Abstract
We investigated the molecular interactions of a cell penetrating peptide (CPP) Pep-1 with model cell membranes using sum frequency generation (SFG) vibrational spectroscopy, supplemented by attenuated total reflectance - Fourier transform infrared spectroscopy (ATR-FTIR). Hydrogenated and deuterated 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG and dDPPG) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG), were used in the experiments to represent gel-phase and fluid-phase lipid bilayers, respectively. Our SFG results indicated that Pep-1 molecules adopted a β-sheet conformation when adsorbed to the surface of gel-phase DPPG lipid bilayers. When interacting with fluid-phase POPG lipid bilayers, Pep-1 adopted a mix of α-helical and β-sheet structures over a broad range of peptide concentrations. The orientation distribution of the α-helical Pep-1 segment associated with the fluid-phase bilayers was found to depend on the peptide concentration. SFG orientation analysis showed that Pep-1 molecules adopted an orientation nearly perpendicular to the plane of the bilayer for peptide concentrations of 0.28 μM and 1.4 μM. When the Pep-1 concentration was increased to 7.0 μM, combined SFG and ATR-FTIR measurements showed that Pep-1 molecules were associated with the bilayer with a broad orientation distribution. Our results demonstrated that lipid bilayer phase and peptide concentration affect the conformation and orientation of Pep-1 molecules associated with model cell membranes, which is crucial to the translocation process of CPPs. A combination of SFG and ATR-FTIR studies can be used to determine the conformation and orientation of CPPs interacting with model cell membranes in situ.
1. Introduction
The cell penetrating peptide (CPP) family has drawn increasing interest in the field of drug delivery because it is one of the most efficient tools for intracellular access.1–7 CPPs are usually short peptides with 11 to 34 amino acids. Being highly hydrophilic and cationic, they are able to translocate across the cell membranes carrying various types of cargos, such as peptides, proteins, plasmid DNAs, oligonucleotides and liposome nanoparticles.8–10 Two main mechanisms for cellular uptake of CPPs have been proposed. One is physically driven to directly interact with and penetrate through the cell membranes and the other is the endocytosis pathway.11 Although numerous studies have been carried out on the therapeutic effects of CPPs, the molecular-level interactions between cell membranes and CPPs remain largely unknown.12
Synthetic peptide carrier Pep-1 is one of the most widely studied peptides in the CPP family. Pep-1 is stable in physiological buffer with high delivery efficiency and low toxicity.13,14 While many other CPPs must be covalently bound to their cargo, Pep-1 can form non-covalent complexes with a broad spectrum of peptides, proteins, and nanoparticles.15 A Pep-1 molecule has three segments: a hydrophobic tryptophan-rich motif (KETWWETWWTEW), a spacer domain (SQP) and a hydrophilic lysine-rich domain (KKKRKV). Previous research using model membranes has shown that Pep-1 appears to directly penetrate through the cell membrane via a physically-driven rather than an endocytosis pathway.16 This peptide has a high affinity for both neutral and negatively charged cell membranes. NMR and CD experiments have shown that the membrane environment can induce the Pep-1 hydrophobic motif to form an α-helical structure.16 By measuring the orientation of Pep-1 in bilayers during the process of translocation, it is possible to understand the molecular mechanism of Pep-1/lipid interactions. However, to date inconsistent orientation distributions have been reported for Pep-1 from studies that use a variety of techniques and model systems.16, 17
Sum frequency generation (SFG) spectroscopy is an intrinsically surface-sensitive technique. It has been widely applied to investigate various types of biointerfaces including those where peptides are associated with model cell membranes.18–40 With the use of SFG, we can observe the process of peptide adsorption onto the lipid bilayer, monitor changes in the lipid bilayer when the peptide interacts, and obtain conformation and orientation information for peptides with a variety of different secondary structures.18,41 We have extensively investigated molecular interactions between model cell membranes and various antimicrobial peptides (AMPs) using SFG. The AMPs investigated include magainin 2,26 MSI-78,42 alamethicin,43 melittin44 and tachyplesin I.27 But to the best of our knowledge, no CPPs have yet been investigated by SFG. CPPs and AMPs are different classes of peptides. AMPs disrupt bacteria cell membranes via one of several modes of action (such as barrel stave, toroidal pore formation or a carpet model) above some threshold concentration, while CPPs usually enter the cell through a physically-driven or endocytosis pathway without disrupting the membranes. The concentration of CPPs required for translocation to occur is usually lower than that required for AMPs to disrupt the cell membranes and therefore a technique with high sensitivity to study CPPs is necessary. In fact, previous research has shown that when increasing the concentration of Pep-1 molecules, they will exhibit toxicity and behave similar to AMPs towards cell membranes.45 Also, it may be challenging to observe the translocation process of CPPs into cells using the simple model cell membranes (e.g., solid supported lipid bilayers) often used for SFG. For example, the endocytosis pathway may require non-lipid components (such as caveolars) to be present in the membrane.46 The transmembrane potential, which is believed to be a driving force for Pep-1 translocation also adds to the difficulty of the model system.45, 47
As a technique with a high sensitivity as well as the ability to obtain orientation information, SFG spectroscopy was applied to study cell penetrating peptide Pep-1 for the first time in this paper. The results revealed that SFG spectroscopy is sensitive enough to detect Pep-1 associated with lipid bilayers and can be used to deduce the orientation of Pep-1 at low concentrations suitable for the study of peptide translocation. The different behaviors of CPPs on gel-phase and liquid-phase lipid bilayers observed explain why the fluidity of the membrane plays an important role in CPP translocation. In addition, attenuated total reflectance - Fourier transform infrared spectroscopy (ATR-FTIR) was used as a supplemental technique to confirm the conclusions drawn from the SFG study. This study is the first step towards fully understanding how CPPs deliver cargo. Studies on the interactions of CPPs with more advanced cell model systems and the translocation process of CPPs with drugs into cells will be carried out in the future.
2. Experimental
Pep-1 (sequence H-KETWWETWWTEWSQPKKKRKV-OH) was purchased from Anaspec with >95% purity. Hydrogenated and deuterated 1,2-dipalmitoyl(d62)-sn-glycero-3-phosphoglycerol (DPPG and dDPPG) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) were purchased from Avanti Polar Lipids Inc (Alabaster, AL).
Lipid bilayers were deposited on CaF2 right angle prisms (Altos Photonics, Bozeman, MT). Langmuir-Blodgett and Langmuir-Schaefer (LB/LS) methods were used to deposit the proximal and then the distal leaflets of the lipid bilayers, respectively, as described in detail previously.26, 44 A KSV2000 LB system and ultrapure water from a Millipore system (Millipore, Bedford, MA) were used throughout the experiments for bilayer preparation. The bilayer was immersed in 50 μM pH=7.2 phosphate buffer inside of a 2 mL reservoir during the experiment. 80, 16 and 3.2 μL of 0.5 mg/mL Pep-1 was injected into the reservoir for concentration-dependent experiments. A magnetic microstirrer was used at a rate of 100 rpm to ensure a homogeneous concentration distribution of peptide molecules in the subphase below the bilayer. The final concentrations of the peptide solutions are 7.0 μM, 1.4 μM and 0.28 μM.
The details of SFG theory, our SFG setup and our experimental design have been described previously.26, 48–62 Spectra were collected from peptides associated with the lipid bilayers in ssp (s-SFG, s-visible, p-IR) and ppp polarization combinations using our previously reported near total reflection geometry.26,44 ATR-FTIR experiments were performed with a Nicolet Magna 550 FTIR spectrometer using a detachable ZnSe total internal reflection crystal (Specac Ltd. RI, U.K.).44 The substrate surface was cleaned with methanol, Contrex AP solution, and deionized water, followed by a treatment in a glow discharge plasma chamber for 2 min to remove residual hydrocarbon contamination. The lipid bilayer was deposited on the crystal surface with a procedure previously reported.44 The appropriate volume of a Pep-1 stock solution (in D2O phosphate buffer) was injected into the subphase of 1.6 mL to achieve the above mentioned concentrations. The s- and p-polarized ATR-FTIR spectra were recorded 1h, followed by a return to the s polarization to ensure that samples were equilibrated and did not change during the timescale of the experiments.
