Abstract
Motor neurons that control limb movements are organized as a neuronal nucleus in the developing ventral horn of the spinal cord called the lateral motor column. Neuronal migration segregates motor neurons into distinct lateral and medial divisions within the lateral motor column that project axons to dorsal or ventral limb targets, respectively. This migratory phase is followed by an aggregation phase whereby motor neurons within a division that project to the same muscle cluster together. These later phases of motor neuron organization depend on limb-regulated differential cadherin expression within motor neurons. Initially, all motor neurons display the same cadherin expression profile, which coincides with the migratory phase of motor neuron segregation. Here, we show that this early, pan-motor neuron cadherin function drives the divisional segregation of spinal motor neurons in the chicken embryo by controlling motor neuron migration. We manipulated pan-motor neuron cadherin function through dissociation of cadherin binding to their intracellular partners. We found that of the major intracellular transducers of cadherin signaling, γ-catenin and α-catenin predominate in the lateral motor column. In vivo manipulations that uncouple cadherin–catenin binding disrupt divisional segregation via deficits in motor neuron migration. Additionally, reduction of the expression of cadherin-7, a cadherin predominantly expressed in motor neurons only during their migration, also perturbs divisional segregation. Our results show that γ-catenin-dependent cadherin function is required for spinal motor neuron migration and divisional segregation and suggest a prolonged role for cadherin expression in all phases of motor neuron organization.
Introduction
Neuronal nuclei are a recurrent organizational scheme within the CNS that clusters functionally related neuronal soma as spatially distinct groups (Cajal, 1995). Despite the critical functions of neuronal nuclei, little is known of the molecular mechanisms that drive their clustering during development, a process termed nucleogenesis (Agarwala and Ragsdale, 2002).
The spinal motor neurons that control limb movement form a neuronal nucleus termed the lateral motor column (LMC) (Jessell, 2000). As with other neuronal nuclei, LMC internal structure is related to the axonal targets of the neurons in the nucleus (Landmesser, 1978). The LMC segregates into lateral (LMCl) and medial (LMCm) divisions related to the dorsal or ventral compartments of the limb to which each division projects. Divisional segregation occurs by inside-out migration of LMCl neurons through the earlier-born LMCm (Hollyday and Hamburger, 1977).
Following motor neuron migration, clusters of motor neurons that project axons to an individual muscle emerge (Whitelaw and Hollyday, 1983). Clustering of these so-called motor neuron pools (Romanes, 1964) results from differential expression of members of the type II family of cadherin cell adhesion molecules (Price et al., 2002). Cadherin expression within the LMC is highly dynamic and encompasses a pan-motor neuron phase during divisional segregation with a later motor pool-specific phase. For example, expression of cadherin-20 is initiated in all motor neurons soon after their generation and is refined via limb-derived signals to a motor pool-specific pattern only after divisional segregation is well underway (Price et al., 2002). Absence of these limb-derived signals perturbs motor pool sorting while leaving divisional segregation intact (Haase et al., 2002; Livet et al., 2002). This suggests that divisional segregation and pool sorting are separable and raises the possibility that early, pan-motor neuron cadherin expression could drive divisional segregation before pool sorting.
The cytoplasmic domain of type II cadherins binds to members of the armadillo family of catenins: β-catenin or γ-catenin (plakoglobin) (Nollet et al., 2000). β-Catenin and γ-catenin bind to α-catenin, which anchors cadherin adhesion to the actin cytoskeleton via the linker protein EPLIN (Abe and Takeichi, 2008). Thus, function of all type II cadherins converges on their interaction with either β- or γ-catenin and absence of β- or γ-catenin abrogates cadherin function (Kintner, 1992; Weis and Nelson, 2006).
We investigated pan-motor neuron cadherin expression in LMC organization through manipulations of catenin–cadherin binding and reduction of a cadherin predominantly expressed during divisional segregation. Expression of a single amino acid mutant of γ-catenin, predicted to uncouple interaction with α-catenin, results in a cell autonomous stalling of motor neuron migration with a concomitant disruption of divisional segregation. Dissociation of cadherin from γ-catenin via expression of an extracellular deleted dominant-negative cadherin also perturbs motor neuron migration and divisional segregation. Finally, knockdown of cadherin-7 perturbs divisional segregation. These data are consistent with a model whereby early pan-motor neuron cadherin function drives the migration of LMC neurons into the ventral horn and suggest a role for cadherin expression throughout nucleogenesis of spinal motor neurons.
Materials and Methods
Chick embryo preparation.
Fertilized Brown Bovan Gold Hen's eggs (Henry Stewart Farms) were incubated in a forced draft incubator at 38°C and staged as in Hamburger and Hamilton (1992). All embryos were treated in accordance with the Animals (Scientific Procedures) Act of 1986, UK. Embryos of both sexes were used in our experiments.
Labeling of migrating neurons.
Horse radish peroxidase (HRP; Roche) 50% solution in PBS with 1% lysolecithin (Sigma) was pressure injected into the dorsal limb essentially as described by Lin et al. (1998) to retrograde label neurons.
BrdU labeling.
BrdU (200 μl, 1 mm; Sigma) was injected directly under the embryo and the eggs sealed and returned to the incubator for the desired length of time. Following cryosectioning, embryo sections were incubated for 5 min in 2 m HCl (Sigma) followed by five washes in PBS, each for 5 min. Primary antibodies to BrdU and subsequent secondary antibody detection was as described in Immunohistochemistry, below.
In situ hybridization histochemistry.
Digoxigenin (DIG)-labeled antisense cRNA probes were used for in situ hybridization histochemistry on 15-μm-thick cryostat sections as in Price et al. (2002). Dual in situ hybridization histochemistry with BrdU labeling was performed by sequential in situ hybridization followed by a 2 m HCl treatment at 20°C for 5 min, three washes in PBS for 5 min, each followed by immunohistochemistry for BrdU with secondary antibodies conjugated to HRP followed by IMPACT DAB staining according to the manufacturers protocol (Vector Labs).
In ovo electroporation.
Expression of cDNAs was achieved by in ovo electroporation using an ECM830 electro-squareporator (BTX). Approximately 0.1 μl of DNA constructs [1–10 μg/μl in H2O with 0.1% Fast Green (Sigma)] was pressure injected into the lumen of the spinal cord. Five 30 volt electrical pulses of 50 ms duration equally spaced over a 5 s period were applied by placing electrodes adjacent to each side of the spinal cord of the embryo. Embryos were electroporated at Hamburger and Hamilton (HH) stages 12–18 and analyzed at HH stages 25–32.
Generation of constructs.
A full-length cDNA for chick γ-catenin was cloned by screening an E3 chick cDNA library. The sequence of chick γ-catenin has been submitted to GenBank (accession number HM102357). Point mutation (L127A) was generated using the QuikChange kit (Stratagene) following the manufacturers protocol. γ-Catenin and γ-catenin (L127A) cDNAs were cloned into a pCAGGS vector containing an internal ribosome entry sequence followed by a cDNA encoding nuclear localization sequence tagged β-galactosidase (pCAGGS inlz). Other constructs used in this work included HA-tagged dominant-negative GSK, HA-tagged β-catenin ΔARM, HA-tagged β-catenin-1-ins, HA-tagged constitutively active GSK, HA-tagged dominant-negative TCF, transposase integrated doxycycline inducible N-cadherinΔ390 (Kawakami and Noda, 2004; Tanabe et al., 2006; Sato et al., 2007; Watanabe et al., 2007), cad-7 shRNA knock-down, control cad-7 shRNA [described, tested and characterized in Barnes et al. (2010), these constructs follow the method described by Das et al. (2006)], pCAGGS inlz, and CMV eGFP (Invitrogen). In situ hybridization probes for cad-20 (MN-cad) and cad-12 were described by Price et al. (2002).
Immunohistochemistry.
Antibodies used in this study were as follows: rabbit (R) anti-GFP (1/1000; Invitrogen), R anti-β-catenin (1/1000; Sigma), R anti-pan-cadherin (1/1000; Sigma), R anti-Hb9 (1/5000), R anti-N-cadherin (1/1000; AbCAM,), Dylight 488-conjugated R anti-HRP (The Jackson Laboratory), rat anti-HA (1/500; Roche), guinea pig (GP) anti-Islet-1(2) (1/20000), GP anti-FoxP1, goat (G) anti-HRP (1/2000; Jackson ImmunoResearch), G anti-β-galactosidase (1/1000), mouse (M) anti-α-Ε-catenin (1/100; Zymed), M anti-γ-catenin (1/100; BD Biosciences), M anti-GFP (1/100; Invitrogen), M anti-BrdU (1/50; Roche), M anti-HA (1/50; Covance). The following mouse monoclonal antibodies were purchased from the Developmental Studies Hybridoma Bank: 745A5 (anti-Nkx2.2) PAX6 (anti-Pax6), 4F2 (anti-Lim1/Lhx1), 2D6 and 4D5 (anti-Islet-1), 5C10 (anti-MNR2/Hb9), A2B11 and EAP3 (anti-transitin), CCD7–1 (anti-cadherin-7). Alkaline phosphatase-conjugated sheep anti-DIG Fab fragments (1/5000; Roche) immunocytochemistry was performed essentially as described previously (Price et al., 2002). Cryostat sections mounted on superfrost plus glass slides were incubated in PBS for 5 min followed by incubation in block solution (PBS with 1% goat serum; Sigma) for 30 min at 20°C. This solution was replaced by antibody diluted in block solution and incubated for 12–16 h at 4°C. Following three washes of 5 min each in PBS, fluorescent-conjugated secondary antibodies were incubated with the sections for 30 min at 20°C in block solution, washed, and mounted with vectashield fluorescent mounting medium (Vector Labs).
Image acquisition and data analysis.
Images were acquired on a Nikon Eclipse E80i fluorescence microscope equipped with a Nikon DS5M and Hamammatsu ORCA ER digital camera or on a Leica SPE confocal microscope. Quantitation of migration lengths was performed using ImageJ software to trace the pathway from ventricular zone to identified motor neurons using knowledge of the curved nature of the pathway following γ-catenin (L127A) expression.
Divisional mixing index.
The divisional mixing index was calculated by focusing on one LMCl cell at a time and counting the number of LMCm that were immediately adjacent to it. The control side of the spinal cord and control electroporations resulted in close to 100% of such cells having no LMCm cells surrounding them. These results were quantitated for at least five different embryos of each phenotype and presented as mean ± SEM percentages in each bin of the mixing index.
