Abstract
Objective
To identify, compare and contrast the microbiota in patients with and without pouchitis after RPC for UC and FAP.
Summary Background Data
Pouchitis is the most common complication following restorative proctocolectomy (RPC). An abnormal host-microbial interaction has been implicated. We investigated the pouch microbiota in patients with and without pouchitis undergoing restorative proctocolectomy for UC and familial adenomatous polyposis (FAP).
Methods
Mucosal pouch biopsies, taken from 16 UC (pouchitis 8) and 8 FAP (pouchitis 3) patients were analysed to the species (or phylotype) level by cloning and sequencing of 3,184 full-length bacterial 16S rRNA genes.
Results
There was a significant increase in Proteobacteria (p= 0.019) and a significant decrease in Bacteroidetes (p= 0.001) and Faecalibacterium prausnitzii (p=0.029) in the total UC compared to the total FAP cohort, but only limited differences were found between the UC non-pouchitis and pouchitis groups and the FAP pouchitis and non-pouchitis groups. Bacterial diversity in the FAP non-pouchitis group was significantly greater than in UC non-pouchitis (p= 0.019) and significantly greater in UC non-pouchitis compared to UC pouchitis (p= 0.009). No individual species or phylotype specifically associated with either UC or FAP pouchitis was found.
Conclusions
UC pouch patients have a different, less diverse, gut microbiota than FAP patients. A further reduction in bacterial diversity but no significant dysbiosis occurs in those with pouchitis. The study suggests that a dysbiosis occurs in the ileal pouch of UC RPC patients which predisposes to, but may not directly cause, pouchitis.
Introduction
Restorative proctocolectomy with ileal pouch-anal anastomosis (RPC) is the procedure of choice in patients with ulcerative colitis (UC) and selected patients with familial adenomatous polyposis (FAP). As pouchitis is a form of inflammatory bowel disease (IBD) and occurs predominantly in patients operated on for UC it may provide a model to study the underlying pathogenesis of IBD.
Whether a dysbiosis (altered gut bacterial composition) or an abnormal host immune response to normal commensal microbiota is the cause of IBD has been the subject of many studies(1). There is evidence from clinical practice to implicate bacteria in pouchitis. Mucosal inflammation is localised to the area of gut with the highest concentration of bacteria(2). Antibiotics have been reported to be effective treatment for both pouchitis and pre-pouch ileitis in up to 87.5% of patients(3;4). Probiotics have been shown to reduce disease relapse(5;6), and reduce the risk of disease onset(7). We have previously demonstrated that the inflammatory response in pouchitis appears to be at the local mucosal level providing indirect evidence to implicate bacteria(8).
However, the microbiology of pouchitis is still poorly understood. Results from early studies of pouch microbiota using culture methods are varied and inconclusive demonstrating no strong evidence that dysbiosis is the cause of pouchitis(9). Since the introduction of molecular techniques for the study of gut microbiology, however, it has been appreciated that culture-based studies fail to identify up to 90% of gut microbiota(10). Studies using molecular techniques have demonstrated changes in the composition of gut microbiota in IBD patients, when compared to non-inflammatory controls. Many investigators report a reduction in bacterial diversity in samples from IBD patients, often with increased Enterobacteriaceae, including E. coli, and a reduction in Firmicutes including Clostridia(11-15). Others have reported no differences in diversity(12;16) or a non-significant increase(17). Both increased(15;18-20) and decreased(21;22) levels of Bacteroidetes have been reported, likewise some studies report differences in microbiota in active and inactive disease(19;21;23), whereas others do not(18).
Two studies have used a molecular technique to compare ileo-anal pouch microbiota in pouchitis and non-pouchitis patients. In the first bacterial DNA from mucosal biopsy samples from 11 patients was sequenced and cloned(24) with apparent significant differences between pouchitis and non-pouchitis groups. The second (from our unit) studied 32 RPC patients using terminal-restriction fragment length polymorphism (TRFLP). No differences between pouchitis and non-pouchitis groups were found(17), Both of these studies, however, had limitations. First, the technique used by Johnson et al (17) was limited to only being able to identify dominant species groups and was not able to identify individual bacterial species. Secondly, in the study of Komanduri et al, samples from groups of patients with or without pouchitis were pooled before cloning and in addition only 8 RPC samples were cloned. It is known however that there is a wide variation in gut microbiota between individuals(25;26), and pooling samples cannot be justified since comparisons can only be made between groups comprised of data from different individuals.
In the present study bacterial 16S rRNA gene cloning and sequencing was used and patient samples were analysed individually to avoid the possible sources of error outlined above. Its aim was to identify, compare and contrast the microbiota in patients with and without pouchitis after RPC for UC and FAP. We also aimed to establish whether a dysbiosis occurs in pouchitis whilst avoiding the limitations of earlier studies. This is an expansion of the previous study(17) from our unit.
MATERIALS AND METHODS
Patients and samples
Ethical permission for the study was granted by the local ethics committee (ethics no. 3238). RPC patients with UC and FAP attending the hospital surgical department either for routine annual review or with symptoms of pouchitis were recruited. All underwent flexible pouchoscopy with biopsy. Chronic pouchitis was defined as three or more episodes of pouchitis per year(27) and active pouchitis was diagnosed when the pouch disease activity index (PDAI)(28) was ≥7.
