Abstract
Myelination by oligodendrocytes in the central nervous system (CNS) is essential for proper brain function, yet the molecular determinants that control this process remain poorly understood. The basic helix-loop-helix transcription factors Olig1 and Olig2 promote myelination, whereas bone morphogenetic protein (BMP) and Wnt/β-catenin signaling inhibit myelination. Here we show that these opposing regulators of myelination are functionally linked by the Olig1/2 common target Smad-interacting protein-1 (Sip1). We demonstrate that Sip1 is an essential modulator of CNS myelination. Sip1 represses differentiation inhibitory signals by antagonizing BMP receptor activated-Smad activity while activating crucial oligodendrocyte-promoting factors. Importantly, a key Sip1-activated target, Smad7, is required for oligodendrocyte differentiation, and partially rescues differentiation defects caused by Sip1 loss. Smad7 promotes myelination by blocking the BMP and β-catenin negative regulatory pathways. Thus, our findings reveal that Sip1-mediated antagonism of inhibitory signaling is critical for promoting CNS myelination and point to new mediators for myelin repair.
Introduction
Myelination in the vertebrate CNS by the unique, compact myelin sheaths produced by oligodendrocytes is required for maximizing the conduction velocity of nerve impulses (Zalc and Colman, 2000) and essential for normal brain function. Demyelinating injury or disease combined with failure of myelin repair impairs rapid propagation of action potential along nerve fibers, leading to nerve degeneration, and is associated with acquired and inherited disorders including devastating multiple sclerosis (MS) and leukodystrophies (Franklin, 2002; Mar and Noetzel, 2010; Trapp et al., 1998). The observation that oligodendrocyte precursor cells (OPCs) are present within demyelinating MS lesions, but fail to differentiate into myelinating oligodendrocytes, suggests that the remyelination process is inhibited at the stage of premyelinating precursors (Chang et al., 2002; Franklin and Ffrench-Constant, 2008).
A major limitation to successful myelin regeneration arises from negative regulatory pathways that operate in the demyelinating environment, such as bone morphogenetic protein (BMP), Wnt and Notch signaling (Emery, 2010; Franklin, 2002; Li et al., 2009). BMPs, members of the TGFβ family, bind to heteromeric complexes of BMP type I (mainly BMPR-Ia or b) and type II (e.g. BMPR-II) serine/threonine kinase receptors (Massague et al., 2005) and activate downstream gene expression including oligodendrocyte differentiation inhibitors Id2 and Id4 mainly through BMP receptor-activated Smads (Smad1/5/8) (Cheng et al., 2007; Samanta and Kessler, 2004). Signaling by BMPs such as BMP4 was shown to block OPC maturation and regulate the timing of myelination (Cheng et al., 2007; Hall and Miller, 2004; Samanta and Kessler, 2004; See et al., 2004). Recently, activation of canonical Wnt signaling by β-catenin stabilization was also found to inhibit oligodendrocyte myelination and remyelination (Fancy et al., 2009; Ye et al., 2009). Finally, Notch signaling activation by its downstream effectors e.g. Hes1 and Hes5 was shown to inhibit the transition of OPCs to mature oligodendrocytes and remyelination (Wang et al., 1998; Wu et al., 2003; Zhang et al., 2009).
As a potential mechanism to counter extrinsic suppressive signaling, a series of cell intrinsic factors, such as the basic helix-loop-helix (bHLH) transcription factors Olig1 and Olig2, have been identified to positively regulate differentiation of oligodendrocytes (Emery et al., 2009; He et al., 2007; Howng et al., 2010; Li et al., 2009; Wegner, 2008; Ye et al., 2009). Olig2 directs early OPC specification and differentiation (Lu et al., 2002; Yue et al., 2006; Zhou and Anderson, 2002), and similarly, Olig1 whose expression is elevated during OPC differentiation promotes oligodendrocyte maturation and is required for repair of demyelinated lesions (Arnett et al., 2004; Li et al., 2007; Xin et al., 2005). This suggests that Olig1 and Olig2 have an overlapping function in regulating myelination in the CNS. However, the underlying mechanisms that balance and coordinate extrinsic with intrinsic inhibitory cues to drive oligodendrocyte myelination are not fully understood.
We hypothesized that the downstream effector(s) regulated by both Olig1 and Olig2 may function to coordinate the inhibitory pathways to promote myelination. By performing whole-genome Chromatin Immunoprecipitation (ChIP)-sequencing and gene profiling analysis, we identified a common target gene of Olig1 and Olig2, encoding Smad-interacting protein-1 (Sip1) [also named zinc finger homeobox protein 1b (Zfhx1b) or Zeb2]. Our present studies reveal a critical role of the transcription factor Sip1 in governing CNS myelination. Sip1 inhibits BMP-Smad negative regulatory pathways while activating the expression of crucial myelination-promoting factors. In addition, we identify Smad7, a member of inhibitory Smads (I-Smads) in the Smad pathway, as a key target induced by Sip1. We show that Smad7 is required for oligodendrocyte differentiation and promotes myelination by blocking BMP and Wnt/β-catenin inhibitory pathways. Thus, by antagonizing activated BMP-Smads while inducing the I-Smad gene Smad7, Sip1 exerts dual-mode regulation of Smad signaling to control oligodendrocyte maturation. Our findings reveal a previously unrecognized role for Sip1 in governing myelination and, in addition, its direct modulation of two Smad pathways, pointing to Sip1 as a nodal point that integrates extrinsic signals and intrinsic regulators to control the myelinogenic program in the CNS.