4. Results
4.1 SFG Results on Pep-1 Interacting with Gel-phase Lipid Bilayers
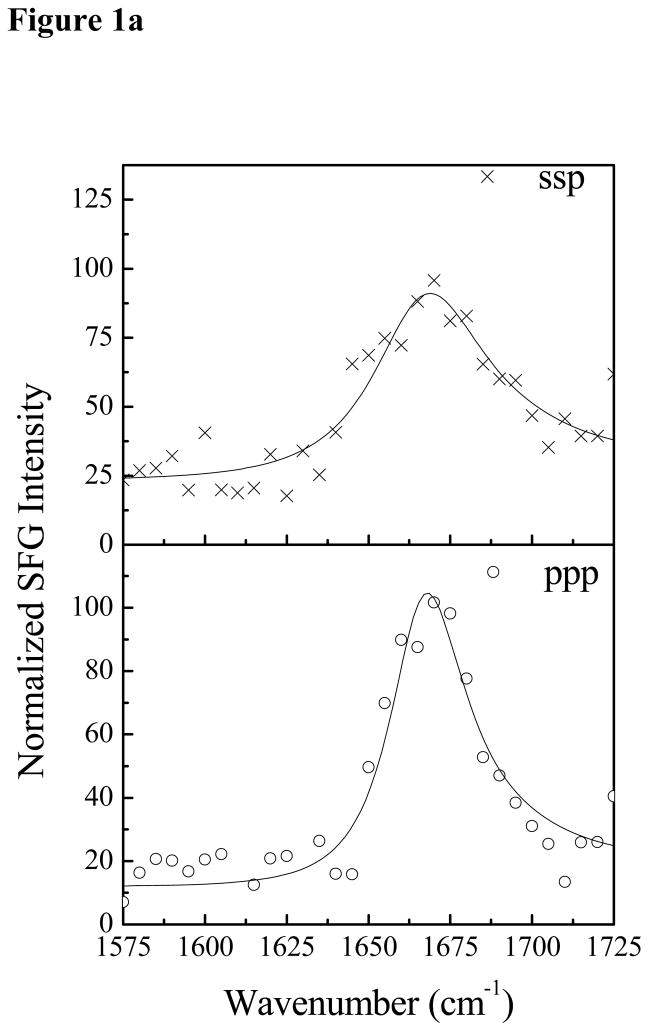
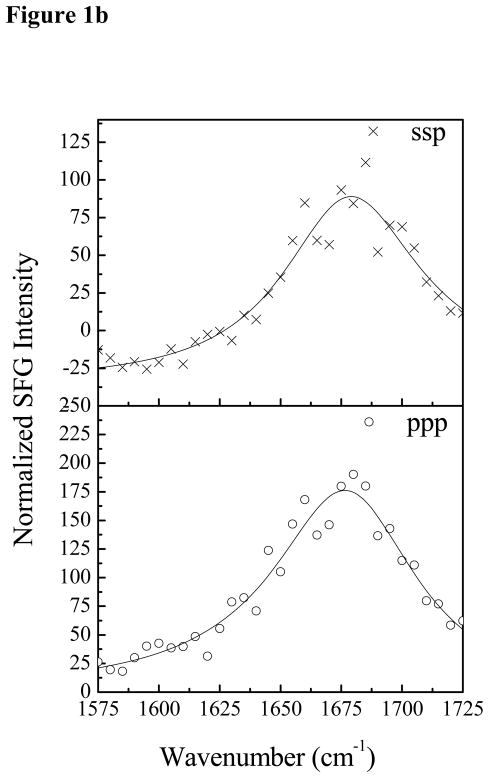
SFG spectra were collected with dDPPG/DPPG bilayers in contact with Pep-1 solutions with different peptide concentrations. At the low Pep-1 concentration of 0.28 μM, no SFG amide I signal from Pep-1 in the lipid bilayer was observed. When the Pep-1 concentration was increased to 1.4 μM, SFG amide I signals centered at 1677 cm−1 was detected from Pep-1 associated with the lipid bilayer, as shown in Fig. 1a. This peak center indicates that Pep-1 likely forms β-sheet type structures on the gel-phase membrane interfaces. The SFG amide signal is quite broad, showing a high degree of structural heterogeneity. This peak center shifted to 1663 cm−1 when the Pep-1 concentration was increased to 7.0 μM (Fig. 1b), which may indicate a change in secondary structure to β-turns and/or disordered structures.
Figure 1.
The ssp and ppp SFG amide I spectra of Pep-1 associated with a dDPPG/dDPPG bilayer at the peptide concentrations of 1.4 μM (a) and 7.0 μM (b).
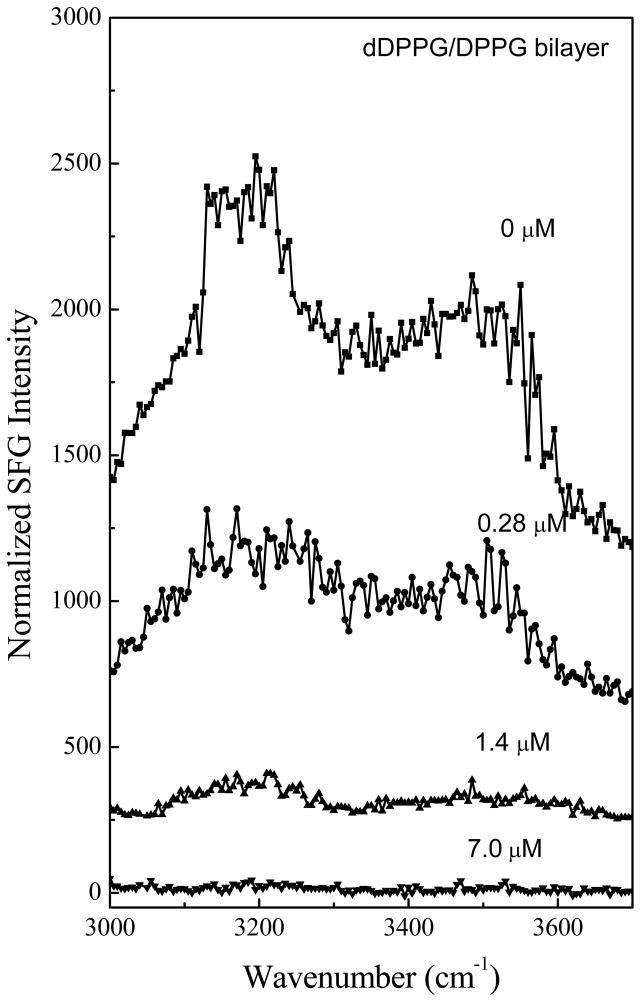
The adsorption and association of Pep-1 to the dDPPG/DPPG bilayer can also be confirmed by the SFG signals collected in the O-H stretching frequency region. Such SFG signals are contributed by ordered water molecules associated with the charged lipid head groups of the dDPPG/DPPG lipid bilayer. As shown in Fig. 2, at the low peptide concentration of 0.28 μM, the detected SFG signal from water decreased upon addition of the peptides, but the spectral feature did not differ substantially. The water SFG signals at 3200 cm−1 and 3500 cm−1 greatly decreased when the peptide concentration was increased to 1.4 μM and completely disappeared at the high concentration of 7.0 μM. We believe that the positively charged Pep-1 molecules interact with and neutralize the negatively charged head group of the dDPPG/DPPG bilayer, therefore disordering the water molecules originally associated with the bilayer. The SFG signals observed in the O-H stretching frequency region indicate that water molecules on the bilayer surface were removed and/or disordered by the adsorption of Pep-1 molecules. When combined with the amide I signals, these results confirm that Pep-1 molecules at a variety of concentrations interact with dDPPG/DPPG bilayers.
