Abstract
Amelogenin is critical for enamel formation and human AMELX gene mutations cause hypoplastic and/or hypomaturation enamel phenotypes. The Amelx null (AKO) mouse has a severe hypoplastic phenotype. This study evaluated the effect of amelogenin loss on enamel formation and crystallite morphology. Enamel from AKO and wild type (WT) mice was used. AKO mice were mated with transgenic mice expressing the most abundant known amelogenin isoform TgM180-87 to rescue (KOM180-87) the enamel crystallite phenotype. Molar enamel was embedded, sectioned with a diamond microtome and photographed using transmission electron microscopy. Crystallite sizes from multiple sections were measured using Image J. Crystallite mean thicknesses were (WT = 26 nm, AKO = 16 nm, KOm180-87 = 25 nm) and the mean widths were (WT = 96 nm, AKO = 59 nm, KOm180-87 = 85 nm). Despite a complete loss of amelogenin in AKO mice, a mineralized enamel layer with well-defined and organized crystallites forms. Enamel crystallites forming in the absence of amelogenin were reduced in thickness and width. For the first time we show that introduction of the m180 amelogenin isoform into the AKO mouse through crossbreeding rescues the crystallite phenotype. We conclude that amelogenin is essential for the development of normal crystallite size.
Keywords: amelogenin, crystallite, transgenic mice, amelogenesis imperfecta
The enamel extracellular matrix (ECM) is known to be essential for the development of enamel that has a normal architecture and composition (1). The exact role the different matrix components play is not fully understood. Amelogenin, the most abundant protein of the enamel extracellular matrix, is thought to function to as a regulator of crystallite growth (2). More specifically it has traditionally been thought that amelogenin and the enamel extracellular matrix help organize crystallite orientation and direction of growth (3). This has been hypothesized to occur by binding and/or organization of the ECM around the developing enamel crystallites (4). Earlier reports have suggested that the ECM serves as a space holder around the developing crystallites and that amelogenin actually inhibits crystallite growth (5-6). Directed processing of the enamel proteins is then thought to regulate crystallite growth specificity by providing control as to face specific growth and defining the ultimate crystallite morphology (7).
The traditional enamel growth paradigm has been challenged recently (8). Several aspects of in vivo observations are cited as being inconsistent with the classic enamel crystal growth theory. As an example, it is noted that enamel crystallites grow in width and thickness during the secretory phase when most of the ECM remains in place and that even when there are proteinase deficiencies, such as occur with the loss of MMP20 or KLK4 mutations, the enamel crystallites still grow in width and thickness (9-10). It has been suggested that the mineralization front provides shape to the enamel crystallites where thin ribbons of mineral develop as the precursor and transition to carbonate substituted hydroxyapatite based enamel crystallites (8). In the absence of enamelin this mineralization front fails to produce these developing ribbons and organized enamel crystallites do not form (11). If the enamel ECM is lacking amelogenin, but other enamel proteins are present, this initial enamel mineral organization does occur (12-13). Amelogenin self assembles into complex three dimensional structures that are believed to contribute to its role in enamel formation (14-15). Given the continued controversy over the function and role of the enamel ECM in enamel crystallite growth, we have undertaken a series of studies to help define the role of amelogenin and its alternatively spliced products on enamel development. The purpose of this study was to evaluate the effect of a loss of amelogenin on crystallite size and shape using the amelogenin knockout mouse as a model.
Materials and Methods
Methods for generating the Amelx null mouse (AKO) and the hypoplastic enamel phenotype have been described previously (12). Mice were housed in an AAALAC accredited facility and treated using procedures approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Male AKO, AKO crossed with TgM180-87 mice, and Amelx (+/y) mice were used for crystallite size studies. Crossing of the AKO and TgM180-87 mice produced offspring lacking the native Amelx gene so no alternative spliced products can be produced but do have an Amelx transgene that produces the most abundant amelogenin isoform (M180). The rescue mouse line is designated as KOM180-87 with the 87 designating a specific founder and registered as Tg(AMELX*M180)87 (Mouse Genome Database, The Jackson Laboratory, Bar Harbor, ME, USA).
