Abstract
The correlation between longevity and stress resistance observed in long-lived mutant animals suggests that the ability to sense and respond to environmental challenges could be important for the regulation of life span. We therefore examined the role of heat shock factor (HSF-1), a master transcriptional regulator of stress-inducible gene expression and protein folding homeostasis, in the regulation of longevity. Down-regulation of hsf-1 by RNA interference suppressed longevity of mutants in an insulin-like signaling (ILS) pathway that functions in the nervous system of Caenorhabditis elegans to influence aging. hsf-1 was also required for temperature-induced dauer larvae formation in an ILS mutant. Using tissue-specific expression of wild-type or dominant negative HSF-1, we demonstrated that HSF-1 acts in multiple tissues to regulate longevity. Down-regulation of individual molecular chaperones, transcriptional targets of HSF-1, also decreased longevity of long-lived mutant but not wild-type animals. However, suppression by individual chaperones was to a lesser extent, suggesting an important role for networks of chaperones. The interaction of ILS with HSF-1 could represent an important molecular strategy to couple the regulation of longevity with an ancient genetic switch that governs the ability of cells to sense and respond to stress.
INTRODUCTION
Studies in a variety of organisms have identified single gene mutations that influence life span (Guarente and Kenyon, 2000). These mutations are pleiotropic and are correlated with resistance to a variety of environmental stresses, including UV radiation, oxidizing conditions, and heat shock (Larsen, 1993; Lithgow et al., 1995; Murakami and Johnson, 1996; Finkel and Holbrook, 2000). We have observed previously that the age-dependent aggregation of polyglutamine proteins was suppressed in long-lived age-1 mutant animals (Morley et al., 2002), suggesting that longevity can be associated with protection not only against exogenously imposed environmental stress but also to biochemical stress associated with expression of misfolded and aggregation-prone proteins. Moreover, these observations suggest that genes that regulate aging are coupled to cellular systems that act to restore protein homeostasis and prevent damage in response to a diverse array of stresses. Alternatively, stress resistance could be a secondary pleiotropic consequence of many cellular processes that are altered in long-lived animals. The observation of hormesis, wherein organisms exposed to sublethal insults become long-lived, however, suggests that activation of stress responses could be sufficient to extend life span (Lithgow et al., 1995; Cypser and Johnson, 2002). Additionally, forward genetic screens to identify stress resistant mutants have been used effectively as strategies to directly isolate long-lived animals (Muänoz and Riddle, 2003). Although cell-protective factors, including antioxidants and molecular chaperones, have been implicated in these processes (Finkel and Holbrook, 2000), how and whether these responses are coordinated and the extent to which chaperones are involved remains unclear.
Among the best-characterized genetic regulatory networks that influence aging is an insulin-like signaling pathway in Caenorhabditis elegans (Gottlieb and Ruvkun, 1994; Dorman et al., 1995; Morris et al., 1996; Kimura et al., 1997). Activation of the insulin/IGF-1-like receptor DAF-2 initiates a kinase cascade that includes a phosphatidylinositol 3-kinase (AGE-1), AKT/PKB, and PDK. This cascade results in phosphorylation and repression of a forkhead family transcription factor (DAF-16) that positively regulates longevity determinants. Consequently, mutations in daf-2 or age-1 that inhibit signaling lead to derepression of DAF-16 resulting in longevity, and these effects are suppressed by loss of DAF-16 function (Lin et al., 1997; Ogg et al., 1997). Mutations that diminish insulin-like signaling also increase life span and stress resistance in Drosophila and mice (Clancy et al., 2001; Holzenberger et al., 2003), suggesting that the effect of this pathway on longevity is conserved. Genetic mosaic analysis and tissue-specific complementation studies indicate that insulin-like signaling functions primarily in the nervous system (although the pathway acts in other tissues—albeit to a lesser extent) of C. elegans to mediate a neuroendocrine pathway regulating longevity (Apfeld and Kenyon, 1998; Wolkow et al., 2000). Presumably, regulatory signals from the nervous system must be transmitted to activate youth preserving factors in other tissues of the body. The recent discovery of a secreted factor, scl-1, that functions downstream of daf-16 is consistent with such a model (Ookuma et al., 2003). However, it is not known what other factors are required to mediate insulin-like signaling (ILS) phenotypes throughout the organism.
Here, we investigate the role of heat shock factor and molecular chaperones, ubiquitous and evolutionarily conserved components of the cellular response to stress (Wu, 1995; Morimoto, 1998, 2002; Nollen and Morimoto, 2002), in the regulation of longevity. Indeed, observations from a number of groups have suggested a direct role for these components the cellular stress response in the regulation of life span. Overexpression of individual molecular chaperones (including members of the Hsp70 and small heat shock protein families) in Drosophila and C. elegans has been shown to extend life span (Tatar et al., 1997; Yokoyama et al., 2002; Walker and Lithgow, 2003). In addition, recent studies have implicated heat shock factor (HSF) itself as a regulator of longevity. For example, it has been shown that inhibition of HSF-1 function leads to decreased life span and an accelerated aging phenotype in C. elegans (Garigan et al., 2002). Conversely, overexpression of HSF-1 in C. elegans has been recently demonstrated to extend life span (Hsu et al., 2003). In this study, we provide new evidence in support of HSF-1 and molecular chaperones as regulators of longevity and extend these observations to provide a better understanding of the newly recognized role of these factors in the regulation of life span.
MATERIALS AND METHODS
DNA Cloning
daf-16/L4440 was created by reverse transcription-polymerase chain reaction (RT-PCR) amplification of the corresponding cDNA from total RNA, digestion with SacI/SalI, and ligation into appropriately digested plasmid L4440 (Timmons et al., 2001) (gift of A. Fire, Carnegie Institute of Washington). hsf-1/L4440 was generated by inserting the EcoRI/XhoI fragment of pBluescript-hsf-1 containing the full-length C. elegans hsf-1 cDNA into EcoRI/XhoI digested L4440.