Results
LMC neurons migrate in an orderly series during divisional segregation
We first characterized the timing of LMC formation as a nucleus in the ventral horn through analysis of the expression of transcription factors that define divisions of the LMC. At lumbar spinal cord levels, expression of the forkhead transcription factor Foxp1 identifies the LMC (Dasen et al., 2008; Rousso et al., 2008). Further, expression of the LIM homeodomain factor Islet-1 identifies neurons of the LMCm (Tsuchida et al., 1994). Neurons of the LMCl express Hb9 (William et al., 2003). The expression of these transcription factors persists through the migratory phase of LMC formation (Tsuchida et al., 1994; William et al., 2003; Dasen et al., 2008; Rousso et al., 2008). Neurons of the medial motor column, which project axons to axial or body wall muscles, coexpress both Islet-1 and Hb9 but do not express Foxp1 (Dasen et al., 2008; Rousso et al., 2008).
More than 95% of LMC neurons are born within the 24 h period before HH (Hamburger and Hamilton, 1992) stage (st) 23, with the peak generation of LMCm in rostral lumbar regions from st18 to st20 and LMCl occurring at st20 and st21 (Hollyday and Hamburger, 1977; Whitelaw and Hollyday, 1983). We thus assessed the time course of motor neuron migration from st23 to st27. Consistent with previous studies, we found that at HH st23, LMCm neurons dominate in the ventral horn with the majority of LMCl neurons found medial to this group (Fig. 1A–C) (Lin et al., 1998; Sockanathan and Jessell, 1998; William et al., 2003). LMCl neurons were first detected in the lateral ventral horn at approximately st24 (data not shown) and divisional segregation continued until approximately st27 (Fig. 1D–I, summarized in Fig. 1J–L). Therefore, ∼24 h appears to be required for the migration of a given LMCl neuron, with LMCl migration extending over a continuous 48 h period after they are generated. Thus, LMCl neurons initiate their migration in an ordered series during divisional segregation.
Figure 1.
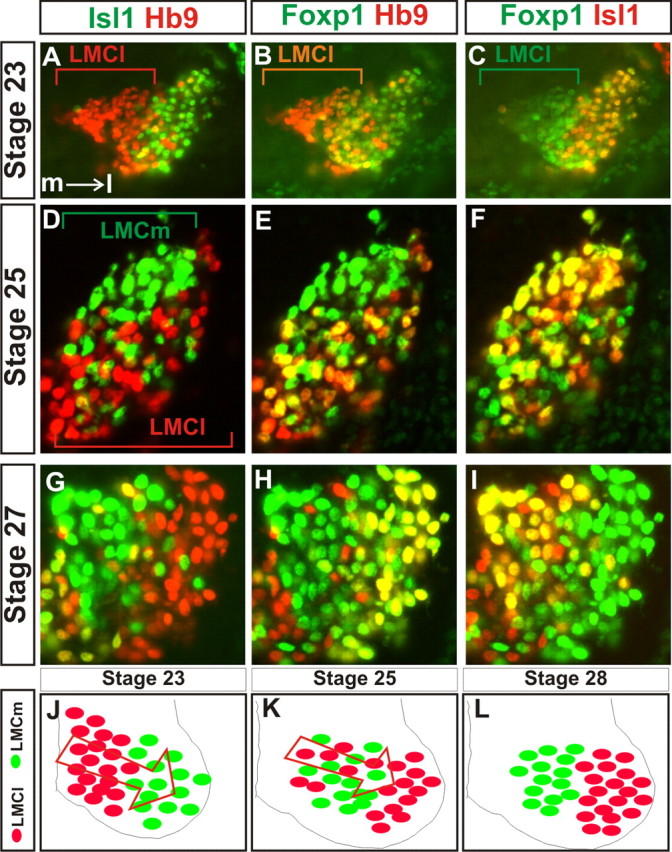
Time course of LMC divisional segregation. A–C, Expression in the ventral horn of a stage 23 embryo of Islet-1 and HB9 (A), Hb9 and Foxp1 (B), and FoxP1 and Islet-1 (C). Foxp1 marks the entire lateral motor column. Hb9 and Islet-1 are LMC divisional markers. Medial is to the left and is illustrated in A. LMCl cells are, at this stage, more medial to the LMCm cells. D–F, Ventral horn expression at stage 25 of the divisional markers Hb9 (D, E) and Islet-1 (D, F) and the LMC marker Foxp1 (E, F). Note that LMCl and LMCm cells are intermingled. G–I, Columnar segregation is largely complete by stage 27 as assessed by ventral horn expression of Foxp1 (H, I), Islet-1 (G, I), and Hb9 (G, H). J–L, Summary of the migration of LMCl cells through LMCm cells during motor neuron divisional segregation at st23 (J), st25 (K), and st28 (L).
γ-Catenin and α-catenin are expressed by the LMC during divisional segregation
We next characterized the expression of classical cadherin signaling components during the stages of motor neuron migration and divisional segregation (Nollet et al., 2000; Zhurinsky et al., 2000a; Hirano et al., 2003; Yamada et al., 2005; Uemura and Takeichi, 2006). We focused our attention on the cytoplasmic binding partners for classical cadherins, which play roles critical to cadherin function, α-catenin, β-catenin, and γ-catenin. We found that α-catenin is expressed in most neurons in the spinal cord and appeared to be expressed in all spinal motor neurons, including the LMC, at all stages analyzed (st20–st32; Fig. 2A,B; data not shown). γ-Catenin was also found in the majority of LMC neurons on both their soma and axons from the time of their first generation to at least HH st32 (Fig. 2C–J; data not shown).
Figure 2.
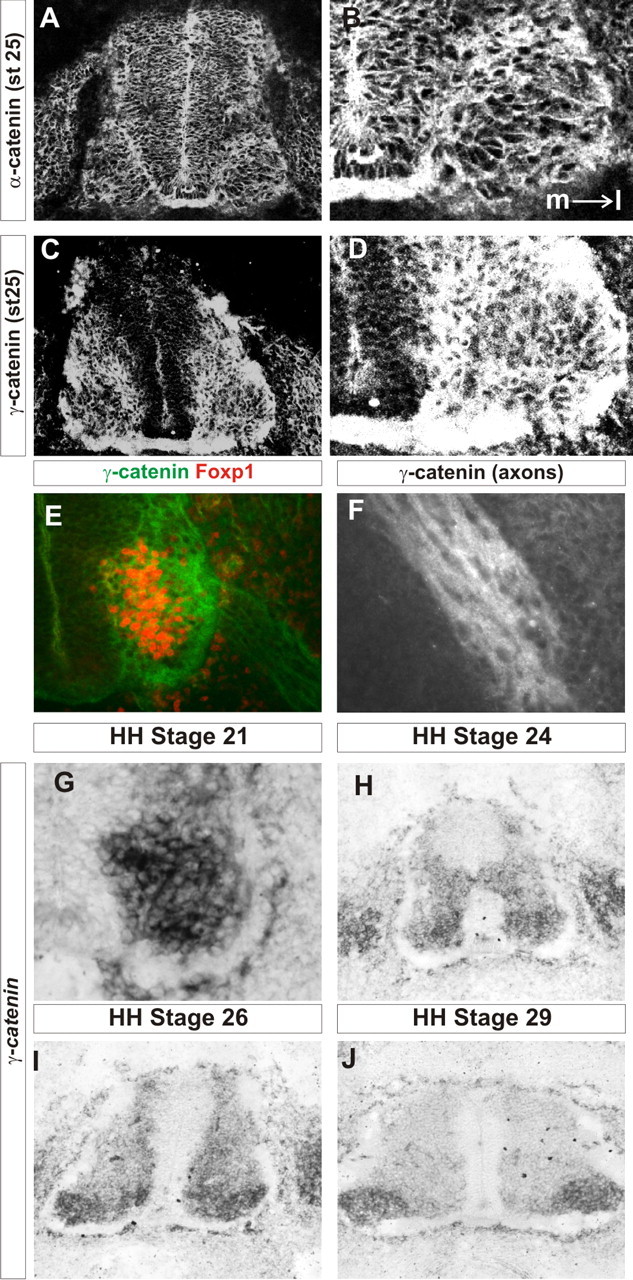
α-Catenin and γ-catenin are expressed in the lateral motor column during motor neuron migration. A, B, α-Catenin immunofluorescence in the lumbar spinal cord at st25. The whole spinal cord is shown in A. The ventral horn is shown in B. Medial is to the left, as shown in B. C, D, γ-Catenin immunofluorescence in the lumbar spinal cord at st25. The whole spinal cord is shown in C. The ventral horn is shown in D. Medial is to the left. E, γ-Catenin is expressed within the LMC, as marked by Foxp1 expression. F, γ-Catenin immunofluorescence in motor axons. G, γ-Catenin transcript expression in the ventral horn of stage 21 lumbar spinal cord. H–J, Lumbar spinal cord expression of γ-catenin transcript at st24 (H), st26 (I), and st29 (J).
β-Catenin is expressed by transitin radial glia in the ventral spinal cord
In contrast to α- and γ-catenin, β-catenin expression was found in a radial pattern within the ventral horn (Fig. 3A). β-Catenin transcript was predominantly found in cells (∼14 per section) in a line approximately parallel to the ventricle within the ventral part of the spinal cord, at the ventricular surface, and in the floor plate (Fig. 3B–E). This expression persisted from st21 to at least st29. Outside of this ventricular zone expression, only low levels of β-catenin could be found in only a small number of cells within the ventral horn; this became more apparent after st26. Within more dorsal regions of the spinal cord, β-catenin was detected in the majority of cells (Fig. 3C–E). Outside the spinal cord, β-catenin expression was also observed at the ventral root exit points (Fig. 3B, arrow).
Figure 3.