Enamel was dissected from first molars of the different mice and embedded in epoxy resin. Multiple ultrathin sections were then cut from each sample and examined using transmission electron microscopy (JEOL TEM 100CXII). Multiple micrographs were taken from each section. Multiple crystallites with clearly delineated margins were then measured in each field using the Image J morphometric software program to establish a mean crystallite width and thickness. A mean crystallite width and thickness score was generated for each sample and these means used to test for size difference in each of the mouse models (n = 7 WT, 6 – AKO and 5 – Rescue mice were evaluated KOM180-87). Differences in crystallite dimensions were evaluated using ANOVA accepting p ≤ 0.05 as significant and Tukey Multiple Mean Test.
Results
Well-delineated crystallites were evident in all the different mouse teeth examined using TEM. There was a range of crystallite sizes seen within each different group of animals and in any given section. Crystallite thicknesses were typically easier to discern in the photomicrographs than were crystallite widths. Results of three independent examiners were similar so the final data set was limited to the observations of SG. The number of crystallites measured for each mouse varied depending on quality of sections and images. For example WT mouse G266 had 71 crystallites measured on 11 different sections while mouse G52 had 11 crystallites measured on two sections. The mean of crystallites measured for each mouse were then used in the final statistical analysis so the final sample size was the same as the number of mice as described in the methods section. This approach provided a stringent statistical evaluation of possible differences in crystallite dimensions for each group of animals.
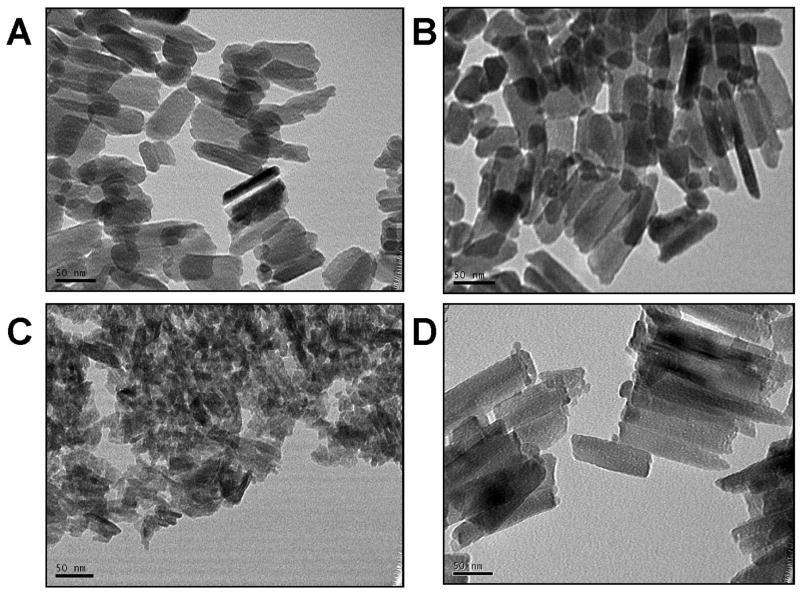
Examples of typical TEM micrographs for each of the animal groups are shown (Fig. 1). The AKO crystallites appeared substantially smaller than the WT and rescue samples (Fig. 1C). While there were areas with crystallites organized parallel to each other in the AKO mice, they did not have the same degree of organized orientation as seen in the WT or KOM180-87 rescue mice. The parallel crystallite organization in the rescue mice appeared similar to that seen in the WT mice as illustrated in Fig. 1.
Figure 1.