Chaperone RNA interference (RNAi) constructs were created by RT-PCR amplification of fragments by using gene-specific oligonucleotides and cloning directly into pGEM-T-Easy. Oligonucleotide sequences used for amplification of trigger sequences were:
hsp-16F 5′-ACTTTACCACTATTTCCGTCCAGC-3′,
hsp-16R 5′-CCTTGAACCGCTTCTTTCTTTG-3′ (381-bp product);
sip-1F 5′-CAACAACATCGTGCCACAACAG-3′,
sip-1R 5′-CGTCCTTTGGAAGAGTGAACTGG-3′ (209-bp product);
T05C3.5F 5′-CACTGAATGAGGCACTTTGCG-3′,
T05C3.5R 5′-CAACTTTTGAGGTAGGGAAGCAGC-3′ (264-bp product);
F39B2.10F 5′-TGTTCTTCCTGGAGAGGTTATTGC-3′,
F39B2.10R 5′-CACCGTGTCCTTCAAAGTCGTC-3′ (287-bp product);
hsp-1F 5′-TGACGCCAAGATGGACAAGAGC-3′,
hsp-1R 5′-TCTCAATACCAAGGGAAAGTGGG-3′ (248-bp product);
C30C11.4F 5′-AGTTCCGTTCTGTTCTCACCTCC-3′,
C30C11.4R 5′-CGTCCAACCATTTCTTCTTGTCC-3′ (300-bp product);
C12C8.1F 5′-ATGTTTTACTCGTTGATGTTGTCCC-3′,
C12C8.1R 5′-CGACTTGTGGAACTCCTCGTGG-3′ (255-bp product); and daf-21F 5′-GCTTTCCGATTTCCTTCGCTAC-3′,
daf-21R 5′-TCCTTGAGTTGTTGGACGCAG-3′ (228-bp product). Feeding RNAi constructs were generated by shuttling trigger sequences from pGEM-T-Easy into L4440 by using appropriate restriction sites.
HSF-1 expression constructs were generated by subcloning the full-length hsf-1 wild-type or mutated cDNA into pPD103.05, pPD30.38 (let-858 promoter and unc-54 promoter, respectively; gift of A. Fire) (Kelly et al., 1997) or vectors containing the F25B3.3 promoter (gift of D. Pilgrim, University of Alberta) (Altun-Gultekin et al., 2001) or vha-6 promoter (gift of J. Wang, Queens College, City University of New York) (Wang et al., 2002). HSF-1 S142A R145A was generated by site-directed mutagenesis by using the QuikChange kit (Stratagene, La Jolla, CA). The N-terminal deletion was generated by PCR amplification of nucleotides encoding residues 290-671 of HSF-1 by using oligonucleotides containing KpnI and EcoRV restriction sites and insertion into pPD103.05. Additional details of oligonucleotide sequences and restriction sites used are available on request. Successful construction was confirmed by DNA sequencing.
RNA Isolation and RT-PCR
Animals were removed from plates and washed two times with M9 buffer. Total RNA was extracted using TRIzol reagent. For RT-PCR, equal amounts of RNA (500 ng) were added to 2× reaction buffer and SS-RT one-step RT-PCR reaction mix (Invitrogen, Carlsbad, CA) with the appropriate forward and reverse gene-specific primers according to the manufacturer's instructions. cDNA was amplified by standard PCR for 20 cycles. Equal amounts of each reaction were then analyzed on 2% TAE agarose gels. Sequences of oligonucleotides used are available upon request.
C. elegans Methods and Generation of Transgenic Lines
Wild-type C. elegans (N2) and strains bearing the alleles age-1(hx546), daf-2(e1370), old-1(zIs3000), and daf-16(mgDf50) were obtained from the Caenorhaditis Genetics Center. Nematodes were handled using standard methods (Brenner, 1974). For generation of transgenic animals, plasmid DNAs were mixed (at 50 μg/ml) with pRF4 (Mello et al., 1991) or pmyo-2::gfp (gift of A. Fire) for a total DNA concentration of 150 μg/ml. Mixtures were microinjected into the gonads of adult hermaphrodite animals by using standard methods (Mello et al., 1991). Transgenic F1 progeny were selected on the basis of roller phenotype or fluorescence in the pharynx. Individual transgenic F2 animals were picked to establish independent lines.
Double-stranded RNAi
Bacterial-mediated RNAi was performed essentially as described (Timmons et al., 2001). Plasmids containing the appropriate trigger sequence or vector L4440 alone were transformed into Escherichia coli strain HT115(DE3) by using standard methods. Individual colonies were inoculated to LB broth containing ampicillin (100 mg/ml) and tetracycline (12.5 mg/ml) and grown overnight at 37°C. Cultures were diluted 1:100 and allowed to grow to OD600 ∼0.4. Isopropyl β-d-thiogalactoside was added to 0.4 mM for initiation of dsRNA synthesis and cultures were induced for 2-4 h at 37°C. Bacteria were then seeded onto NGM plates containing ampicillin, tetracycline, and isopropyl β-d-thiogalactoside and allowed to dry overnight.
Life Span Assays
To assess whether HSF-1 has a role in genetic pathways that influence aging, we first tested whether inactivation of the single C. elegans hsf-1 ortholog by bacterial RNAi would affect the life span extension of age-1(hx546) animals. In longevity experiments, animals were allowed to grow to adulthood (3 d old) before exposure to RNAi to eliminate the contribution of developmental effects of hsf-1(RNAi) on life span. This was necessary because microinjection of hsf-1 dsRNA resulted in larval arrest and bacterial RNAi initiated in embryos (by growth of L4 animals on RNAi bacteria and examining progeny) gave rise to small sterile adult animals (our unpublished data). Similarly, RNAi for the various molecular chaperones was not initiated until adulthood, because exposure to RNAi for C30C11.4, hsp-1, and daf-21 resulted in developmental phenotypes (our unpublished data).
Life span assays were performed essentially as described previously (Dillin et al., 2002). Embryos were synchronized by treatment of adult hermaphrodites with alkaline hypochlorite and allowed to develop for 3 d. Adult animals were then transferred to new plates and scored every other day. Animals were judged as dead when they ceased pharyngeal pumping and did not respond to prodding with a platinum wire. Assays were performed on NGM RNAi plates containing 5′-fluorodeoxyuridine (5 mg/l, to inhibit progeny development) at either 20 or 25°C as indicated. Animals were transferred to fresh RNAi plates every 7-10 d. hsf-1(RNAi) also inhibited age-1(hx546) longevity when experiments were carried out on standard NGM plates (our unpublished data), indicating that FUDR did not influence the effect of HSF-1 on longevity. Life span time = 0 was the first day of fertile adulthood. Although no difference in life span has been reported between animals grown on HT115(DE3) versus standard OP50, experiments on HSF-1-overexpressing animals were performed on RNAi plates seeded with L4440 transformed HT115(DE3) to maintain consistency. Statistical significance between groups was determined using one-way analysis of variance. Data were analyzed and survival curves generated using the Prism 3 statistical analysis software package.
Heat Shock Reporter Assays
Animals bearing the C12C8.1::gfp reporter fusion (and other transgenes as indicated) were exposed to control (growth at ambient temperature) or heat shock (33°C for 1 h) conditions on NGM agar plates seeded with OP50. Heat-shocked animals were allowed to recover for 6-8 h before analysis. Adult transgenic animals were assayed for GFP expression at 100× magnification by using a Leica MZFLIII stereomicroscope equipped for epifluorescence. Images were acquired using OPENLAB software (Improvision, Lexington, MA) and a charge-coupled device camera.
Dauer Formation Assays
Assessment of dauer formation was performed as described previously (Ailion and Thomas, 2000). Adult hermaphrodites were allowed to lay embryos at room temperature for 4-6 h to generate a synchronized population. Plates were then shifted for growth at 25°C or 27°C as indicated. Animals were scored for dauer formation (by morphology and resistance to 1% SDS) after 44 h (27°C) or 48 h (25°C).