Four groups of patients were studied:
UC RPC non-pouchitis patients (n=8)
UC RPC patients with active pouchitis (PDAI ≥7) and a history of chronic pouchitis (n=8)
FAP RPC non-pouchitis patients (n=5)
FAP RPC patients with active pouchitis (PDAI ≥7) (n=3)
Patients with pouchitis were treated with four weeks of antibiotic treatment (500mg ciprofloxacin and metronidazole 400mg twice daily) after which they underwent a second clinical and endoscopic assessment. Biopsies were taken before repeat pouchoscopy and discarded in those where follow-up pouchoscopy failed to demonstrate mucosal healing in order to exclude antibiotic-resistant pouchitis cases from this study (Figure 1). Inclusion criteria for UC and FAP non-pouchitis included patients with good pouch function, no previous history of pouchitis and PDAI < 7. FAP pouchitis included patients with active pouchitis (PDAI ≥7). Patients with complications including retained ano-rectal cuff inflammation, stricture, anastomotic leakage, fistula, a history of non-steroidal anti-inflammatory drug use, immunomodulator or other IBD therapy in the previous two months and those on antibiotic or probiotic therapy within the preceding two weeks were excluded from the study.
Figure 1.
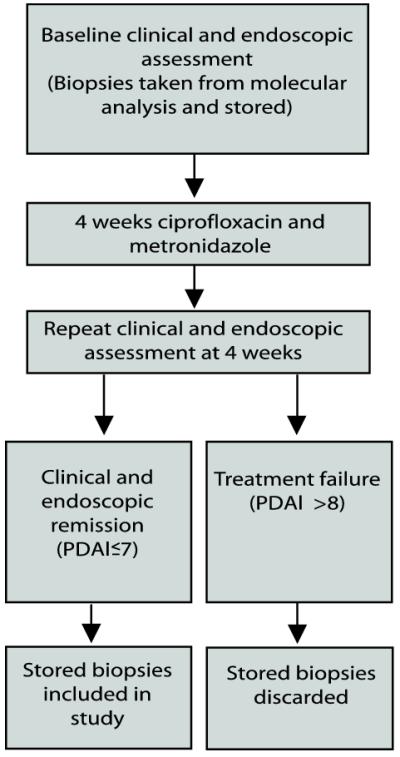
Selection of patient samples from pouchitis group
Mucosal biopsy sampling
Each patient received a phosphate enema (Forest, UK) prior to the procedure. Two mucosal biopsies (each approximately 1 × 2 mm) were collected during pouchoscopy approximately 10cm from the anal verge away from suture or staple lines. Each sample was placed in a sterile cryovial without preservative, snap frozen in liquid nitrogen and stored at −70°C until analysis. Four biopsies for routine histological examination were taken and examined by a GI histopathologist. A medical history was taken, hospital records were reviewed and the PDAI calculated for each patient.
DNA extraction and PCR amplification of bacterial 16S rRNA genes
DNA extraction was performed on single biopsy specimens using the DNeasy blood and tissue kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions.
The broad-range bacterial primers Bact-7F (5′-AGAGTTTGATYMTGGCTCAG-3″) and Bact-1510R (5′-ACGGYTACCTTGTTACGACTT-3′) were used to amplify community 16S rRNA genes. Each 100μl PCR mixture contained 20μl of Go-Taq Buffer (Promega, UK), 3mM MgCl2, 200μM dNTPs, 0.4μM primer F, 0.4μM primer R, 0.5μl Go-Taq DNA polymerase (Promega, UK), 47.5μl nuclease free water and 2μl of the sample DNA solution. The control contained 2μl of nuclease free water in place of sample DNA.
PCR amplification was performed using a Hybaid Px2 Thermal Cycler (Thermo Scientific, Waltham, USA) with one denaturation step at 95°C for 5min followed by 30 cycles of 95°C for 30 seconds, 58°C for 30 seconds and 72°C for 2 minutes with a final elongation step at 72°C for 10 minutes.
Clone library construction and sequence analysis
Clone library construction and sequencing were performed as described previously(29). 192 colonies were randomly selected for sequencing from agar plates. Sequences were aligned using the NAST aligner(27) and extensive manual curation of alignments was performed using the ARB package(30). Sequences were tested for chimeras using Mallard(31). Bellerophon(27), and Pintail(32) and chimeric sequences were removed. After removal of chimeras and other suspect sequences an average of 133 sequences per sample remained (3184 full-length sequences in total). These sequences (deposited in GenBank under GQ156578-GQ159761) were given a broad classification at the phylum and family levels using the Classifier tool at the RDPII website(33). To obtain more detailed taxonomic information the sequences were divided into phylotypes. Distance matrices were generated with ARB using the Olsen correction and entered into the DOTUR program(34) set to the furthest neighbour and 99%-similarity setting. Resulting phylotypes were then assigned similarities to nearest neighbours using MegaBLAST (35). The Shannon diversity index (SDI) for each individual sample was calculated using DOTUR(34).
Statistical analysis
SPSS version 15 (SPSS inc. Chicago, USA) was used for all statistical analysis. For the description of data, the median and range were calculated. The Mann-Whitney U test was used to compare groups. A two-tailed p-value <0.05 was considered significant.
RESULTS
Biopsy specimens were obtained from eight UC RPC non-pouchitis patients, eight UC RPC patients with active chronic pouchitis who later entered clinical and endoscopic remission following antibiotic treatment, five FAP RPC non-pouchitis patients and three FAP RPC patients with active pouchitis. Clinical and demographic details are shown in Table 1.