Figure 2.
The ssp SFG spectra in the O-H stretching frequency range detected when the dDPPG/dDPPG bilayer is in contact with Pep-1 solutions with different concentrations.
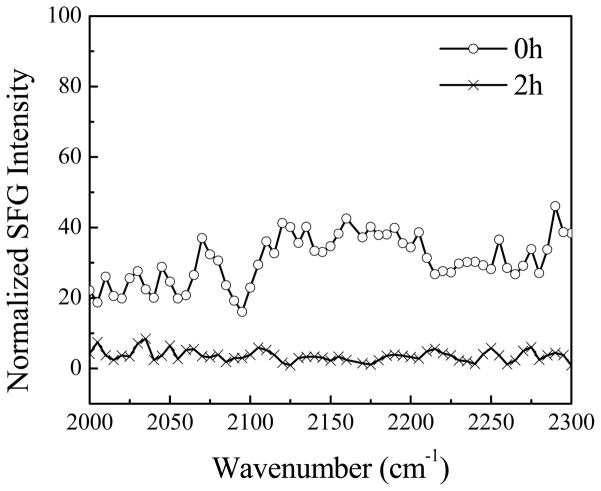
We also studied the behavior of the lipid bilayer when interacting with Pep-1. To avoid a potential overlap of signals from the peptide and the lipids in the C-H stretching frequency region, we also used a fully deuterated lipid bilayer (dDPPG/dDPPG). Amide I spectra from the peptide were found to be the same as when dDPPG/DPPG bilayers were used. No C-D stretching signal was observed from the lipids before the addition of Pep-1 to the subphase, showing that the dDPPG/dDPPG bilayer was symmetric (as expected). After the introduction of Pep-1 into the subphase, at the concentration of 7.0 μM, no SFG C-D stretching signal was detected (Fig. 3). This implies that Pep-1 binds to the lipid headgroups rather than inserting into the gel- phase lipid bilayers. This observation is different from the peptides that disrupt the gel-phase lipid bilayers we investigated previously.63
Figure 3.
The ssp SFG spectra in the C–D stretching frequency range detected before, and two hours after, the dDPPG/DPPG bilayer is in contact with the Pep-1 solution with a concentration of 7.0 μM.
4.2 SFG Results on Pep-1 Interacting with Liquid-phase Lipid Bilayers
Concentration-dependent Pep-1 experiments were also performed using liquid-phase lipid bilayers (POPG/POPG), and the results were compared to those from gel-phase bilayers. Unlike the asymmetric dDPPG/DPPG bilayer, the disruption of the POPG bilayer could not be monitored directly by SFG. This is because POPG bilayers are prone to rapid flip-flop, and this rapid exchange of lipids between leaflets prevents the use of deuterated lipids to create asymmetry. Thus in this section we mainly focus on the SFG signals generated from the peptides. It is found that the SFG spectra collected from Pep-1 in fluid phase bilayers were significantly different from the gel phase bilayer results.
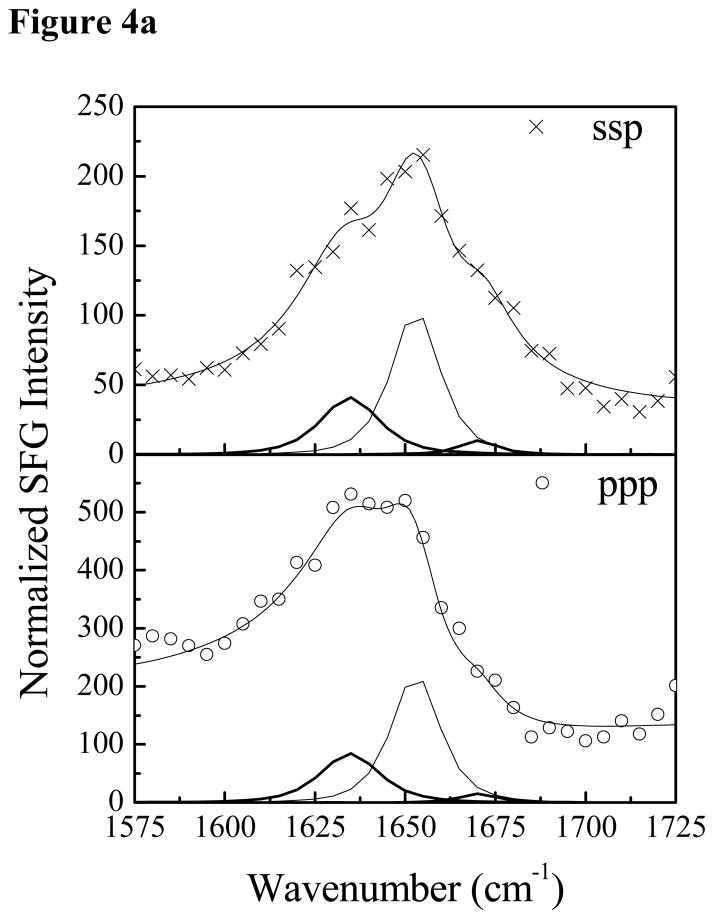
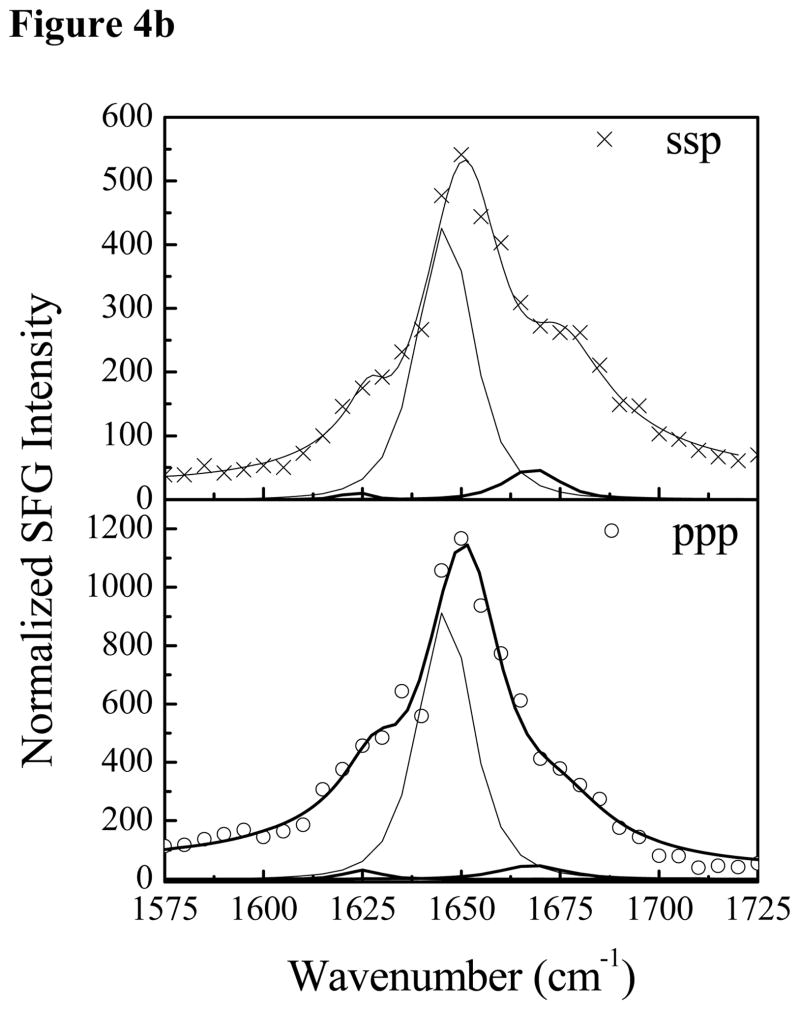
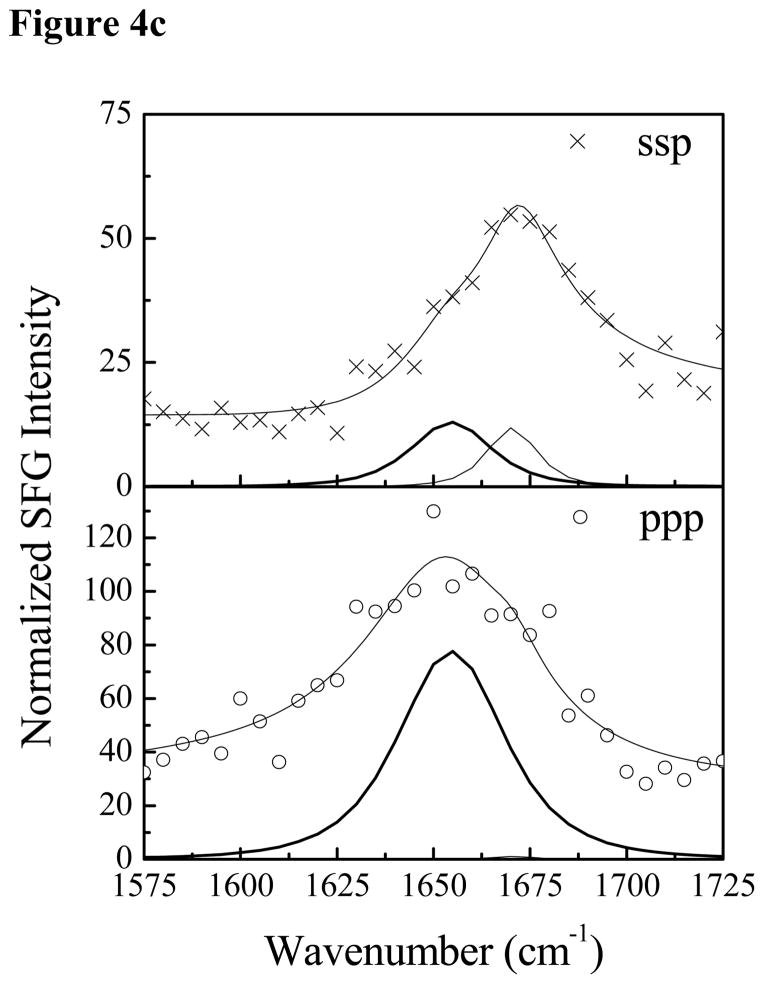
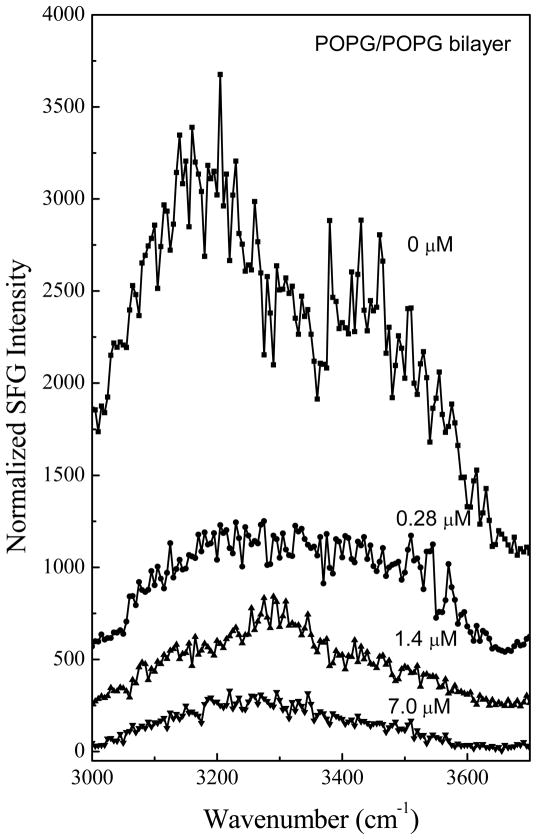
Fig. 4 shows the SFG amide I signals collected from Pep-1 interacting with a POPG/POPG bilayer with the same peptide concentrations as used previously. At the low concentration of 0.28 μM, a prominent peak at 1653 cm−1 was detected in both the ssp and ppp spectra, suggesting that some peptide molecules associated with the POPG/POPG bilayer adopted an α-helical structure. Additional peak shoulders at 1634 cm−1 and 1670 cm−1 