Abstract
Background
Chromosome rearrangements are caused by many mutational mechanisms; of these, recurrent rearrangements can be particularly informative for teasing apart DNA sequence-specific factors. Some recurrent translocations are mediated by homologous recombination between large blocks of segmental duplications on different chromosomes. Here we describe a recurrent unbalanced translocation casued by recombination between shorter homologous regions on chromosomes 4 and 18 in two unrelated children with intellectual disability.
Results
Array CGH resolved the breakpoints of the 6.97-Megabase (Mb) loss of 18q and the 7.30-Mb gain of 4q. Sequencing across the translocation breakpoints revealed that both translocations occurred between 92%-identical human endogenous retrovirus (HERV) elements in the same orientation on chromosomes 4 and 18. In addition, we find sequence variation in the chromosome 4 HERV that makes one allele more like the chromosome 18 HERV.
Conclusions
Homologous recombination between HERVs on the same chromosome is known to cause chromosome deletions, but this is the first report of interchromosomal HERV-HERV recombination leading to a translocation. It is possible that normal sequence variation in substrates of non-allelic homologous recombination (NAHR) affects the alignment of recombining segments and influences the propensity to chromosome rearrangement.
Keywords: HERV-H, HERV, NAHR, translocation, t(4;18), recurrent translocation, 4q35.1, 18q22.3, 18q
Background
Chromosome rearrangements play a major role in intellectual disability, birth defects, and autism [1-3], and many heterogeneous mechanisms have been implicated in the formation of chromosome rearrangements. Analyses of breakpoint junctions from a host of chromosome rearrangements have revealed signatures of homologous recombination (HR), nonhomologous end-joining (NHEJ), microhomology-mediated end joining (MMEJ), and break-induced replication (BIR) [4-6].
Recurrent chromosome abnormalities give us a unique opportunity to unravel specific factors involved in rearrangement, as their frequency and shared breakpoints indicate a rearrangement-prone genomic architecture. Deletions and duplications mediated by non-allelic homologous recombination (NAHR) are the most common class of recurrent constitutional chromosome rearrangements and are responsible for several genetic syndromes [7-9]. Such rearrangements are relatively easy to identify by paralogous genomic segments that are highly identical and typically hundreds of kilobases in size flanking breakpoint junctions [9-11]. Analysis of translocation breakpoints has shown that some recurrent translocations are also mediated by NAHR. For example, recurrent translocations between chromosomes 4p and 8p [12], 4q and 10q [13], and 4p and 11p [14,15] are known to be mediated by homologous recombination between large blocks of segmental duplications, whereas recurrent translocations between 11q and 22q [16] and 8q and 22q [17] are initiated by palindromic AT-rich sequences at breakpoints. However, most constitutional translocations are not recurrent, and their breakpoints lack significant sequence homology that would suggest NAHR [18-20].
NAHR between homologous interspersed repeats like LINE, Alu, and human endogenous retrovirus (HERV) elements can also lead to genomic rearrangements [21,22]. Alu-Alu recombination events have been described at multiple loci, giving rise to pathogenic deletions, duplications, and translocations [6,23-26]. Homologous recombination between HERV15 elements underlies the recurrent Y chromosome microdeletion that removes the azoospermia factor a (AZFa) region and causes male infertility [27-29]. Recently, a HERV-H-mediated deletion of chromosome 8q13.3 has been described in a child with heterozygous loss of the EYA1 gene and branchio-oto-renal syndrome [30]. Nevertheless, homologous recombination between HERV elements on different chromosomes has not been previously described as a mechanism for recurrent translocations.
HERVs make up ~3% of the human genome [31,32]; however, most copies have mutations and/or deletions that disrupt one or more of the ORFs, rendering the retrovirus inactive [33-35]. Though most HERVs are inactive as retrotransposons, they may spread through the genome via ectopic recombination processes. Phylogenetic studies of HERV-K elements have revealed signatures of intraelement gene conversion and recombination [36,37]. Thus, the density of HERVs and the sequence homogenization between copies make them ideal substrates for NAHR in the human genome.
