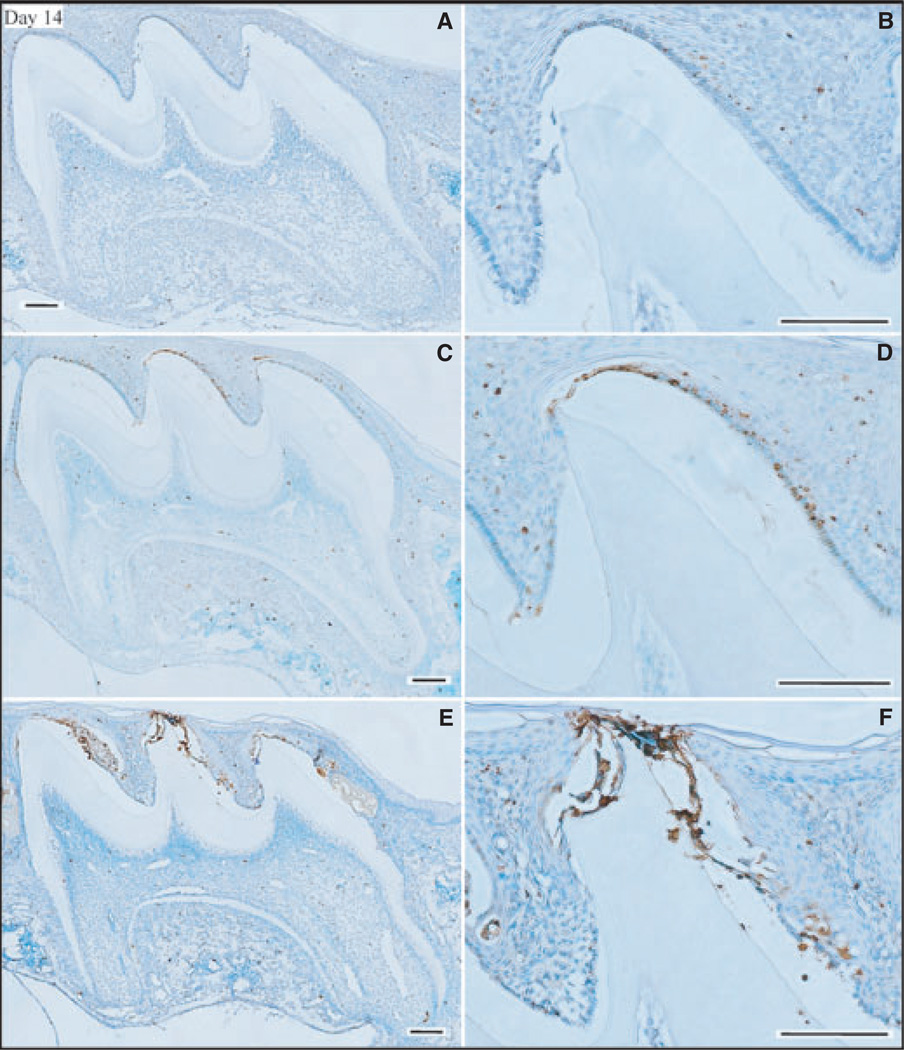
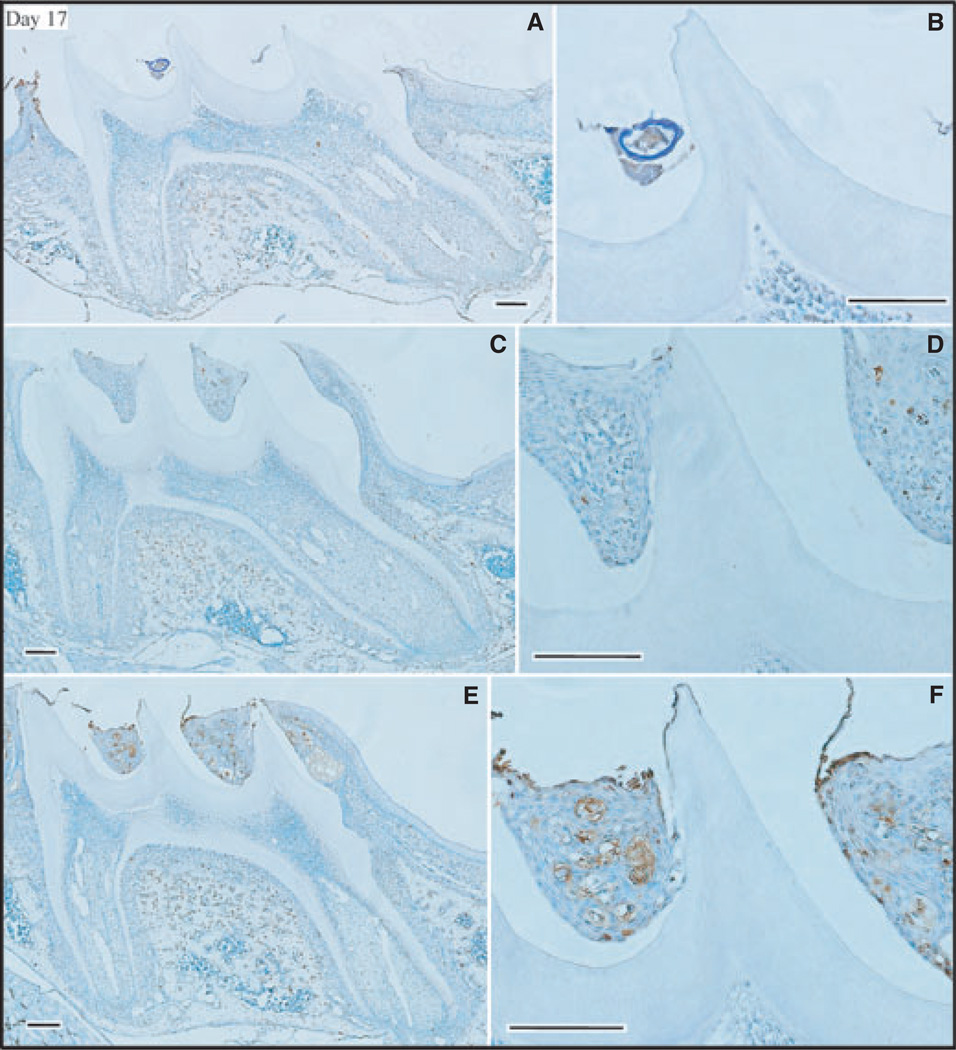
By day 14 the ameloblasts were reduced to cuboidal or low columnar in form. Positive TUNEL staining in the day-14 maxillary first molars of wild-type mice (Fig. 4A,B) was similar to that observed in the day-9 molars of wild-type mice. In contrast, apoptosis staining was elevated in both the Enam heterozygous (Fig. 4C,D) and null mice (Fig. 4E,F). The surprising finding at day 14 was the high level of apoptosis in the Enam heterozygous mice. This is the first evidence that Enam haploinsufficiency causes pathology in maturation-stage ameloblasts. On day 17 the maxillary first molars erupt into the oral cavity and small differences in the amount of eruption translate into large differences in the amount of reduced enamel epithelium still associated with the crown. In the Enam null mouse there appeared to be high levels of apoptosis within the wedge-shaped remnants of enamel organ between the cusps (Fig. 5). These remnants were not retained in the molars of wild-type mice (Fig. 5A,B), which showed slightly more advanced eruption than the molars of Enam heterozygous (Fig. 5C,D) and Enam null (Fig. 5E,F) mice.
Fig. 4.
Apoptosis [determined using terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL)] and assessment of day-14 mouse maxillary first molars from wild-type (A, B), enamelin (Enam) heterozygous (C, D), and Enam null (E, F) mice. Panels on the right (B, D, F) detail the apoptotic staining in the ameloblast layer of the central cusp tip. Bars = 100 µm. Ameloblasts are now reduced enamel epithelium.
