Abstract
The methylenetetrahydrofolate reductase (MTHFR) genes and folate in one-carbon metabolism are essential for DNA methylation and synthesis. However, their role in carcinogen DNA damage in target lung tissue, a dosimeter for cancer risk, is not known. Our study aimed to investigate the association between genetic and nutritional one-carbon metabolism factors and DNA adducts in target lung. Data on 135 lung cancer cases from the Massachusetts General Hospital were studied. Genotyping was completed for MTHFR C677T (rs1801133) and A1298C (rs1801131). Information on dietary intake for one-carbon related micronutrients, folate and other B vitamin, was derived from a validated food frequency questionnaire. DNA adducts in lung were measured by 32P-postlabeling. After adjusting for potential confounders, DNA adduct levels in lung significantly increased by 69.2% [95% confidence interval (CI), 5.5% to 171.5%] for the MTHFR 1298AC+CC genotype. The high risk group, combining the A1298C (AC+CC) plus C677T (CT+TT) genotypes, had significantly enhanced levels of lung adducts by 210.7% (95% CI, 21.4% to 695.2%) in contrast to the A1298C (AA) plus C677T (CC) genotypes. Elevation of DNA adduct was pronounced - 111.3% (95% CI, −3.0 to 360.5%) among 1298AC+CC patients who consumed the lowest level of folate intake as compared with 1298AA individuals with highest tertile of intake. These results indicate that DNA adducts levels are influenced by MTHFR polymorphisms and low folate consumption, suggesting an important role of genetic and nutritional factors in protecting DNA damage from lung carcinogen in at-risk populations.
Keywords: MTHFR, folate, genetic polymorphisms, DNA adducts, one carbon metabolism
INTRODUCTION
DNA adducts serve as a reliable marker of tobacco-associated exposure and cancer 1. DNA adducts caused by tobacco smoking carcinogen such as polycyclic aromatic hydrocarbons (PAHs), listed as group 1 (carcinogenic to human) by International Agency for Research on Cancer (IARC), are formed by covalently binding to DNA. If the DNA adducts are left unrepaired, the resultant DNA damage can result in mutation that may ultimately lead to development of lung cancer. The methylenetetrahydrofolate reductase (MTHFR)genes and folate in one-carbon metabolism are essential for DNA synthesis, repair, and methylation. A few human studies have examined their role in DNA damage in non-target tissue, peripheral blood 2,3, but none have explored in carcinogen DNA damage in the target tissue, lung, as a molecular dosimeter for lung cancer.
Folate, a water soluble vitamin B9, has been considered as an ‘essential vitamin’ because it modulates potential DNA damage and the risk of developing cancer, although not consistently 4. In vivo and in vitro evidences suggest that folate deficiency results in DNA damage and instability, and altered DNA methylation, and eventually result in cell death via apoptosis, all of which may promote tumor initiation 5–9. The MTHFR protein, a central enzyme in folate metabolism, has been implicated with lung cancer risk 10–13, because it is involved in methyl group synthesis process by catalyzing the irreversible conversion of 5, 10-methylenetetrahydrofolate (THF) to 5-methyl THF, which serves as methyl donor for the remethylation of homocystein to methionine and the precursor of S-adenosylmethionine (SAM). The polymorphisms in the MTHFR gene: C677T and A1298C are known to have functional relevance and thus variation of MTHFR in the folate metabolic pathway may be associated with variable folate levels 14 and is suspected of influencing the risk of lung cancer.
In this study, we aimed to investigate independent and combined effects of two polymorphisms of a key one-carbon pathway gene, MTHFR C677T and A1298C, and as well as dietary folate intake on DNA damage in target lung tissue. As DNA adducts in target lung tissue are mainly formed by cigarette smoking, the main source of exposure that contribute to the increased risk of lung cancer in smokers 15, DNA adducts were analyzed in uninvolved lung tissues in lung cancer patients with exposed to cigarette smoking.
MATERIALS AND METHODS
Study population
The study population consisted of 142 lung cancer patients at Massachusetts General Hospital(MGH, Boston, MA), as described previously 16, 17. Among the participants, nonsmokers (n = 7) were excluded. Surgically resected non-involved lung tissue was sampled from the same lobe, distal to tumor in patients. Lung tissue specimens were frozen immediately on dry ice and stored deep-frozen at −80 until DNA adduct analysis. Blood samples for genotyping, socio-demographic information(including age, gender, and smoking status), and dietary intake were collected at the time of recruitment by trained personnel. Informed consent was obtained from all study participants. This study was approved by the Committees on the Use of human Subjects in Research at the MGH and the Harvard School of Public Health.