Table 1.
Demographic details of study patients.
| UC | FAP | |||
|---|---|---|---|---|
| Non-pouchitis n=8 |
Pouchitis n=8 |
Non-pouchitis n=5 |
Pouchitis n=3 |
|
| Age in years | 51 (19-63) | 39 (19-64) | 40 (25-72) | 32 (30-54) |
| Median interval since RPC in months |
103 (35-325) | 119 (10-1234) | 124 (42-203) | 50 (25-53) |
| Sex | 6 males | 5 males | 4 males | 1 male |
| Pouch configuration |
4 ‘W’ 3 ‘J’ 1 ‘S’ |
1 ‘W’ 7 ‘J’ |
1 ‘W’ 4 ‘J’ |
3 ‘J’ |
| 24hr Stool frequency |
5 (2-9) | 12( 8-16) | 4 (3-7) | 12 (6-21) |
| PDAI | 0 (0-3) | 11 (8-14) | 0 (0-1) | 12 (9-12) |
Values shown are medians with range in parentheses
Sequence analysis
In total, 3184 full-length sequences were generated from the twenty-four clone libraries. In common with other gut bacterial surveys(22;26) the majority of the sequences (99.8%) corresponded to just four bacterial phyla: Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria. A very small number of sequences corresponded to Verrucomicrobia and Fusobacteria. As has been repeatedly shown in other studies of the gut microbiota(22;24) we also found a large inter-individual variation.
Phylum level analysis between groups
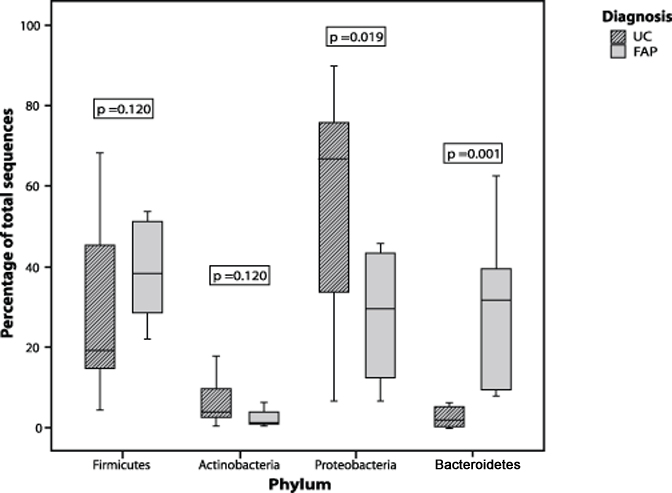
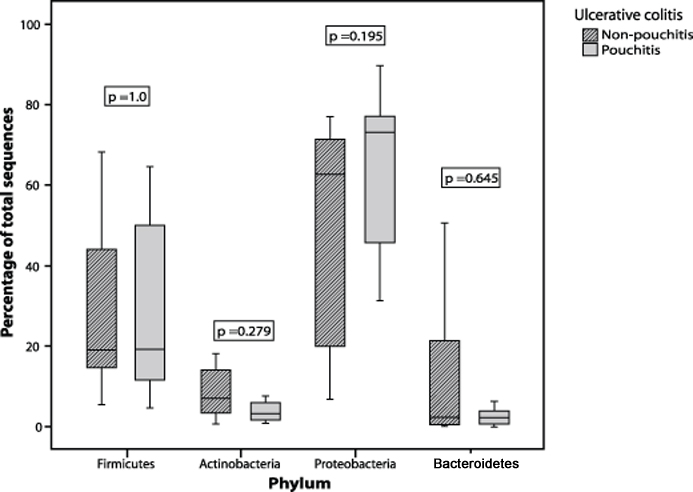
Although the four phyla above were predominant in our samples it is clear that the pouch microbiota, particularly in patients with UC, is drastically different from that typically encountered in the colon. Normally the gut microbiota is dominated by Bacteroidetes and Firmicutes. In contrast, samples taken from UC RPC patients were marked by unusually high proportions of Proteobacteria (mean of around 60% of total clones) while Bacteroidetes and the major Firmicutes families Lachnospiraceae and Ruminococcaceae were, for the most part, greatly reduced. The FAP RPC samples, while still harbouring relatively high Proteobacteria proportions, generally appeared to be composed of microbial communities more typical of a normal colon. When comparing the UC and FAP cohorts, there was a significant increase in the proportion of sequences in the Proteobacteria (p= 0.019) and a significant decrease in the proportion of Bacteroidetes (p= 0.001) phyla in the total UC compared with the total FAP patient cohort (Figure 2). Similar differences were identified when the UC pouchitis group was compared to the FAP pouchitis group, with increased Proteobacteria (p= 0.041) and reduced Bacteroidetes (p= 0.014). When the UC non-pouchitis group was compared with the UC pouchitis group, however, there were no significant differences in the proportion of sequences from the any of the phyla (Figure 3). There was also no significant difference between FAP pouchitis and FAP non-pouchitis groups (Table 2). Therefore, at the phylum level, although we could demonstrate differences between the two different patient cohorts we were unable to demonstrate a dysbiosis within each of the two disease groups.
Figure 2.
Box plot comparing the percentage of sequences identified from the four predominant bacterial phyla in UC patient samples compared with FAP patient samples
Figure 3.