RESULTS
Down-Regulation of HSF-1 Suppresses Longevity of ILS Mutants
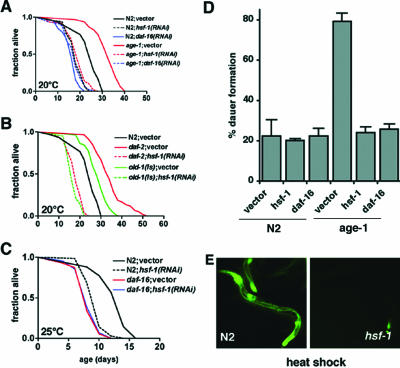
To examine a role for HSF-1 in aging, we first asked whether hsf-1 was required for longevity of age-1(hx546) mutants. Functional inactivation of HSF-1 by RNAi was demonstrated by inhibition of heat-inducible expression of a heat shock promoter::gfp transgene in hsf-1(RNAi) animals (Figure 1E). Down-regulation of hsf-1 in adulthood suppressed longevity of age-1(hx546) animals to an extent similar to RNAi for daf-16 (p < 0.001; Figure 1A and Table 1). hsf-1(RNAi) also suppressed longevity associated with other components of the insulin-like signaling pathway including daf-2(e1370) mutants and OLD-1 (Murakami and Johnson, 2001) overexpressing animals (p < 0.0001; Figure 1B and Table 1). Similar suppression of age-1(hx546) longevity by hsf-1(RNAi) was observed at 20 or 25°C (Table 1), and subsequent life span analysis was conducted at 25°C. These results corroborate previous observations that HSF-1 levels influence the rate of aging (Garigan et al., 2002) and that hsf-1(RNAi) inhibits daf-2 mediated longevity (Hsu et al., 2003). Our epistasis analysis further shows that HSF-1 is required for longevity of other ILS mutants and indicates that HSF-1 may act downstream of these known components. Because RNAi was not initiated until adult stages, these data also demonstrate for the first time that HSF-1 functions in adulthood to regulate longevity as has been shown previously for components of ILS (Dillin et al., 2002).
Figure 1.
Suppression of longevity associated with insulin-like signaling mutants by hsf-1(RNAi). (A) Life span measurements of wild-type (N2) and age-1(hx546) mutant animals grown on bacteria transformed with RNAi vectors for daf-16, hsf-1, or empty vector as indicated. (B) Life spans of daf-2(e1370) and old-1(zIs3000) animals with or without hsf-1 RNAi. (C) Comparison of shortened life span observed in daf-16(mgDf50) null mutants grown on control or hsf-1 RNAi bacteria. For statistical analysis (see Table 1). (D) Dauer formation of wild-type (N2) or age-1(hx546) animals grown at 27°C on the indicated RNAi bacteria. Data are mean ± SEM of duplicate plates from one experiment. Three independent trials gave similar results. (E) Epifluorescence images demonstrating expression of a C12C8.1(hsp70)::gfp reporter gene in wild-type(N2) or hsf-1(RNAi) animals after a 1-h heat shock at 33°C followed by 8 h of recovery.
Table 1.
Effects of hsf-1(RNAi) on life span of wild-type (N2) or ILS mutant animals at 20°C or 25°C
| Adult lifespan (d) mean ± SEM | N | Control lifespan, % | P value | |
|---|---|---|---|---|
| 20 C | ||||
| N2; vector | 23.2±0.8 | 123 | ||
| N2;hsf-1(RNAi) | 17.9±0.5 | 121 | 77 | <0.001 |
| N2;daf-16(RNAi) | 16.9±0.8 | 86 | 73 | <0.001 |
| age-1(hx546) ;vector | 32.1±0.7 | 142 | ||
| age-1(hx546);hsf-1(RNAi) | 18.5±0.6 | 135 | 58 | <0.001 |
| age-1(hx546);daf-16(RNAi) | 17.9±0.6 | 140 | 56 | <0.001 |
| daf-2(e1370) ;vector | 34.3±0.9 | 128 | ||
| daf-2(e1370);hsf-1(RNAi) | 17.8±0.5 | 112 | 52 | <0.001 |
| old-1(zls3000) ;vector | 28.0±0.7 | 127 | ||
| old-1(zls3000);hsf-1(RNAi) | 16.8±0.4 | 131 | 60 | <0.001 |
| 25 C | ||||
| N2;vector | 13.5±0.3 | 46 | ||
| N2;hsf-1(RNAi) | 9.6±0.2 | 70 | 71 | <0.001 |
| age-1(hx546); vector | 25.5±0.5 | 50 | ||
| age-1(hx546);hsf-1(RNAi) | 12.2±0.3 | 42 | 48 | <0.001 |
| daf-16(mgDf50); vector | 8.4±0.2 | 71 | ||
| daf-16(mgDf50);hsf-1(RNAi) | 8.5±0.2 | 70 | 101 | >0.05 |
Data are shown as days ± SEM.
HSF-1 Is Required for Dauer Formation Mediated by ILS
In addition to its role in longevity, ILS in C. elegans regulates a developmental checkpoint mediating entry into the alternative dauer larval stage in response to starvation, stress, or hormonal signals (Gottlieb and Ruvkun, 1994). To further investigate the relationship between hsf-1 and ILS, we asked whether HSF-1 influences dauer formation in wild-type and age-1 animals. Growth at 27°C resulted in induced dauer formation in wild-type (22%) and age-1(hx546) (79%) animals as has been reported previously (Figure 1D; Malone et al., 1996; Ailion and Thomas, 2000). Both daf-16 and hsf-1 were required for this effect in age-1 mutant animals because RNAi for either of these factors reduced dauer formation to wild-type levels (Figure 1D). Neither daf-16(RNAi) nor hsf-1(RNAi) affected dauer formation at 27°C in wild-type animals (Figure 1D), suggesting that these genes were specifically required for enhanced dauer formation resulting from the age-1 mutation.
Combination of daf-16 and hsf-1 Loss of Function Does Not Further Decrease Life Span
Our observations indicate that HSF-1 could function downstream from all of these components of insulin-like signaling or in an independent pathway to influence life span and dauer formation. RNAi for either hsf-1 or daf-16 shortened wild-type life span by 23 and 27%, respectively (Figure 1A and Table 1). In fact, life spans of hsf-1(RNAi) animals were nearly identical to that of daf-16(RNAi) animals (p > 0.05) in the different genetic backgrounds tested (Figure 1A and Table 1; our unpublished data). To address whether HSF-1 and DAF-16 function in a common pathway to regulate aging, we asked whether the life span of daf-16(mgDf50) null animals was affected by hsf-1 down-regulation (Ogg et al., 1997). Combination of daf-16(mgDf50) and hsf-1(RNAi) did not lead to a further decrease in life span (p > 0.05; Figure 1C and Table 1), suggesting that daf-16 and hsf-1 may function in a common pathway to regulate longevity.