DNA adduct analysis
We used previously reported data on DNA adducts in lung samples determined by the 32P-postlabeling assay 18, 19. These DNA adducts are considered primarily to represent tobacco-derived aromatic hydrophobic adducts, mainly polycyclic aromatic hydrocarbon(PAH)-DNA adducts 18, 19. The half-life of DNA adduct in the lung tissue of lung cancer patients has been reported to be approximately 1.7 years, indicating that DNA adduct persist longer in lung tissues than other tissues 20. Total relative DNA adducts were measured in the diagonal reactive zone plus discrete adducts as in prior studies 18, 19. Each sample was repeated at least twice as a validation analysis and average adducts levels were obtained from the combination of all experiments of the relative adduct levels. The coefficient of variation for the repeated measurements was 14% for the positive control sample 19.
Genotyping
DNA was extracted from peripheral blood samples using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN). Of the 135 patients, the MTHFR polymorphisms, C677T (rs1801133) and A1298C (rs1801131), were genotyped in 96 and 102 patients using the TaqMan method with an ABI7900HT sequence detection system (Applied Biosystems, Foster City, CA) and a random 5% of the samples were repeated for the validation of genotyping procedures. Genotypes were verified by two independent readers.
Assessment of Dietary Intake
Dietary folate intake was assessed from the Harvard-Willet validated food frequency questionnaire (FFQ) of each patient at the time of recruitment 21, 22, asking how often on average the subject have used the amount specified during the past one year. The FFQ, designed to estimate an individual’s habitual intake over a defined period of time (e.g., a year), has been used extensively as the standard tool for dietary assessment in epidemiologic studies. The 126-food item FFQ, developed by the Nutrition Department at the Harvard School of Public Health, has been validated in a group of female Caucasian nurses 22 and male health professionals 21 living in Boston. In the FFQ, a commonly used unit or portion size was specified for each food item or supplement, and subjects were asked about their average consumption over the past year before enrollment. Estimated average intakes for each specific food were obtained and nutrient intake was computed using the Harvard database, which is a modification of the U.S. Department of Agriculture Nutrition Composition Laboratory’s food composition database.
Statistical analysis
The dependent variable, DNA lung adduct per 1010 nucleotides, was transformed using natural logarithm to improve normality and to stabilize the variance. Genotypes were coded as wild type (major-allele homozygote) and variant genotype (minor-allele homozygote + heterozygote). Dietary folate intake values were fist adjusted for total energy intake using multivariate nutrient-density model based on an established method 23. Because nutrient intake highly depends on total energy intake, adjustment for total energy intake is necessary. Energy-adjusted folate categorized into tertile with the highest tertile as the reference group. Potential confounders including age at diagnosis(continuous), gender(male and female), smoking status (ex-smoker and current smoker), and pack-years of smoking (continuous)were adjusted in the multivariate analysis. We estimated the percent change in DNA lung adduct levels for the risk genotype compared with the common allele as [eβ − 1]×100%, with 95% CI [e(β ± 1.96×SE) −1]× 100%, where β and SE are the estimated regression coefficient and its standard error from multiple regression analysis. To examine the combined effects of C677T and A1298C, we constructed a model that included all possible combinations between C677T and A1298C polymorphisms, with the low-risk combination of the homozygous wild-type genotype, 677CC plus 1298AA, as a reference category. To analyze whether DNA adduct level changed with the tertile of folate intake, trend tests were conducted by treating each category as a continuous variable in a regression model. We also assessed the nonlinear relationship by fitting the energy-adjusted folate intake using a natural cubic spline with 2 degrees of freedom. All statistical analyses were performed using SAS version 9.2 (SAS Institute Inc., Carry, NC, USA) and R (version 2.11.0; The R Foundation for Statistical Computing 2010).
RESULTS
Details of demographic and clinical characteristic of the study population are presented in Table 1. The geometric mean (GM) of DNA adduct levels was 86.2 adducts per 1010 nucleotides in lung tissue(mean, 172 adducts per 1010 nucleotides). The mean daily intake was 487.5μg for folate and 2,207 kcal for total energy, respectively. To check for possible selection bias due to lack of MTHFR genotyping data on 25% of the population, we analyzed whether differences exist between the groups, classified by cases with MTHFR genotypes (n = 94) and cases without MTHFR genotype (n = 41). We found no significant differences in variables including age, gender, smoking status, histology, pack-years, energy-adjusted folate, and lung DNA adduct levels.
Table 1.