Fig. 5.
Apoptosis [determined using terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL)] and assessment of day-17 mouse maxillary first molars from wild-type (A, B), enamelin (Enam) heterozygous (C, D), and Enam null (E, F) mice. Panels on the right (B, D, F) detail the apoptotic staining in the ameloblast layer of the central cusp tip. Bars = 100 µm. These molars are at the point of eruption into the oral cavity.
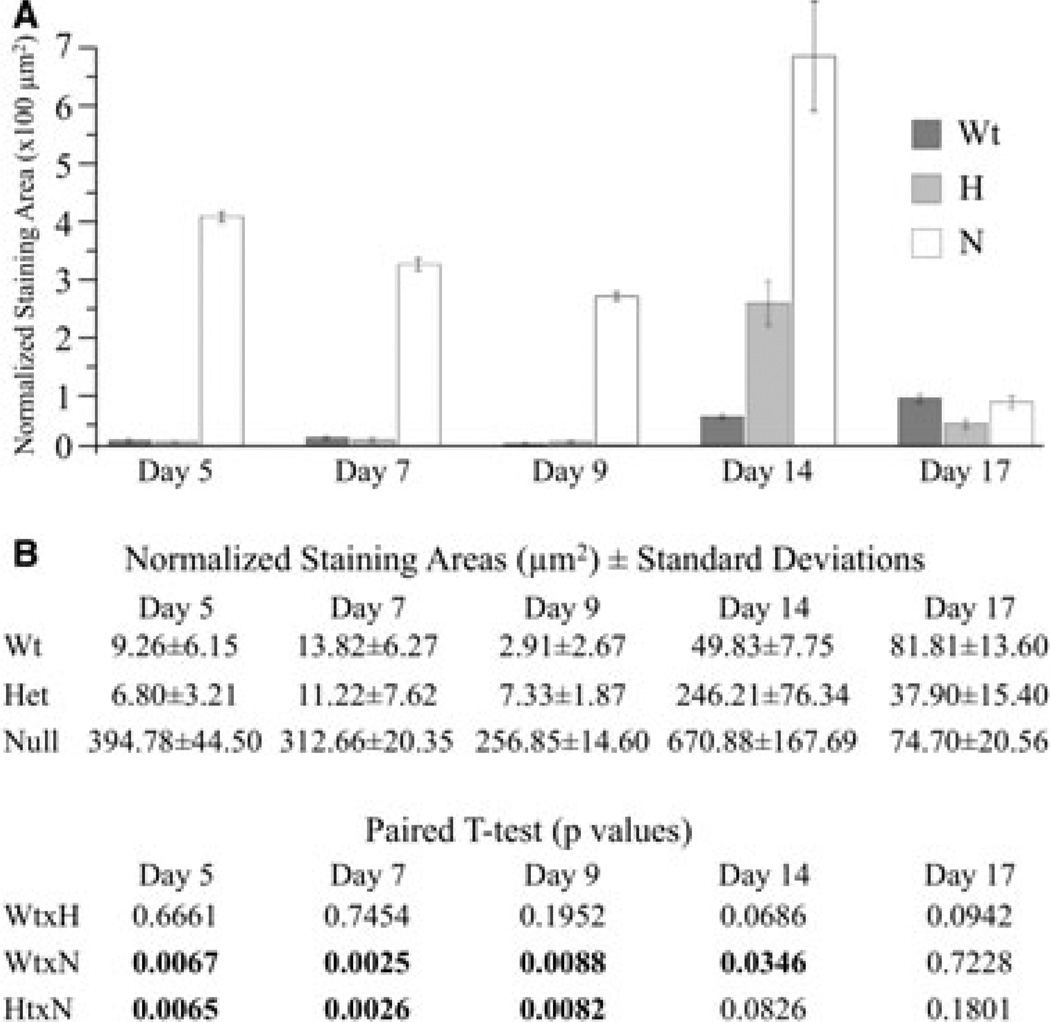
The combined ameloblast and stratum intermedium layers (as it was not always possible to resolve these layers) were manually outlined, as were the TUNEL staining areas within these layers. The averaged areas of apoptotic staining are plotted in Fig. 6. Apoptosis staining in secretory-stage ameloblasts (day 5) and in modulating ameloblasts (day 7 and 9) in the maturation stage was dramatically higher in the maxillary first molars of the Enam null mice than in Enam heterozygous mice and wild-type controls, and these differences were statistically significant. Apoptosis staining in the reduced enamel epithelium (days 14 and 17) was higher in the maxillary first molars of the Enam null mice compared with wild-type mice and was statistically significant. Apoptosis was also higher in the molars of Enam heterozygous mice relative to the molars of wild-type mice in the reduced enamel epithelium (day 14); however, these differences were not statistically significant because of a large SD and a small sample size, but the effect seemed real.