Here we describe a recurrent translocation mediated by NAHR between HERVs on chromosomes 4q and 18q. Sequencing the breakpoint junctions in two unrelated individuals with similar translocations [38,39] revealed breakpoints within a few hundred basepairs (bp) of each other. Both sets of translocation breakpoints are located in HERV-H elements, and the orientation and sequence homology between recombining segments on 4q and 18q are consistent with a NAHR rearrangement mechanism.
Results and discussion
In an earlier study, we identified two individuals (18q-82C and 18q-146C) with unbalanced translocations between the ends of the long arms of chromosomes 4 and 18. Both carry a derivative chromosome missing the end of chromosome 18q, with an additional copy of the end of 4q, as shown previously by array comparative genomic hybridization (CGH) [38,39]. Although two children are not enough for us to evaluate the phenotype associated with this unbalanced translocation comprehensively, some shared clinical features are worth noting. Patients 18q-82C and 18q-146C both exhibited developmental delays, auditory canal atresia, midface hypoplasia, microcephaly, and a broad nasal bridge. Parental studies revealed that 18q-82C carries a de novo translocation, derived from paternal chromosomes 4 and 18 [39]. Parents of 18q-146C were not available for study.
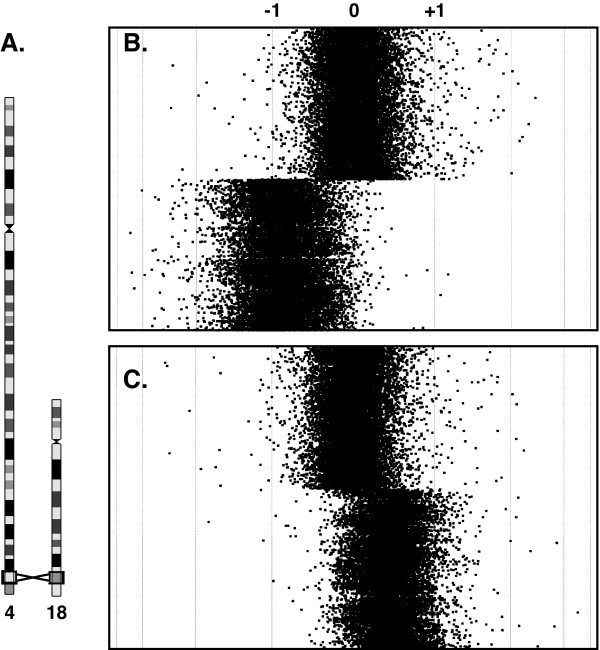
Using high-resolution array CGH, we resolved the 4q and 18q breakpoints in both subjects. We designed a custom oligonucleotide array targeting the 5 Mb spanning the 4q and 18q breakpoints with a mean probe spacing of one oligonucleotide per 100 bp. Array CGH revealed the same breakpoints in both individuals: a 6.97-Mb loss of 18q and a 7.30-Mb gain of 4q (Figure 1).
Figure 1.
High-resolution array CGH reveals the breakpoints of the unbalanced translocation from patient 18q-146C. (a) The recurrent translocation occurs between 4q35.1 and 18q22.3, as indicated by the boxed chromosome bands. (b) The 6.97-Mb loss of 18q and (c) the 7.30-Mb gain of the end of 4q are shown. Averaged log2 ratios of probe signal intensities are shown (black dots). Dashed lines indicate log2 ratios of -1, 0, and +1. Similar array CGH results were obtained from patient 18q-82C's translocation.
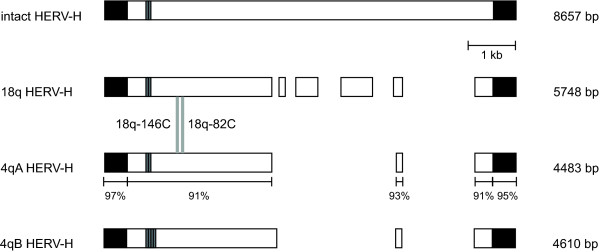
Based on the array CGH data, we designed PCR primers to amplify across the breakpoint junctions of the derivative chromosomes 18 from patients 18q-82C and 18q-146C. We cloned and sequenced the breakpoint junctions to generate complete sequence across the two independent junction fragments (GenBank sequences 18q82C_junction and 18q146C_junction). In both translocations, the junction between chromosomes 18 and 4 lies in a HERV-H element (Figure 2). As represented in the reference genome (Build 36.1, hg18), the HERV-H elements on chromosomes 4q and 18q are 4.6 kilobases (kb) and 5.7 kb, respectively, and are ~92% identical overall.