General and clinical characteristics of patients of lung cancer (N=135)
| N (%) or mean ±SD | |
|---|---|
| Age at diagnosis, y | 66.3 ± 10.6 |
| Gender (male) | 78 (57.8) |
| Smoking | |
| Current | 56 (41.5) |
| Former | 79 (58.5) |
| Histology | |
| Adenocarcinoma | 65 (48.1) |
| Squamous | 44 (32.6) |
| Others | 26 (19.3) |
| Pack-years, y | 63.5 ± 40.6 |
| Folate, μg/d | 487.5 ± 259.5 |
| Total energy intake, kcal/d | 2,207.7 ± 738.5 |
| Lung DNA adduct levels, adducts per 1010 nucleotides† | 86.2 ± 4.7† |
| MTHFR C677T (rs1801133)‡ | |
| CT+TT | 47 (46.1) |
| CC | 55 (53.9) |
| MTHFR A1298C(rs1801131)‡ | |
| AC+CC | 58 (60.4) |
| AA | 38 (39.6) |
Geometric mean ± geometric SD
The reference SNP identification numbers (ref SNP ID) are from the NCBI SNP database (http://www.ncbi.nlm.nih.gov/SNP).
Genotype distribution of the MTHFR polymorphisms and their association with DNA lung adduct levels are given in Table 2. The DNA adduct levels in lung tissue significantly increased by 69.2% (95% CI, 5.5% to 171.5%) for A1298C AC+CC genotypes, but no association was founded with C667T CT+TT genotypes. In addition to the independent effect of each MTHFR genes, we also examined combined effect of A1298C and C677T genotype, in which the low-risk combination of the homozygous wild-type genotype, 677CC plus 1298AA, as the reference group in comparison. The high risk group, 677CT+TT plus 1298AC+CC, was highly significantly associated with an enhanced DNA adduct level in the lung of 210.7% (95% CI, 21.4 % to 695.2%, P = 0.02) compared with the low-risk reference group. There was a trend towards increased level of DNA lung adducts for increased risk combination of MTHFR genotypes (P for trend = 0.012).
Table 2.
Adjusted geometric mean (GM)† and estimated percent changes (95% CIs)† in DNA adducts in lung by MTHFR polymorphisms
| N (%) | Adjusted GM | % Change (95% CI)† | |
|---|---|---|---|
| MTHFR A1298C | |||
| AC+CC | 47 (46.1) | 336 | 69.2 (5.5 to 171.5)* |
| AA | 55 (53.9) | 199 | Reference |
| MTHFR C677T | |||
| CT+TT | 58 (60.4) | 240 | 1.2 (−41.5 to 75.2) |
| CC | 38 (39.6) | 237 | Reference |
| Combined MTHFR | |||
| A1298C (AC+CC) and C677T (CT+TT) | 18 (19.2) | 376 | 210.7 (21.4 to 695.2)* |
| A1298C (AC+CC) and C677T (CC) | 27 (28.7) | 313 | 159.0 (6.4 to 530.7)* |
| A1298C (AA) and C677T (CT+TT) | 38 (40.4) | 216 | 78.5 (−23.8 to 318.0) |
| A1298C (AA) and C677T (CC) | 11 (11.7) | 121 | Reference |
| P for trend | 0.012 | ||
Adjusted for age, gender, smoking status, pack-years, and total energy adjusted-folate intake.
P < 0.05
Table 3 reports joint associations of MTHFR genotypes and dietary folate intake with DNA adduct levels. The DNA adducts levels in the lung significantly decreased by −92.6% (95% CI, −99.2% to −30.3%) with increased level of dietary folate intake (μg/kcal) among patients with 1298AC+CC genotypes, but no associations were found with C677T genotypes. The DNA adduct levels were elevated by 111.3% (95% CI, −3.0 to 360.5%) among 1298AC+CC patients who consumed the lowest tertile of folate intake compared with 1298AA individuals with highest tertile of intake (P for trend = 0.067), whereas no associations were found with C677T polymorphism. The interaction terms between folate intake and MTHFR polymorphisms in relation to DNA adduct in the lung were not statistically significant. When stratifying by smoking status, similar and consistent decreases in DNA adduct levels in the lung were observed among former (−90.2%, 95% CI −99.8 to 423.2), and current smokers (−91.5%, 95% CI −99.7 to 122.8) with increased level of folate among cases with 1298AC+CC genotypes, but our analysis with small sample size suffers from a lack of statistical power to find associations. We further analyzed the effects of vitamin B6 and B12 on DNA adduct levels in the lung. No associations were found in overall subjects. A weak association was found with vitamin B6 among patients with 1298 A+CC genotypes(−74.9%, 95% CI, −92.4 to −17.4), but no significant trend was observed (P for trend = 0.68).
Table 3.