Fig. 6.
Apoptotic activities of the ameloblast + statum intermedium layer of mouse maxillary first molars from days 5, 7, 9, 14, and 17. Apoptosis [determined using terminal deoxynucleotidyl transferase (TdT)-mediated biotin–dUTP nick-end labelling (TUNEL)] in the enamelin (Enam) null mice was dramatically higher than in both the Enam heterozygous (H) mice and the wild-type controls, and these differences were statistically significant (numbers in bold) for days 5, 7, and 9 (secretory-stage and maturation-stage ameloblasts). Apoptosis staining in the Enam null and heterozygous mice was higher in the reduced enamel epithelium (day 14) relative to the wild-type mice. The lack of statistical significance when comparing Enam heterozygous mice with the wild-type and Enam null mice was attributed to a large SD and a small sample size, as the observed differences were evident histologically.
Enamelin is normally expressed during the secretory stage, and the trend towards a lowered normalized TUNEL staining area from day 5 to day 9 suggests that cell pathology correlates with the times of greatest enamelin expression. The plots of normalized positive staining increase at day 14; however, the reduced enamel epithelium has very little volume relative to the earlier stages when ameloblasts are columnar and associated with the stratum intermedium. Reducing the area of the cell layer increases the normalized TUNEL staining area because it is normalized to the area of the cells. Although the day-14 TUNEL staining increases, the difference between the wild-type and the Enam heterozygote is not statistically significant and the statistical significance of the difference between the wild-type and the Enam null mouse is not as robust as in the earlier stages.
Because the failure to express Enam at normal levels early in tooth development might have induced an increase in cell proliferation, the developing maxillary first molars from wild-type, Enam heterozygous (+/−), and Enam null (−/−) mouse at postnatal days 4 and 5 were analyzed for increased DNA synthesis by BrdU staining (Fig. S2). Positive BrdU staining appeared to be comparable in all three genetic backgrounds and was concentrated mainly in the region of the cervical loop.
Discussion
Early tooth development is regulated by a series of reciprocal epithelial–mesenchymal interactions between cells that have migrated from the cranial neural crest and the oral epithelium along the future alveolar ridge (28, 29). These interactions are later superseded by cell–matrix interactions as dentin and enamel layers develop between the epithelium and the mesenchyme. The transition from epithelial–mesenchymal interactions to cell–matrix interactions involves the fenestration and reabsorption of the basement membrane and establishment of a mineralization front along the secretory surface of the ameloblast distal membrane (30). This transition is a critical event and easily disrupted. Defects in the genes encoding the pre- and post-transition membrane-associated complexes result in ameloblast pathologies that interfere with the onset of dental enamel formation and can abort it altogether. Hemidesmosomes are crucial in maintaining the integrity of the ameloblast layer. Laminin-332 (formerly laminin V), an essential basal lamina component in skin and developing teeth, is an assembly of three gene products (LAMB3, LAMA3, and LAMC2). Genetic defects in these genes are the major cause of junctional epidermolysis bullosa with enamel defects (31–33). Although laminin is expressed throughout early tooth formation, presecretory ameloblasts in the Lama3 null mice are indistinguishable from those of wild-type mice. The pathology in tooth formation appears to initiate at the time of removal of the basal lamina, when, instead of progressing to secretory ameloblasts, the pre-ameloblasts become progressively smaller in size relative to the wild-type (34). On the other side of the transition from epithelial–mesenchymal interactions to cell–matrix interactions are the enamel proteins, most notably enamelin and ameloblastin, without which enamel does not form (19).