Figure 2.
HERV-H elements from chromosomes 4q and 18q, as compared to the intact HERV-H consensus sequence. The HERV-H consensus is 8657 bp as represented in RepeatMasker [40]. HERVs and long terminal repeats (LTRs) are depicted as open and filled black rectangles, respectively. The 18q HERV-H sequence is derived from Build 36.1 (hg18) of the human genome assembly. HERV-H sequences from the 4qA and 4qB alleles were derived from 18q-82P. Sequence identity between the 4q and 18q HERV-Hs are shown across aligning segments. The 61-bp tandem repeats are represented as dark grey rectangles within the HERV-Hs. Sites of recombination between 4q and 18q that gave rise to translocations in 18q-82C and 18q-146C are shown as vertical grey lines.
We aligned our junction sequences from patients 18q-82C and 18q-146C to sequence from the 4q and 18q HERV-H elements in the human genome assembly (Build 36.1, hg18). Recombination for both translocations occurred in an ~three-kb region that is 91% identical between the 4q and 18q HERV-Hs (Figure 2). Though the HERV-Hs are highly identical, chromosome-specific SNPs distinguish the 18q and 4q sides of the translocation junctions and allow us to further resolve the sites of recombination. The sites of recombination mediating the translocations in patients 18q-82C and 18q-146C are ~150 bp apart (Figure 3).
Figure 3.
Sequence alignment of the translocation junction sequences from patients 18q-82C and 18q-146C. Sequences for chromosomes 18 (chr 18) and 4 (chr 4) were derived from Build 36.1 (hg18) of the human genome assembly. SNPs that are specific to chromosome 4 or chromosome 18 are shown in bold and marked by asterisks. The sites of recombination between chromosomes 18q and 4q are highlighted in grey.
It is possible that some copies of the 4q and 18q HERV-Hs share more homology than represented in the human genome assembly, which may affect the propensity of the two chromosomes to recombine. There are reports of sequence variation in HERVs at several loci, which may occur via gene conversion or transposition processes [29,36,37]. To capture the actual HERV-H sequences that recombined to form the translocations in patients 18q-82C and 18q-146C, it was necessary to sequence the parental 4q and 18q alleles. Microsatellite analysis of 18q-82C and his parents revealed that the de novo translocation was paternal in origin [39]. We sequenced both alleles of the HERV-H elements on 4q and 18q in 18q-82C's father (18q-82P). The two 18q HERV-H alleles in 18q-82P were 99.5% identical to the 18q HERV-H in the reference genome, with no significant differences between the two alleles. However, 18q-82P has two different 4q HERV-H alleles, described here as 4qA (GenBank sequence 82Pchr4HERVHA) and 4qB (GenBank sequence 82Pchr4HERVHB).
The HERV-H elements on 4qA and 4qB are 99.0% identical, but 4qB has a 122-bp duplication not present in 4qA (Figure 2). The duplication is made up of a 61-bp tandem repeat present in two copies and four copies on the 4qA and 4qB alleles, respectively. The human reference genome (Build 36.1, hg18) corresponds to the 4qB allele, including the 122-bp duplication. The 122-bp duplication is not present in the 18q HERV-H or in the HERV-H consensus sequence [40], consistent with the duplication being a new event that arose on the 4qB allele.
To determine the frequency of the 4qA and 4qB alleles in the human population, we designed a genotyping assay that distinguishes the two alleles. We performed a nested PCR that specifically amplifies a six-kb region, including the entire 4q HERV-H in the first PCR, followed by a second PCR that amplifies the region around the 122-bp duplication. This results in a 326-bp band for the 4qA allele and a 446-bp band for the 4qB allele. We genotyped DNA from 99 Caucasians obtained from the Coriell Cell Repository (Human Variation Panel HD100CAU), representing 198 4q alleles, of which 37 (18.7%) were 4qA and 161 (81.3%) were 4qB (Additional file 1). We also genotyped 62 samples from 10 populations included in the Human Genome Diversity Panel (HGDP). Allele frequencies were not significantly different between geographic populations, and as a group, there were 26 (21.0%) 4qA alleles and 98 (79.0%) 4qB alleles (Additional file 1). Thus, the 4qA allele carried by 18q-82P is the minor allele, and the 4qB allele in the reference genome is the major allele.