Estimated percent changes (95% CIs)† in DNA lung adducts associated with folate intake by MTHFR polymorphisms
| Overall‡ | MTHFR A1298C | MTHFR C677T | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| AC+CC | AA | CT+TT | CC | |||||||
|
| ||||||||||
| Adjusted GM | % Change (95% CI) | Adjusted GM | % Change (95% CI) | Adjusted GM | % Change (95% CI) | Adjusted GM(95% CI) | % Change (95% CI) | Adjusted GM | % Change (95% CI) | |
| Folate, μg/kcal | −77.0 (−97.0 to 75.1) | −92.6 (−99.2 to −30.3)* | −65.9 (−98.6 to 726.7) | −77.8 (−98.7 to 287.2) | −91.8 (−99.8 to 176.8) | |||||
| Folate, μg/kcal, tertile | ||||||||||
| Low | 356 | 76.7 (−8.9 to 242.7) | 495 | 111.3 (−3.0 to 360.5) | 180 | 81.9 (−29.5 to 369.4) | 327 | 72.3 (−32.5 to 340.0) | 448 | 167.9 (−19.2 to 788.4) |
| Medium | 212 | 8.3 (−42.9 to 105.3) | 290 | 24.5 (−43.2 to 173.1) | 170 | 13.7 (−55.1 to 187.7) | 232 | 31.3 (−50.4 to 247.7) | 177 | 5.9 (−61.4 to 190.6) |
| High | 199 | Reference | 235 | Reference | 153 | Reference | 182 | Reference | 170 | Reference |
| P for trend | 0.094 | 0.067 | 0.214 | 0.188 | 0.103 | |||||
Note: Percentchange represents per unit change in DNA adduct levels (DNA adduct in lung/1010 nucleotides) with per unit increase in folate level (μg/kcal)or decreasing tertile of folate level (μg/kcal).
Adjusted for age, gender, smoking status, and pack-years.
Adjusted for age, gender, smoking status, pack-years, and MTHFR A1298C and C677T.
P < 0.05
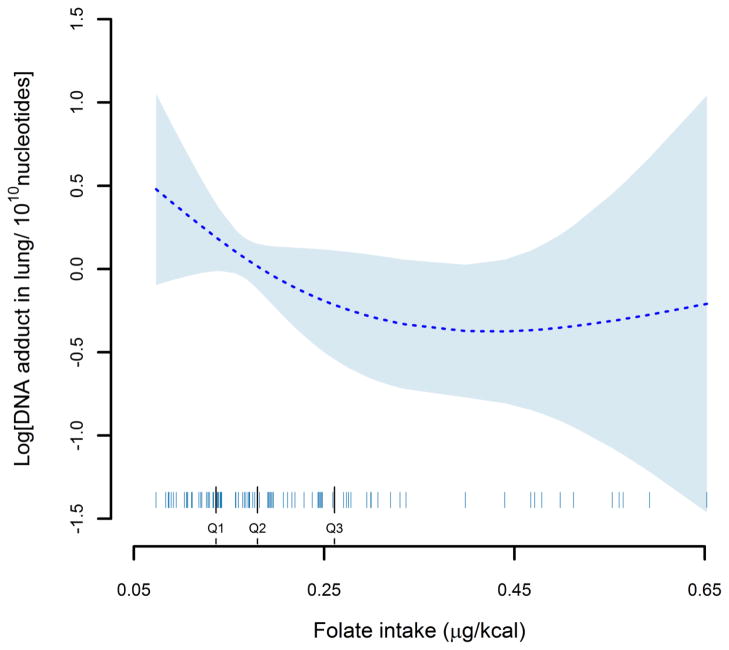
We examined the assumption of the nonlinear association between folate intake and DNA adduct in the lung, as shown in Figure 1. The DNA adducts levels in lung decreased with increasing dietary folate consumption through 0.4 μg/kcal, but it plateaued with wide 95% CIs after 0.4 μg/kcal.
Figure 1.
Hypothesized nonlinear association between folate intake and the changes in DNA lung adduct level adjusting for age, gender, smoking status, pack-years, total energy and MTHFR A1298C polymorphism. The predicted values are indicated by the dashed line and their 95% confidence intervals by the blue shades. Folate intakes of all individual subjects are indicated by short vertical lines on the abscissa.
DISCUSSION
In our study, increased levels of lung DNA adducts were associated with the MTHFR risk genotypes, A1298C variantor in combination with C677T variant. This suggests that a clustering of MTHFR risk genotypes increases DNA adduct levels in lung. In addition, an inverse association between dietary folate intake and lung adduct levels was observed among individuals with the A1298C variant genotype, implying that the combination of low folate intake and impaired folate metabolic polymorphisms may be implicated in DNA damage in target lung tissue.