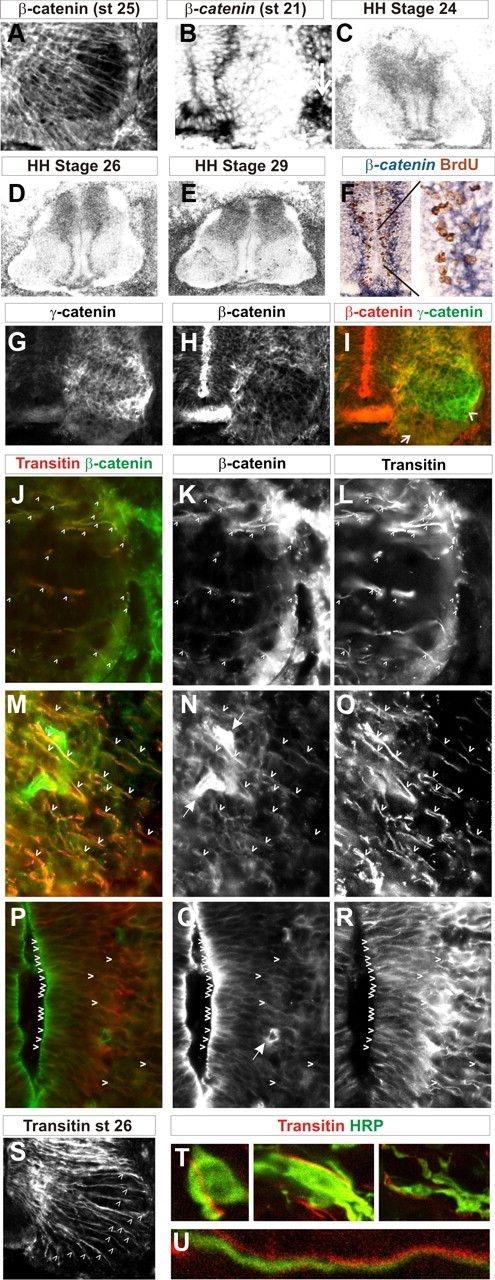
β-Catenin is expressed in transitin radial glia. A, β-Catenin immunoreactivity in the ventral horn of a stage 25 embryo. B–E, β-Catenin transcript expression in stage 21 ventral spinal cord (B), stage 24 (C), stage 26 (D), and stage 29 (E) lumbar spinal cords. F, BrdU expression (brown) with β-catenin transcript (blue) following a 1 h pulse application of BrdU to a stage 23 embryo. Right, Magnification of the area shown by the black lines. G–I, Double immunofluorescence of γ-catenin (G, I) and β-catenin (H, I) at stage 25. I, Arrow, β-Catenin expression in radial glia; arrowhead, γ-catenin expression in motor neurons. J–R, β-Catenin (J, K, M, N, P, Q) and transitin (J, L, M, O, P, R) localization at stage 26 in the ventral horn (J–L), intermediate part of the ventral spinal cord (M–O), and ventral ventricular zone (P–R). Arrowheads, Regions of colocalization in radial glia. N, Q, Arrows, Blood vessel expression of β-catenin. S, Transitin immunofluorescence in the ventral spainal cord of a st26 embryo. Arrowheads, Individual transitin radial glia processes. T, U, Confocal sections (0.15 μm) showing HRP labeling of migrating motor neurons (green) and transitin (red) expression following retrograde tracing of LMCl cells. T, Cell bodies of five different motor neurons are shown. Note the close apposition of the motor neurons to the transitin fibers. U, Motor axons could also be observed in close apposition to transitin radial glial processes.
We asked whether the ventral line of cells that express β-catenin progress through the cell cycle. A 1 h pulse of BrdU was applied to embryos at HH st22 and subsequent immunohistochemistry for BrdU and in situ hybridization for β-catenin was performed. We found that ∼30% of BrdU+ve cells were colabeled with β-catenin, suggesting that β-catenin-expressing cells synthesize DNA but that only a subset of progenitor cells express detectable levels of β-catenin (Fig. 3F). We also confirmed that γ-catenin is predominantly expressed within motor neurons while β-catenin is excluded from motor neurons by double immunofluorescence of the two proteins (Fig. 3G–I).
The expression of β-catenin within a subset of progenitor cells suggested that the radial staining of β-catenin could be within radial glia. Within the chick spinal cord, radial glia express the intermediate filament protein transitin (Cole and Lee, 1997). Double immunofluorescence staining indicated a colocalization of transitin-expressing radial fibers and β-catenin within the ventral spinal cord (Fig. 3J–R). Within the ventral horn, transitin immunofluorescence at HH st26 showed an average of 14 (modal value; range 12–15) glial fibers (Fig. 3S), similar to the number of β-catenin cells observed in the ventral ventricular zone. Together, this suggests that β-catenin is predominantly expressed in transitin radial glia in the ventral spinal cord and that these radial glia represent a subset of ventral progenitor cells.
Transitin radial glia mark pathways of motor neuron migration
We further characterized the transitin/β-catenin radial glia by asking whether the pathways of migration of LMC neurons coincide with them. We identified subsets of migrating LMCl motor neurons through injection of the retrograde axonal tracer, HRP, into the dorsal limb mesenchyme at st25. Migrating motor neurons were identified by their location within the LMCm domain at the time of analysis. Confocal analysis of 0.15-μm-thin optical sections revealed that in all cases (50/50 HRP+ve neurons analyzed within the LMCm), transitin fibers and migrating motor neurons were closely juxtaposed (Fig. 3T). Additionally, we found evidence of motor axons following the paths of transitin radial glia (Fig. 3U). This suggests that transitin/β-catenin radial glia mark the pathways of LMC migration during LMC divisional segregation.
Overexpression of γ-catenin leaves LMC organization unperturbed
We focused our attention on γ-catenin as motor neurons express it predominantly. We cloned a full-length cDNA of chick γ-catenin and found that the transcript was 86% identical to that of human γ-catenin and its amino acid sequence was 89% identical (94% similar) to that of human γ-catenin. Additionally, chick γ-catenin was 69% identical (89% similar) to chick β-catenin, illustrating the high level of conservation between these two armadillo family members (data not shown). We expressed wild-type γ-catenin by in ovo electroporation (Momose et al., 1999) and confirmed expression of the protein, noting in particular its presence at the apical surface of the ventricular zone (data not shown). Overexpression of γ-catenin (marked by nuclear β-galactosidase immunoreactivity in Fig. 4A) or the empty DNA vector had no observable effect on the total number of motor neurons or the position of those motor neurons in the ventral horn or the segregation of LMCl and LMCm divisions (Fig. 4A,B; data not shown). This suggests that the levels of γ-catenin are saturating with regard to a role in motor neuron migration.
Figure 4.
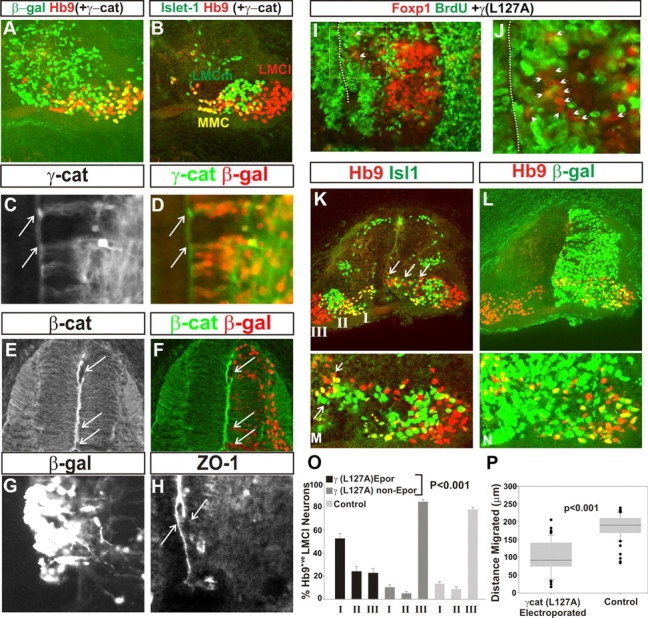
γ-Catenin (L127A) expression disrupts LMC migration and divisional segregation. A, B, Lack of effect of γ-catenin overexpression on divisional segregation at stage 29. Cells that had been electroporated are marked by β-gal immunoreactivity (A, green). Hb9 (A, B) and Islet-1 (B) mark the medial motor column (MMC), LMCl, and LMCm in the ventral horn. C, D, Expression of γ-catenin (L127A) viewed by γ-catenin immunoreactivity. Electroporated cells are marked by β-gal immunofluorescence (D, red). Arrows, Apical expression of γ-catenin at the ventricle surface. E, F, Disruption of β-catenin apical expression following γ-catenin(L127A) expression. Arrows, Regions of disruption. G, H, Disruption of ZO1 apical expression following γ-catenin(L127A) expression. H, Left arrow, ZO1 apical expression on the contralateral spinal cord; right arrow, disruption on the electroporated side of the spinal cord. I, J, The ventral spinal cord after electroporation with γ-catenin (L127A) at st18. Foxp1 (red) and BrdU (green) expression after BrdU application from st23 to st28. No colocalization of motor nuclei with BrdU was observed, indicating that all motor neurons had been born before st23 following γ-catenin(L127A) expression. J, Magnification of boxed area in I. I, J, Dotted lines, Midline; arrows, Foxp1 nuclei stalled in their migration and are unlabeled by BrdU. K–N, Effects of γ-catenin(L127A) expression on divisional segregation in st29 lumbar spinal cords. K, L, The left side of the spinal cord is unelectroporated (β-gal immunofluorescence is absent; L). K, LMCm and LMCl segregate normally on the left side, as viewed with Hb9 (red) and Islet-1 (green) immunofluorescence. Following γ-catenin(L127A) expression on the right side of the spinal cord, LMCl and LMCm segregation was perturbed. The LMC spread over a much larger area than the control, with LMC cells found close to the ventricle and LMCl and LMCm cells intermingled. K, Arrows, Some of the LMC cells that stalled in their migration. M, N, Ventral horn of a different embryo electroporated by γ-catenin(L127A) showing that both LMCm and LMCl were affected; the midline as at the left side of the panel. M, Arrows, Some of the LMCm and LMCl cells close to the ventricle surface. K, I, II, III, Regions quantitated in O. O, Quantitation of LMCl neuron position in bins illustrated in K. P, Quantitation of distance migrated by γ-catenin(L127A)-expressing LMCl neurons compared with those on the electroporated (Epor) side of the spinal cord that did not express γ-catenin(L127A). Error bars are SEM.