Conclusions
Recurrent copy number variations (e.g., deletions, duplications, and translocations) provide mechanistic insight into the etiology of chromosome rearrangements. We sequenced the breakpoint junctions of two independent translocations with nearly identical breakpoints on chromosomes 4q and 18q. Analysis of the recombining segments revealed that the breakpoints lie in HERV-H elements that are 92% identical. NAHR between other HERVs has been found to underlie interstitial deletions of the Y chromosome and chromosome 8; in both cases, the recombining HERVs were ~94% identical [27-30].
NAHR between larger segmental duplications causes the most common microdeletion and microduplication syndromes. In these cases, recombining segmental duplications range from 10 kb to hundreds of kilobases in size and are typically 95% identical or greater [10,41-43]. NAHR between smaller substrates with greater sequence divergence, such as the HERV-Hs described here, is probably less frequent than NAHR between large segmental duplications. However, sequence variation in recombining segments could also impact rearrangement frequency. Polymorphism in the HERV15 elements that mediate the recurrent AZFa microdeletion leads to better sequence alignment between the recombining regions on the Y chromosome and is predicted to predispose to microdeletion [29]. It is possible that sequence variation in the 4q HERV-H also affects its propensity to recombine with the 18q HERV-H. Our study of two recurrent translocations is not comprehensive enough to draw conclusions about HERV-H sequence variation and recombination frequency. However, the lack of the 122-bp duplication in the 4qA HERV-H makes it more similar to the 18q HERV-H, which may be important for recombination between the two chromosomes. Furthermore, the fact that 18q-82P carries a 4qA allele is consistent with recombination between the 4qA HERV-H and the 18q HERV-H to give rise to patient 18q-82C's translocation. Translocations in patients 18q-82C and 18q-146C both occurred in the same region of the HERV-H that is polymorphic between the 4q alleles (Figure 2), and the 122-bp duplication alters the sequence alignment between this part of chromosomes 4q and 18q.
It is important to point out that we only recognized the signatures of HERV-HERV NAHR at the translocation breakpoints of patients 18q-82C and 18q-146C after sequencing the breakpoint junctions. Inferring chromosomal rearrangement mechanisms from lower-resolution approaches (e.g., array CGH only) is likely to underestimate the frequency of NAHR-mediated events between shorter homologous segments and only detect NAHR between large blocks of segmental duplication. Furthermore, NAHR between homologous interspersed repeats, such as Alus, LINEs, and HERVs, would be overlooked by strategies focusing on recombination between segmental duplications [15]. We have previously detected a translocation that is the product of interchromosomal LINE-LINE recombination between L1PA2s on chromosomes 6 and 16 that are 96% identical over ~six kb by sequencing across the translocation breakpoint junction [6]. In addition, sequencing of normal copy number variation breakpoints has uncovered signatures of intrachromosomal NAHR [44,45]. Future sequencing-based studies of other chromosome rearrangements will likely capture more NAHR events between shorter homologous segments, which would give us a better understanding of the requirements for interchromosomal and intrachromosomal NAHR in the human genome.
Methods
Participants
We obtained informed consent from individuals with chromosome 18 abnormalities and their families. The human subjects protocol was approved by the Institutional Review Board of the University of Texas Health Science Center at San Antonio.
Array CGH
Using a 244K platform from Agilent Technologies (Santa Clara, CA), we designed a custom two-plex array covering the five-Mb regions spanning the previously described breakpoints on chromosomes 4q and 18q [39], with a mean probe spacing of one oligonucleotide per 100 basepairs (bp). Oligonucleotides were designed using Agilent's eArray program (https://earray.chem.agilent.com/earray/). To minimize non-unique oligonucleotides that would not be informative in array CGH, we performed a high definition (HD) probe search to prefer existing "catalog probes" and used the most stringent "similarity score filter" designed to select probes that hybridize to only one genomic location. The unique identifier (AMADID) for the array design is 021748; this design is available upon request.