Enamel crystallites in the WT mice (A and B) were well-developed and often showed a parallel organization with respect to adjacent crystallites. In contrast the AKO enamel crystallites (C) were smaller and less organized than the WT crystallites. The KOM180-87 mouse (D) showed a return to normal crystallite development in both size and with an often parallel orientation similar to that seen in the WT enamel. (TEMs all exposed at 270K magnification) (scale bar = 50 nm).
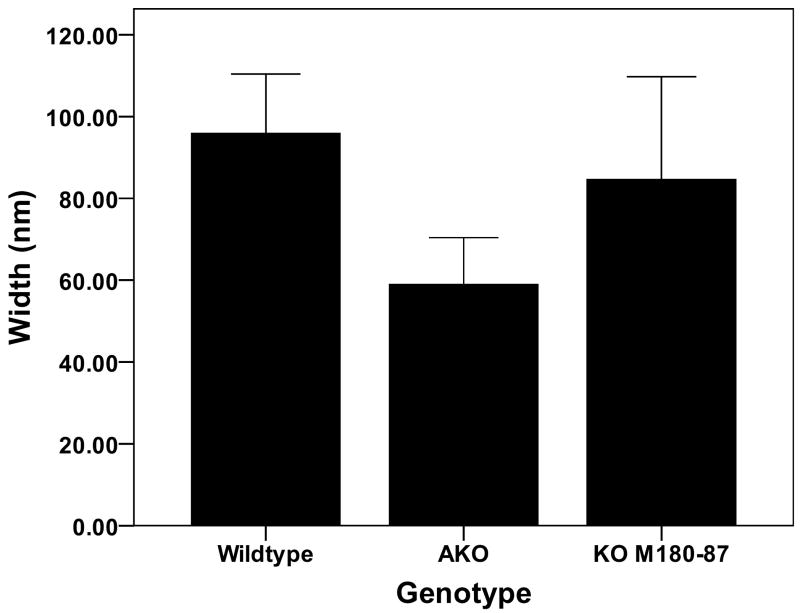
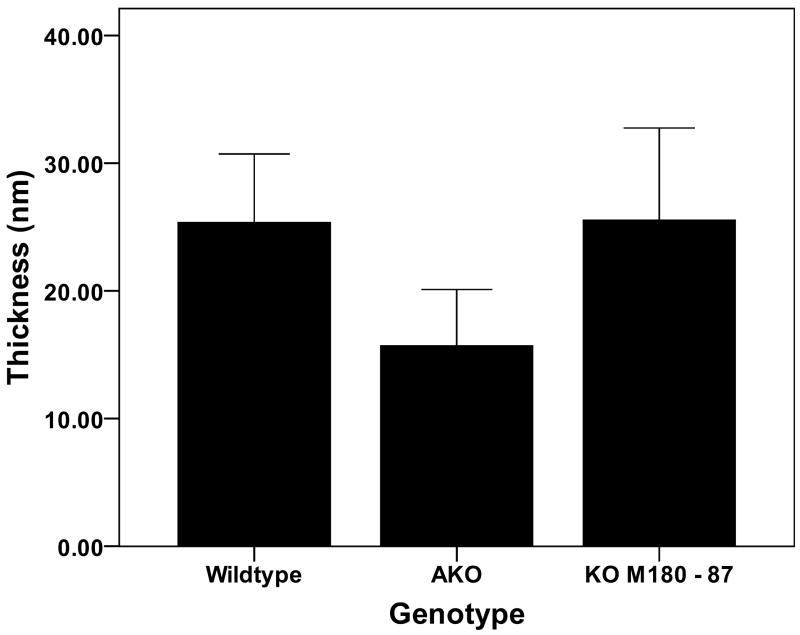
The mean enamel widths and thicknesses for each genotype are illustrated in Figs. 2 and 3. The enamel crystallite widths were significantly greater (p = 0.005) in the WT (96 ± 14 nm) compared with the AKO enamel crystallites (59 ± 11 nm). The rescue mice, KOM180-87, had enamel crystallite widths (85 ± 25 nm) that were similar to the WT mice and were not significantly different from the AKO mice (p = 0.079). The crystallite thicknesses were significantly larger comparing WT (25 ± 5 nm) and KOM180-87 (26 ± 7 nm) with the AKO enamel crystallites (mean 16 nm ± 4) (p = 0.023 and p = 0.035, respectively). The crystallite width to thickness ratios were similar for the three groups of animals (WT = 3.8. AKO = 3.7, KOM180-87 = 3.3)
Figure 2.