Overexpression of HSF-1 Increases Life Span
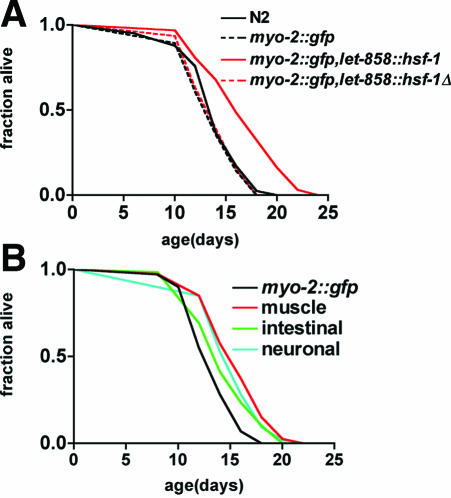
If HSF-1 is a regulator of longevity, we would predict that increasing HSF-1 levels could extend life span. Overexpression of HSF-1 ubiquitously in somatic cells under the control of the let-858 promoter resulted in a 22% increase in life span, whereas expression of a mutant HSF-1 lacking the amino terminal DNA binding domain had no effect (Figure 2A and Table 2). Overexpression of WT and mutant HSF-1 were confirmed using RT-PCR (our unpublished data). Life span extension in HSF-1-overexpressing animals was not associated with delayed larval development (wild-type: 98% L4/young adult animals after 48 h at 25°C, n = 424; let-858::hsf-1 WT: 96%, n = 113) or constitutive dauer formation (our unpublished data). The extent of life span extension achieved by HSF-1 overexpression was less than observed in age-1 or daf-2 mutants, although similar to that previously observed for DAF-16 overexpression (Lin et al., 2001). Both HSF-1 and DAF-16 are transcription factors subject to posttranslational negative regulation, consequently increasing expression levels is likely insufficient to achieve maximal activation of downstream targets. Consistent with this, ubiquitous HSF-1 overexpression resulted in constitutive activation of an hsp-70::gfp reporter but to a lesser extent than heat shock induction (Table 3).
Figure 2.
HSF-1 acts in multiple tissues to regulate life span. (A) Life span of wild-type (N2) or transgenic animals expressing marker gene (myo-2::gfp) alone or in combination with let-858::hsf-1 or let-858::hsf-1Δ (encoding residues 290-671 of C. elegans HSF-1). (B) Life spans of transgenic animals expressing marker alone (myo-2::gfp) or in combination with hsf-1 in body-wall muscle (unc-54::hsf-1), intestine (vha-6::hsf-1), or nervous system (F25B5.3::hsf-1). Data are representative of experiments with two independent transgenic lines. No significant differences were noted between the duplicate lines. The data reported represent two independent trials that yielded similar results. In each case, the appropriate control was tested in parallel in the same experiment. For statistical analysis, see Table 2.
Table 2.
Effect of wild-type or dominant negative expression on life span of wild-type or age-1(hx546) animals at 25°C
| Adult lifespan (d) mean ± SEM | N | Control lifespan, % | P value | |
|---|---|---|---|---|
| Wild-type | ||||
| myo-2::gfp | 13.8 ± 0.5 | 99 | 99 | |
| myo-2::gfp, let-858::hsf-1 | 16.8 ± 0.5 | 64 | 122 | < 0.001 |
| myo-2::gfp, let858::hsf-1 Δ | 14.2 ± 0.5 | 60 | 102 | > 0.05 |
| myo-2::gfp, unc-54::hsf-1 | 15.9 ± 0.3 | 70 | 115 | < 0.001 |
| myo-2::gfp, F25B3.3::hsf-1 | 15.9 ± 0.3 | 83 | 115 | < 0.001 |
| myo-2::gfp, vha-6::hsf-1 | 14.7 ± 0.2 | 111 | 107 | < 0.05 |
| Age-1(hx546) | ||||
| myo-2::gfp | 27.8 ± 0.7 | 48 | ||
| myo-2::gfp, unc-54::hsf-1DN | 23.2 ± 0.4 | 65 | 84 | < 0.001 |
| myo-2::gfp, F25B3.3::hsf-1DN | 24.3 ± 0.4 | 78 | 87 | < 0.001 |
| myo-2::gfp,vha-6::hsf-1DN | 26.7 ± 0.3 | 53 | 96 | > 0.05 |
Table 3.
Effect of tissue-specific expression of wild-type (WT) or dominant negative (DN) HSF-1 on heat shock promoter (pC12C8.1::gfp) activation in neuronal, body-wall muscle (BWM), and intestinal cells
| Control (% transgenic animals with GFP in each tissue)
|
||||
|---|---|---|---|---|
| Neuronal | BWM | Intestine | N | |
| let-858::hsf-1 WT | 25 | 18 | 10 | 59 |
| let-858::hsf-1 Δ | 4 | 2 | 0 | 48 |
| F25B3.3::hsf-1 WT | 23 | 4 | 0 | 70 |
| unc-54::hsf-1 WT | 2 | 20 | 0 | 51 |
| vha-6::hsf-1 WT | 2 | 2 | 13 | 45 |
| Heat Shock (% transgenic animals with GFP in each tissue)
|
||||
|---|---|---|---|---|
| Neuronal | BWM | Intestine | N | |
| let-858::hsf-1 WT | 91 | 97 | 97 | 75 |
| let-858::hsf-1 Δ | 89 | 97 | 98 | 62 |
| F25B3.3::hsf-1 DN | 10 | 94 | 98 | 50 |
| unc-54::hsf-1 DN | 93 | 12 | 97 | 58 |
| vha-6::hsf-1 DN | 98 | 96 | 8 | 52 |
HSF-1 Functions in Multiple Tissues to Increase Life Span
Our observation that HSF-1 overexpression is sufficient to extend life span is consistent with a recent finding by Kenyon's group (Hsu et al., 2003). However, it is unclear whether the effect of HSF-1 on longevity is specific to the nervous system, similar to insulin-like signaling, or is required in multiple tissues to influence longevity. To examine this question, we tested the effect of HSF-1 on life span of wild-type animals when overexpressed in neuronal, body-wall muscle, or intestinal cells by using tissue-specific promoters. Intestinal expression of HSF-1 under control of the vha-6 promoter resulted in a 7% life span increase (p < 0.05; Figure 2B and Table 2) of wild-type animals. Expression of HSF-1 in neuronal (F25B3.3 promoter) or body-wall muscle cells (unc-54 promoter) each resulted in a significant (15%, p < 0.001) extension of wild-type life span (Figure 2B). Previous studies have described that green fluorescent protein (GFP) expression from these promoters is restricted to specific tissues (Altun-Gultekin et al., 2001; Morley et al., 2002; Wang et al., 2002). To further establish the tissue-specificity of these promoters in our system, we examined activation of an hsp-70::gfp reporter in animals expressing HSF-1 in different tissues (Table 3). Within the limits of GFP detection, we did not observe any promiscuous expression from these promoters (Table 3). Together, these results suggest that HSF-1 has significant beneficial effects in multiple tissues to influence longevity.