Box-plot comparing the percentage of sequences identified from the four predominant bacterial phyla in UC pouchitis patient samples compared with UC non-pouchitis patient samples.
Table 2.
Table to demonstrate difference in median proportion of clones belonging to each phylum
| UC | FAP | UC | FAP | UC Pouchitis FAP pouchitis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Non- pouchitis |
Pouchitis | P- value |
Non- pouchitis |
Pouchitis | P- value |
All | All | P- value |
P-value |
| Firmicutes | 18.9 | 18.9 | 1.0 | 29.9 | 50.0 | 0.393 | 18.9 | 38.3 | 0.120 | 0.307 |
| Actinobacteria | 6.8 | 3.1 | 0.279 | 2.1 | 1.2 | 0.786 | 4.0 | 1.5 | 0.120 | 0.220 |
| Proteobacteria | 62.6 | 73.2 | 0.195 | 41.7 | 18.4 | 0.786 | 66.6 | 29.5 | 0.019 | 0.041 |
| Bacteroidetes | 2.1 | 2.1 | 0.645 | 29.4 | 31.0 | 0.786 | 2.1 | 31.7 | 0.001 | 0.014 |
Family level analysis
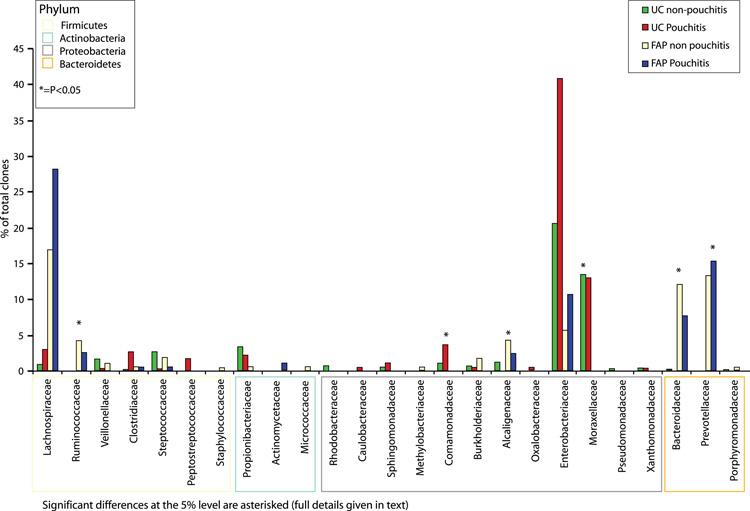
We then attempted to pinpoint the significant differences between the samples by examining the sequence data at the family level (Figure 4). These results showed that the differences at the phylum level between the UC and FAP cohorts corresponded to significant increases in the levels of the proteobacterial families Comamonadaceae (p= 0.007), Moraxellaceae (p= 0.027) and Alcaligenaceae (p= 0.03) in tandem with a significant reduction in the Bacteroidetes families Bacteroidaceae (p= 0.013) and Prevotellaceae (p= 0.023) and the Firmicutes family Ruminococcaceae (p= 0.007) in UC. The other families identified are illustrated in Tables 5-7 (supplementary material).
Figure 4.
Median percentage of clones identified from each bacterial family in samples from UC pouchitis, UC non-pouchitis, FAP pouchitis and FAP non-pouchitis patients.
Significant differences at the 5% level are asterisked (full details given in text).
When comparing the UC pouchitis to the UC non-pouchitis groups we found that streptococci and Alcaligenaceae were reduced in patients with pouchitis (p= 0.04 and p= 0.026 respectively). Enterobacteriaceae, including E. coli, accounted for the highest proportion of proteobacterial sequences and were increased in both UC pouchitis versus UC non-pouchitis and FAP pouchitis versus FAP non-pouchitis. Due to the large degree of inter-individual variation between patients, however, these differences did not reach significance.
Species level analysis
Each patient sample was analysed at the species level by splitting the sequences into phylotypes comprised of >99%-identical sequences using DOTUR(34) (complete details of each patient sample and the species present are provided in Tables 8-31, supplementary data). When comparing the UC and FAP cohorts, Faecalibacterium prausnitzii, which has been postulated to have anti-inflammatory properties and may be reduced in IBD patients, was detected in 6 out of 8 FAP patients but only 4 out of 16 UC patients (p=0.029). Bacteroides vulgatus was also significantly increased in the FAP group compared to the UC group (p=0.031).
There were, however, no individual species or phylotypes that significantly differed between the UC pouchitis and UC non-pouchitis cohorts. This included both F. prausnitzii and B. vulgatus as well as other species that have previously been implicated in IBD such as Bacteroides fragilis(14) and E. coli (Table 3). Further bacteria that have been implicated in the pathogenesis of IBD such as Mycobacterium avium subspecies paratuberculosis, Listeria and Yersinia spp.(27) were not detected in any of the samples. Sulphate-reducing bacteria (SRB) have also been linked to IBD pathogenesis, however we only detected the Desulfobulbaceae family of SRB in one out of eight UC pouchitis patients, while the Desulfovibrionaceae family of SRB was only detected in one FAP non-pouchitis patient.
Table 3.