indicate the coexistence of a β-sheet structure. At the intermediate concentration of 1.4 μM, in addition to the dominant 1651 cm−1 peak, shoulders at 1630 cm−1 and 1673 cm−1 were also detected. Therefore, at these two concentrations, Pep-1 adopts a mix of α-helical and β-sheet structures when associated with the POPG/POPG bilayer. SFG spectra were also collected from Pep-1 molecules associated with the POPG/POPG bilayer when the peptide concentration was increased to 7.0 μM. Interestingly, for a high solution concentration of Pep-1, the observed peak intensities in the SFG spectra were much weaker than signals detected at lower peptide concentrations. It is well known that the SFG intensity is affected by molecular ordering/orientation as well as the number of molecules, and a drop in signal as peptide concentration increases suggests that the Pep-1 molecules were either lying down on the surface or else adopted a more random orientation distribution.
Figure 4.
The ssp and ppp SFG amide I spectra of Pep-1 associated with a POPG/POPG bilayer at the peptide concentrations of 0.28 μM (a), 1.4 μM (b) and 7.0 μM (c).
The orientation information could be further quantified with the methodology our group has recently developed.26, 65 We want to emphasize here that the 1653 cm−1 peak is solely due to the α-helical structure based on the following reasons. (a) Due to the lack of a high-resolution three-dimensional structure, we were unable to calculate the SFG signal contributed by the random coil section of Pep-1. However, in other cases such as cytochrome-b5,71 we found that the random coil part of the peptide would contribute less than 2% of the SFG signal generated from the α-helical components, even if they could have a somewhat ordered structure. This means that unlike linear vibrational spectroscopic techniques (e.g., FTIR), SFG is much more sensitive to α-helices than random components. (b) It was reported in ref. 64 that the structure of Pep-1 calculated by DYANA has free C and free N termini (referred to as PepW in the paper). The random coil parts in this structure are very dynamic and have no preferred ordering. Therefore even if the random coil part contributes a small SFG signal from one peptide molecule, those signals would be averaged out as an ensemble. (c) The width for the SFG α-helical peak (e.g., at 1.4 μM peptide concentration) is 12 cm−1 with a peak center of 1653cm−1. These parameters are similar to those of the purely α-helical peptide magainin-2.26 If random coil signals contributed to the overall lineshape, we would expect to see a larger peak width and a lower peak center frequency.