Expression of a point mutation in γ-catenin
Previous work identified a 29 amino acid region of γ-catenin that is both necessary and sufficient for binding to α-catenin (Aberle et al., 1996). This region is highly conserved between different species with only one replacement (to a similar amino acid) between chicken and human γ-catenin. Additionally, single amino acid mutations in this region can reduce binding to α-catenin to background levels (Aberle et al., 1996). We reasoned that expression of γ-catenin containing such a mutation might uncouple cadherin–catenin interaction, thus abrogating cadherin function. We generated a single amino acid substitution of chick γ-catenin of L127-A, γ(L127A), which is located in the α-catenin binding domain of γ-catenin and reduces this binding to <2% of the wild-type γ-catenin for the human protein (Aberle et al., 1996). We expressed γ(L127A) by in ovo electroporation, confirming its misexpression by immunofluorescence (Fig. 4C,D; electroporated cells are marked by nuclear β-gal staining in D). γ(L127A) expression should result in a delocalization of adherens junctional complexes, which depend on interaction with the actin cytoskeleton via α-catenin. We found that the apical expression of β-catenin in radial glia in the dorsal spinal cord was disrupted following γ(L127A) expression (Fig. 4E,F), consistent with its predicted mode of action. α-Catenin binds to the tight junction protein ZO1 and disruption of cadherin function follows disruption of ZO1 binding to the cadherin complex (Itoh et al., 1997; Imamura et al., 1999). We thus followed the localization of ZO1 protein expression following γ(L127A) expression and again found that its localization to the apical end-feet of radial glia was disrupted (Fig. 4G,H). Together, these data suggest that γ(L127A) disrupts cadherin function most likely through uncoupling the cadherin–catenin complex.
Following γ(L127A) expression, the total number of LMCm and LMCl motor neurons (56 ± 5 and 58 ± 3 motor neurons per section, respectively) was not significantly different to the control side of the spinal cord (62 ± 6 and 55 ± 5 per section, respectively; p > 0.05, Student's t test), indicating that the general differentiation of LMC divisions was not perturbed. We did, however, observe an ∼50% decrease in the number of medial motor column neurons when γ(L127A) was expressed (25 ± 3 vs 15 ± 2; control vs experimental; p < 0.05, Student's t test). Application of BrdU to embryos from st23 to st27, following γ(L127A) expression, revealed that LMC neurons were born before stage 23, as is found in wild-type embryos (Fig. 4I,J). Thus, the major program of LMC differentiation in terms of neuron number within each division and the timing of motor neuron generation is not perturbed by γ(L127A) expression. This finding allowed us to investigate the role of γ(L127A) expression on LMC divisional segregation.
LMC neuron position and divisional segregation is perturbed by γ(L127A) cell autonomously
LMC divisional segregation is normally complete before st29, 4 d after the first generation of LMC neurons. We thus investigated the effect of γ(L127A) expression on divisional segregation at st29. Following γ(L127A) expression, we observed a striking perturbation of LMC neuron positioning. In contrast to the control, nonelectroporated side of the spinal cord (Fig. 4K–N), the area encompassing the LMC was greatly expanded (over twofold) with neurons of both LMCl and LMCm found close to the ventricle. Divisional segregation was also severely disrupted with many of the Hb9+ve/Islet-1−ve LMCl neurons located in a position medial to the Hb9−ve/Islet-1+ve LMCm neurons (Fig. 4K, arrows). We also observed mixing of LMCm and LMCl with LMCl neurons found within the domain normally occupied by LMCm neurons.
We quantitated LMCl cell body position by considering the percentage of neurons that were located in three defined mediolateral bins in the ventral spinal cord, bin I being most medial and bin III the most lateral (Fig. 4K). Expression of γ(L127A) results in a mosaic misexpression due to stochastic incorporation of the electroporated construct (indicated by nuclear-localized β-galactosidase). We could thus quantify the cell autonomy of mispositioning of LMC cells in relation to their expression of γ(L127A) (Fig. 4O). In bin I, 54 ± 3% of LMCl neurons and 20% of LMCm neurons that had acquired γ(L127A) were found, compared with 12% and 3% respectively on the control side of the spinal cord (p < 0.001, Student's t test). In contrast, 85 ± 4% of Hb9+ve/Islet-1−ve/ β-gal−ve LMCl neurons were located in bin III, similar to that found in the control ventral horn (p > 0.1, Student's t test; Fig. 2K). Additionally, ∼25% of Hb9+ve/Islet-1−ve/β-gal+ve LMCl neurons were found within the LMCm, indicating that divisional mixing also occurs following γ(L127A) expression. These data are consistent with a cell autonomous perturbation of LMC divisional segregation and motor neuron positioning in response to γ(L127A) expression.
We next quantitated the distance migrated by either Hb9+ve/Islet-1−ve/β-gal+ve or Hb9+ve/Islet-1−ve/β-gal−ve cells. We found that LMCl cells expressing γ(L127-A) had migrated approximately half the distance of controls (Fig. 4P). This suggests that LMC cell body positioning defects are due to a perturbation in the distance migrated by the cells.
Stalled LMCl neurons project axons into the limb normally
The perturbation of LMC neuron positioning prompted us to ask whether motor axon trajectory proceeded normally following γ(L127A) expression. To address this, we injected HRP into the dorsal limb mesenchyme at HH st28 to trace LMCl cell body position following coexpression of γ(L127A) and GFP, the latter to trace motor neuron cell body and axon position. We found that following γ(L127A)/GFP coexpression, the region of the spinal cord occupied by HRP+ve cells was increased compared with controls (Fig. 5A–C). Particularly notable was the presence of HRP labeling in neurons located close to the ventricle (Fig. 5B, arrows). This indicated that the mispositioned LMCl neurons still project axons to the dorsal limb. In contrast, axonal GFP was not restricted to the dorsal limb mesenchyme, suggesting normal axon trajectories of LMCm and LMCl cells following γ(L127-A) expression (Fig. 5D–F, arrows). Additionally, we noted that within the ventral spinal cord, GFP fluorescence was enhanced medially compared with control expression of GFP alone (Fig. 5G), consistent with the cell autonomous perturbation of LMCl position following γ(L127A) expression. This further suggests that the normal program of LMC differentiation occurs following expression of γ(L127A) despite aberrant motor neuron positioning.
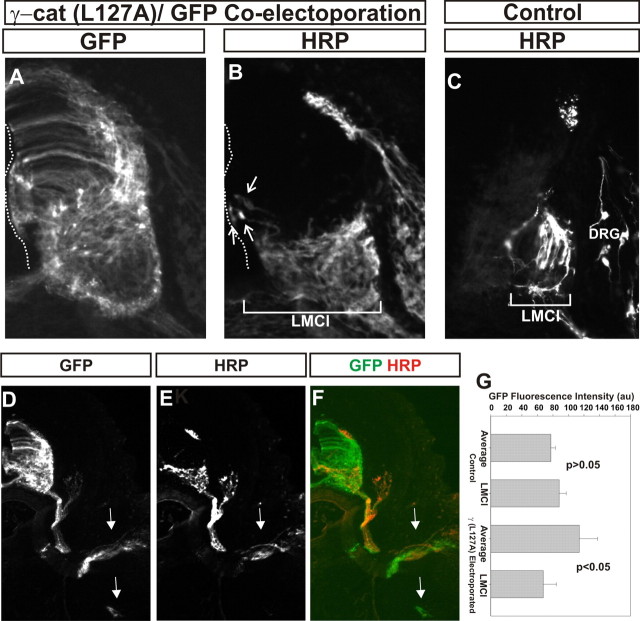
Figure 5.
Motor neuron projections appear normal following γ-catenin (L127A) expression. GFP and γ-catenin(L127A) were coelectroporated at stage 18 and HRP was injected into the dorsal limb at stage 29. A, B, GFP (A) and HRP (B) immunofluorescence in one section. B, Arrows, Motor neurons close the ventricular surface that projected into the dorsal limb. A, B, Dotted lines, Ventricle surface. C, HRP retrograde labeling in control embryos. B, C, Bars, Mediolateral extent of retrogradely labeled motor neurons. Note that the bar in B is more than twice as long as the one in C. D–G, GFP immunofluorescence is more intense medially in the spinal cord than in controls following γ-catenin(L127A)/GFP coelectroporation. D, F, GFP immunofluorescence. E, F, HRP immunofluorescence. G, Quantitation of average GFP immunofluorescence in the spinal cord versus that found in the lateral LMC following GFP or γ-catenin(L127A)/GFP coelectroporation. GFP fluorescence is lower in the lateral ventral horn following γ-catenin(L127A) expression, consistent with a defect in motor neuron migration. D–F, Arrows, Motor axon tracks to dorsal and ventral limb. Note that GFP is present in both, whereas HRP immunofluorescence is exclusively in the dorsal limb tracks. Error bars are SEM.
γ-Catenin (L127A) expression does not perturb dorsal interneuron positioning
The predominant expression of γ-catenin is within the ventral horn. We thus asked whether the observed migration defect following γ(L127A) expression is specific to the ventral spinal cord. We investigated the effect of γ(L127A) expression on the positioning of both interneurons and motor neurons within the spinal cord at st29 through Lhx1 expression, a LIM homeodomain transcription factor expressed within the LMCl and also within the majority of interneurons within the spinal cord. We also assessed the positioning of the subset of dorsal commissural interneurons that express Islet-1. Consistent with previous results, following γ(L127A) expression, we detected a large number of Lhx1+ve cells adjacent to the ventral ventricular surface with a concomitant reduction in the number of Lhx1+ve cells in the lateral ventral horn (Fig. 6A–C). However, we saw no defect in positioning of either Lhx1+ve or Islet-1+ve dorsal interneurons following γ(L127A) expression (Fig. 6B,C, right arrows). These data suggest that the mispositioning of neurons following γ(L127A) expression is specific to the ventral region of the spinal cord and does not result in a general perturbation of cell body positioning in the spinal cord.
Figure 6.