Table to demonstrate difference in median proportion of clones for particular bacterial species in each study group
| UC | FAP | UC | FAP | UC Pouchitis FAP pouchitis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | Non- pouchitis |
Pouchitis | P- value |
Non- pouchitis |
Pouchitis | P- value |
All | All | P- value |
P-value |
|
Bacteroides
vulgatus |
0 | 0 | 0.538 | 3.52 | 0 | 0.093 | 0 | 1.00 | 0.031 | 0.545 |
|
Bacteroides
fragilis |
0 | 0 | 0.144 | 0 | 0 | 1.0 | 0 | 0 | 0.307 | 0.364 |
|
Faecalibacterium
prausnitzii |
0 | 0 | 0.783 | 1.69 | 0 | 0.453 | 0 | 1.56 | 0.029 | 0.604 |
| Escherichia coli | 17.37 | 8.47 | 1.0 | 5.65 | 1.84 | 0.655 | 12.70 | 0 | 0.118 | 0.414 |
Shannon Diversity Index (SDI)
Using the phylotypes generated by DOTUR we calculated the SDI,(27) which is a measure of the number of different species and their relative abundance in a given environment, for each sample and for each patient group (Figures 4-6). These are given in Table 4 (median and range shown). The median SDI for all UC RPC patients was 2.61 compared to 3.2 for all FAP RPC patients and this difference was statistically significant (p= 0.004). The median SDI in the FAP non-pouchitis group was significantly higher than in the UC non-pouchitis group (p= 0.019) however no difference was observed between the FAP pouchitis and UC pouchitis group (p= 0.066), overall indicating that a less diverse bacterial community exists in UC RPC patients than in FAP RPC patients. Comparison within disease groups showed the median SDI in the UC non-pouchitis group was 2.70 and in the UC pouchitis group 2.32. This difference was statistically significant (p= 0.009). The median SDIs in the FAP non-pouchitis and pouchitis groups were 3.19 and 3.34(p= 0.18). These results demonstrate that, overall, there was a simpler, less diverse bacterial community in the UC group in comparison to the FAP group, and that a further reduction in diversity of the bacterial community occurs in patients with pouchitis.
Figure 5.
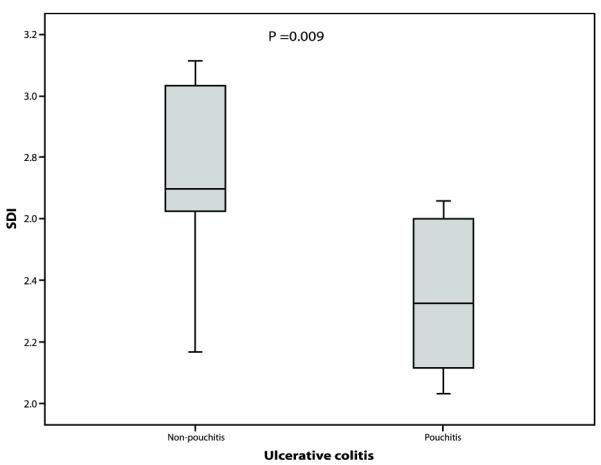
Boxplot comparing the Shannon Diversity index in samples from UC pouchitis patients compared to UC non-pouchitis patient samples.
Figure 6.
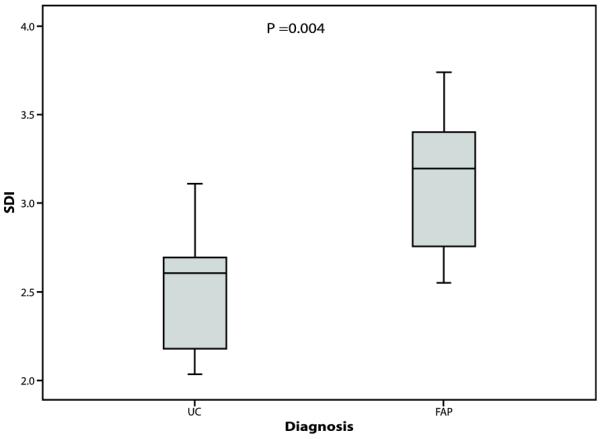
Box plot comparing the Shannon Diversity index in the total UC cohort and total FAP cohort
Table 4.
Shannon diversity index (SDI) by patient group
| Shannon diversity index | ||||
|---|---|---|---|---|
| Patient group | Pouchitis | Non-pouchitis | P-value | All patients |
| UC | 2.32 (2.10-2.66) |
2.70 (2.17-3.11) |
0.009 | 2.61 (2.10-3.09) |
| FAP | 2.79 (2.55-3.22) |
3.34 (2.72-3.46) |
0.18 | 3.20 (2.72-3.46) |
| P-value | 0.066 | 0.019 | 0.004 | |
Values shown are medians with range in parentheses
DISCUSSION
It has been suggested that the term pouchitis describes a spectrum of diseases(36). In this study we attempted to reduce heterogeneity by studying only UC pouchitis patients with chronic pouchitis rather than all types of pouchitis. We repeated clinical and endoscopic assessment following treatment with standard combination antibiotic therapy to ensure the study group entered both clinical and endoscopic remission and to exclude patients with antibiotic resistant pouchitis. In addition, in all patients at pouchoscopy there was diffuse pouch inflammation as opposed to inflammation confined to one area of the pouch. No patient had risk factors for mesenteric ischaemia. Therefore we believe that no patient had an ischaemic component to their pouchitis. The inclusion of FAP pouchitis and FAP non-pouchitis patients is novel and allows comparison between patients with a previous history of IBD and those without. Pouchitis in FAP patients has not been well studied and its incidence is about ten times lower than in UC patients(37), the reasons for this are unclear.