The enamel crystallite widths measured from TEM photomicrographs were significantly reduced in the AKO mice compared with the WT mice. The KOM180-87 mice had widths that were similar to the WT mice.
Figure 3.
The enamel crystallite thickness was markedly reduced in the AKO mice compared with the WT enamel. Replacement of amelogenin in the KOM180-87 mice rescued the crystallite phenotype and allowed development of crystallites with normal thicknesses.
Discussion
Although the exact role of the enamel extracellular matrix molecules during enamel formation remains controversial, results of the present in vivo study using a transgenic mouse approach further illustrates the important relationship between the presence of amelogenin and the enamel crystallite size and orientation. Humans with AMELX associated amelogenesis imperfecta also have abnormal enamel formation that is in many instances similar to those changes seen in the various AmelX transgenic mouse models. Collectively these studies provide ample support for an important role of amelogenin during enamel formation (12, 16). The specific amelogenin protein alteration seen in these human conditions ultimately determines the effect on enamel formation and the clinical phenotype observed, providing further evidence as to critical functional domains in the amelogenin protein (17). As previously noted, enamel crystallites do form in the absence of amelogenin producing a thin layer of enamel that is approximately 20 μm thick (12). This is not the case in the enamelin null mouse (11) and only a very thin mineralized layer is produced when full length ameloblastin is not secreted by the ameloblasts (18-19). The ability of ameloblasts in the AmelX KO mouse to generate a thin layer of organized enamel crystallites shows that amelogenin is not essential to the initial process of enamel mineralization and crystallite organization while other enamel extracellular matrix proteins such as enamelin are critical.
The present in-vivo study assessing the role of amelogenin in the formation of enamel crystallites demonstrates that crystallite growth is significantly diminished in the absence of amelogenin. The diminution in size of the enamel crystallite in the Amelx null mouse involves all three of its axes causing a reduction in crystallite width, thickness and length (crystallite length is considered same as enamel thickness from dentin-enamel junction to enamel surface). Given that the crystallite width to thickness ratio remains the same in WT and AKO mice it appears that the loss of amelogenin does not alter appositional growth of the crystallite with any face specificity. The width to thickness ratio of enamel crystallites is reported to be 2.6 with the mean width of human crystallites being 68 and the thickness 26 nm (20-21). Crystallite measurements in the present study were similar to those of previous reports regarding thickness but the width dimension was larger than previously described. In the present study measurements of the width dimension were more variable than thickness likely due to the variance in sectioning plane that does not consistently give a true cross section of the crystallite. Despite this variation in width measurement, the similarity of width to thickness ratio for crystallites growing in the presence or absence of amelogenin indicate this abundant ECM protein does not regulate the width to thickness growth ratio. This observation suggests that binding and regulation of crystallite growth at specific crystallite faces by amelogenin is not critical for defining the crystallite aspect ratio. It has been proposed, and in-vitro studies indicate, that amelogenin can help shape crystallite growth (22-23). The presence of well-delineated crystallites in the AmelX KO mouse indicates that amelogenin is not critical to prevent crystallite fusion to a major extent as has also been hypothesized. It also has been proposed that amelogenin helps prevent random proliferation of mineralization nuclei and help promote the direction of crystallite growth (24). In the AKO mouse that is completely lacking amelogenin we did not observe the development of random foci of mineral nucleation. Enamel mineralization appeared to be initiated at the dentin-enamel junction. It is possible that random foci of mineralization did occur in the AKO mice at a greater rate than normal after the initial enamel formation but this is difficult to assess with the methods used in the present study. While initial crystallite orientation at the dentin enamel junction was ordered, there was a more random order and disorganization of crystallite directionality just a few microns from the dentin-enamel junction. There was no evidence of crystallites being organized into a prismatic architecture in the absence of amelogenin.