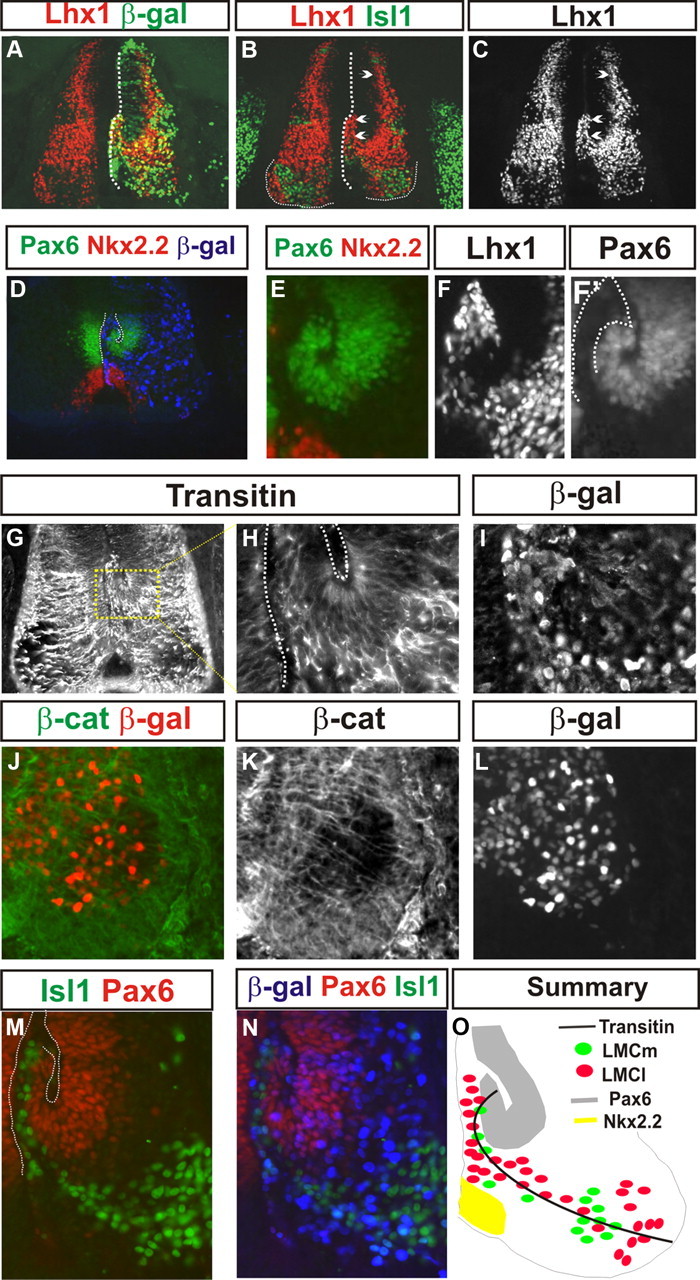
Stalled motor neuron migration results in bucking of the ventral Pax6 domain following γ-catenin(L127A) expression. A–C, Lhx1-expressing ventral interneurons and LMCl neurons (B, C, left-pointing arrowheads) and islet-1 cells (I) are found close to the ventricle (A, B, dotted lines). Electroporated neurons are marked by β-gal immunofluorescence (A, green). B, C, Right-pointing arrowheads, Absence of effect on dorsal interneuron populations. D, E, The ventral Pax 6 (green) progenitor domain buckles following γ-catenin(L127A) expression but the Nkx2.2 (red) domain is unperturbed. Electroporation is marked by β-gal immunofluorescence (D, blue). D, Dotted lines, Ventricle surfaces. E, Magnified image of the ventral progenitor domains of the right side of the spinal cord in D. Note that the electroporated cells (stalled LMC neurons) are close to the ventricle, whereas the majority of the buckled ventral Pax6 domain is not electroporated. F, Lhx1 immunofluorescence focusing on the ventricular zone of the adjacent section to that shown in D and E. F′, Position of the Lhx1 cells (outlined by the dotted lines) in relation to the Pax6 domain of E. G–I, Transitin immunofluorescence (G, H) following electroporation of γ-catenin(L127A). The right side of the spinal cord in G was electroporated. H, I, Magnified images of the dotted box in G. H, Dotted lines, Ventricle surfaces. I, β-Gal immunoreactivity showing electroporated cells. J–L, β-Catenin (β-cat; J, K) immunofluorescence in radial glia in the ventral horn at stage 25 after γ(L127A) expression marked by β-gal immunoreactivity in J and L. M, N, Islet-1 (green) and Pax6 (red) immunofluorescence at stage 26 following γ-catenin(L127A) expression, showing that motor neurons stalled in their migration are found adjacent to the bucked Pax6 domain (as in F, F′). Motor neuron position relative to the Pax6 domain is therefore normal. β-gal immunoreactivity (N, blue) shows electroporated neurons. O, Summary of the data in this figure showing the relative positions of transitin radial glia, progenitor domains, and LMC neurons following γ-catenin(L127A) expression.
Stalled LMC migration causes the motor neuron progenitor domain to buckle
The location of Lhx-1+ve, Islet-1+ve/Hb9−ve, and Islet-1−ve/Hb9+ve cells close to the ventricle following γ(L127A) expression prompted us to investigate the integrity of the spinal progenitor cells within the ventricular zone at these later stages of spinal cord development. We analyzed the expression at st28 of homeodomain transcription factors expressed within ventral progenitor domains, specifically Pax6 and Nkx2.2, which label adjacent domains (Briscoe et al., 2000). On the γ(L127A) electroporated side of the spinal cord, the v3 (Nkx2.2) progenitor domain was found in a normal ventral position, adjacent to the floor plate. (Fig. 6D,E). In contrast, the ventral part of the Pax6 domain folded medially and dorsally such that its more ventrolateral aspect was closer to the ventricle (Fig. 6D,E). We noted that in many embryos it was only the stalled motor neurons and not the ventral Pax6 domain that was electroporated. This suggested that the buckling of the Pax6 domain is a noncellautonomous effect most likely mediated by γ(L127A) expression in motor neurons. The position of stalled ventral Lhx1 cells fits between the Pax6 domain and the ventricle surface (Fig. 6F,F′). The length of the Pax6 domain as a coherent group of cells was, however, similar to the control side of the spinal cord (239.6 ± 8.4 μm vs 240.6 ± 7.8 μm; p > 0.1, Student's t test). Additionally, the dorsal part of the Pax6 domain resided in a normal position. At this stage, a small number of Pax6 cells were found in the mantle zone of the spinal cord, presumably having migrated from the ventricular zone. We observed that following γ(L127A) expression, the total number of these cells was not significantly different from the control side of the spinal cord (13 ± 1 vs 13 ± 3 cells per section; p > 0.1, χ2 test). This suggests that the general integrity of the Pax6 progenitor domain is not affected by γ(L127A) expression.
Transitin/β-catenin radial glia still traverse to the ventral horn following γ(L127A) expression
Because of the bucking of the ventral Pax6 progenitor domain, we investigated the morphology of transitin radial glia fibers in the vicinity of stalled motor neurons. We found that in the ventral spinal cord, transitin/β-catenin fibers still course from the ventricular zone to the pial surface within the ventral horn (Fig. 6G–L). However, the folded nature of the ventral Pax-6 domain resulted in these fibers first taking a ventral and then lateral route (Fig. 6G,H). Related to the Pax6 domain, the position of stalled LMC neurons located close to the ventricle were, however, normal (Fig. 6M,N). This indicates that motor neurons adjacent to the ventricular surface are not within the ventricular zone itself, as defined by the expression of Pax6. Thus, following γ(L127A) expression, the pathway of migration of spinal motor neurons still follows the course outlined by transitin+ve fibers (Summarized in Fig. 6O).
Cadherin dominant-negative expression stalls migration of LMC neurons
The effects of γ(L127A) expression suggests a cadherin dependence of γ-catenin function in LMC neuron migration and divisional segregation. We therefore asked whether uncoupling cadherin function from intracellular binding to γ-catenin also perturbed motor neuron divisional segregation. Expression of a truncation of the extracellular domain of N-cadherin, NΔ390, has been shown to act in a cadherin dominant-negative fashion by sequestering endogenous β- or γ-catenin (Fujimori and Takeichi, 1993). Expression of NΔ390 from st18 caused a phenotype similar to γ(L127A) expression, albeit at a reduced level (Fig. 7A–F). In contrast to γ(L127A) expression, we did, however, observe a small decrease (∼20%) in the total number of motor neurons cells (64 ± 2 Hb9 cells vs 73 ± 2; p < 0.05, χ2 test; 39 ± 2 vs 50 ± 2 Islet-1 cells per section; p < 0.05, χ2 test). Similar to γ(L127A) expression, NΔ390 expression left the LMC spread over a greater area, approximately double that of controls. Additionally, we found ∼5% of both LMCl and LMCm cells adjacent to the ventricular surface (Fig. 7G). We quantitated divisional segregation of LMCl and LMCm and found that LMCl cells mingled throughout the LMCm division (Fig. 7H), consistent with a perturbation of LMC migration.
Figure 7.
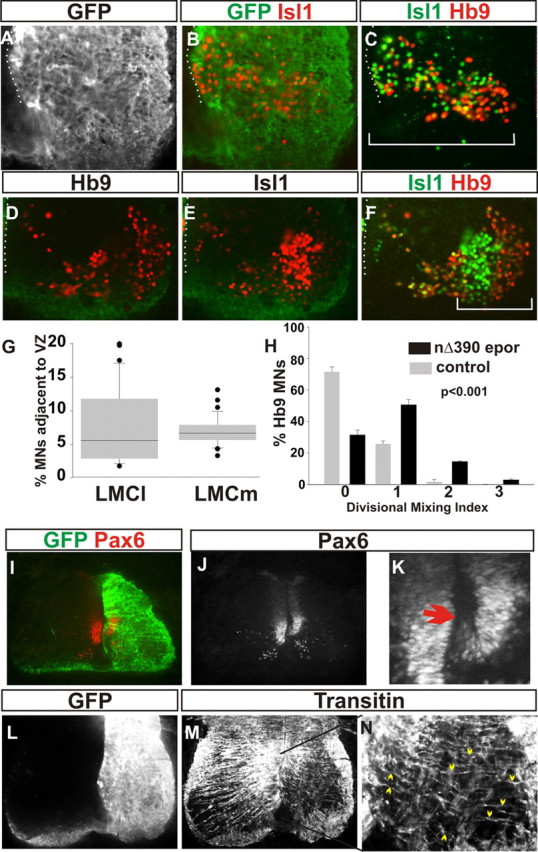
NΔ390 dominant-negative cadherin expression results in a similar phenotype to γ-catenin(L127A) expression in disruption of LMC divisional segregation. A–C, GFP, Islet-1, and HB9 immunoreactivity following NΔ390 cadherin expression shows stalled motor neurons (MNs) and perturbed divisional segregation of the LMC. D–F, Hb9 and Islet-1 immunoreactivity on the contralateral LMC. A–F, Dotted lines, Ventricular surface. C, F, Solid lines, Mediolateral extent of the LMC. Note that following NΔ390 cadherin expression, the LMC spans approximately twice the extent of the contralateral LMC. G, Percentage of Hb9 and Islet-1 cells adjacent to the ventricular zone (VZ) following NΔ390 cadherin expression. Contralateral spinal cords have no cells in similar positions. H, Divisional mixing index following NΔ390 cadherin expression compared with contralateral spinal cord (Student's t test of bins 0, 1, and 2, p < 0.001; χ2 test, p < 0.001 of the entire distribution 2df). Error bars are SEM. I–K, Pax6 expression following NΔ390 cadherin expression, marked by GFP in I. K, Magnification of the buckled Pax 6 domain (arrow) in J. L–N, Transitin immunoreactivity (M, N) following NΔ390 cadherin expression marked by GFP in L. N, Magnification of area indicated by black line in M. N, Arrowheads, Transitin fibers coursing from the ventricle surface to the pial surface of the spinal cord.