As in other studies we analysed the mucosal-adherent microbiota since these are likely to be more important than luminal microbiota in the pathogenesis of IBD(15;19;22). This mucosal-adherent microbiota, which is in close contact with the gut mucosa, has been shown(38;39) to be distinct from the luminal or faecal microbiota, which is comprised of free-living or particle-attached cells. The difference in community structure is likely driven by a number of factors such as differential substrate availability (e.g. mucus vs undigested dietary residues), oxygen levels and host-microbe interactions. The close proximity of the mucosally-adherent microbiota to the gut epithelium means that these bacteria are presumed to be more important than luminal microbiota in the pathogenesis of IBD since they, and their excreted products, are considered more likely to have direct contact with the host(22).
The particular strengths of the present study are that we have studied pouchitis in both IBD and non-IBD RPC patients and that each sample was cloned individually and sequenced to the species/phylotype level. To the best of our knowledge this is the first investigation in which this has been undertaken. We performed a power calculation of the study data which has shown that an estimated sample size of eight patients per group was required to demonstrate a 5% statistical significance and 80% power in SDI between the UC pouchitis (2.35±0.26) versus UC non-pouchitis groups (2.75 ±0.31).
Similarly an estimated sample size of seven patients per group was required to demonstrate a 5% statistical significance and 80% power in SDI between all UC patients (2.55±0.35) versus FAP patients (3.12 ±0.41).
There are, however, limitations to this present study. First, 16S rRNA gene sequencing results represent gene copy number, not true bacterial counts, and may also be biased by differential DNA extraction and PCR amplification rates. The methodology is currently, however, the best available and regarded as the “gold standard” for the analysis of gut-associated microbiota(10). Secondly, the study included small numbers of patients. This is due to difficulty in accrual since patients with chronic antibiotic-dependent pouchitis are uncommon and represent about 5% of all patients. FAP RPC patients are uncommon and FAP pouchitis, particularly, is rare(40-44). The patient group was recruited from the largest European centre and although it may have been possible to include patients from other centres this may have increased heterogeneity into the study population due to differences in the diagnostic criteria of pouchitis. Thirdly, we included patients with chronic pouchitis who had not received antibiotic therapy for a minimum of two weeks. This might have influenced the gut microbiota but this cut-off was chosen for practical and ethical reasons. Others have done the same, for example in the study by Komanduri et al one patient had been treated with antibiotics two weeks prior to sampling. These authors reported that there was no difference in the microbiota identified in this patient when compared with those who had not received an antibiotic for four weeks and concluded that a two week wash-out period was sufficient(25).
Around 99% of gut microbiota are contained in four phyla; Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria(22;26). At the species level however, each individual has his or her own unique gut microbiota(24). This causes difficulty in studying gut microbiota and also demonstrates the importance of cloning and sequencing individual samples rather than analysing pooled samples. The present study has shown that the ileal pouch microbiota is different from the normal large intestine. UC pouches in particular, with or without pouchitis, appear to harbour more unusual microbiota than FAP pouches. Proteobacteria, which normally account for only a small proportion of the microbiota in the healthy colon(39) and up to 20% of the microbiota in IBD patients(22) comprised up to 90% (median = 66.6%) of the microbiota in the UC RPC patients in the study. There were also lower than normal proportions of Bacteroidetes, Lachnospiraceae and Ruminococcaceae. Comparison of the two study cohorts showed that UC RPC patients have increased proportions of the phylum Proteobacteria and decreased levels of Bacteroidetes compared with FAP RPC patients. A similar pattern was reported in a recent study in which surgical specimens from IBD (UC and Crohn’s disease) and non-IBD patient controls were compared. There was a reduction in the numbers of Bacteroidetes and Lachnospiraceae and an increase in Proteobacteria in a subset of IBD patients(22). In other studies increases in Proteobacteria in IBD patients have also been demonstrated(12;16) and the Enterobacteriaceae family of Proteobacteria have often been shown to be increased in IBD patients compared with controls(14;45).
Bacterial diversity was significantly lower in UC RPC patients, with or without pouchitis, than for FAP RPC patients. Furthermore, diversity was significantly reduced in UC pouchitis patients compared to those without. It has previously been shown that VSL#3 increases bacterial diversity in pouchitis(46) and perhaps this may account for the reduced risk of relapse. A reduction in bacterial diversity has also been reported in both CD(47) and UC(48;49). The results of the present study therefore are further evidence of the importance of bacterial diversity in maintaining normal gut homeostasis.
Although the study aimed to establish whether a dysbiosis might be associated with pouchitis only minor differences between UC pouchitis and UC non-pouchitis were found; comparisons revealed only borderline significance between a very small number of bacterial groups. We recognise that when comparing multiple groups, significance may occur in a limited number simply by chance and have taken care not to overstate the importance of these observations. There was, however, a difference in the microbiota between the total UC RPC and FAP RPC cohorts with a reduction in F. prausnitzii and B. vulgatus in the UC RPC patient group. This is an interesting finding, given that F. prausnitzii has previously been postulated to have anti-inflammatory properties and may be reduced in IBD patients(50;51).
