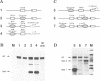
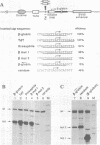
Abstract
Mammalian gene promoters for transcription by RNA polymerase II are typically organized in the following order: upstream sequence motif(s)/TATA box/initiation site. Here we report studies in which the order, orientation and DNA sequences of these three elements are varied to determine how these affect polarity of transcription. We have constructed promoters with an 'octamer' upstream sequence ATTTGCAT (or its complement ATGCAAAT) in combination with several different TATA boxes and initiation (cap) sites, and tested these promoters in transfection experiments with cultured cells. TATA boxes derived from the adenovirus major late promoter (TATAAAA), immunoglobulin kappa light chain (TTATATA) and heavy chain (TAAATATA) promoter functioned equally well or even better when inverted. Only the beta-globin TATA box (CATAAAA) was poorly active when inverted. In addition, a symmetrical TATA box (TATATATA) derived from a casein gene was very active. Our results suggest that the asymmetry of most TATA boxes (consensus TATAAAA) is not a primary determinant of the polarity of transcription. We also found that the initiation (cap) site, which usually consists of an adenine embedded in a pyrimidine-rich region (PyPyCAPyPyPyPyPy), was permissive towards sequence alterations; even a randomly composed sequence worked well. However, an inverted, hence purine-rich, cap site reduced transcript levels to 1/7th, as did an oligo G sequence. Irrespective of the presence of a cap site, the configuration: 'TATA box/octamer' yielded a strong leftward, rather than rightward transcription. From this, we conclude that the polarity of transcription is primarily determined by the linear order of an upstream sequence relative to a TATA box, rather than by the individual orientations of either of these two elements.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballard D. W., Bothwell A. Mutational analysis of the immunoglobulin heavy chain promoter region. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9626–9630. doi: 10.1073/pnas.83.24.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Sharp P. A., Guarente L. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature. 1988 Jul 7;334(6177):37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- Carcamo J., Maldonado E., Cortes P., Ahn M. H., Ha I., Kasai Y., Flint J., Reinberg D. A TATA-like sequence located downstream of the transcription initiation site is required for expression of an RNA polymerase II transcribed gene. Genes Dev. 1990 Sep;4(9):1611–1622. doi: 10.1101/gad.4.9.1611. [DOI] [PubMed] [Google Scholar]
- Cavallini B., Faus I., Matthes H., Chipoulet J. M., Winsor B., Egly J. M., Chambon P. Cloning of the gene encoding the yeast protein BTF1Y, which can substitute for the human TATA box-binding factor. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concino M. F., Lee R. F., Merryweather J. P., Weinmann R. The adenovirus major late promoter TATA box and initiation site are both necessary for transcription in vitro. Nucleic Acids Res. 1984 Oct 11;12(19):7423–7433. doi: 10.1093/nar/12.19.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Strubin M., Ponticelli A. S., Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991 Apr 19;65(2):341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Dollard C., Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989 Sep 22;58(6):1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Gidoni D., Kadonaga J. T., Barrera-Saldaña H., Takahashi K., Chambon P., Tjian R. Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science. 1985 Nov 1;230(4725):511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- Gill G., Tjian R. A highly conserved domain of TFIID displays species specificity in vivo. Cell. 1991 Apr 19;65(2):333–340. doi: 10.1016/0092-8674(91)90166-v. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989 Sep 22;58(6):1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- Henco K., Brosius J., Fujisawa A., Fujisawa J. I., Haynes J. R., Hochstadt J., Kovacic T., Pasek M., Schamböck A., Schmid J. Structural relationship of human interferon alpha genes and pseudogenes. J Mol Biol. 1985 Sep 20;185(2):227–260. doi: 10.1016/0022-2836(85)90401-2. [DOI] [PubMed] [Google Scholar]
- Heuchel R., Matthias P., Schaffner W. Two closely spaced promoters are equally activated by a remote enhancer: evidence against a scanning model for enhancer action. Nucleic Acids Res. 1989 Nov 25;17(22):8931–8947. doi: 10.1093/nar/17.22.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T., Dynlacht B. D., Peterson M. G., Pugh B. F., Tjian R. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell. 1990 Jun 29;61(7):1179–1186. doi: 10.1016/0092-8674(90)90682-5. [DOI] [PubMed] [Google Scholar]