Lymphoblastoid cell lines derived from 18q-82C, 18q-82P, and 18q-146C were established previously [39]. We extracted genomic DNA from cell lines using the Gentra Puregene DNA Extraction Kit (Qiagen, Valencia, CA). Subject DNA was co-hybridized with reference DNA from the GM15510 cell line (Coriell Cell Repositories, Camden, NJ). Arrays were scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA), and signal intensities were evaluated using Feature Extraction Version 9.5.1.1 software (Agilent Technologies, Santa Clara, CA). We used DNA Analytics Version 4.0 software (Agilent Technologies, Santa Clara, CA) to analyze the array data and call breakpoints.
Breakpoint PCR
Starting with breakpoints identified by array CGH, we designed PCR primers to amplify across the translocation breakpoint junctions in 18q-82C and 18q-146C. The chromosome 18-specific primer is 5'-TCAACTGTAGAAGAGGTCAGAGCTCCCCTA-3', and the chromosome 4-specific primer is 5'-GGTCAATGATCCGGAGGGTTCTGGATG-3'. We performed PCR using TaKaRa Ex Taq polymerase (Clontech Laboratories, Inc., Madison, WI) with 1× PCR buffer, 0.2 mM dNTP, 8 pmol of each primer, and 50-100 ng of DNA template. PCR conditions were as follows. 94°C for 1 min; 10 cycles at 94°C for 30 s, 66°C for 1 min, decreasing 0.5°C per cycle, and 72°C for 7 min; 20 cycles at 94°C for 30 s, 61°C for 1 min, and 72°C for 7 min; and a final extension at 72°C for 10 min. Bands were visualized via gel electrophoresis on a 1% agarose gel.
Nested PCR
We designed a nested PCR to specifically amplify the HERV-Hs on 4qA and 4qB. The first PCR amplifies a 6-kb region larger than the 4q HERV-Hs using the following primers: 5'-GATCATTTTGTCAATGAAATCTCACAAGAGGGC-3' and 5'-GGTCAATGATCCGGAGGGTTCTGGATG-3'. The PCR reagents were the same as described above, except for the addition of betaine (0.7 M final concentration in 50 μl PCR). Conditions for the first PCR were: 94°C for 1 min; 10 cycles at 94°C for 30 s, 65°C for 1 min, decreasing 0.5°C per cycle, and 72°C for 7.5 min; 30 cycles at 94°C for 30 s, 60°C for 1 min, and 72°C for 7.5 min; and a final extension at 72°C for 10 min.
We diluted amplicons from the first PCR 1:1000 to use as template in the second PCR. The second PCR amplifies a 326-bp or a 446-bp product from the 4qA and 4qB alleles, respectively, using the following primers: 5'-CACCTGCTTTGGTCCTTCAC-3' and 5'-ACTTTCCCCTCTCCCAGAAA-3'. Conditions for the second PCR were: 94°C for 1 min; 35 cycles at 94°C for 20 s, 55°C for 10 s, and 72°C for 10 s; and a final extension at 72°C for 1 min. Bands were visualized via gel electrophoresis on a 1% agarose gel.
Sequence analysis
We purified PCR products from agarose gels using the QIAquick gel extraction kit (Qiagen, Valencia, CA), and cloned them into a TOPO-TA vector following the manufacturer's protocol (Invitrogen, Carlsbad, CA). We transformed the ligated construct into SURE 2 Supercompetent Cells (Agilent Technologies, Cedar Creek, TX) following the manufacturer's protocol. We propagated plasmids in recombination-deficient SURE 2 Escherichia coli to prevent rearrangement of the cloned insert.