HSF-1 Is Required in Multiple Tissues for Age-1(hx546) Longevity
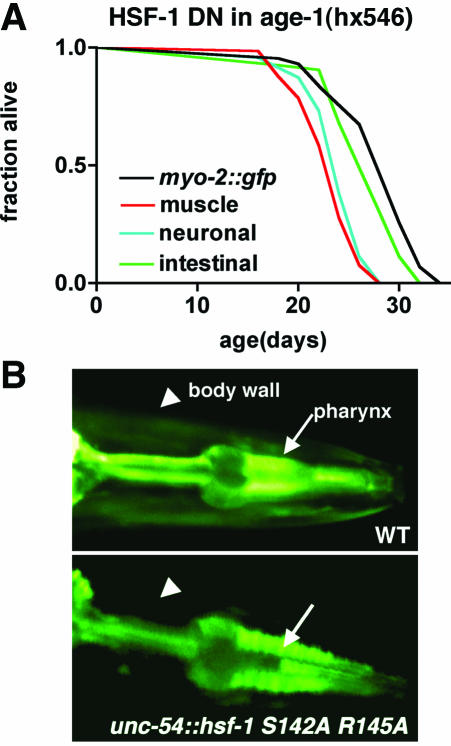
To further explore this hypothesis, we expressed a mutant HSF-1 carrying point mutations in the DNA binding domain (S142A R145A), which should form nonfunctional HSF-1 trimers defective in binding to the heat shock elements in age-1(hx546) animals. Expression of this dominant negative HSF-1 (HSF-1 DN) under control of the unc-54 promoter resulted in muscle specific loss of heat shock promoter inducible GFP reporter expression (Figure 3B and Table 3). Both intestinal and neuronal expression of HSF-1 DN also resulted in tissue-specific inhibition of the heat shock response (Table 3). Tissue-specific expression of the dominant-negative HSF-1, by using the promoters described above for wild-type HSF-1 overexpression, demonstrated that inhibition of endogenous HSF-1 activity in neuronal or body-wall muscle cells suppressed age-1(hx546) life span by 13 and 16%, respectively (Figure 3A and Table 2). Intestinal expression of the dominant negative HSF-1 resulted in a 4% decrease in age-1(hx546) life span that was not statistically significant. We were unable to recover transgenic lines of animals ubiquitously (let-858 promoter) expressing HSF-1 DN, presumably due to the developmental toxicity resulting from functional loss of HSF-1 activity. Additionally, unlike our experiments in which animals were allowed to grow to adulthood before being exposed to RNAi, by using tissue-specific promoters to express wild-type or HSF-1 DN does not allow us to distinguish between effects during larval development and adulthood. Although we have not examined the effects of HSF-1 gain and loss of function in all cell types, the results observed for muscle and neuronal cells are intriguing given previous observations that preservation of the nervous system is important in the regulation of longevity and that muscle cells of C. elegans are also particularly susceptible to aging-related decline (Wolkow et al., 2000; Herndon et al., 2002).
Figure 3.
HSF-1 is required in multiple tissues for longevity of age-1(hx546) animals. (A) Life spans of age-1(hx546) expressing marker alone (myo-2::gfp) or in combination with hsf-1 dominant negative (HSF-1 DN) in body-wall muscle, intestine, or nervous system of age-1(hx546) animals. Data shown are representative of results from two independent transgenic lines expressing each construct. No significant differences were noted between the duplicate lines. Experiments were performed in two independent trials with similar results. For statistical analysis, see Table 2. (B) Epifluorescence images illustrating expression of a C. elegans hsp-70::gfp reporter after heat shock in wild-type or transgenic animals expressing HSF-1 dominant negative in body-wall muscle cells. Arrowheads, body-wall muscle cells. Arrows, pharynx.
Molecular Chaperones Contribute to age-1(hx546) Longevity
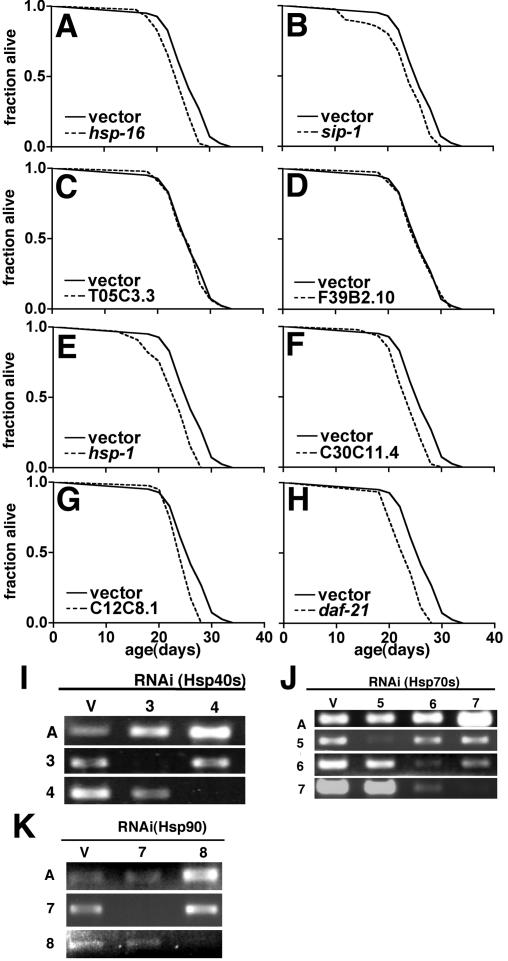
How does HSF-1 act to influence life span? Among the best-characterized cellular functions of HSF-1 is in the transcriptional control of heat shock genes, many of which encode molecular chaperones and components of degradative machineries. Elevated expression of multiple heat shock genes (including hsp-16 and Hsp90) has been observed in long-lived daf-2 and age-1 mutants (Hsu et al., 2003; McElwee et al., 2003; Murphy et al., 2003). Additionally, transgenic overexpression of Hsp16 or Hsp70 in Drosophila and C. elegans can increase life span (Tatar et al., 1997; Tower, 2000; Yokoyama et al., 2002; Walker and Lithgow, 2003). These observations have implicated a link between aging and molecular chaperones; however, overexpression of chaperones inevitably has pleiotropic effects on many signaling pathways influencing cell growth and death (Beere et al., 2000; Song et al., 2001; Nollen and Morimoto, 2002). To test whether chaperones are required for longevity of age-1 animals, we generated a series of feeding RNAi constructs to target eight molecular chaperones representing four major classes of heat shock proteins: small HSPs (hsp-16.48, sip-1; Lund et al., 2002), DnaJ/Hsp40s (T05C3.5, F39B2.10), Hsc/Hsp70s (hsp-1 [Snutch et al., 1988], C30C11.4, C12C8.1), and Hsp90 (daf-21 [Birnby et al., 2000]). This list of candidate genes is not exhaustive, as there are 18 small HSPs, 23 DnaJ/Hsp40s, and 12 Hsp70s in the predicted C. elegans proteome (www.wormbase.org Release WS100). This set of genes is however representative of constitutively and inducibly expressed Hsps, including genes shown previously to be transcriptionally regulated during aging based on microarray analysis (Lund et al., 2002).