To date, there is no clinical and epidemiological evidence regarding the effects of MTHFR polymorphisms and/or dietary folate intake on DNA adducts in lung. A few human studies have evaluated their influence on DNA adducts in peripheral leukocyte 2, 3 used as a surrogate for the target lung tissue 17. In the European Prospective Investigation into Cancer and Nutrition (EPIC)-Italy cross-sectional study, reduced DNA adduct levels in peripheral white blood cells were associated with the frequent consumption of fresh fruit and vegetables, which are the major sources of one-carbon nutrients including folate and the intake of antioxidants. No associations were seen with folic acid 2 and MTHFR C677T polymorphisms 3. Several studies have shown the association of an increased DNA adducts levels and the risk of lung cancer 24–28, and MTHFR genetic polymorphisms and dietary folate in one-carbon metabolism factors may modulate their link. MTHFR, a fundamental enzyme in one-carbon (methyl group) metabolism, balances the folate pool involved in DNA synthesis and methylation. The MTHFR A1298C polymorphism is associated with an increased risk of lung cancer in women, but not in men 11, and with diminished DNA methylation 29. No association was also observed between the MTHFR C677T and A1298C polymorphisms and risk of lung cancer 13. Our study found the association between DNA adducts in lung tissue with A1298C polymorphisms, but not with C677T polymorphisms, which is consistent with the previous study 3. It is still unclear why and how MTHFR C677T and A1298C genes play a role in DNA damage in the lung. This may probably be due to complex MTHFR polymorphisms that many other factors such as environmental exposure to chemicals and diet, and other genes in the same pathway, may contribute to these inconsistent results in smaller data sets 11. More extensive studies with large sample size are needed to confirm and interpret of these findings.
Folate deficiency can lead to DNA damage by causing chromosomal breaks, oxidative lesions, or both, which could contribute to the increased risk of cancer 30. Experimental studies have shown that dietary folate deficiency increases DNA damage and initiates tumor development in mice, and that MTHFR mutations play a role in this phenomenon 5. An in vitro study showed that folate depletion induces DNA instability 8 and increases chromosomal breakage (measured by micronuclei frequency) and abnormalities in human lymphocytes 31. Recent epidemiologic evidence showed that increasing serum folate levels were inversely associated with the risk of lung cancer among former and current smokers, but not among nonsmokers 32. Although low dietary folate intake was weakly correlated with lung DNA adducts levels (P = 0.096 in Table 3)among overall subjects, enhanced DNA adducts was observed with increased levels of dietary folate among individuals with MTHFR A1298C variant genotypes (P = 0.029). Marginally significant trend of increased levels of lung DNA adducts with decreased dietary folate was also observed (P for trend = 0.067).
Although the biological mechanism remains unclear for the observed associations, possible mechanisms may include the alteration of DNA methylation in one-carbon (methyl group) metabolism pathways. The disruption of homeostasis in one-carbon metabolism affects the risk of cancer 6. PAHs such as benzo[a]pyrene are metabolized in vivo to form highly genotoxic and tumorigenic diol epoxides that bind to DNA at the guanine residues forming adducts. These PAH-DNA adducts have been served as markers of biologically effective doses from exposure to tobacco smoke. The state of methylation enhances carcinogen DNA adduct formation for a particular mutation site in the p53 gene in human lung cancer 33, 34. In coke oven workers, DNA methylation was associated not only with PAHs exposure, assessed by urinary 1-hydroxypyrene, but also with anti-benzo[a]pyrene diolepoxide (BPDE)-DNA adduct levels in peripheral blood leukocytes, suggesting that the epigenetic effect of PAHs exposure involve changes in DNA methylation status 35. Folate as a methyl donor can lead to elevated DNA damage and altered DNA methylation. MTHFR is a critical gene in folate-mediated one-carbon metabolism that is responsible for DNA remethylation and DNA synthesis pathways. DNA methylation might be impaired in individuals with carrying the variant MTHFR genotypes, particularly with low folate status 6, 36, providing biological plausible link between low-folate diet and MTHFR genotypes involved in DNA methylation 37, which may modulate adduct formation 35.
A significant gene-gene and gene-nutrient interactions between MTHFR A1298C and C677T and between A1298C polymorphism and folate intake were observed in the Poison regression model (P for interaction < 0.0001) 18. The Recommended Dietary Allowance (RDA) of folate for US adults aged ≥ 19 years is 400 μg dietary folate equivalent per day 38. In this study, 48% of the subjects had folate below RDA. As a sensitivity analysis, when we re-analyzed the data using this criterion, the subjects with folate levels below RDA had significantly increased levels lung adducts by 88% (95% CI, 3.3% to 243%) as compared with individuals with folate above RDA, taking MTHFR polymorphisms into account as well as other covariates.