The use of the cross matching of AKO and M180 transgenic mice to reintroduce the amelogenin protein back into the system provides a powerful model to substantiate the potential role of amelogenin on enamel development and crystallite growth. We show that replacing the full length amelogenin using this approach essentially fully rescues the enamel crystallite morphology. This strongly supports that the alteration in crystallite growth is a direct result of amelogenin protein loss and not from other factors. Crystallite growth was normalized in the width, thickness and largely in its length with the enamel thickness being rescued as well (see Gibson et al., this issue). Interestingly, the prism structure was not typically rescued in the KOM180-87 suggesting that factors other than just the full length amelogenin could be required to routinely develop this architecture. For example, there could be alternatively spliced amelogenins that were not replaced in the present model that are critical for organizing crystallites into prisms. Previous studies suggest that the LRAP amelogenin isoform could potentially play a role in prism formation (25). Alternatively, rescue with only the full amelogenin may alter ameloblast – matrix interactions or other critical processes essential for development of the prismatic architecture.
The present study shows that normal appositional growth of the enamel crystallite is diminished in the absence of amelogenin. The mechanism for this could be altered crystallite spacing during the initial development of the first mineral phase adjacent to the dentin-enamel junction. The absence of amelogenin as a space holder between the developing enamel mineral ribbons could alter the spatial relationship of the crystallites leading ultimately to a lack of space to allow for continued crystallite growth. The distance between mineralizing ribbons of enamel at the dentin-enamel junction is 20 nm (26). Numerous studies also indicate this is similar in size to the self-assembled amelogenin protein that forms nanospheres (27). Further TEM analysis of developing enamel crystallites at the dentin-enamel junction during early secretory stage could help provide answers as to the possible function of amelogenin in the separation and organization of early developing crystallites.
The present study shows that the nascent crystallites orient in a parallel fashion in the absence of amelogenin and then display a reduced organization as the crystallites grow in length away from the dentin enamel junction compared with the WT enamel. This indicates that a parallel orientation of enamel crystallites is not solely mediated by the presence of amelogenin and that other mechanisms must play a role in crystallite orientation as well. Studies on the ameloblast enamel matrix interactions continue to suggest a critical role for normal enamel formation. Even in the absence of amelogenin, a structure similar to that seen left by the ameloblast Tomes’ process can be seen decorating the enamel surface area of the teeth (28). Studies suggest ameloblast – matrix interactions are important and new knowledge in this field continues to emerge such as the identification of a fibronectin binding domain in ameloblastin (29-30).
In summary, the present study shows well-developed and distinct crystallites are formed in the complete absence of amelogenin. The complete loss of amelogenin during enamel development results in a reduction in crystallite growth during initial enamel formation and crystallites that have the same width to thickness aspect ratio as normal enamel but that are smaller in dimension. This suggests that amelogenin is a critical determinant of the ultimate crystallite size but not essential for establishing or maintaining the width to thickness aspect ratio. The lack of prismatic structure and greatly diminished thickness of enamel in the AKO mouse shows that amelogenin is essential for the sustained growth in the length of the crystallites and that it appears to play a critical role in helping to form the complex prismatic architecture of normal murine enamel.