Using these RNAi constructs, we tested the effect of down-regulating molecular chaperones on life span in wild-type and age-1(hx546) animals. To eliminate the influence of developmental effects of chaperone gene inactivation, animals were allowed to develop to adulthood before transfer to RNAi bacteria as for hsf-1 RNAi experiments. None of the genes targeted had significant effects on life span of wild-type animals (p > 0.05; Table 4). This did not reflect ineffectiveness of RNAi, because down-regulation of target mRNA levels was observed using RT-PCR (Figure 4, I and J) and RNAi for hsp-1 and daf-21 initiated in embryos resulted in strong developmental phenotypes (our unpublished data). Down-regulation of hsp-16, sip-1, hsp-1, C30C11.4, C12C8.1, and daf-21, but not either Hsp40 homolog tested, resulted in small (8-13%) but reproducible reductions in age-1(hx546) life span (p < 0.05; Figure 4, A-H, and Table 4).
Table 4.
Effect of chaperone RNAi on life span of wild-type (N2) and age-1(hx546) animals at 25°C
| Family | Adult lifespan (d) mean ± SEM | N | Control lifespan, % | P value | |
|---|---|---|---|---|---|
| N2 | |||||
| vector | 14.1 ± 0.3 | 65 | |||
| hsp-16 | sHsp | 14.3 ± 0.3 | 69 | 101 | > 0.05 |
| sip-1 | 14.2 ± 0.3 | 68 | 100 | > 0.05 | |
| T05C3.5 | Hsp40/DnaJ | 14.6 ± 0.2 | 76 | 103 | >0.05 |
| F39B10.2 | 14.6 ± 0.2 | 79 | 103 | > 0.05 | |
| hsp-1 | 14.6 ± 0.3 | 75 | 103 | > 0.05 | |
| C30C11.4 | Hsp70 | 14.3 ± 0.2 | 85 | 101 | > 0.05 |
| C12C8.1 | 14.2 ± 0.3 | 79 | 100 | > 0.05 | |
| daf-21 | Hsp90 | 14.6 ± 0.2 | 84 | 103 | > 0.05 |
| Age-1(hx546) | |||||
| vector | 26.7 ± 0.4 | 83 | |||
| hsp-16 | sHsp | 24.5 ± 0.4 | 107 | 92 | < 0.05 |
| sip-1 | 24.1 ± 0.4 | 102 | 90 | < 0.01 | |
| T05C3.5 | Hsp40/DnaJ | 26.3 ± 0.5 | 95 | 99 | > 0.05 |
| F39B10.2 | 26.5 ± 0.5 | 82 | 100 | > 0.05 | |
| hsp-1 | 23.4 ± 0.6 | 91 | 88 | < 0.001 | |
| C30C11.4 | Hsp70 | 23.6 ± 0.3 | 112 | 88 | < 0.001 |
| C12C8.1 | 24.1 ± 0.4 | 111 | 90 | < 0.001 | |
| daf-21 | Hsp90 | 23.1 ± 0.4 | 104 | 87 | <0.001 |
Figure 4.
Molecular chaperones are required for age-1(hx546) longevity. (A-H) Longevity curves for age-1(hx546) animals grown from young adulthood on bacteria expressing control vector or the indicated RNAi bacteria. All life spans were measured concurrently and vector control data shown is the same in each panel. The data reported represent two independent trials that yielded similar results. In each case, the appropriate control was tested in parallel in the same experiment. For statistical analysis, see Table 4. (I-K) Analysis of RNAi inactivation of Hsp40 (I, 3 = T05C3.5, 4 = F39B10.2) and Hsp70 (J, 5 = hsp-1, 6 = C30C11.4, 7 = C12C8.1) and Hsp90 (daf-21) genes by using RT-PCR. Total RNA was extracted from 3-d-old animals using TRIzol reagent after a 1-h heat shock at 33°C. Amplification of actin (A) was used as a control. Consistent with the high degree of sequence identity between Hsp70 family members, some cross-interference with expression of C12C8.1 expression is observed in C30C11.4 RNAi.
DISCUSSION
Heat shock factor and molecular chaperones are well-known mediators of stress resistance. Thus, the finding that HSF-1 and molecular chaperones influence life span provides a molecular link between genes that regulate longevity and those that protect cells from stress. A large body of evidence accumulated in cellular and animal models of senescence has consistently indicated that heat shock gene expression is induced poorly in aged animals (Fargnoli et al., 1990; Fawcett et al., 1994). In instances where it has been tested, lack of heat shock gene induction corresponds with deficient oligomerization and DNA binding of HSF-1 without changes in the absolute levels of the protein (Fawcett et al., 1994). However, it has been difficult to distinguish whether diminished activation of the heat shock response simply represents one of many cellular processes that decline during aging or whether HSF-1 itself participates in the regulation of cellular senescence and aging. Our present results support a direct role for HSF-1 as a regulator of longevity.
One potential concern is that the influence of HSF-1 inhibition on life span is a nonspecific effect resulting from loss of essential cellular functions. However, multiple lines of evidence support a central role for hsf-1 in the genetic regulation of aging. HSF-1 gain and loss of function have reciprocal effects on life span. Whereas loss of many essential genes might shorten life span, the set of factors that can extend life span is likely to be more specific. Suppression of dauer formation in age-1 mutants indicates that HSF-1 influences not only longevity but also a specific developmental checkpoint regulated by ILS. The failure of hsf-1(RNAi) to further shorten life span of daf-16 null animals is also consistent with the suggestion that these factors function, at least in part, through a similar mechanism. In support of this, a recent report found similarly that HSF-1 participates in the regulation of life span through cooperation with ILS (Hsu et al., 2003). Importantly, these workers found that hsf-1(RNAi) suppressed longevity of age-1 animals but not isp-1 or eat-2 mutants that function through a different mechanism (Hsu et al., 2003). Together, all of these observations strongly support the idea that HSF-1 is a bona fide regulator of aging through a mechanism similar to ILS.