This study demonstrates that ameloblast apoptosis is a consequence of failure to express an enamel protein. The most surprising finding was not detecting apoptosis, however, but that the TUNEL staining in the Enam null mice was restricted in its localization to near the cusp tips and on the mesial cusp slopes. Ameloblasts near the cervical margin did not seem to be as susceptible to apoptosis in the absence of enamelin expression. Ameloblasts associated with irregular mineralizing nodules were most severely affected, and apoptotic nuclei were embedded in the mineral, as though the cells had become entrapped. Our explanation for this pathology envisions ameloblasts as continuously monitoring the extracellular matrix and adjusting their activities accordingly. Ameloblast cell pathology might relate less to a loss of attachment than to a loss of feedback/stimulation from the unmineralized extracellular matrix. Our model suggests that in the absence of enamelin or ameloblastin, the mineralization front fails to initiate enamel crystal formation, a mineralized enamel matrix does not form, and extracellular matrix receptors that may or may not include enamelin or ameloblastin as components fail to stimulate ameloblasts, which degenerate and undergo apoptosis. Continual secretion of calcium and phosphate ions results in aberrant and ectopic mineral deposition that can trap cells.
Cell attachment to the matrix is not emphasized in this model because secretory-stage ameloblasts seem to be only weakly attached to the mineralizing matrix. When we surgically extract secretory-stage mouse molars, it is a common experience for the enamel organ (and pulp) to slide right off the crown, particularly for a second molar at day 5, allowing the clean extraction of the mineralizing crown shell without the surrounding soft tissue. During the secretory stage, ameloblasts are constantly retreating from an expanding enamel surface, so they are probably only weakly attached. In contrast, the enamel organ must be torn away from crowns in the maturation stage, when the enamel organ has regenerated a basal lamina (35).
During development of normal teeth the forming enamel matrix rapidly expands in thickness. In the Enam null mice, the enamel matrix does not expand, or expands only slightly, so the ameloblast layer rests upon a smaller surface than normal. This may cause crowding and stresses that could contribute to ameloblast pathology and apoptosis. We assayed cell proliferation, by BrdU staining, in mice at postnatal days 4 and 5, but did not observe any apparent increase or decrease in cell proliferation among the wild-type, Enam heterozygous, or Enam null mice. There does not appear to be a feedback loop that reduces cell proliferation in the cervical loop to compensate for secretory ameloblasts that are not moving away from the dentin and thereby expanding the outer coronal surface area.
We have demonstrated ameloblast apoptosis at all stages of enamel development in Enam null mice. The direct cause of this cell pathology is not understood, but its existence suggests that ameloblasts must normally monitor and react to conditions in the extracellular environment to generate dental enamel, and genetic disturbances that arrest the normal mechanism of enamel deposition lead to a degeneration of the ameloblast layer. A better understanding of the situation will require better characterization of the onset of pathology and the extracellular matrix-sensing mechanisms that stimulate ameloblasts to continue the process of amelogenesis. The finding of extensive apoptosis in Enam null mice suggests that the enamel phenotype in subjects with ENAM mutations may be explained in part by cell pathology secondary to a primary mineralization deficit.
Supplementary Material
Acknowledgments
This investigation was supported by US-PHS Research Grant DE011301 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 29892, USA.
Footnotes
Conflicts of interest – All authors declare that there are no potential conflicts of interest.
Supporting Information
Additional Supporting information may be found in the online version of this article:
Fig. S1. TUNEL staining in maxillary first molars from days 5, 7, 9, 14 and 17.