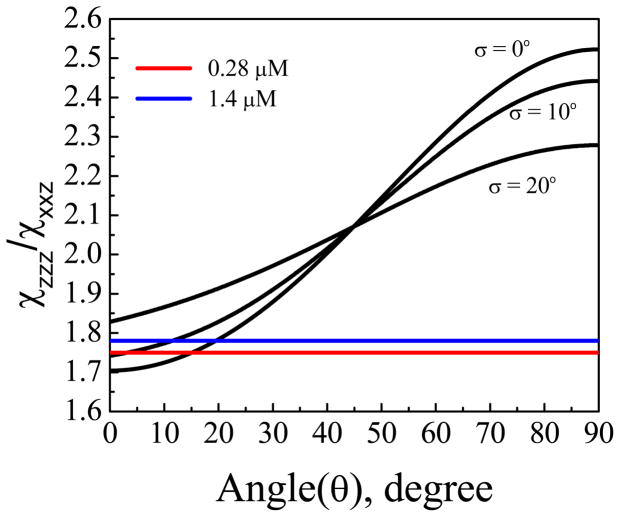
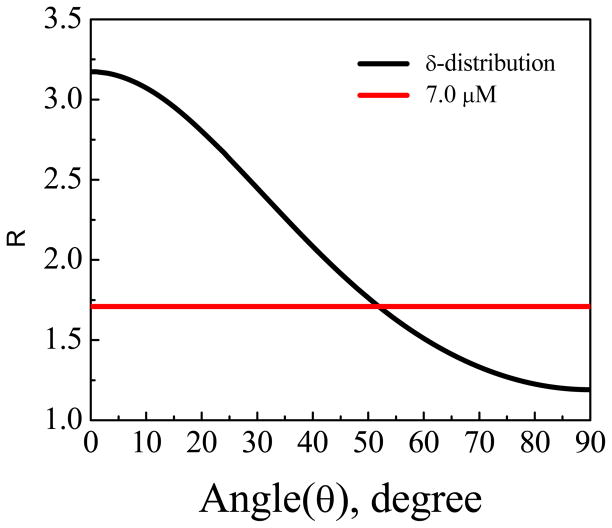
To relate the expected signal intensities to molecular orientation, we first generated a theoretical curve by plotting the ratio between the two susceptibility tensor elements for the α-helix peak as a function of the tilt angle of α-helical component (residues 4–13)16, 64 of the Pep-1 molecules associated with the lipid bilayer.65 Here we define the molecular c axis as along the α-helical backbone and the tilt angle θ is the angle between the backbone and the surface normal of the bilayer. Then we calculate the experimental value of from the fitted signal strength ratio , which includes a correction for the Fresnel coefficients. Lastly, we find the corresponding orientation information of the experimental from the generated theoretical curve. If we assume that the molecules adopt a Gaussian orientation distribution, the relationship between the ratio and the tilt angle θ of the α-helix relative to the bilayer normal is plotted in Fig. 5, for various distribution widths. For Pep-1 in fluid-phase lipid bilayers, the ratio was found to depend on peptide concentration: 1.81±0.03 for Pep-1 in the lipid bilayer at the intermediate peptide concentration of 1.4 μM, and 1.75 ± 0.07 at the low concentration of 0.28 μM. The larger error bar in the measurement from the low peptide concentration is because the lower SFG signal induces weaker signal to noise ratio. The deduced tilt angles (relative to the membrane normal) for the two peptide concentrations are ~15° and ~18°, respectively, if we assume that all molecules adopt the same orientation (σ is 0°, a δ-distribution). If the distribution width is assumed to be 10 degrees, the tilt angles for the two concentrations are ~3° and ~12°, respectively. Based on the experimental data and the curves in Figure 5, a Gaussian distribution of 20° or greater would be unlikely. This implies that for both concentrations, the helical components in the Pep-1 molecules in the POPG/POPG orient perpendicular to the membrane surface with a narrow distribution. If we assume that the molecules at both concentrations adopt a δ-distribution, we can further deduce from the fitted SFG signal strengths that the ratio of the number of Pep-1 molecules in lipid bilayers for 0.28 μM and 1.4 μM cases is about 1:2 (although the ratio of the numbers of peptide molecules in the bulk solutions is 1:5). At higher concentration 7.0 μM, although the overall spectral lineshape was reproducible, the reduced signal intensity hindered efforts to reliably determine molecular orientation.
Figure 5.
Dependence of the measured SFG ratio of a 10-residue α-helix on the helix tilt angle relative to the surface normal.
Helices interacting with lipid membranes sometimes unravel in the end. In a previous publication, we reported the calculated SFG responses of alpha helices with different numbers of amino acids65 (11, 10 and 9 residues), but these curves are quite similar especially in the region of interest. Therefore a slight unraveling would not affect the conclusion that the peptides mainly adopt a perpendicular orientation.
The SFG spectra collected in the O-H stretching frequency region show that the O-H stretching signals decreased after the Pep-1 molecules were introduced to the subphase of the fluid-phase bilayer (Fig. 6). This is similar to what was observed when a gel-phase dDPPG/DPPG bilayer was used. However, for the fluid phase bilayer, the drop in signal intensity is accompanied by a change in the overall lineshape the O-H stretching frequency region. We believe that this change is due to the SFG signal generated from the Pep-1 N-H stretching mode at 3300 cm−1. SFG N-H signals have been used to study interfacial peptides previously.38,70,72 No peak at 3300 cm−1 was seen for Pep-1 in gel-phase lipid bilayers, suggesting that lipid bilayer phase has an effect on Pep-1-lipid bilayer interactions, and Pep-1 interacts with the gel-phase and liquid-phase bilayers differently. For the intermediate Pep-1 concentration of 1.4 μM, the 3300 cm−1 peak became more distinct due to the further decrease in the water O-H stretching signal. For the higher peptide concentration of 7.0 μM, the signal in the O-H stretching region decreased further, indicating the bilayer associated water molecules were even more disordered. However, no N-H stretching signals were observed at this higher concentration, suggesting that the Pep-1 molecules interact with the lipid bilayer differently at the high peptide concentration compared to that at the intermediate concentration. Again, this agrees with the results obtained from studies on the amide I frequency region.
Figure 6.
The ssp SFG spectra in the O-H stretching frequency range detected when the POPG/POPG bilayer is in contact with Pep-1 solutions with different concentrations.
4.3 ATR-FTIR Results on Pep-1 Interacting with Liquid-phase Lipid Bilayers
We also performed ATR-FTIR experiments to supplement our SFG studies on Pep-1 interacting with lipid bilayers. Whereas SFG is more sensitive to α-helices than β-sheets or random coils due to the dependence of SFG signals on molecular ordering, ATR-FTIR can readily detect amide I signals from many different secondary structures, but it is not able to detect very low concentrations of peptides.
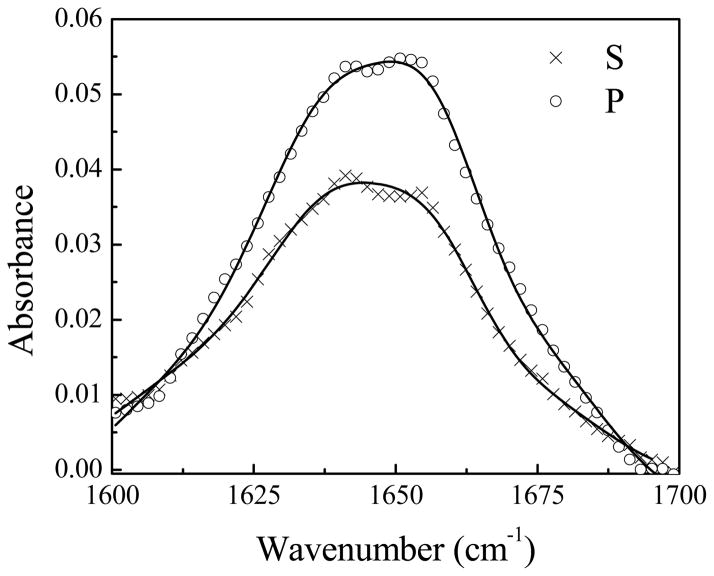
No discernible ATR-FTIR signal was detected from Pep-1 associated with the POPG/POPG bilayer at low (0.28 μM) and intermediate (1.4 μM) peptide concentrations. At the high peptide concentration of 7.0 μM, ATR-FTIR signals were observed (Fig. 7). The fitting results for the ATR-FTIR spectra collected using the s- and p-polarized light are shown in Table 1. From the signal strength ratio of the s and p polarized spectra, the tilt angle of the α-helical component with respect to the membrane normal was determined to be 52°, assuming a δ orientation distribution (Fig. 8). However, as we discussed in Section 4.2, very weak SFG signals were detected from 7.0 μM Pep-1 in the POPG/POPG lipid bilayer, suggesting that the δ-distribution is not a good assumption here. In fact, this orientation angle deduced by polarized ATR-FTIR is close to the “magic” angle (54.7°) that would be predicted for a random orientation of molecules. From the combination of SFG and ATR-FTIR we believe that the helical sections of Pep-1 molecules adopt a random orientation distribution.