The influence of ILS on life span is thought to be mediated through a neuroendocrine signaling pathway. This hypothesis is based on two major findings. First, genetic mosaic experiments have demonstrated daf-2 can function cell nonautonomously (Apfeld and Kenyon, 1998). Second, the longevity of age-1 mutants could be reversed completely by expression of wild-type AGE-1 only in neurons, whereas expression of the rescuing constructs in muscle or intestinal cells had only minor effects on life span (Wolkow et al., 2000). In contrast, expression of wild-type or HSF-1 DN had nearly identical effects whether they were expressed in neuronal or muscle cells (and to a lesser extent in intestinal cells). These observations suggest that HSF-1 exerts beneficial effects in multiple tissues throughout the organism to influence life span. Thus, these findings raise the intriguing possibility that regulatory signals from ILS in the nervous system could be conveyed to HSF-1 and molecular chaperones to mediate longevity in other tissues of the body, although this idea remains to be tested directly.
In contrast to the striking effects of hsf-1(RNAi), suppression of age-1(hx546) longevity by inactivation of individual chaperones was relatively small. Given the redundancy of chaperone function (Werner-Washburne et al., 1987), it is somewhat surprising to observe any effects resulting from down-regulation of a single chaperone. However, the incremental effects of chaperones on age-1(hx546) longevity and the essential role of HSF-1 in development may explain why these factors have not been identified in screens for genetic suppressors of daf-2/age-1 mutant phenotypes. Additionally, when RNAi for small heat shock proteins was initiated in embryos (this was possible because none of the genes they examined caused developmental phenotypes), suppression of age-1(hx546) longevity was greater (∼25%) than we observed by initiating RNAi in adults (Hsu et al., 2003). However, in both of our studies, suppression resulting from down-regulation of any single chaperone was much less than that observed for hsf-1(RNAi). An incremental role for many different molecular chaperones is also consistent with observations where overexpression of a single chaperone has only small effects on life span (Tower, 2000; Yokoyama et al., 2002; Walker and Lithgow, 2003). Additionally, a recent study found that RNAi-mediated inhibition of individual DAF-16 targets identified by microarray analysis had much smaller effects in life span than inhibition of DAF-16 itself (Murphy et al., 2003). Thus, the incremental role of each individual chaperone may reflect a general principle in genetic networks influencing longevity where upstream regulators, such as DAF-16 and HSF-1, have large effects on life span by orchestrating the activities of numerous effector molecules that each exert a small or specific form of protective influence.
In addition to its role in longevity, we provide evidence in this study that HSF-1 is required for dauer formation of age-1 mutants. This finding is intriguing because it implicates HSF-1 in a developmental switch that regulates the body plan of the organism. A remaining question of considerable interest is whether HSF-1 might be required for dauer formation mediated through pathways other than ILS, such as, transforming growth factor-β or cGMP signaling. At least part of the answer is provided by a recent report demonstrating that HSF-1 is required for both ILS and TGF-β-mediated dauer formation—although dauer formation of ILS mutants were considerably more sensitive to the inhibitory effects of hsf-1(RNAi) (Walker et al., 2003). Thus, it seems possible that HSF-1 may serve as a common modulator of dauer formation signaled by diverse regulatory pathways. Although the requirement for HSF-1 in dauer formation is unexpected, it is consistent with phenotypes of HSF-deficient Drosophila suggesting HSF-1 regulates genes important for normal developmental progression and fertility (Jedlicka et al., 1997). Additionally, many of the environmental conditions that induce dauer formation, including elevated temperature, high osmolarity, and starvation are themselves stressful. Whether HSF-1 acts by sensing these homeostatic challenges and activating genes necessary to facilitate and integrate stress induced dauer formation mediated by multiple pathways remains to be determined.
We have previously demonstrated that the reduced rate of aging in age-1 mutant animals is associated with delayed aggregation and toxicity of polyglutamine expansions that underlie Huntington's Disease and at least eight other mature-onset neurodegenerative disorders (Morley et al., 2002). Here, we demonstrate that HSF-1 and molecular chaperones, factors essential for the maintenance of protein folding homeostasis, are themselves regulators of longevity. Additionally, inactivation of daf-16, hsf-1, or small heat shock proteins accelerated the aggregation of polyglutamine expansion proteins (Hsu et al., 2003), supporting the idea that ILS could coordinately influence aging and protein aggregation through the action of DAF-16, HSF-1, and molecular chaperones. Together, these studies have revealed a common set of factors that link the genetic regulation of protein homeostasis, stress responsiveness, and longevity. This suggests that longevity and fitness may be, at least in part, a consequence of the efficient detection, capture, and resolution of misfolded and aggregation-prone proteins. Thus, these studies provide a new perspective on the way in which decreased ability to sense and respond to stress in old animals may contribute to declining cellular function and, in particular, increased susceptibility to diseases of protein misfolding observed during aging.
Acknowledgments
We acknowledge Greg Beitel and members of the Morimoto laboratory for discussion and comments of the manuscript. We thank A. Fire, D. Pilgrim, and J. Wang for plasmids. Some of the strains used in this study were provided by the Caenorhabditis Genetics Center, which is supported by a grant from the National Institutes of Health. J.F.M. is supported by an individual National Research Service Award from the National Institute of Neurological Disease and Stroke. R.I.M. is supported by a grant from the National Institute for General Medical Science, the Huntington's Disease Society of America Coalition for the Cure, and the Daniel F. and Ada L. Rice Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0532. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0532.