Fig. S2. BrdU staining of day 4 and 5 maxillary first molars of wild-type, Enam heterozygous and Enam null mice.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki K, Suzuki T, Weiss KM. Genetic basis for the evolution of vertebrate mineralized tissue. Proc Natl Acad Sci USA. 2004;101:11356–11361. doi: 10.1073/pnas.0404279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP. Cloning and characterization of porcine enamelin mRNAs. J Dent Res. 1997;76:1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- 4.Yamakoshi Y. Carbohydrate moieties of porcine 32 kDa enamelin. Calcif Tissue Int. 1995;56:323–330. doi: 10.1007/BF00318054. [DOI] [PubMed] [Google Scholar]
- 5.Yamakoshi Y, Pinheiro FH, Tanabe T, Fukae M, Shimizu M. Sites of asparagine-linked oligosaccharides in porcine 32 kDa enamelin. Connect Tissue Res. 1998;39:39–46. doi: 10.3109/03008209809023910. [DOI] [PubMed] [Google Scholar]
- 6.Hu JC, Yamakoshi Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14:387–398. doi: 10.1177/154411130301400602. [DOI] [PubMed] [Google Scholar]
- 7.Wright JT. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet A. 2006;140:2547–2555. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JW, Simmer JP, Lin BP, Seymen F, Bartlett JD, Hu JC. Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci. 2006;114 Suppl. 1:3–12. doi: 10.1111/j.1600-0722.2006.00278.x. [DOI] [PubMed] [Google Scholar]
- 9.Chan HC, Mai L, Oikonomopoulou A, Chan HL, Richardson AS, Wang SK, Simmer JP, Hu JC. Altered enamelin phosphorylation site causes amelogenesis imperfecta. J Dent Res. 2010;89:695–699. doi: 10.1177/0022034510365662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang HY, Seymen F, Lee SK, Yildirim M, Tuna EB, Patir A, Lee KE, Kim JW. Candidate gene strategy reveals ENAM mutations. J Dent Res. 2009;88:266–269. doi: 10.1177/0022034509333180. [DOI] [PubMed] [Google Scholar]
- 11.Hart TC, Hart PS, Gorry MC, Michalec MD, Ryu OH, Uygur C, Ozdemir D, Firatli S, Aren G, Firatli E. Novel ENAM mutation responsible for autosomal recessive amelogenesis imperfecta and localised enamel defects. J Med Genet. 2003;40:900–906. doi: 10.1136/jmg.40.12.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mårdh CK, Backman B, Holmgren G, Hu JC, Simmer JP, Forsman-Semb K. A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2) Hum Mol Genet. 2002;11:1069–1074. doi: 10.1093/hmg/11.9.1069. [DOI] [PubMed] [Google Scholar]
- 13.Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, e-namelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- 14.Al-Hashimi N, Lafont AG, Delgado S, Kawasaki K, Sire JY. The enamelin genes in lizard, crocodile, and frog and the pseudogene in the chicken provide new insights on enamelin evolution in tetrapods. Mol Biol Evol. 2010;27:2078–2094. doi: 10.1093/molbev/msq098. [DOI] [PubMed] [Google Scholar]
- 15.Sire JY, Delgado SC, Girondot M. Hen’s teeth with enamel cap: from dream to impossibility. BMC Evol Biol. 2008;8:246. doi: 10.1186/1471-2148-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meredith RW, Gatesy J, Cheng J, Springer MS. Pseudogenization of the tooth gene enamelysin (MMP20) in the common ancestor of extant baleen whales. Proc Biol Sci. 2011;278:993–1002. doi: 10.1098/rspb.2010.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meredith RW, Gatesy J, Murphy WJ, Ryder OA, Springer MS. Molecular decay of the tooth gene Enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000634. e1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu JC, Hu Y, Smith CE, Mckee MD, Wright JT, Yamakoshi Y, Papagerakis P, Hunter GK, Feng JQ, Yamakoshi F, Simmer JP. Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith CE, Wazen R, Hu Y, Zalzal SF, Nanci A, Simmer JP, Hu JC. Consequences for enamel development and mineralization resulting from loss of function of ameloblastin or enamelin. Eur J Oral Sci. 2009;117:485–497. doi: 10.1111/j.1600-0722.2009.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawada T, Sekiguchi H, Uchida T, Yamashita H, Shintani S, Yanagisawa T. Histological and immunohistochemical analyses of molar tooth germ in enamelin-deficient mouse. Acta Histochem. 2011;113:542–546. doi: 10.1016/j.acthis.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A, Arakaki M, Harada H, Nonaka K, Nakamura S, Yamada Y, Fukumoto S. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J Biol Chem. 2009;284:27176–27184. doi: 10.1074/jbc.M109.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beyeler M, Schild C, Lutz R, Chiquet M, Trueb B. Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp Cell Res. 2010;316:1202–1212. doi: 10.1016/j.yexcr.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Addison WHF, Appleton JL. On the development of the a-meloblasts of the molars of the albino rat, with special reference to the enamel-free areas. Anat Rec. 1921;21:43. [Google Scholar]
- 25.Yamamoto H, Ishizeki K, Sasaki J, Nawa T. Ultrastructural and histochemical changes and apoptosis of inner enamel epithelium in rat enamel-free area. J Craniofac Genet Dev Biol. 1998;18:44–50. [PubMed] [Google Scholar]
- 26.Shibata S, Suzuki S, Tengan T, Yamashita Y. A histochemical study of apoptosis in the reduced ameloblasts of erupting mouse molars. Arch Oral Biol. 1995;40:677–680. doi: 10.1016/0003-9969(95)00021-g. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa S, Sasaki F. DNA localization in nuclear fragments of apoptotic ameloblasts using anti-DNA immunoelectron microscopy: programmed cell death of ameloblasts. Histochem Cell Biol. 1995;104:151–159. doi: 10.1007/BF01451574. [DOI] [PubMed] [Google Scholar]
- 28.Lumsden AG. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103 Suppl:155–169. doi: 10.1242/dev.103.Supplement.155. [DOI] [PubMed] [Google Scholar]
- 29.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch D, Soriano P, Mcmahon A, Sucov H. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 30.Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89:1024–1038. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchroithner B, Klausegger A, Ebschner U, Anton-Lamprecht I, Pohla-Gubo G, Lanschuetzer CM, Laimer M, Hintner H, Bauer JW. Analysis of the LAMB3 gene in a junctional epidermolysis bullosa patient reveals exonic splicing and allele-specific nonsense-mediated mRNA decay. Lab Invest. 2004;84:1279–1288. doi: 10.1038/labinvest.3700164. [DOI] [PubMed] [Google Scholar]
- 32.Mclean WH, Irvine AD, Hamill KJ, Whittock NV, Coleman-Campbell CM, Mellerio JE, Ashton GS, Dopping-Hepenstal PJ, Eady RA, Jamil T, Phillips RJ, Shabbir SG, Haroon TS, Khurshid K, Moore JE, Page B, Darling J, Atherton DJ, Van Steensel MA, Munro CS, Smith FJ, Mcgrath JA. An unusual N-terminal deletion of the laminin alpha3a isoform leads to the chronic granulation tissue disorder laryngo-onycho-cutaneous syndrome. Hum Mol Genet. 2003;12:2395–2409. doi: 10.1093/hmg/ddg234. [DOI] [PubMed] [Google Scholar]
- 33.Pfendner EG, Lucky AW. Pagon RA, editor. Junctional epidermolysis bullosa. [accessed on 10 September 2011];Gene Reviews. 2008 Available online at: http://www.ncbi.nlm.nih.gov/books/NBK1125/
- 34.Ryan MC, Lee K, Miyashita Y, Carter WG. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145:1309–1323. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al Kawas S, Warshawsky H. Ultrastructure and composition of basement membrane separating mature ameloblasts from enamel. Arch Oral Biol. 2008;53:310–317. doi: 10.1016/j.archoralbio.2007.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.