Figure 7.
Polarized ATR-FTIR amide I spectra of Pep-1 in a POPG/POPG bilayer in contact with the peptide solution with a concentration of 7.0 μM.
Table 1.
Fitting parameters for s and p polarized ATR-FTIR spectra.
| Frequency | Assignment | Peak Width | A (S polarization) | A (P polarization) |
|---|---|---|---|---|
| 1673 | Turn and β-sheet | 9.72 | 0.007 | 0.014 |
| 1656 | α-helix | 6.46 | 0.0205 | 0.0308 |
| 1640 | Random coil | 6.97 | 0.0229 | 0.033 |
| 1628 | Intermolecular β-sheet | 6.87 | 0.0065 | 0.0091 |
| 1613 | Side chains | 10.59 | 0.012 | 0.020 |
Figure 8.
Dependence of the ATR-FTIR measured p to s spectral intensity ratio of an α-helix on the helix tilt angle vs. the surface normal.
5. Further Discussion and Conclusion
Previous studies using surface plasmon resonance (SPR) showed that Pep-1 - lipid interactions are modulated by membrane fluidity.66 When the fluidity increases, more Pep-1 molecules bind and insert into the membrane. This is in agreement with our results from SFG and ATR-FTIR, but our results provide a way to reveal molecular level information about Pep-1 conformation and orientation while interacting with lipid bilayers.
While ATR-FTIR has been proven to be a powerful tool to study the conformations of peptides associated with membrane lipids, previous work focused on samples that were semi-dehydrated. As a result, Pep-1 molecules not inserted into the membrane could precipitate as aggregates and contribute to the ATR-FTIR spectrum.17 By contrast, SFG is uniquely sensitive to interfaces, and therefore can selectively monitor the structures of peptides associated with the lipid bilayer without contributions from peptide molecules in the bulk environment (e.g., in solution or as aggregates). Also, compared to ATR-FTIR, SFG is more sensitive. Whereas no ATR-FTIR signals were observed from Pep-1 in a POPG/POPG bilayer at peptide concentrations of 0.28 and 1.4 μM, SFG signals were detected at those concentrations. By using different polarization combinations of the laser beams, we are able to deduce the orientation of the α-helical component of Pep-1. Our results clearly show that Pep-1 can interact with both gel-phase (DPPG) and fluid-phase (POPG) lipid bilayers, as indicated by a decrease in the water O-H stretching signal from water molecules at the lipid-water interface. In gel-phase lipid bilayers, Pep-1 generated very weak signals centered around 1670 cm−1 in the amide I frequency range, suggesting a random coil or β-sheet conformation. By contrast, for fluid-phase bilayers at low and intermediate Pep-1 concentrations, a strong peak around 1653 cm−1 could be detected from, indicating an α-helical conformation.
5.1 Orientation Information
Elucidating the orientation of the α-helical component in Pep-1 is essential to understand the process of membrane translocation. Previous studies on this process have produced various results. For example, based on fluorescence results, Heitz et al. first proposed that translocation involves the construction of a transient transmembrane pore-like structure.16 They concluded that the tryptophan residues in the α-helix are embedded in a hydrophobic environment, which would be consistent with Pep-1/ membrane interactions that place the helical axis perpendicular to the membrane plane. Further support for this claim came from electrophysiological measurements.67 However, spin-label studies by Weller and coworkers revealed a three-amino acid periodicity in signal attenuation, leading them to conclude that CPP lies parallel with the surface of DPC/SDS micelles.64 ATR-FTIR has also been applied to measure the in situ orientation of Pep-1 with respect to the membrane normal in various types of lipids, and without the need for exogenous labels.17 In POPC and mixed POPC/Cholesterol multilayers, the angles were measured to be 46.5° and 44.5°, respectively. These values are not very different from the average orientation angle expected for randomly oriented peptides. It was suggested from such measurements that the cyto-toxicity of Pep-1 is due to a “carpet-like” mechanism. Their study17 used high concentrations of Pep-1. Translocation activity at lower peptide concentrations was not studied, possibly due to the limited sensitivity of ATR-FTIR spectroscopy.
The combined SFG and ATR-FTIR studies reported in this paper showed that the behavior of Pep-1 associated with POPG/POPG bilayers is concentration-dependent (schematic in Fig. 9). At the highest concentration of 7.0 μM, results lead to a random orientation of Pep-1 helical component, in agreement with the previous ATR-FTIR studies.17 At the low and intermediate peptide concentrations, SFG results showed that the Pep-1 helical component is more or less perpendicular to the lipid bilayer surface, indicating that Pep-1 inserts into the membrane in this concentration range.
Figure 9.
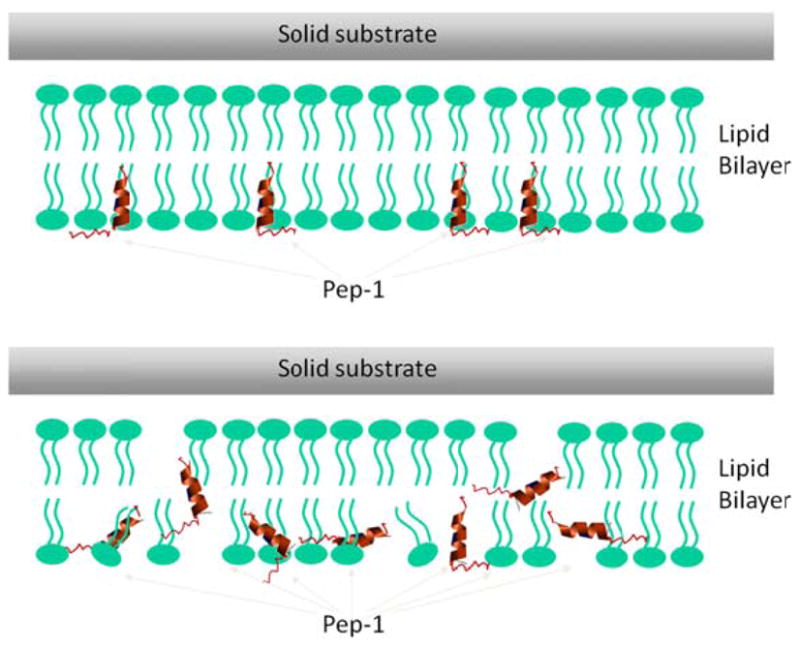
Schematics showing the interactions between lipid bilayers and Pep-1 with low or intermediate (a) and high (b) peptide concentrations.