References
- Ailion, M., and Thomas, J.H. (2000). Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156, 1047-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin, Z., Andachi, Y., Tsalik, E. L., Pilgrim, D., Kohara, Y., and Hobert, O. (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951-1969. [DOI] [PubMed] [Google Scholar]
- Apfeld, J., and Kenyon, C. (1998). Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95, 199-210. [DOI] [PubMed] [Google Scholar]
- Beere, H.M., Wolf, B.B., Cain, K., Mosser, D.D., Mahboubi, A., Kuwana, T., Tailor, P., Morimoto, R.I., Cohen, G.M., and Green, D.R. (2000). Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2, 469-475. [DOI] [PubMed] [Google Scholar]
- Birnby, D.A., Link, E.M., Vowels, J.J., Tian, H., Colacurcio, P.L., and Thomas, J.H. (2000). A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155, 85-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, D.J., Gems, D., Harshman, L.G., Oldham, S., Stocker, H., Hafen, E., Leevers, S.J., and Partridge, L. (2001). Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104-106. [DOI] [PubMed] [Google Scholar]
- Cypser, J.R., and Johnson, T.E. (2002). Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J. Gerontol. A Biol. Sci. Med. Sci. 57, B109-B114. [DOI] [PubMed] [Google Scholar]
- Dillin, A., Crawford, D.K., and Kenyon, C. (2002). Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830-834. [DOI] [PubMed] [Google Scholar]
- Dorman, J.B., Albinder, B., Shroyer, T., and Kenyon, C. (1995). The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141, 1399-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargnoli, J., Kunisada, T., Fornace, A.J., Jr., Schneider, E.L., and Holbrook, N.J. (1990). Decreased expression of heat shock protein 70 mRNA and protein after heat treatment in cells of aged rats. Proc. Natl. Acad. Sci. USA 87, 846-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett, T.W., Sylvester, S.L., Sarge, K.D., Morimoto, R.I., and Holbrook, N.J. (1994). Effects of neurohormonal stress and aging on the activation of mammalian heat shock factor 1. J. Biol. Chem. 269, 32272-32278. [PubMed] [Google Scholar]
- Finkel, T., and Holbrook, N.J. (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408, 239-247. [DOI] [PubMed] [Google Scholar]
- Garigan, D., Hsu, A.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Kenyon, C. (2002). Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161, 1101-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb, S., and Ruvkun, G. (1994). daf-2, daf-16 and daf-23, genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics 137, 107-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente, L., and Kenyon, C. (2000). Genetic pathways that regulate ageing in model organisms. Nature 408, 255-262. [DOI] [PubMed] [Google Scholar]
- Herndon, L.A., Schmeissner, P.J., Dudaronek, J.M., Brown, P.A., Listner, K.M., Sakano, Y., Paupard, M.C., Hall, D.H., and Driscoll, M. (2002). Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419, 808-814. [DOI] [PubMed] [Google Scholar]
- Holzenberger, M., Dupont, J., Ducos, B., Leneuve, P., Gâeloèen, A., Even, P.C., Cervera, P., and Le Bouc, Y. (2003). IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182-187. [DOI] [PubMed] [Google Scholar]
- Hsu, A.L., Murphy, C.T., and Kenyon, C. (2003). Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142-1145. [DOI] [PubMed] [Google Scholar]
- Jedlicka, P., Mortin, M.A., and Wu, C. (1997). Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 16, 2452-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W.G., Xu, S., Montgomery, M.K., and Fire, A. (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K.D., Tissenbaum, H.A., Liu, Y., and Ruvkun, G. (1997). daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942-946. [DOI] [PubMed] [Google Scholar]
- Larsen, P.L. (1993). Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 90, 8905-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K., Dorman, J.B., Rodan, A., and Kenyon, C. (1997). daf-16, An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319-1322. [DOI] [PubMed] [Google Scholar]
- Lin, K., Hsin, H., Libina, N., and Kenyon, C. (2001). Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28, 139-145. [DOI] [PubMed] [Google Scholar]
- Lithgow, G.J., White, T.M., Melov, S., and Johnson, T.E. (1995). Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. USA 92, 7540-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, J., Tedesco, P., Duke, K., Wang, J., Kim, S.K., and Johnson, T.E. (2002). Transcriptional profile of aging in C. elegans. Curr. Biol. 12, 1566-1573. [DOI] [PubMed] [Google Scholar]
- Malone, E.A., Inoue, T., and Thomas, J.H. (1996). Genetic analysis of the roles of daf-28 and age-1 in regulating Caenorhabditis elegans dauer formation. Genetics 143, 1193-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee, J., Bubb, K., and Thomas, J.H. (2003). Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2, 111-121. [DOI] [PubMed] [Google Scholar]
- Mello, C.C., Kramer, J.M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, R.I. (1998). Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12, 3788-3796. [DOI] [PubMed] [Google Scholar]
- Morimoto, R.I. (2002). Dynamic remodeling of transcription complexes by molecular chaperones. Cell 110, 281-284. [DOI] [PubMed] [Google Scholar]
- Morley, J.F., Brignull, H.R., Weyers, J.J., and Morimoto, R.I. (2002). The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99, 10417-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.Z., Tissenbaum, H.A., and Ruvkun, G. (1996). A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382, 536-539. [DOI] [PubMed] [Google Scholar]
- Muänoz, M.J., and Riddle, D.L. (2003). Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163, 171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, S., and Johnson, T.E. (1996). A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143, 1207-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, S., and Johnson, T.E. (2001). The OLD-1 positive regulator of longevity and stress resistance is under DAF-16 regulation in Caenorhabditis elegans. Curr. Biol. 11, 1517-1523. [DOI] [PubMed] [Google Scholar]
- Murphy, C.T., McCarroll, S.A., Bargmann, C.I., Fraser, A., Kamath, R.S., Ahringer, J., Li, H., and Kenyon, C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277-283. [DOI] [PubMed] [Google Scholar]
- Nollen, E.A., and Morimoto, R.I. (2002). Chaperoning signaling pathways: molecular chaperones as stress-sensing `heat shock' proteins. J. Cell Sci. 115, 2809-2816. [DOI] [PubMed] [Google Scholar]
- Ogg, S., Paradis, S., Gottlieb, S., Patterson, G.I., Lee, L., Tissenbaum, H.A., and Ruvkun, G. (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994-999. [DOI] [PubMed] [Google Scholar]
- Ookuma, S., Fukuda, M., and Nishida, E. (2003). Identification of a DAF-16 transcriptional target Gene, scl-1, that regulates longevity and stress resistance in Caenorhabditis elegans. Curr. Biol. 13, 427-431. [DOI] [PubMed] [Google Scholar]
- Snutch, T.P., Heschl, M.F., and Baillie, D.L. (1988). The Caenorhabditis elegans hsp70 gene family: a molecular genetic characterization. Gene 64, 241-255. [DOI] [PubMed] [Google Scholar]
- Song, J., Takeda, M., and Morimoto, R.I. (2001). Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 3, 276-282. [DOI] [PubMed] [Google Scholar]
- Tatar, M., Khazaeli, A.A., and Curtsinger, J.W. (1997). Chaperoning extended life. Nature 390, 30. [DOI] [PubMed] [Google Scholar]
- Timmons, L., Court, D.L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. [DOI] [PubMed] [Google Scholar]
- Tower, J. (2000). Transgenic methods for increasing Drosophila life span. Mech. Ageing Dev. 118, 1-14. [DOI] [PubMed] [Google Scholar]
- Walker, G.A., and Lithgow, G.J. (2003). Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2, 131. [DOI] [PubMed] [Google Scholar]
- Walker, G.A., Thompson, F.J., Brawley, A., Scanlon, T., and Devaney, E. (2003). Heat shock factor functions at the convergence of the stress response and developmental pathways in Caenorhabditis elegans. FASEB J 17, 1960-1962. [DOI] [PubMed] [Google Scholar]
- Wang, J., Tokarz, R., and Savage-Dunn, C. (2002). The expression of TGFbeta signal transducers in the hypodermis regulates body size in C. elegans. Development 129, 4989-4998. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne, M., Stone, D.E., and Craig, E.A. (1987). Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow, C.A., Kimura, K.D., Lee, M.S., and Ruvkun, G. (2000). Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290, 147-150. [DOI] [PubMed] [Google Scholar]
- Wu, C. (1995). Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11, 441-469. [DOI] [PubMed] [Google Scholar]
- Yokoyama, K., Fukumoto, K., Murakami, T., Harada, S., Hosono, R., Wadhwa, R., Mitsui, Y., and Ohkuma, S. (2002). Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. 516, 53-57. [DOI] [PubMed] [Google Scholar]