We purified plasmid DNA (Qiagen Miniprep kit, Valencia, CA) and submitted plasmids for Sanger sequencing (Beckman Coulter Genomics, Danvers, MA). DNA sequences were analyzed by comparing reads to the human genome reference assembly (NCBI36/hg18) using the BLAT tool [46] on the UCSC Genome Browser (http://genome.ucsc.edu/). Junction sequences from 18q-82C (18q82C_junction) and 18q-146C (18q146C_junction) have been deposited in GenBank. The breakpoint junctions on chromosome 4 correspond to positions chr4:183974152-183974212 (18q-146C) and chr4:183974285-183974380 (18q-82C) and the breakpoint junctions on chromosome 18 correspond to positions chr18:69144350-69144410 (18q-146C) and chr18:69144483-69144578 (18q-82C) of the NCBI36/hg18 build of the human genome.
We also cloned and sequenced the products of the first round of nested PCR from 82P to characterize the entire 6-kb HERV-H regions from the 4qA and 4qB alleles. These sequences have been deposited in GenBank as 82Pchr4HERVHA and 82Pchr4HERVHB. We aligned the intact HERV-H sequences from 4qA and 4qB to the HERV-H consensus as represented in RepeatMasker [40] and analyzed the 61-bp tandem repeat within the HERV-H using Tandem Repeats Finder (TRF), http://tandem.bu.edu/trf/trf.html[47].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KEH performed the array CGH, breakpoint sequencing, and genotyping experiments. KEH and MKR performed the sequence alignments and analyzed the data. MKR drafted the manuscritpt. US and JDC participated in the design of the study and the manuscript. All authors have read and approved the manuscript.
Supplementary Material
Gentoype results of the 4qA and 4qB alleles in the population.
Contributor Information
Karen E Hermetz, Email: khermet@emory.edu.
Urvashi Surti, Email: surtus@mail.magee.edu.
Jannine D Cody, Email: CODY@uthscsa.edu.
M Katharine Rudd, Email: katie.rudd@emory.edu.
Acknowledgements
The authors wish to thank families and members of the Chromosome 18 Registry for their participation in this study. This study was supported by a grant from the National Institute of Mental Health (1R01MH092902 to MKR). We thank Madhuri Hegde for comments on the manuscript and Cheryl Strauss for editorial assistance.
References
- Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, Ballif BC, Bejjani BA. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V. et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST. et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in medicine. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A. et al. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. [DOI] [PubMed] [Google Scholar]
- Luo Y, Hermetz KE, Jackson JM, Mulle JG, Dodd A, Tsuchiya KD, Ballif BC, Shaffer LG, Cody JD, Ledbetter DH. et al. Diverse mutational mechanisms cause pathogenic subtelomeric rearrangements. Hum Mol Genet. 2011;20:3769–3778. doi: 10.1093/hmg/ddr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC. et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Keene J, Bunke B, Kaminsky EB, Adam MP, Mulle JG, Ledbetter DH, Martin CL. Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Hum Mol Genet. 2009;18:2957–2962. doi: 10.1093/hmg/ddp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/S0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M. et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maarel SM, Deidda G, Lemmers RJ, van Overveld PG, van der Wielen M, Hewitt JE, Sandkuijl L, Bakker B, van Ommen GJ, Padberg GW, Frants RR. De novo facioscapulohumeral muscular dystrophy: frequent somatic mosaicism, sex-dependent phenotype, and the role of mitotic transchromosomal repeat interaction between chromosomes 4 and 10. Am J Hum Genet. 2000;66:26–35. doi: 10.1086/302730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Whitby H, Maxwell T, Aston E, Brothman AR, Carey JC. Co-occurrence of 4p16.3 deletions with both paternal and maternal duplications of 11p15: modification of the Wolf-Hirschhorn syndrome phenotype by genetic alterations predicted to result in either a Beckwith-Wiedemann or Russell-Silver phenotype. Am J Med Genet A. 2008;146A:2691–2697. doi: 10.1002/ajmg.a.32516. [DOI] [PubMed] [Google Scholar]