Acknowledgments
The work was supported by NIH grant DE011089
References
- 1.ROBINSON C, KIRKHAM J, BRIGGS HD, ATKINSON PJ. Enamel proteins from secretion to maturation. J Dent Res. 1982;61:1490–1495. [PubMed] [Google Scholar]
- 2.FINCHAM AG, BELCOURT AB. Amelogenin biochemistry:current concepts. In: Butler WT, editor. The Chemistry and Biology of Mineralized Tissues. Birmingham: Ebsco Media; 1985. pp. 240–247. [Google Scholar]
- 3.WANG L, GUAN X, DU C, MORADIAN-OLDAK J, NANCOLLAS GH. Amelogenin Promotes the Formation of Elongated Apatite Microstructures in a Controlled Crystallization System. J Phys Chem C Nanomater Interfaces. 2007;111:6398–6404. doi: 10.1021/jp0675429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LAKSHMINARAYANAN R, YOON I, HEGDE BG, FAN D, DU C, MORADIAN-OLDAK J. Analysis of secondary structure and self-assembly of amelogenin by variable temperature circular dichroism and isothermal titration calorimetry. Proteins. 2009;76:560–569. doi: 10.1002/prot.22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AOBA T, TANABE T, MORENO E. Function of amelogenins in porcine enamel mineralization during the secretory stage of amelogenesis. Adv Dental Res. 1987;2:252–260. doi: 10.1177/08959374870010021401. [DOI] [PubMed] [Google Scholar]
- 6.AOBA T, FUKAE M, TANABE T, SHIMIZU M, MORENO EC. Selective adsorption of porcine-amelogenins onto hydroxyapatite and their inhibitory activity on hydroxyapatite growth in supersaturated solutions. Calcif Tissue Int. 1987;41:281–289. doi: 10.1007/BF02555230. [DOI] [PubMed] [Google Scholar]
- 7.SIMMER JP, HU JC. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43:441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- 8.SIMMER JP, PAPAGERAKIS P, SMITH CE, FISHER DC, ROUNTREY AN, ZHENG L, HU JC. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89:1024–1038. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WRIGHT JT, DALY B, SIMMONS D, HONG S, HART SP, HART TC, ATSAWASUWAN P, YAMAUCHI M. Human enamel phenotype associated with amelogenesis imperfecta and a kallikrein-4 (g.2142G>A) proteinase mutation. Eur J Oral Sci. 2006;114(Suppl 1):13–17. doi: 10.1111/j.1600-0722.2006.00291.x. discussion 39-41, 379. [DOI] [PubMed] [Google Scholar]
- 10.CATERINA JJ, SKOBE Z, YANLI DING J-S, SIMMER JP, BIRKEDAL-HANSEN H, BARTLETT JD. Enamelysin (Matrix Metalloproteinase 20)-deficient Mice Display an Amelogenesis Imperfecta Phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 11.HU JCC, HU Y, SMITH CE, MCKEE MD, WRIGHT JT, YAMAKOSHI Y, PAPAGERAKIS P, HUNTER GK, FENG JQ, YAMAKOSHI F, SIMMER JP. Enamel Defects and Ameloblast-Specific Expression in Enamelin Knockout/LacZ Knockin mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GIBSON CW, YUAN ZA, HALL B, LONGENECKER G, CHEN E, THYAGARAJAN T, SREENATH T, WRIGHT JT, DECKER S, PIDDINGTON R, HARRISON G, KULKARNI AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 13.WRIGHT JT, HART TC, HART PS, SIMMONS D, SUGGS C, DALEY B, SIMMER J, HU J, BARTLETT JD, LI Y, YUAN ZA, SEOW WK, GIBSON CW. Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4. Cells Tissues Organs. 2009;189:224–229. doi: 10.1159/000151378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FINCHAM AG, MORADIAN-OLDAK J, SIMMER JP, SARTE P, LAU EC, DIEKWISCH T, SLAVKIN HC. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol. 1994;112:103–109. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 15.MORADIAN-OLDAK J, GOLDBERG M. Amelogenin supra-molecular assembly in vitro compared with the architecture of the forming enamel matrix. Cells Tissues Organs. 2005;181:202–218. doi: 10.1159/000091382. [DOI] [PubMed] [Google Scholar]