Our study limitations include its relatively small sample size and the inclusion of Caucasian population which limited generalizability across other populations, not unlike many other molecular epidemiologic studies. In addition, we did not see associations with other B vitamins, such as vitamin B6 and B12 involved in one-carbon metabolism in overall subjects, but only a weak association with vitamin B6 among patients with 1298 A+CC genotypes. We speculate that this is probably due to 90% and 76% of the total subjects who met current recommendations intake for B12 (2.4 μg/d)and B6 [1.3 mg/d for ages 19 to 50, 1.7 mg/d (men) and 1.3 mg/d (women) for age ≥ 51), respectively. DNA adducts may lead to lesions that are expressed as micronuclei (MN), which reflects chromosome breakage and abnormal chromosome segregation 39. Although we found increased DNA adduct levels in target lung with decreased dietary folate in small dataset, further studies using prospective designs with measures of DNA adducts, as a marker of biological effective dose, and MN frequency, as an indicator of early biological effect, are needed to confirm our findings. Further studies with large sample size are needed to determine other micronutrients that can affect the one-carbon metabolic pathways underlying the role of gene and nutrient and their interactions on DNA damage.
In summary, our results provide evidence that DNA adduct levels in target lung tissue are influenced by MTHFR polymorphisms and low folate intake, implying a significant role of genetic and nutritional factors in prevention of DNA damage from lung carcinogen in at-risk populations.
Acknowledgments
Grant Support: National Institutes of Health(CA074386, CA092824, CA090578)
The authors gratefully acknowledge the patients and physicians from the Massachusetts General Hospital in Boston and Dr. John Wiencke for technical assistance.
Abbreviations
- FFQ
food frequency questionnaire
- GM
geometric mean
- MTHFR
methylenetetrahydrofolate reductase
- PAHs
polycyclic aromatic hydrocarbons
- RDA
recommended dietary allowance
- SAM
S-adenosylmethionine
- THF
5, 10-methylenetetrahydrofolate
Footnotes
Disclosure of Potential Conflicts of Interest: None.
References
- 1.Wiencke JK. DNA adduct burden and tobacco carcinogenesis. Oncogene. 2002;21:7376–91. doi: 10.1038/sj.onc.1205799. [DOI] [PubMed] [Google Scholar]
- 2.Palli D, Vineis P, Russo A, Berrino F, Krogh V, Masala G, Munnia A, Panico S, Taioli E, Tumino R, Garte S, Peluso M. Diet, metabolic polymorphisms and dna adducts: the EPIC-Italy cross-sectional study. Int J Cancer. 2000;87:444–51. doi: 10.1002/1097-0215(20000801)87:3<444::aid-ijc21>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Palli D, Masala G, Vineis P, Garte S, Saieva C, Krogh V, Panico S, Tumino R, Munnia A, Riboli E, Peluso M. Biomarkers of dietary intake of micronutrients modulate DNA adduct levels in healthy adults. Carcinogenesis. 2003;24:739–46. doi: 10.1093/carcin/bgg003. [DOI] [PubMed] [Google Scholar]
- 4.Kim YI. Folate and carcinogenesis: evidence, mechanisms, and implications. J Nutr Biochem. 1999;10:66–88. doi: 10.1016/s0955-2863(98)00074-6. [DOI] [PubMed] [Google Scholar]
- 5.Knock E, Deng L, Wu Q, Leclerc D, Wang XL, Rozen R. Low dietary folate initiates intestinal tumors in mice, with altered expression of G2-M checkpoint regulators polo-like kinase 1 and cell division cycle 25c. Cancer Res. 2006;66:10349–56. doi: 10.1158/0008-5472.CAN-06-2477. [DOI] [PubMed] [Google Scholar]
- 6.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–42. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Fenech M. A comparison of folic acid and 5-methyltetrahydrofolate for prevention of DNA damage and cell death in human lymphocytes in vitro. Mutagenesis. 2003;18:81–6. doi: 10.1093/mutage/18.1.81. [DOI] [PubMed] [Google Scholar]
- 8.Duthie SJ, Hawdon A. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. FASEB J. 1998;12:1491–7. [PubMed] [Google Scholar]