Further, similar to γ(L127A) expression, we observed a dorsomedial folding of the ventral Pax6 progenitor domain. However, we noted that the extent of folding of the ventricular zone following NΔ390 expression was smaller than that following γ(L127A) expression, consistent with the reduced effect of NΔ390 compared with γ(L127A) expression (Fig. 7I–K). Additionally, similar to γ(L127A) expression, the pathway of transitin+ve radial glia mirrored the perturbed LMC neuron position and were still associated with the ventricle surface (Fig. 7L–N). Thus, the essential features of the phenotype of γ(L127A) expression on LMC migration were captured by expression of a cadherin dominant-negative. Together, these results suggest that catenin-dependent cadherin function is necessary for the correct radial migration of spinal motor neurons.
Canonical Wnt signaling does not influence LMC migration
The perturbations of cadherin and catenin function described here have the potential to alter canonical Wnt signaling (Nelson and Nusse, 2004). We thus asked whether direct perturbations of Wnt signaling alter motor neuron migration and LMC divisional segregation. We expressed constructs in vivo that have been shown to result in the upregulation or downregulation of canonical Wnt signaling (Roose et al., 1999; Krylova et al., 2000; Zhurinsky et al., 2000b). Expression of either wild-type β-catenin (Fig. 8A–D) or a truncated version of β-catenin [β-cat-1-ins (Zhurinsky et al., 2000b); Fig. 8E–H] or a dominant-negative GSK3β construct (Fig. 8I–L) resulted in no observable difference in motor neuron migration compared with the control, unelectroporated side of the spinal cord. Thus, constructs that upregulate Wnt signaling have no effect on LMC divisional segregation.
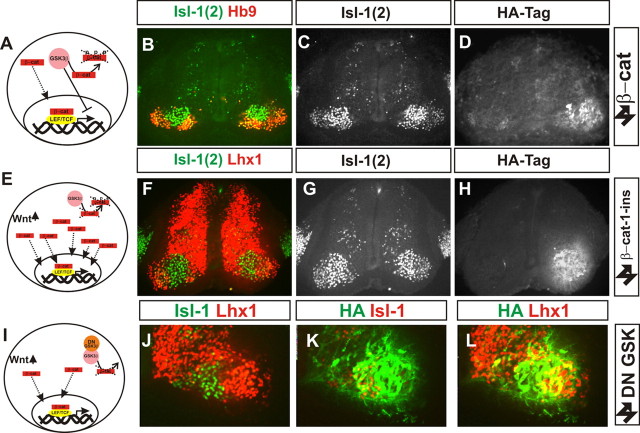
Figure 8.
Upregulation of canonical Wnt signaling does not disrupt LMC neuron organization. A–D, Overexpression of β-catenin. A, Summary of Wnt signaling. B–D, Hb9 and Islet-1(2) (B, C) immunohistochemistry after β-catenin expression marked by HA immunoreactivity (D). E–H, Expression of β-catenin-1-ins construct. E, Summary of effect to increase Wnt signaling. F–H, Lhx1 and islet-1(2) (F, G) immunoreactivity after β-cat-1-ins expression marked by HA immunoreactivity (H). I–L, Lack of effect on LMC organization of expression of a dominant-negative GSK3β construct. I, Schematic of action of the construct. J–L, HA-tag (K, L, green), Islet-1 (J, green; K, red), and Lhx1 (J, L, red) immunoreactivity following electroporation of the construct.
Additionally, we sought to downregulate canonical Wnt signaling pathway through the expression of wild-type GSK3β (Fig. 9A–D), constitutively active GSK3β (Fig. 9E–H), or a dominant-negative TCF construct (Fig. 9I–L) (Ciani et al., 2004). Again, we observed no differences in spinal motor neuron migration following expression of these constructs. These data suggest that the observed migration and divisional segregation defects following γ(L127A) or NΔ390 expression are not consistent with a perturbation of canonical Wnt signaling.
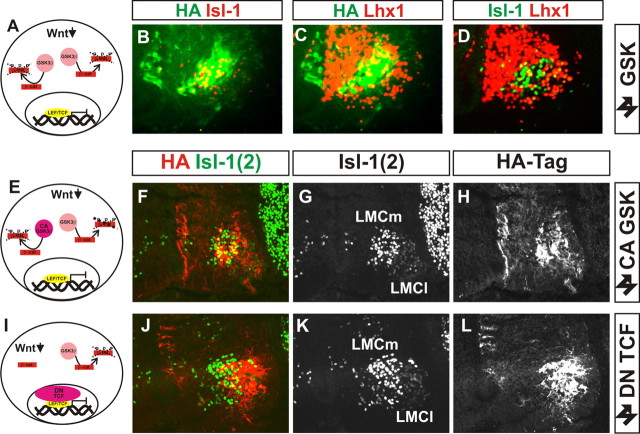
Figure 9.
Downregulation of canonical Wnt signaling does not disrupt LMC neuron organization. A–D, Lack of effect on LMC organization of expression of wild-type GSK3β. A, Schematic of action of the construct. B–D, HA-tag (B, C, green), Islet-1 (B, red; D, green) and Lhx1 (C, D, red) immunoreactivity following electroporation of the construct. E–L, Downregulation of Wnt signaling pathway by expression of constitutively active GSK3β construct (E–H) or dominant-negative TCF transcription factor expression (I–L). E, I, Summary of action of each construct. Islet-1(2) immunohistochemistry (F, G, J, K) reveals normal motor neuron migration and divisional segregation following expression of the constructs, marked by HA immunoreactivity (F, H, J, L). Islet1(2) immunofluorescence is lower in LMCl than LMCm.
Cadherin-7 is required for LMC migration and divisional segregation
We have shown that uncoupling general cadherin function intracellularly disrupts divisional segregation by neuronal migration. We sought to manipulate the expression of a single cadherin that was predominantly expressed during LMC migration with little or no expression later in development. To begin to address this, we screened expression of type I and type II cadherin family members for such an expression profile. We found that type I cadherins, including N-cadherin, appear excluded from LMC neurons, and instead were found in radial fibers in the ventral horn (Fig. 10A–D). In contrast, type II cadherins, including cad-7, cad-12, and cad-20, were expressed in the majority of the LMC at early stages of development (Fig. 10E–L) (Luo et al., 2006). Both cadherin-12 and cadherin-20 also have prominent expression within a number of motor neuron pools after divisional segregation (Fig. 10F,H) (Price et al., 2002). In contrast, cad-7 was predominantly expressed early during LMC divisional segregation and is downregulated in the vast majority of LMC neurons after divisional segregation is complete (st28; Fig. 10I–L) (Price et al., 2002). Therefore, cad-7 is expressed within the majority of the LMC neurons during their migration; we thus focused our attention on its function during LMC divisional segregation (Luo et al., 2006).
Figure 10.
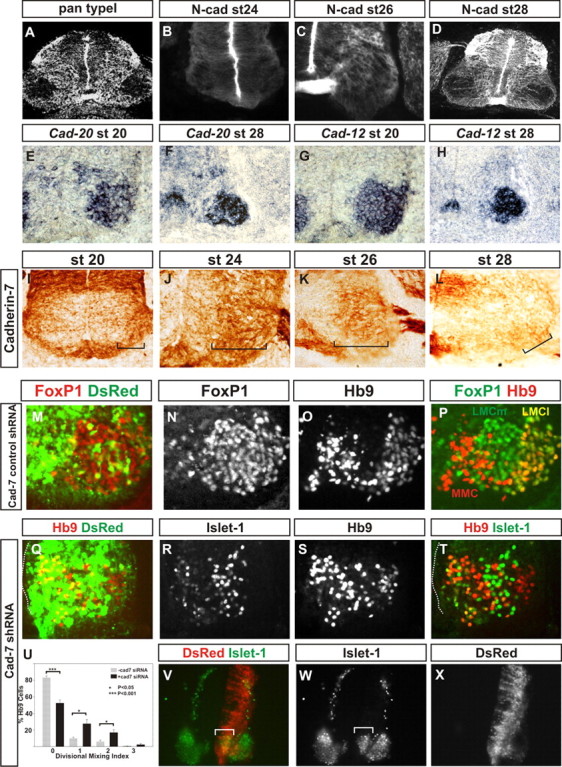
Pan-motor neuron type II cadherin expression and cadherin-7 siRNA restricts LMC neuron migration and divisional segregation. A, Pan type I cadherin immunolabeling at st28. B–D, N-cadherin immunoreactivity at st24 (B), st26 (C), and st28 (D). E, F, Cad-20 in situ hybridization in the ventral horn at st20 (E) and at caudal lumbar regions of st28 (F). G, H, Cad-12 in situ hybridization in the ventral horn at st20 (G) and at caudal lumbar regions of st28 (H). I–L, Cad-7 immunohistochemistry in the ventral spinal cord at st20 (I) and in the ventral horn at HH st24 (J), st26 (K), and st28 (L). Cad-7 appears to be expressed in the majority of LMC neurons during their migration (I–K) and is weakly expressed in only a small subset of motor neurons subsequently (L). M–P, Cad-7 control shRNA expression. Foxp1 (M, N, P), a pan-LMC marker, and Hb9 immunoreactivity (O, P) show normal segregation and positioning of LMCl, LMCm, and medial motor column following control shRNA expression, marked by dsRed fluorescence in M. Q–U, Cad-7 shRNA perturbs LMC divisional segregation. Q–T, HB9 (Q, S, T) and Islet-1 (R, T) immunohistochemistry at st28 following expression of cad-7 shRNA revealed by dsRed fluorescence (Q). Q, T, Dotted lines, Ventricle surface. Note that there are motor neurons close to the ventricle surface. U, Divisional mixing index following cad-7 shRNA expression compared with cells that had not acquired the construct (p < 0.001 for the 0 bin, p < 0.05 for the 1 and 2 bins, Student's t test; χ2 analysis, p < 0.001 2df). V–X, Islet-1 immunohistochemistry (V, W) at st24 reveals a perturbation of LMCm migration following cad-7 shRNA expression (V, W, bracket) revealed by dsRed fluorescence (V, X); note the paucity of dsRed fluorescence in the lateral LMC.