5.2 The effect of lipid bilayer phase
SFG results show that on gel-phase lipid bilayers, Pep-1 molecules are loosely adsorbed on the surface with random or β-sheet type structures. On fluid-phase lipid bilayers, new peaks around 1653 cm−1 indicate the existence of α-helices. Previous CD studies have shown that in the range between 0.1 and 0.3 mg/mL, Pep-1 molecules in aqueous solution are poorly ordered,16 but that range is three orders of magnitude larger than the concentrations studied in our experiments. Interestingly, here even at the low concentration of 0.28 μM, β-sheet type structures were detected on the POPG/POPG lipid bilayers. This implies that these β-sheet type structures are not a consequence of peptide aggregation, but rather perhaps an intermediate species in the absorption process. It is the high fluidity of the lipid bilayer rather than the headgroups that will induce the formation and insertion of α-helices. Our results demonstrate that with SFG, it is possible to examine the translocation process from a molecular level.
5.3 Difference between CPPs and AMPs
CPPs and AMPs are both membrane-active peptides. The two classes of peptides share several characteristics such as charge, amphipathicity, helicity and length. CPPs have the capability to translocate biological membranes in a non-disruptive way, whereas AMPs can induce membrane permeabilization. The molecular mechanisms that underlie these differences in membrane interactions remain unclear.68, 69 Our SFG studies on both types of peptides can shed light on how they differ in their interactions with membranes.
MSI-78 is a synthetic analog of magainin 2 with high antimicrobial activity. SFG results42 showed that at the low concentration of 400 nM, MSI-78 molecules lie down on the surface of the negatively-charged gel-phase DPPG bilayer with ~70° deviation from the membrane surface normal. When the concentration is increased to 600 nM, MSI-78 inserts into the membrane with a ~25° tilt from the lipid bilayer. Multiple orientations were observed for an even higher peptide concentration, possibly indicating a toroidal-pore mechanism. This is an interesting contrast to studies on Pep-1, for which even at a low concentrations of 280 nM, the molecules were observed to insert into the fluid-phase POPG bilayer with α-helical structure. In gel-phase DPPG bilayers, Pep-1 does not form α-helices. This different performance from MSI-78 suggested that formation of α-helical structure in Pep-1 is induced by the fluid lipid chains in the hydrophobic interior of the bilayer rather than by interactions with the charged PG lipid headgroups. This agrees with the claim from previous research that CPPs are generally less amphipathic than AMPs.12
Alamethicin is an antibiotic peptide that can form voltage-gated ion channels in membranes. It interacts with cell membranes through the barrel-stave mode. SFG results43 indicated that alamethicin molecules lie down on gel-phase bilayers but adopt a mixed α-helical and 310-helical structure in the fluid-phase bilayers. The α-helical component at the N-terminus tilts ~63° while 310-helical component at the C-terminus tilts ~43° versus the surface normal. Similar to Pep-1, alamethicin selectively forms α-helices in fluid-phase lipid bilayers, but not gel-phase lipid bilayers. However, there are also key differences. For Pep-1, signals from α-helical and β-sheet structures were observed simultaneously, although the magnitude of the β-sheet signals decreased as we increased the concentration of Pep-1. This shows that there is an equilibrium from the β-sheet type to the α-helical component in the Pep-1-membrane interface. However for alamethicin, such coexistence has not been observed.
In this study we expand the application of SFG studies on AMPs to CPPs. This work is our first step towards understanding the molecular interactions between CPPs and cell membranes. The effect of a membrane potential on CPP translocation will be investigated using SFG in the future. In addition, molecular interactions between cell membranes and CPPs with various molecular cargos (including small molecules, nanoparticles, proteins and DNAs) and lipid bilayers will be examined.
Acknowledgments
This work is supported by the National Institute of Health (1R01GM081655). B. D. acknowledges the Rackham International Student Fellowship from the University of Michigan. We thank Dr. Pei Yang for technical support and insightful discussion.
References
- 1.Fernández-Carneado J, Kogan MJ, Pujals S, Giralt E. Biopolymers. 2004;76:196–203. doi: 10.1002/bip.10585. [DOI] [PubMed] [Google Scholar]
- 2.Magzoub M, Gräslund A. Quart Rev Biophys. 2004;37:147–195. doi: 10.1017/s0033583505004014. [DOI] [PubMed] [Google Scholar]
- 3.Patel LN, Zaro JL, Shen W-C. Pharmaceut Res. 2007;24:1977–1992. doi: 10.1007/s11095-007-9303-7. [DOI] [PubMed] [Google Scholar]
- 4.Vivès E, Schmidt J, Pèlegrin A. Biochim Biophys Acta. 2008;1786:126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Heitz F, Morris MC, Divita G. Brit J Pharmacol. 2009:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fonseca SB, Pereira MP, Kelley SO. Adv Drug Deliver Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Chugh A, Eudes F, Shim Y-S. IUBMB Life. 2010;62:183–193. doi: 10.1002/iub.297. [DOI] [PubMed] [Google Scholar]
- 8.Moore MJ, Rosbash M. Adv Sci. 2001;294:1841–1842. doi: 10.1126/science.1067676. [DOI] [PubMed] [Google Scholar]
- 9.Kumar P, Wu H, McBride JL, Jung K-E, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 10.Muñoz-Morris MA, Heitz F, Divita G, Morris MC. Biochem Biophys Res Commun. 2007;355:877–882. doi: 10.1016/j.bbrc.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 11.Räägel H, Säälik P, Pooga M. Biochim Biophys Acta. 2010;1798:2240–2248. doi: 10.1016/j.bbamem.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Herce HD, Garcia AE. J Biol Chem. 2007;33:345–356. doi: 10.1007/s10867-008-9074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris MC, Depollier J, Mery J, Heitz F, Divita G. Nature Biotech. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 14.Deshayes S, Plénat T, Charnet P, Divita G, Molle G, Heitz F. Biochim Biophys Acta. 2006;1758:1846–1851. doi: 10.1016/j.bbamem.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Gros E, Deshayes S, Morris MC, Aldrian-Herrada G, Depollier J, Heitz F, Divita G. Biochim Biophys Acta. 2006;1758:384–393. doi: 10.1016/j.bbamem.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Deshayes S, Heitz A, Morris MC, Charnet P, Divita G, Heitz F. Biochemistry. 2004;43:1449–1457. doi: 10.1021/bi035682s. [DOI] [PubMed] [Google Scholar]
- 17.Henriques ST, Quintas A, Bagatolli LA, Homblé F, MARB Mol Membr Biol. 2007;24:282–293. doi: 10.1080/09687860601142936. [DOI] [PubMed] [Google Scholar]
- 18.Ye S, Nguyen KT, Le Clair SV, Chen Z. J Struct Biol. 2009;168:61–77. doi: 10.1016/j.jsb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Buck SM, Even MA, Chen Z. J Am Chem Soc. 2002;124:13302–13305. doi: 10.1021/ja026881m. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Clarke ML, Zhang Y, Chen X, Chen Z. Langmuir. 2003;19:7862–7866. [Google Scholar]