- 9.Kim YI, Pogribny IP, Basnakian AG, Miller JW, Selhub J, James SJ, Mason JB. Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am J Clin Nutr. 1997;65:46–52. doi: 10.1093/ajcn/65.1.46. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki T, Matsuo K, Hiraki A, Saito T, Sato S, Yatabe Y, Mitsudomi T, Hida T, Ueda R, Tajima K. Impact of one-carbon metabolism-related gene polymorphisms on risk of lung cancer in Japan: a case control study. Carcinogenesis. 2007;28:1718–25. doi: 10.1093/carcin/bgm104. [DOI] [PubMed] [Google Scholar]
- 11.Shi Q, Zhang Z, Li G, Pillow PC, Hernandez LM, Spitz MR, Wei Q. Sex differences in risk of lung cancer associated with methylene-tetrahydrofolate reductase polymorphisms. Cancer Epidemiol Biomarkers Prev. 2005;14:1477–84. doi: 10.1158/1055-9965.EPI-04-0905. [DOI] [PubMed] [Google Scholar]
- 12.Boccia S, Boffetta P, Brennan P, Ricciardi G, Gianfagna F, Matsuo K, van Duijn CM, Hung RJ. Meta-analyses of the methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and risk of head and neck and lung cancer. Cancer Lett. 2009;273:55–61. doi: 10.1016/j.canlet.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 13.Shen H, Spitz MR, Wang LE, Hong WK, Wei Q. Polymorphisms of methylene-tetrahydrofolate reductase and risk of lung cancer: a case-control study. Cancer Epidemiol Biomarkers Prev. 2001;10:397–401. [PubMed] [Google Scholar]
- 14.Parle-McDermott A, Mills JL, Molloy AM, Carroll N, Kirke PN, Cox C, Conley MR, Pangilinan FJ, Brody LC, Scott JM. The MTHFR 1298CC and 677TT genotypes have opposite associations with red cell folate levels. Mol Genet Metab. 2006;88:290–4. doi: 10.1016/j.ymgme.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Godschalk R, Nair J, van Schooten FJ, Risch A, Drings P, Kayser K, Dienemann H, Bartsch H. Comparison of multiple DNA adduct types in tumor adjacent human lung tissue: effect of cigarette smoking. Carcinogenesis. 2002;23:2081–6. doi: 10.1093/carcin/23.12.2081. [DOI] [PubMed] [Google Scholar]
- 16.Lee MS, Su L, Christiani DC. Synergistic Effects of NAT2 slow and GSTM1 null Genotypes on Carcinogen DNA Damage in the Lung. Cancer Epidemiol Biomarkers Prev. 2010 doi: 10.1158/1055-9965.EPI-09-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MS, Su L, Mark EJ, Wain JC, Christiani DC. Genetic modifiers of carcinogen DNA adducts in target lung and peripheral blood mononuclear cells. Carcinogenesis. 2010;31:2091–6. doi: 10.1093/carcin/bgq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiencke JK, Kelsey KT, Varkonyi A, Semey K, Wain JC, Mark E, Christiani DC. Correlation of DNA adducts in blood mononuclear cells with tobacco carcinogen-induced damage in human lung. Cancer Res. 1995;55:4910–4. [PubMed] [Google Scholar]
- 19.Wiencke JK, Thurston SW, Kelsey KT, Varkonyi A, Wain JC, Mark EJ, Christiani DC. Early age at smoking initiation and tobacco carcinogen DNA damage in the lung. J Natl Cancer Inst. 1999;91:614–9. doi: 10.1093/jnci/91.7.614. [DOI] [PubMed] [Google Scholar]
- 20.Schoket B, Phillips DH, Kostic S, Vincze I. Smoking-associated bulky DNA adducts in bronchial tissue related to CYP1A1 MspI and GSTM1 genotypes in lung patients. Carcinogenesis. 1998;19:841–6. doi: 10.1093/carcin/19.5.841. [DOI] [PubMed] [Google Scholar]
- 21.Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC. Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 1992;135:1114–26. doi: 10.1093/oxfordjournals.aje.a116211. discussion 27–36. [DOI] [PubMed] [Google Scholar]
- 22.Willett WC, Sampson L, Stampfer MJ, Rosner B, Bain C, Witschi J, Hennekens CH, Speizer FE. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol. 1985;122:51–65. doi: 10.1093/oxfordjournals.aje.a114086. [DOI] [PubMed] [Google Scholar]
- 23.Willett WC, Howe GR, Kushi LH. Adjustment for total energy intake in epidemiologic studies. Am J Clin Nutr. 1997;65:1220S–8S. doi: 10.1093/ajcn/65.4.1220S. discussion 9S–31S. [DOI] [PubMed] [Google Scholar]