In the previous study from our group using culture and T-RFLP (17), the failure to find any differences between UC and FAP patients whether with pouchitis or not, is a reflection of the methodology which permits only cultured bacteria to be studied and is less specific and sensitive in the identification of species. The present study did find differences which not only added new information regarding the microbiota but also obtained results which are different than those of Komanduri et al (24). These authors compared the microbiota after RPC for UC only (not FAP) in patients with and without pouchitis but there are methodological objections to the study. Length heterogeneity polymerase chain reaction (LH-PCR) was used and because this technique provides limited information about the bacterial differences at a species/phylotype level, the LH-PCR products from three non-IBD controls, five UC non-pouchitis and three UC pouchitis patients were pooled before cloning and data were then filtered. As a result only phylotypes representing more than 5% of the total clone library were analysed and only 73% of the microbiota were identified. The cloning and sequencing of these pooled samples identified an increase in the proportion of Enterobacteriaceae and Fusobacteria, a reduction in streptococci and a difference in the Ruminococcus species associated with pouchitis (R. obeum) and non-pouchitis (R. gnavus). In our study we studied individual patients and did not pool samples or filter our data. Using this methodology we were able to study individual species/phylotypes.
The differing methodological approaches may explain the different results between the study of Komanduri et al, and ours in which Fusobacteria were not detected in the UC RPC samples and no difference was found in the proportions of Clostridium paraputrificum or Ruminococcus species in pouchitis and non-pouchitis. In our study, there was a doubling in Enterobacteriaceae in UC and FAP patients with pouchitis but this was not statistically significant owing to the high individual variation between patients, further indicating the danger of pooling samples. Indeed individual variation was so great that the numbers of patients required to detect any statistically significant difference in microbiota within UC patients would be too large to be practicable. In agreement with Komanduri et al, however, we did observe a reduction in streptococci in the UC pouchitis patients compared with non-pouchitis.
This study has demonstrated that a dysbiosis occurs in UC RPC patients when compared with a non-IBD (FAP) population. There was a reduction in diversity but only minor compositional differences between the microbiota of UC patients with active pouchitis and those with no history of pouchitis. This suggests that either this dysbiosis predisposes UC patients to pouchitis by increasing the likelihood of immune system stimulation or that the reduction in diversity is sufficient to stimulate the immune system and lead to mucosal inflammation. The failure to identify a particular bacterial species associated with pouchitis is in keeping with clinical experience where antibiotics with very different spectra of antimicrobial activity are equally effective in pouchitis. We have recently shown that many patients with pouchitis refractory to empirical antibiotic treatment have antibiotic resistant coliforms and microbiological testing is able to predict an effective antibiotic regime(52). This, taken with the findings of the present study, suggests that antibiotic therapy is effective in pouchitis by reducing the total gut microbial load and therefore the stimulus to the immune system rather than the elimination of a specific disease-activating bacterial species.
CONCLUSION
This is the first study to compare the microbiota in individual patients having RPC for UC and FAP RPC using 16S rRNA gene sequencing. UC patients have a different and less diverse gut microbiota than FAP. A further reduction in bacterial diversity but only minor changes occurs in active pouchitis. The study suggests that a dysbiosis occurs in UC RPC patients which predisposes to, but does not directly cause, pouchitis.
Supplementary Material
Mini abstract.
We investigated the microbiota in ulcerative colitis and familial adenomatous polyposis restorative proctocolectomy patients using 16S rRNA gene cloning and sequencing. We identified significant differences in Proteobacteria, Bacteroidetes and Faecalibacterium prausnitzii in the total UC compared with the total FAP cohort, but only limited differences between non-pouchitis and pouchitis groups.
Acknowledgments
Grant support Funding for SM was provided by The St. Mark’s Hospital Foundation and The Broad Medical Research Programme
Funding for LP was provided by the FAIR Foundation and The Broad Medical Research Programme
Funding for AW, CC, JP, GD and sequencing was provided by The Wellcome Trust [Grant no. WT076964]
Footnotes
All authors declare no conflicts of interest exist. There are no financial disclosures for any author.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- (1).Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- (2).Schultz M, Sartor RB. Probiotics and inflammatory bowel diseases. Am J Gastroenterol. 2000;95(1 Suppl):S19–S21. doi: 10.1016/s0002-9270(99)00812-6. [DOI] [PubMed] [Google Scholar]
- (3).Shen B. Diagnosis and treatment of patients with pouchitis. Drugs. 2003;63:453–61. doi: 10.2165/00003495-200363050-00002. [DOI] [PubMed] [Google Scholar]
- (4).McLaughlin SD, Clark SK, Bell AJ, et al. An open study of antibiotics for the treatment of pre-pouch ileitis following restorative proctocolectomy with ileal pouch-anal anastomosis. Aliment Pharmacol Ther. 2008 doi: 10.1111/j.1365-2036.2008.03858.x. [DOI] [PubMed] [Google Scholar]
- (5).Gionchetti P, Rizzello F, Venturi A, et al. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology. 2000;119:305–9. doi: 10.1053/gast.2000.9370. [DOI] [PubMed] [Google Scholar]
- (6).Mimura T, Rizzello F, Helwig U, et al. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108–14. doi: 10.1136/gut.53.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gionchetti P, Rizzello F, Helwig U, et al. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–9. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- (8).Bell AJ, Nicholls RJ, Forbes A, et al. Human lymphocyte stimulation with pouchitis flora is greater than with flora from a healthy pouch but is suppressed by metronidazole. Gut. 2004;53:1801–5. doi: 10.1136/gut.2003.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lim M, Sagar P, Finan P, et al. Dysbiosis and pouchitis. Br J Surg. 2006;93:1325–34. doi: 10.1002/bjs.5602. [DOI] [PubMed] [Google Scholar]