To address a role for cad-7 expression in spinal motor neuron migration, we reduced its expression using a previously characterized and successful shRNA approach (Barnes et al., 2010). Following expression of a control cad-7 shRNA construct, visualized by dsRed fluorescence, both migration of LMCm and LMCl cells and their segregation into divisions appeared normal (Fig. 10M–P). DsRed fluorescence was also observed laterally as well as medially in the ventral horn (Fig. 10M). In contrast, following expression of the cad-7 shRNA construct, cells that had acquired the shRNA were concentrated more medially (Fig. 10Q). These data suggest that the cells expressing cad-7 shRNA were perturbed in their migration within the lateral motor column. We next characterized the position of LMCm and LMCl cells following cad-7 shRNA expression. At st27, cad-7 shRNA expression resulted in HB9+ve Islet-1−ve LMCl cells located medial to the LMCm (Fig. 10R–T). Quantitation of the effect of cad-7 shRNA at st27 showed divisional segregation of LMCm and LMCl was compromised (Fig. 10U). Further, at earlier stages (st25), we observed an increase in the number of Islet-1+ve LMCm cells located medially close to the ventricular zone (Fig. 10V–X). Together, these data suggest that knockdown of cad-7 stalls the migration of LMC motor neurons and perturbs LMC divisional segregation.
Discussion
Spinal motor neuron migration to the ventral horn is highly organized. More than 95% of motor neurons are born rapidly over ∼24 h, whereas their migration occurs over a substantially longer time course (Hollyday and Hamburger, 1977; Whitelaw and Hollyday, 1983; Lin et al., 1998; Sockanathan and Jessell, 1998; William et al., 2003). Thus, because of their rapid generation, a backlog of motor neurons waiting to migrate is formed. Our data (schematized in Fig. 11) suggest that γ-catenin-dependent cadherin function is required for the migration of spinal motor neurons.
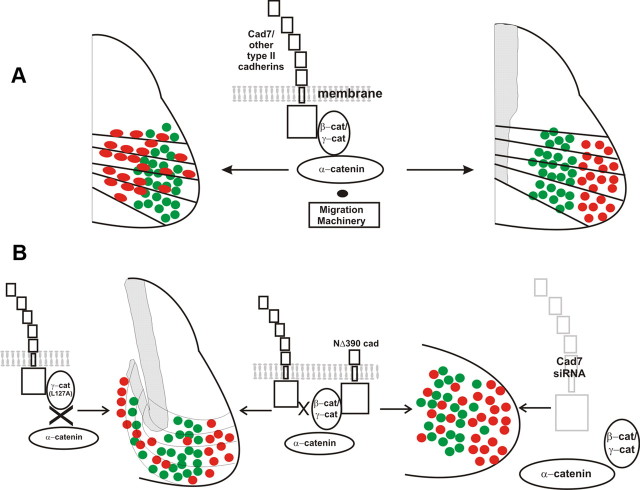
Figure 11.
Schematic summary of results. A, Schematic of motor neuron migration along pathways of transitin radial glia and divisional segregation and schematic of cadherin–catenin interactions. B, Schematic of the effects of γ(L127A) on migration and divisional segregation. Cad-7 shRNA disrupts divisional segregation and NΔ390 cadherin expression results in migration and divisional segregation effects similar to γ(L127A) and the cad-7 shRNA.
Transitin radial glia as a scaffold for motor neuron migration
Within the ventral spinal cord, coexpression of transitin and β-catenin identifies a subset of progenitor cells with characteristics of radial glia. Expression of β-catenin within radial glia demarcates two domains of ventral progenitor cells: apical cells that do not express β-catenin and more basal cells that do express it. Radial migration of spinal motor neurons has previously been inferred (Barron, 1946; Wentworth, 1984; Leber et al., 1990; Leber and Sanes, 1995; Eide and Glover, 1996) and we suggest that this occurs on transitin radial glia for two reasons. First, retrograde labeling of migrating motor neurons indicated that their migration paths follow transitin radial glia. There are relatively few transitin glia (∼14) in the ventral horn and their juxtaposition to migrating motor neurons seems unlikely to result from chance alone. More tellingly, stalled motor neurons were still found along pathways defined by transitin glia following γ(L127A) expression. This would not be predicted by motor neurons taking the most direct route in their migration and strongly suggests that spinal motor neuron migration follows routes labeled by transitin glia.
Motor neuron migration requires γ-catenin function
Both β- and γ-catenin bind to classical cadherins and link their extracellular interactions to intracellular signaling (Hirano et al., 2003), particularly via α-catenin, which provides a bridge for cadherin function to the actin cytoskeleton (Abe and Takeichi, 2008). Generally, it is believed that β-catenin is the major transducer of cadherin signaling intracellularly. We were thus surprised to find that β-catenin is absent from spinal motor neurons. Instead, α-catenin and γ-catenin are the predominant catenins expressed in the LMC. Thus, within the chick LMC, γ-catenin is the major transducer of cadherin function.
Expression of either γ-catenin or γ(L127A) leaves the total number and timing of generation of LMCm and LMCl cells unaffected. Additionally, motor neurons were labeled by retrograde tracing from the limb following γ(L127A) expression, suggesting that the time required for axon growth to the limb [>48 h (Tosney and Landmesser, 1985)] had occurred despite motor neuron migration being retarded. Thus, γ(L127A) does not perturb the generation of LMC neurons.
Expression of γ(L127A) resulted in a cell autonomous perturbation of LMC motor neuron position: ∼60% of γ(L127A)+ve LMCl MNs were located close to the ventricle with a further ∼25% found within the domain occupied by the LMCm. We found that the distances migrated were less than half those compared with control neurons, suggesting that migration of LMC neurons was severely compromised by γ(L127A). Importantly, LMC neurons on the experimental side of the spinal cord that had not acquired γ(L127A) behaved similarly to the control side of the spinal cord. While we cannot rule out an additional role for perturbed β-catenin expression in transitin radial glia in contributing to this migration phenotype, the cell autonomous nature of the effect of γ(L127A) strongly suggests that the predominant role for γ(L127A) is within the motor neurons themselves.
Stalled LMC migration causes buckling of the ventral pax6 progenitor domain
The majority of ventral Pax6 cells express neither β-catenin nor γ-catenin. Instead, transitin radial glia represent ∼30% of these progenitor cells and express detectable levels of β-catenin. The general integrity of this progenitor domain and position of generation of motor neurons from it appears normal following NΔ390 or γ(L127A) manipulations. The ventral buckling of the Pax6 domain was often seen in embryos where the Pax6 cells were not electroporated but was never observed when motor neurons were not electroporated. Further, motor neurons that had not acquired the γ(L127A) manipulation reached a normal settling position. Thus, the motor neurons stalled in their migration probably contribute most to the buckling of the Pax6 domain. We suggest that the force required for the buckling of the progenitor domain could arise from the continued mitotic activity of the ventral Pax6 domain cells. BrdU labeling following γ(L127A) expression demonstrated that cell division is not halted. Additionally, the number of Lhx1 cells arrested close to the ventricle surface was greater than the number of Hb9+ve/Islet-1−ve LMCl cells. Thus, the ventral Pax6 domain continues to generate postmitotic Lhx1 interneurons. This cell division might exert a force rearward away from stalled LMC neurons which would result in the progenitor domain buckling at its ventral extent.
Cadherin involvement in motor neuron migration
Cadherin dominant-negative expression (Fujimori and Takeichi., 1993) caused a similar phenotype to that of γ(L127A). We found that type I cadherins were not expressed in LMC motor neurons. Of the type II classical cadherins, cad-7 was expressed predominantly in most LMC neurons during divisional segregation and downregulated in the vast majority of the LMC thereafter. Downregulation of cad-7 by shRNA resulted in a similar migration and divisional mixing phenotype to that observed in NΔ390 expression: motor neurons were found close to the ventricle and LMCm and LMCl divisions were mixed. Cad-7 appears not to be expressed in ventral transitin radial glia. This further suggests that the predominant role for cadherins in motor neuron migration is within the motor neurons themselves. Demireva et al. (2011) reported perturbed LMC organization following conditional ablation of both β- and γ-catenin from mouse motor neurons. These findings are in broad agreement with those presented here. However, this conditional ablation approach in mouse revealed a much milder migration phenotype than we show for the chicken. Differences in gene expression, rate of motor neuron generation, and migration between chicken and mouse may underlie differences in severity of catenin and cadherin manipulations in these different species.
How could cadherin and catenin function facilitate motor neuron migration? A model has recently been proposed whereby α-catenin acts as a molecular clutch that links substrate adhesion to retrograde actin flow during cell migration (Bard et al., 2008). Our results are consistent with a model where both γ(L127A) and NΔ390 constructs perturb γ-catenin function intracellularly within motor neurons, disrupting cell migration, likely mediated at least in part through cad-7-based linkage to the migration machinery. Whether retrograde actin flow is a dominant force in motor neuron migration as it is in other cell types will require further characterization of the dynamics of cadherin and catenin linkage (Drees et al., 2005; Yamada et al., 2005) to the actin cytoskeleton during motor neuron migration.
Neuronal nucleus formation through regulated cadherin function
The LMC is organized as a neuronal nucleus. Neuronal nuclei are generally found in more evolutionarily ancient regions of the CNS. It seems likely that the mechanisms of nucleogenesis will reveal similarities and differences with organizational mechanisms in evolutionarily newer regions of the CNS, such as the lamination of the cortex (Rakic, 2006; Lui et al., 2011). Radial glia provide a scaffold for excitatory projection neuronal migration in the cortex and are a major progenitor cell type of the ventricular zone. Additional progenitor cells, generated from radial glia, are found more basally in the subventricular zone (Noctor et al., 2004). Within the spinal cord, radial glia delineate pathways of spinal motor neuron migration, although, in contrast to the cortex, spinal transitin glia represent only a small proportion of the ventral progenitor cells. Additionally, these radial glia are basal to the ventricular progenitor cells. One major difference in the generation of motor neurons from that observed to drive lamination of the cortex is the rate at which LMC neurons are generated. Rapid LMC generation results in a backlog of neurons waiting to migrate. In contrast, neurons in the cortex destined to populate more superficial layers are born sequentially and migrate in an ordered fashion to generate the layers of the laminated cortex (Noctor et al., 2004). It is likely that the availability of spinal radial glia represents a rate limiting step for motor neuron migration.