- Ou Z, Stankiewicz P, Xia Z, Breman AM, Dawson B, Wiszniewska J, Szafranski P, Cooper ML, Rao M, Shao L. et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21:33–46. doi: 10.1101/gr.111609.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- Sheridan MB, Kato T, Haldeman-Englert C, Jalali GR, Milunsky JM, Zou Y, Klaes R, Gimelli G, Gimelli S, Gemmill RM. et al. A palindrome-mediated recurrent translocation with 3:1 meiotic nondisjunction: the t(8;22)(q24.13;q11.21) Am J Hum Genet. 2010;87:209–218. doi: 10.1016/j.ajhg.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajecka M, Gentles AJ, Tsai A, Chitayat D, Mackay KL, Glotzbach CD, Lieber MR, Shaffer LG. Unexpected complexity at breakpoint junctions in phenotypically normal individuals and mechanisms involved in generating balanced translocations t(1;22)(p36;q13) Genome Res. 2008;18:1733–1742. doi: 10.1101/gr.077453.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, Donovan DJ, Eisenman R, Fan Y, Farra CG, Ferguson HL. et al. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82:712–722. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kalscheuer V, Tzschach A, Menzel C, Ullmann R, Schulz MH, Erdogan F, Li N, Kijas Z, Arkesteijn G. et al. Mapping translocation breakpoints by next-generation sequencing. Genome Res. 2008;18:1143–1149. doi: 10.1101/gr.076166.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Human mutation. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Rouyer F, Simmler MC, Page DC, Weissenbach J. A sex chromosome rearrangement in a human XX male caused by Alu-Alu recombination. Cell. 1987;51:417–425. doi: 10.1016/0092-8674(87)90637-4. [DOI] [PubMed] [Google Scholar]
- Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pousi B, Hautala T, Heikkinen J, Pajunen L, Kivirikko KI, Myllyla R. Alu-Alu recombination results in a duplication of seven exons in the lysyl hydroxylase gene in a patient with the type VI variant of Ehlers-Danlos syndrome. Am J Hum Genet. 1994;55:899–906. [PMC free article] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, Page DC. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- Blanco P, Shlumukova M, Sargent CA, Jobling MA, Affara N, Hurles ME. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J Med Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Valle A, Wang X, Potocki L, Xia Z, Kang SH, Carlin ME, Michel D, Williams P, Cabrera-Meza G, Brundage EK. et al. HERV-mediated genomic rearrangement of EYA1 in an individual with branchio-oto-renal syndrome. Am J Med Genet A. 2010;152A:2854–2860. doi: 10.1002/ajmg.a.33686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Paces J, Pavlicek A, Paces V. HERVd: database of human endogenous retroviruses. Nucleic Acids Res. 2002;30:205–206. doi: 10.1093/nar/30.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patience C, Wilkinson DA, Weiss RA. Our retroviral heritage. Trends Genet. 1997;13:116–120. doi: 10.1016/S0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/S0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat Genet. 2001;29:487–489. doi: 10.1038/ng775. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics. 2005;171:1183–1194. doi: 10.1534/genetics.105.043976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn SR, Mohammed M, Reveles XT, Viskochil DH, Palumbos JC, Johnson-Pais TL, Hale DE, Lancaster JL, Hardies LJ, Boespflug-Tanguy O. et al. Molecular characterization of a patient with central nervous system dysmyelination and cryptic unbalanced translocation between chromosomes 4q and 18q. Am J Med Genet A. 2003;120A:127–135. doi: 10.1002/ajmg.a.20026. [DOI] [PubMed] [Google Scholar]
- Horbinski C, Carter EM, Heard PL, Sathanoori M, Hu J, Vockley J, Gunn S, Hale DE, Surti U, Cody JD. Molecular and clinical characterization of a recurrent cryptic unbalanced t(4q;18q) resulting in an 18q deletion and 4q duplication. Am J Med Genet A. 2008. [DOI] [PMC free article] [PubMed]
- Hirose Y, Takamatsu M, Harada F. Presence of env genes in members of the RTVL-H family of human endogenous retrovirus-like elements. Virology. 1993;192:52–61. doi: 10.1006/viro.1993.1007. [DOI] [PubMed] [Google Scholar]
- Naylor JA, Buck D, Green P, Williamson H, Bentley D, Giannelli F. Investigation of the factor VIII intron 22 repeated region (int22h) and the associated inversion junctions. Hum Mol Genet. 1995;4:1217–1224. doi: 10.1093/hmg/4.7.1217. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR. The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet. 1997;6:1595–1603. doi: 10.1093/hmg/6.9.1595. [DOI] [PubMed] [Google Scholar]
- Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gentoype results of the 4qA and 4qB alleles in the population.