- (10).Zoetendal EG, Rajilic-Stojanovic M, De Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–15. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- (11).Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- (12).Gophna U, Sommerfeld K, Gophna S, et al. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–41. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, et al. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–45. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- (15).Swidsinski A, Weber J, Loening-Baucke V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–9. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Seksik P, Rigottier-Gois L, Gramet G, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–42. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Johnson MW, Rogers GB, Bruce KD, et al. Bacterial community diversity in cultures derived from healthy and inflamed ileal pouches after restorative proctocolectomy. Inflamm Bowel Dis. 2009 doi: 10.1002/ibd.21022. [DOI] [PubMed] [Google Scholar]
- (18).Bibiloni R, Mangold M, Madsen KL, et al. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn’s disease and ulcerative colitis patients. J Med Microbiol. 2006;55:1141–9. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- (19).Swidsinski A, Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- (20).Kleessen B, Kroesen AJ, Buhr HJ, et al. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034–41. doi: 10.1080/003655202320378220. [DOI] [PubMed] [Google Scholar]
- (21).Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–18. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- (22).Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- (24).Komanduri S, Gillevet PM, Sikaroodi M, et al. Dysbiosis in pouchitis: evidence of unique microfloral patterns in pouch inflammation. Clin Gastroenterol Hepatol. 2007;5:352–60. doi: 10.1016/j.cgh.2007.01.001. [DOI] [PubMed] [Google Scholar]
- (25).Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–9. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Eckburg PB, Relman DA. The role of microbes in Crohn’s disease. Clin Infect Dis. 2007;44:256–62. doi: 10.1086/510385. [DOI] [PubMed] [Google Scholar]
- (27).DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sandborn WJ, Tremaine WJ, Batts KP, et al. Pouchitis after ileal pouch-anal anastomosis: a Pouchitis Disease Activity Index. Mayo Clin Proc. 1994;69:409–15. doi: 10.1016/s0025-6196(12)61634-6. [DOI] [PubMed] [Google Scholar]
- (29).Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ludwig W, Strunk O, Westram R, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ashelford KE, Chuzhanova NA, Fry JC, et al. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–41. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ashelford KE, Chuzhanova NA, Fry JC, et al. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–36. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–6. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Johnson M, Zaretskaya I, Raytselis Y, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(Web Server issue):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Shen B, Lashner BA. Pouchitis: a spectrum of diseases. Curr Gastroenterol Rep. 2005;7:404–11. doi: 10.1007/s11894-005-0011-3. [DOI] [PubMed] [Google Scholar]
- (37).McLaughlin SD, Clark SK, Tekkis PP, et al. Review Article: restorative proctocolectomy, indications, management of complications and follow-up: a guide for gastroenterologists. Aliment Pharmacol Ther. 2008 doi: 10.1111/j.1365-2036.2008.03643.x. [DOI] [PubMed] [Google Scholar]
- (38).Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68(7):3401–7. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Romanos J, Samarasekera DN, Stebbing JF, et al. Outcome of 200 restorative proctocolectomy operations: the John Radcliffe Hospital experience. Br J Surg. 1997;84:814–8. [PubMed] [Google Scholar]
- (41).Stahlberg D, Gullberg K, Liljeqvist L, et al. Pouchitis following pelvic pouch operation for ulcerative colitis. Incidence, cumulative risk, and risk factors. Dis Colon Rectum. 1996;39:1012–8. doi: 10.1007/BF02054692. [DOI] [PubMed] [Google Scholar]
- (42).Salemans JM, Nagengast FM, Lubbers EJ, et al. Postoperative and long-term results of ileal pouch-anal anastomosis for ulcerative colitis and familial polyposis coli. Dig Dis Sci. 1992;37:1882–9. doi: 10.1007/BF01308083. [DOI] [PubMed] [Google Scholar]
- (43).Hurst RD, Molinari M, Chung TP, et al. Prospective study of the incidence, timing and treatment of pouchitis in 104 consecutive patients after restorative proctocolectomy. Arch Surg. 1996;131:497–500. doi: 10.1001/archsurg.1996.01430170043007. [DOI] [PubMed] [Google Scholar]
- (44).Lovegrove RE, Tilney HS, Heriot AG, et al. A comparison of adverse events and functional outcomes after restorative proctocolectomy for familial adenomatous polyposis and ulcerative colitis. Dis Colon Rectum. 2006;49:1293–306. doi: 10.1007/s10350-006-0608-0. [DOI] [PubMed] [Google Scholar]
- (45).Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–40. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- (46).Kuhbacher T, Ott SJ, Helwig U, et al. Bacterial and fungal microbiota in relation to probiotic therapy (VSL#3) in pouchitis. Gut. 2006;55:833–41. doi: 10.1136/gut.2005.078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Andoh A, Sakata S, Koizumi Y, et al. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955–62. doi: 10.1002/ibd.20151. [DOI] [PubMed] [Google Scholar]
- (49).Sokol H, Lepage P, Seksik P, et al. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol. 2006;44:3172–7. doi: 10.1128/JCM.02600-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–9. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- (52).McLaughlin SD, Clark SK, Shafi S, et al. Fecal coliform testing to identify effective antibiotic therapies for patients with antibiotic-resistant pouchitis. Clin Gastroenterol Hepatol. 2009;7:545–8. doi: 10.1016/j.cgh.2009.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.