- 16.PAINE ML, LUO W, ZHU DH, BRINGAS P, JR, SNEAD ML. Functional domains for amelogenin revealed by compound genetic defects. J Bone Miner Res. 2003;18:466–472. doi: 10.1359/jbmr.2003.18.3.466. [DOI] [PubMed] [Google Scholar]
- 17.WRIGHT JT, HART PS, ALDRED MJ, SEOW WK, CRAWFORD PJM, HONG SP, GIBSON C, HART TC. Relationship of phenotype and genotype in X-linked amelogenesis imperfecta. Connect Tissue Res. 2003;44(suppl):72–78. [PubMed] [Google Scholar]
- 18.FUKUMOTO S, KIBA T, HALL B, IEHARA N, NAKAMURA T, LONGENECKER G, KREBSBACH PH, NANCI A, KULKARNI AB, YAMADA Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WAZEN RM, MOFFATT P, ZALZAL SF, YAMADA Y, NANCI A. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 2009;28:292–303. doi: 10.1016/j.matbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DACULSI G, KEREBEL B. High-resolution electron microscope study of human enamel crystallites: size, shape, and growth. J Ultrastruct Res. 1978;65:163–172. doi: 10.1016/s0022-5320(78)90053-9. [DOI] [PubMed] [Google Scholar]
- 21.KEREBEL B, DACULSI G, KEREBEL LM. Ultrastructural studies of enamel crystallites. J Dent Res. 1979;58:844–851. doi: 10.1177/00220345790580023701. [DOI] [PubMed] [Google Scholar]
- 22.IIJIMA M, MORADIAN-OLDAK J. Interactions of amelogenins with octacalcium phosphate crystal faces are dose dependent. Calcif Tissue Int. 2004;74:522–531. doi: 10.1007/s00223-002-0011-3. [DOI] [PubMed] [Google Scholar]
- 23.MORADIAN-OLDAK J. Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- 24.AOBA T. Recent observations on enamel crystal formation during mammalian amelogenesis. Anat Rec. 1996;245:208–218. doi: 10.1002/(SICI)1097-0185(199606)245:2<208::AID-AR8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 25.GIBSON CW, LI Y, DALY B, SUGGS C, YUAN ZA, FONG H, SIMMONS D, ARAGON M, KULKARNI AB, WRIGHT JT. The leucine-rich amelogenin peptide alters the amelogenin null enamel phenotype. Cells Tissues Organs. 2009;189:169–174. doi: 10.1159/000151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DIEKWISCH TG, BERMAN BJ, GENTNER S, SLAVKIN HC. Initial enamel crystals are not spatially associated with mineralized dentine. Cell Tissue Res. 1995;279:149–167. doi: 10.1007/BF00300701. [DOI] [PubMed] [Google Scholar]
- 27.MORADIAN-OLDAK J, PAINE ML, LEI YP, FINCHAM AG, SNEAD ML. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J Struct Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 28.PRAKASH SK, GIBSON CW, WRIGHT JT, BOYD C, CORMIER T, SIERRA R, LI Y, ABRAMS WR, ARAGON MA, YUAN ZA, VAN DEN VEYVER IB. Tooth enamel defects in mice with a deletion at the Arhgap 6/Amel X locus. Calcif Tissue Int. 2005;77:23–29. doi: 10.1007/s00223-004-1213-7. [DOI] [PubMed] [Google Scholar]
- 29.FUKUMOTO S, YAMADA A, NONAKA K, YAMADA Y. Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation. Cells Tissues Organs. 2005;181:189–195. doi: 10.1159/000091380. [DOI] [PubMed] [Google Scholar]
- 30.BEYELER M, SCHILD C, LUTZ R, CHIQUET M, TRUEB B. Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp Cell Res. 2010;316:1202–1212. doi: 10.1016/j.yexcr.2009.12.019. [DOI] [PubMed] [Google Scholar]