- 21.Wang J, Even Ma, Chen X, Schmaier AH, Waite JH, Chen Z. J Am Chem Soc. 2003;125:9914–9915. doi: 10.1021/ja036373s. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Wang J, Sniadecki JJ, Even MA, Chen Z. Langmuir. 2005:2662–2664. doi: 10.1021/la050048w. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Clarke M, Chen LX, Even M, Johnson AW, Chen CZ. Surf Sci. 2005;587:1–11. [Google Scholar]
- 24.Wang J, Paszti Z, Clarke ML, Chen X, Chen Z. J Phys Chem B. 2007;111:6088–6095. doi: 10.1021/jp070383o. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Lee S-H, Chen Z. J Phys Chem B. 2008;112:2281–2290. doi: 10.1021/jp077556u. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen KT, Le Clair SV, Ye S, Chen Z. J Phys Chem B. 2009;113:12358–12363. doi: 10.1021/jp904154w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen KT, King JT, Chen Z. J Phys Chem B. 2010;114:8291–8300. doi: 10.1021/jp102343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boughton AP, Andricioaei I, Chen Z. Langmuir. 2010;26:16031–16036. doi: 10.1021/la1024394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Ward R, Tian Y, Malizia F, Gracias DH, Shen YR, Somorjai GA. J Biomed Mater Res. 2002;62:254–264. doi: 10.1002/jbm.10075. [DOI] [PubMed] [Google Scholar]
- 30.Mermut O, Phillips DC, York RL, McCrea K, Ward RRS, Somorjai GA. J Am Chem Soc. 2006;128:3598–3607. doi: 10.1021/ja056031h. [DOI] [PubMed] [Google Scholar]
- 31.Phillips DC, York RL, Mermut O, McCrea KR, Ward RS, Somorjai GA. J Phys Chem C. 2007;111:255–261. [Google Scholar]
- 32.York RL, Browne WK, Geissler PL, Somorjai GA. Isr J Chem. 2007;47:51–58. [Google Scholar]
- 33.Weidner T, Apte JS, Gamble LJ, Castner DG. Langmuir. 2010;26:3433–3440. doi: 10.1021/la903267x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baugh L, Weidner T, Baio JE, Nguyen PC, Gamble LJ, Stayton PS, Castner DG. Langmuir. 2010;26:16434–16441. doi: 10.1021/la1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu L, Ma G, Yan EC. J Am Chem Soc. 2010;132:5405–5412. doi: 10.1021/ja909546b. [DOI] [PubMed] [Google Scholar]
- 36.Anglin TC, Liu J, Conboy JC. Biophys J. 2007;92:L01–L03. doi: 10.1529/biophysj.106.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anglin TC, Brown KL, Conboy JC. J Struct Biol. 2009;168:37–52. doi: 10.1016/j.jsb.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung S-Y, Lim S-M, Albertorio F, Kim G, Gurau MC, Yang RD, Holden MA, Cremer PS. J Am Chem Soc. 2003;125:12782–12786. doi: 10.1021/ja037263o. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Sagle LB, Cremer PS. J Am Chem Soc. 2007;129:15104–15105. doi: 10.1021/ja075034m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall SA, Jena KC, Trudeau TG, Hore DK. J Phys Chem C. 2011;113:15364–15372. [Google Scholar]
- 41.Chen X, Chen Z. Biochim Biophys Acta. 2006;1758:1257–1273. doi: 10.1016/j.bbamem.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Yang P, Ramamoorthy A, Chen Z. Langmuir. 2011;27:7760–7767. doi: 10.1021/la201048t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye S, Nguyen KT, Chen Z. J Phys Chem B. 2010;114:3334–3340. doi: 10.1021/jp911174d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Wang J, Boughton AP, Kristalyn CB, Chen Z. J Am Chem Soc. 2007;129:1420–1427. doi: 10.1021/ja067446l. [DOI] [PubMed] [Google Scholar]
- 45.Henriques T, Castanho MARB. J Pept Sci. 2008;1:482–487. doi: 10.1002/psc.1003. [DOI] [PubMed] [Google Scholar]
- 46.Fittipaldi A, Ferrari A, Zoppé M, Arcangeli C, Pellegrini V, Beltram F, Giacca M. J Biol Chem. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 47.Henriques ST, Castanho MARB. Biochemistry. 2004;43:9716–9724. doi: 10.1021/bi036325k. [DOI] [PubMed] [Google Scholar]
- 48.Shen Y. Nature. 1989;337:519–525. [Google Scholar]
- 49.Zhuang X, Miranda PB, Kim D, Shen YR. Phys Rev B. 1999;59:12632–12640. [Google Scholar]
- 50.Chen Z, Shen YR, Somorjai GA. Ann Rev Phys Chem. 2002;53:437–465. doi: 10.1146/annurev.physchem.53.091801.115126. [DOI] [PubMed] [Google Scholar]
- 51.Eisenthal KB. Chem Rev. 1996;96:1343–1360. doi: 10.1021/cr9502211. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z. Prog Polym Sci. 2010;35:1376–1402. doi: 10.1016/j.progpolymsci.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Z. Polym Inter. 2007;56:577–587. [Google Scholar]
- 54.Li G, Ye S, Morita S, Nishida T, Osawa M. J Am Chem Soc. 2004;126:12198–12199. doi: 10.1021/ja046183x. [DOI] [PubMed] [Google Scholar]
- 55.Voges AB, Al-Abadleh HA, Musorrariti MJ, Bertin PA, Nguyen ST, Geiger FM. J Phys Chem B. 2004;108:18675–18682. [Google Scholar]
- 56.Li QF, Hua R, Chea IJ, Chou KC. J Phys Chem B. 2008;112:694–697. doi: 10.1021/jp072147j. [DOI] [PubMed] [Google Scholar]
- 57.Ye HK, Gu ZY, Gracias DH. Langmuir. 2006;22:1863–1868. doi: 10.1021/la052030r. [DOI] [PubMed] [Google Scholar]
- 58.Yatawara AK, Tiruchinapally G, Bordenyuk AN, Andreana PR, Benderskii AV. Langmuir. 2009;25:1901–1904. doi: 10.1021/la803113h. [DOI] [PubMed] [Google Scholar]
- 59.Moad AJ, Simpson GJ. J Phys Chem. 2004;108:3548–3562. [Google Scholar]
- 60.Moad AJ, Moad CW, Perry JM, Wampler RD, Goeken GS, Begue NJ, Shen T, Heiland R, Simpson GJ. J Comp Chem. 2007;28:1996–2002. doi: 10.1002/jcc.20706. [DOI] [PubMed] [Google Scholar]
- 61.Tong YJ, Li N, Liu HJ, Ge AL, Osawa M, Ye S. Angew Chem Int Ed. 2010;49:2319–2323. doi: 10.1002/anie.200904950. [DOI] [PubMed] [Google Scholar]
- 62.Paszti Z, Guczi L. Vib Spectro. 2009;50:48–56. [Google Scholar]
- 63.Chen X, Wang J, Kristalyn CB, Chen Z. Biophys J. 2007;93:866–875. doi: 10.1529/biophysj.106.099739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weller K, Lauber S, Lerch M, Renaud a, Merkle HP, Zerbe O. Biochemistry. 2005;44:15799–15811. doi: 10.1021/bi051535d. [DOI] [PubMed] [Google Scholar]
- 65.Nguyen KT, Le Clair SV, Ye S, Chen Z. J Phys Chem B. 2009;113:12169–12180. doi: 10.1021/jp904153z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henriques ST, Castanho MARB, Pattenden LK, Aguilar M-I. J Pept Sci. 2010;94:314–322. doi: 10.1002/bip.21367. [DOI] [PubMed] [Google Scholar]
- 67.Deshayes S, Morris MC, Divita G, Heitz F. Biochim Biophys Acta. 2006;1758:328–335. doi: 10.1016/j.bbamem.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 68.Henriques ST, Melo MN, Castanho MARB. Biochem J. 2006;399:1–7. doi: 10.1042/BJ20061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bobone S, Piazzon A, Orioni B, Pedersen JZ, Nan YH, Hahm K-S, Shin SY, Stella L. J Pept Sci. 2011:335–341. doi: 10.1002/psc.1340. [DOI] [PubMed] [Google Scholar]
- 70.Weidner T, Breen NF, Drobny GP, Castner DG. J Phys Chem B. 2009;113:15423–6. doi: 10.1021/jp908773c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen KT, Soong R, Lm S-C, Waskell L, Ramamoorthy A, Chen Z. J Am Chem Soc. 2010;132:15112–5. doi: 10.1021/ja106508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu L, Liu J, Yan ECY. J Am Chem Soc. 2011;133:8094–7. doi: 10.1021/ja201575e. [DOI] [PubMed] [Google Scholar]