We have demonstrated that early, pan-motor neuron cadherin function is required for LMC divisional segregation through neuronal migration. Subsequently, differential type II cadherin function is required for motor neuron pool formation (Price et al., 2002; Patel et al., 2006). Our results therefore suggest a prolonged requirement for cadherin function in all phases of motor neuron cell body organization. How could cadherin-dependent radial LMC migration and later motor pool sorting be coupled? Recent work has suggested that perturbation of reelin signals disrupts later phases of motor neuron organization (Palmesino et al., 2010). Reelin can regulate cadherin function in the control of neuronal migration in the cortex (Franco et al., 2011). Thus, reelin signaling may provide a mechanism for dissociation of cadherin- and catenin-mediated radial migration from a later involvement of cadherin expression in motor pool sorting.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council of the UK (Grant BBS/B/06512) and the Wellcome Trust (GR072914 and 094399/B/10/Z). S.M.B. was supported by a scholarship from the Commonwealth Scholarship Commission in the UK. We thank Thomas Jessell for his support during this project, which was started while S.R.P. was a postdoctoral fellow in his laboratory. We are grateful to T. Jessell, M. Takeichi, P. Salinas, and S. Nakagawa for constructs and antibodies. Additionally, we thank Elena Demireva, Thomas Jessell, Artur Kania, Ivo Lieberam, Ben Novitch, Patricia Salinas, and Niccolo Zampieri for critical comments on the manuscript and P Salinas, S. Wilson, C. Stern, and J. Parnavelas for advice. The hybridomas obtained from the Developmental Studies Hybridoma Bank were developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, Iowa.
References
- Abe K, Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci U S A. 2008;105:13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberle H, Schwartz H, Hoschuetzky H, Kemler R. Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to alpha-catenin. J Biol Chem. 1996;271:1520–1526. doi: 10.1074/jbc.271.3.1520. [DOI] [PubMed] [Google Scholar]
- Agarwala S, Ragsdale CW. A role for midbrain arcs in nucleogenesis. Development. 2002;129:5779–5788. doi: 10.1242/dev.00179. [DOI] [PubMed] [Google Scholar]
- Bard L, Boscher C, Lambert M, Mège RM, Choquet D, Thoumine O. A molecular clutch between the actin flow and N-cadherin adhesions drives growth cone migration. J Neurosci. 2008;28:5879–5890. doi: 10.1523/JNEUROSCI.5331-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SH, Price SR, Wentzel C, Guthrie SC. Cadherin-7 and cadherin-6B differentially regulate the growth, branching and guidance of cranial motor axons. Development. 2010;137:805–814. doi: 10.1242/dev.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron DH. Observations on the early differentiation of the motor neuroblasts in the spinal cord of the chick. J Comp Neurol. 1946;85:149–169. doi: 10.1002/cne.900850202. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Cajal SR. Histology of the nervous system of man and vertebrates. New York: Oxford UP; 1995. [Google Scholar]
- Ciani L, Krylova O, Smalley MJ, Dale TC, Salinas PC. A divergent canonical WNT-signaling pathway regulates microtubule dynamics: dishevelled signals locally to stabilize microtubules. J Cell Biol. 2004;164:243–253. doi: 10.1083/jcb.200309096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole GJ, Lee JA. Immunocytochemical localization of a novel radial glial intermediate filament protein. Brain Res Dev Brain Res. 1997;101:225–238. doi: 10.1016/s0165-3806(97)00068-0. [DOI] [PubMed] [Google Scholar]
- Das RM, Van Hateren NJ, Howell GR, Farrell ER, Bangs FK, Porteous VC, Manning EM, McGrew MJ, Ohyama K, Sacco MA, Halley PA, Sang HM, Storey KG, Placzek M, Tickle C, Nair VK, Wilson SA. A robust system for RNA interference in the chicken using a modified microRNA operon. Dev Biol. 2006;294:554–563. doi: 10.1016/j.ydbio.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Demireva EY, Shapiro LS, Jessell TM, Zampieri N. Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell. 2011;147:641–652. doi: 10.1016/j.cell.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide AL, Glover JC. Development of an identified spinal commissural interneuron population in an amniote: neurons of the avian Hofmann nuclei. J Neurosci. 1996;16:5749–5761. doi: 10.1523/JNEUROSCI.16-18-05749.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Müller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Takeichi M. Disruption of epithelial cell–cell adhesion by exogenous expression of a mutated nonfunctional N-cadherin. Mol Biol Cell. 1993;4:37–47. doi: 10.1091/mbc.4.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase G, Dessaud E, Garcès A, de Bovis B, Birling M, Filippi P, Schmalbruch H, Arber S, deLapeyrière O. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35:893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hirano S, Suzuki ST, Redies C. The cadherin superfamily in neural development: diversity, function and interaction with other molecules. Front Biosci. 2003;8:d306–d355. doi: 10.2741/972. [DOI] [PubMed] [Google Scholar]
- Hollyday M, Hamburger V. An autoradiographic study of the formation of the lateral motor column in the chick embryo. Brain Res. 1977;132:197–208. doi: 10.1016/0006-8993(77)90416-4. [DOI] [PubMed] [Google Scholar]
- Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. Functional domains of alpha-catenin required for the strong state of cadherin-based cell adhesion. J Cell Biol. 1999;144:1311–1322. doi: 10.1083/jcb.144.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166:895–899. doi: 10.1534/genetics.166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner C. Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell. 1992;69:225–236. doi: 10.1016/0092-8674(92)90404-z. [DOI] [PubMed] [Google Scholar]
- Krylova O, Messenger MJ, Salinas PC. Dishevelled-1 regulates microtubule stability: a new function mediated by glycogen synthase kinase-3β. J Cell Biol. 2000;151:83–94. doi: 10.1083/jcb.151.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L. The distribution of motoneurones supplying chick hind limb muscles. J Physiol. 1978;284:371–389. doi: 10.1113/jphysiol.1978.sp012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber SM, Sanes JR. Migratory pathways of neurons and glia in the embryonic chick spinal cord. J Neurosci. 1995;15:1236–1248. doi: 10.1523/JNEUROSCI.15-02-01236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber SM, Breedlove SM, Sanes JR. Lineage, arrangement, and death of clonally related motoneurons in the chick spinal cord. J Neurosci. 1990;10:2451–2462. doi: 10.1523/JNEUROSCI.10-07-02451.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95:393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, Arber S. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Ju MJ, Redies C. Regionalized cadherin-7 expression by radial glia is regulated by Shh and Pax7 during chicken spinal cord development. Neuroscience. 2006;142:1133–1143. doi: 10.1016/j.neuroscience.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Momose T, Tonegawa A, Takeuchi J, Ogawa H, Umesono K, Yasuda K. Efficient targeting of gene expression in chick embryos by microelectroporation. Dev Growth Differ. 1999;41:335–344. doi: 10.1046/j.1440-169x.1999.413437.x. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Palmesino E, Rousso DL, Kao TJ, Klar A, Laufer E, Uemura O, Okamoto H, Novitch BG, Kania A. Foxp1 and lhx1 coordinate motor neuron migration with axon trajectory choice by gating reelin signalling. PLoS Biol. 2010;8:e1000446. doi: 10.1371/journal.pbio.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109:205–216. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- Rakic P. A century of progress in corticoneurogenesis: from silver impregnation to genetic engineering. Cereb Cortex. 2006;16(Suppl 1):i3–i17. doi: 10.1093/cercor/bhk036. [DOI] [PubMed] [Google Scholar]
- Romanes GJ. The motor pools of the spinal cord. Prog Brain Res. 1964;11:93–119. doi: 10.1016/s0079-6123(08)64045-5. [DOI] [PubMed] [Google Scholar]
- Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the β-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Kasai T, Nakagawa S, Tanabe K, Watanabe T, Kawakami K, Takahashi Y. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev Biol. 2007;305:616–624. doi: 10.1016/j.ydbio.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94:503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Takahashi Y, Sato Y, Kawakami K, Takeichi M, Nakagawa S. Cadherin is required for dendritic morphogenesis and synaptic terminal organization of retinal horizontal cells. Development. 2006;133:4085–4096. doi: 10.1242/dev.02566. [DOI] [PubMed] [Google Scholar]
- Tosney KW, Landmesser LT. Development of the major pathways for neurite outgrowth in the chick limb. Dev Biol. 1985;109:193–214. doi: 10.1016/0012-1606(85)90360-4. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Uemura M, Takeichi M. Alpha N-catenin deficiency causes defects in axon migration and nuclear organization in restricted regions of the mouse brain. Dev Dyn. 2006;235:2559–2566. doi: 10.1002/dvdy.20841. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Saito D, Tanabe K, Suetsugu R, Nakaya Y, Nakagawa S, Takahashi Y. Tet-on inducible system combined with in ovo electroporation dissects multiple roles of genes in somitogenesis of chicken embryos. Dev Biol. 2007;305:625–636. doi: 10.1016/j.ydbio.2007.01.042. [DOI] [PubMed] [Google Scholar]
- Weis WI, Nelson WJ. Re-solving the cadherin-catenin-actin conundrum. J Biol Chem. 2006;281:35593–35597. doi: 10.1074/jbc.R600027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth LE. The development of the cervical spinal cord of the mouse embryo. I. A Golgi analysis of ventral root neuron differentiation. J Comp Neurol. 1984;222:81–95. doi: 10.1002/cne.902220108. [DOI] [PubMed] [Google Scholar]
- Whitelaw V, Hollyday M. Thigh and calf discrimination in the motor innervation of the chick hindlimb following deletions of limb segments. J Neurosci. 1983;3:1199–1215. doi: 10.1523/JNEUROSCI.03-06-01199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William CM, Tanabe Y, Jessell TM. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130:1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J Cell Sci. 2000a;113:3127–3139. doi: 10.1242/jcs.113.18.3127. [DOI] [PubMed] [Google Scholar]
- Zhurinsky J, Shtutman M, Ben-Ze'ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by β-catenin and plakoglobin. Mol Cell Biol. 2000b;20:4238–4252. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]