- Hoffman A., Sinn E., Yamamoto T., Wang J., Roy A., Horikoshi M., Roeder R. G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature. 1990 Jul 26;346(6282):387–390. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989 Sep 28;341(6240):299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Luciw P. A., Duchange N. Structural arrangements of transcription control domains within the 5'-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988 Sep;2(9):1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- Kao C. C., Lieberman P. M., Schmidt M. C., Zhou Q., Pei R., Berk A. J. Cloning of a transcriptionally active human TATA binding factor. Science. 1990 Jun 29;248(4963):1646–1650. doi: 10.1126/science.2194289. [DOI] [PubMed] [Google Scholar]
- Kelley K. A., Pitha P. M. Characterization of a mouse interferon gene locus II. Differential expression of alpha-interferon genes. Nucleic Acids Res. 1985 Feb 11;13(3):825–839. doi: 10.1093/nar/13.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Müller H. P., Schaffner W. Transcriptional enhancers can act in trans. Trends Genet. 1990 Sep;6(9):300–304. doi: 10.1016/0168-9525(90)90236-y. [DOI] [PubMed] [Google Scholar]
- Müller M. M., Gerster T., Schaffner W. Enhancer sequences and the regulation of gene transcription. Eur J Biochem. 1988 Oct 1;176(3):485–495. doi: 10.1111/j.1432-1033.1988.tb14306.x. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Blair D. L., Murphy W. J., Granner D. K. Structure of the 5' ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984 May;81(9):2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Berk A. J. Multiple transcription factor binding sites mediate adenovirus E1A transactivation. J Virol. 1989 Aug;63(8):3499–3506. doi: 10.1128/jvi.63.8.3499-3506.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. G., Tanese N., Pugh B. F., Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990 Jun 29;248(4963):1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatt M. D., Rusconi S., Schaffner W. A single DNA-binding transcription factor is sufficient for activation from a distant enhancer and/or from a promoter position. EMBO J. 1990 Feb;9(2):481–487. doi: 10.1002/j.1460-2075.1990.tb08134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A., Patil N., Chao M. A constitutive promoter directs expression of the nerve growth factor receptor gene. Mol Cell Biol. 1988 Aug;8(8):3160–3167. doi: 10.1128/mcb.8.8.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman J. G., Max E. E., Leder P. A kappa-immunoglobulin gene is formed by site-specific recombination without further somatic mutation. Nature. 1979 Aug 2;280(5721):370–375. doi: 10.1038/280370a0. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Baltimore D. The "initiator" as a transcription control element. Cell. 1989 Apr 7;57(1):103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Schmidt M. C., Berk A. J., Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Hunkapiller M., Yuen D., Silvert D., Fristrom J., Davidson N. Cuticle protein genes of Drosophila: structure, organization and evolution of four clustered genes. Cell. 1982 Jul;29(3):1027–1040. doi: 10.1016/0092-8674(82)90466-4. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Stuve L. L., Myers R. M. A directly repeated sequence in the beta-globin promoter regulates transcription in murine erythroleukemia cells. Mol Cell Biol. 1990 Mar;10(3):972–981. doi: 10.1128/mcb.10.3.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington C. A., Leder P. Rescuing the in vitro function of a globin pseudogene promoter. Nature. 1982 Jul 8;298(5870):192–195. doi: 10.1038/298192a0. [DOI] [PubMed] [Google Scholar]
- Tamura T., Sumita K., Hirose S., Mikoshiba K. Core promoter of the mouse myelin basic protein gene governs brain-specific transcription in vitro. EMBO J. 1990 Oct;9(10):3101–3108. doi: 10.1002/j.1460-2075.1990.tb07507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K., Hirose S., Suzuki Y. In monkey COS cells only the TATA box and the cap site region are required for faithful and efficient initiation of the fibroin gene transcription. Nucleic Acids Res. 1984 Feb 10;12(3):1543–1558. doi: 10.1093/nar/12.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C., Weber H. The interferon genes. Prog Nucleic Acid Res Mol Biol. 1986;33:251–300. doi: 10.1016/s0079-6603(08)60026-4. [DOI] [PubMed] [Google Scholar]
- Westin G., Gerster T., Müller M. M., Schaffner G., Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987 Sep 11;15(17):6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland S., Schatt M. D., Rusconi S. Role of TATA-element in transcription from glucocorticoid receptor-responsive model promoters. Nucleic Acids Res. 1990 Sep 11;18(17):5113–5118. doi: 10.1093/nar/18.17.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Staudt L., Baltimore D. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature. 1987 Sep 10;329(6135):174–178. doi: 10.1038/329174a0. [DOI] [PubMed] [Google Scholar]
- Yoshimura M., Oka T. Isolation and structural analysis of the mouse beta-casein gene. Gene. 1989 May 30;78(2):267–275. doi: 10.1016/0378-1119(89)90229-1. [DOI] [PubMed] [Google Scholar]