- 24.Bak H, Autrup H, Thomsen BL, Tjonneland A, Overvad K, Vogel U, Raaschou-Nielsen O, Loft S. Bulky DNA adducts as risk indicator of lung cancer in a Danish case-cohort study. Int J Cancer. 2006;118:1618–22. doi: 10.1002/ijc.21551. [DOI] [PubMed] [Google Scholar]
- 25.Peluso M, Munnia A, Hoek G, Krzyzanowski M, Veglia F, Airoldi L, Autrup H, Dunning A, Garte S, Hainaut P, Malaveille C, Gormally E, et al. DNA adducts and lung cancer risk: a prospective study. Cancer Res. 2005;65:8042–8. doi: 10.1158/0008-5472.CAN-04-3488. [DOI] [PubMed] [Google Scholar]
- 26.Perera FP, Mooney LA, Stampfer M, Phillips DH, Bell DA, Rundle A, Cho S, Tsai WY, Ma J, Blackwood A, Tang D. Associations between carcinogen-DNA damage, glutathione S-transferase genotypes, and risk of lung cancer in the prospective Physicians’ Health Cohort Study. Carcinogenesis. 2002;23:1641–6. doi: 10.1093/carcin/23.10.1641. [DOI] [PubMed] [Google Scholar]
- 27.Tang D, Phillips DH, Stampfer M, Mooney LA, Hsu Y, Cho S, Tsai WY, Ma J, Cole KJ, She MN, Perera FP. Association between carcinogen-DNA adducts in white blood cells and lung cancer risk in the physicians health study. Cancer Res. 2001;61:6708–12. [PubMed] [Google Scholar]
- 28.Cheng YW, Chen CY, Lin P, Huang KH, Lin TS, Wu MH, Lee H. DNA adduct level in lung tissue may act as a risk biomarker of lung cancer. Eur J Cancer. 2000;36:1381–8. doi: 10.1016/s0959-8049(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 29.Friso S, Girelli D, Trabetti E, Olivieri O, Guarini P, Pignatti PF, Corrocher R, Choi SW. The MTHFR 1298A>C polymorphism and genomic DNA methylation in human lymphocytes. Cancer Epidemiol Biomarkers Prev. 2005;14:938–43. doi: 10.1158/1055-9965.EPI-04-0601. [DOI] [PubMed] [Google Scholar]
- 30.Ames BN. DNA damage from micronutrient deficiencies is likely to be a major cause of cancer. Mutat Res. 2001;475:7–20. doi: 10.1016/s0027-5107(01)00070-7. [DOI] [PubMed] [Google Scholar]
- 31.Narayanan S, McConnell J, Little J, Sharp L, Piyathilake CJ, Powers H, Basten G, Duthie SJ. Associations between two common variants C677T and A1298C in the methylenetetrahydrofolate reductase gene and measures of folate metabolism and DNA stability (strand breaks, misincorporated uracil, and DNA methylation status) in human lymphocytes in vivo. Cancer Epidemiol Biomarkers Prev. 2004;13:1436–43. [PubMed] [Google Scholar]
- 32.Johansson M, Relton C, Ueland PM, Vollset SE, Midttun O, Nygard O, Slimani N, Boffetta P, Jenab M, Clavel-Chapelon F, Boutron-Ruault MC, Fagherazzi G, et al. Serum B vitamin levels and risk of lung cancer. JAMA. 2010;303:2377–85. doi: 10.1001/jama.2010.808. [DOI] [PubMed] [Google Scholar]
- 33.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–2. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 34.Denissenko MF, Chen JX, Tang MS, Pfeifer GP. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc Natl Acad Sci U S A. 1997;94:3893–8. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, Baccarelli A. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125:1692–7. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 36.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–11. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friso S, Choi SW. Gene-nutrient interactions and DNA methylation. J Nutr. 2002;132:2382S–7S. doi: 10.1093/jn/132.8.2382S. [DOI] [PubMed] [Google Scholar]
- 38.Institute of Medicine. Dietary reference intakes for thiamin, riboflavin, niacin, vitamin B6, folate, vitamin B12, pantothenic acid, biotin, and cholineed. Washington DC: National Academy Press; 1998. [PubMed] [Google Scholar]
- 39.Pavanello S, Kapka L, Siwinska E, Mielzynska D, Bolognesi C, Clonfero E. Micronuclei related to anti-B[a]PDE-DNA adduct in peripheral blood lymphocytes of heavily polycyclic aromatic hydrocarbon-exposed nonsmoking coke-oven workers and controls. Cancer Epidemiol Biomarkers Prev. 2008;17:2795–9. doi: 10.1158/1055-9965.EPI-08-0346. [DOI] [PubMed] [Google Scholar]