Results
Identification of the oligodendrocyte-enriched transcription factor Sip1 as a common target of Olig1 and Olig2
To identify the target genes directly regulated by Olig2, we carried out whole-genome ChIP sequencing using purified rat oligodendrocytes. Due to the lack of ChIP-grade anti-Olig1 antibody, gene-chip microarray transcriptome analysis was used, in this case, to screen for Olig1-regulated genes. ChIP-sequencing data revealed 5,439 genes carrying candidate Olig2 binding sites with four-fold enrichment over control (Figure S1A). We compared these candidates with Olig1-regulated genes that are downregulated in the optic nerve of Olig1 null mutants (Chen et al., 2009b) and identified 398 genes (Figure S1A) as common candidate targets of Olig2 and Olig1 (Table S1). The majority of them are involved in biological processes that connect to myelination (Figure S1B). By focusing on oligodendrocyte-enriched transcriptional regulators regulated by both Olig1 and Olig2, we identified the zinc finger homeobox transcription factor Sip1/Zfhx1b. Olig2 was found to bind strongly to multiple sites around and within the Sip1 gene that are highly conserved in vertebrates (Figure S1C). The Sip1 transcript is highly enriched in the spinal white matter, and substantially downregulated in Olig2 and Olig1 null mice at embryonic day (E) E18.5 and postnatal day (P) P14, respectively (Figure 1A,B). In addition, overexpression of Olig1 and Olig2, individually or in combination, was found to activate Sip1 expression in adult rat hippocampus-derived early oligodendrocyte progenitor cells (Figure 1C) (Chen et al., 2009b; Hsieh et al., 2004). Collectively, these data suggest that the Sip1 gene is a common downstream target regulated by both Olig1 and Olig2.
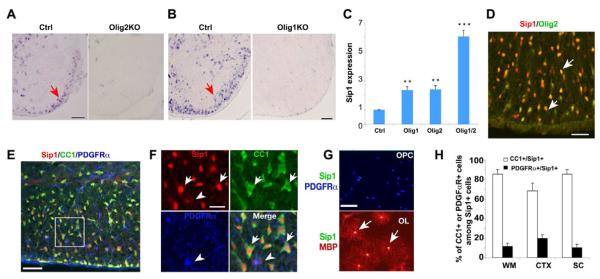
Figure 1. Identification of Olig1/2-regulated Oligodendrocyte-enriched Transcription Factor Sip1.
(A-B) Expression of Sip1 was examined in spinal cord of control and E18.5 Olig2 null (A) and P14 Olig1 null (B) mice by in situ hybridization. Arrows indicate the labeled cells.
(C) qRT-PCR analysis of Sip1 in neural progenitor cells transfected with pCS2MT-nls-Olig1, Olig2, both Olig1/2, or control vector. The fold change was present from cells transfected with Olig1 or/and Olig2 versus control (** p < 0.01, *** p < 0.001, Student’s t-test).
(D-F) The spinal cord of wildtype mice at P14 was immunostained for Sip1, Olig2, CC1, PDGFRα, respectively. Panel F showing a high magnification of panel E. Arrows indicate co-labeled cells. Arrowhead indicates a PDGFRα+ OPC.
(G) Primary rat OPCs and differentiated oligodendrocytes (OL) were immunostained for Sip1, MBP and PDGFRα. Arrows indicate the co-labeled cells.
(H) Quantification of the percentage of CC1 or PDGFRα positive cells among Sip1-positive cells in the cerebral white matter (WM), cortex (CTX) and spinal cord (SC) from P14 WT mice (n = 3).
Scale bars, in A, D, E; 50 μm; B, 100 μm; F-G, 30 μm.
To identify Sip1-expressing cell types, we performed immunohistochemistry analysis of Sip1 and co-stained for the oligodendrocyte lineage marker Olig2. Sip1 was detected in the majority, if not all, of Olig2-positive (+) cells in the white matter of the spinal cord at P14 (Figure 1D). We determined the developmental state of Sip1+ cells in the oligodendrocyte lineage, by co-labeling Sip1 with the stage-specific markers for differentiated oligodendrocytes (CC1+ or MBP+) or their precursors (PDGFRα+) in the spinal cord and in cultured oligodendrocytes. High Sip1 protein levels were detected in mature oligodendrocytes, in contrast to low levels in OPCs (Figure 1E-G). In addition, the majority of Sip1+ cells in the oligodendrocyte lineage were differentiated oligodendrocytes in the corpus callosum, cortex and spinal cord (Figure 1H). The proportions of CC1+ and Olig2+ cells among the Sip1+ cells in the spinal white matter at P14 are 82.5±5.8% and 96.0±4.0%, respectively (> 500 cell count; n=3). We did not observe Sip1 expression in GFAP+ astrocytes in white matter tracts of the CNS (data not shown). These observations suggest that Sip1 is largely confined to oligodendrocytes in the developing white matter.
Sip1 is required for oligodendrocyte maturation and myelination
To assess the functional role of Sip1 in oligodendrocyte development in vivo, we generated oligodendrocyte-lineage specific Sip1 knockout mice. Conditional Sip1flox/flox mice (Higashi et al., 2002) were bred with an Olig1-Cre line, in which Cre recombinase is produced in the oligodendrocyte lineage (Xin et al., 2005; Ye et al., 2009) (Figure 2A). We observed that all resulting mutant Sip1flox/flox;Olig1Cre+/− mice (referred to as Sip1cKO), but not their control littermates, developed generalized tremors, hindlimb paralysis and seizures from postnatal week 2 (Figure 2B, upper panel), although they were born at a normal Mendelian ratio. Sip1cKO mice exhibited the phenotypes reminiscent of myelin-deficient mice (Nave, 1994), and died around postnatal week 3, in contrast to the normal lifespan of wild-type and Sip1 conditional heterozygous control (Sip1flox/+;Olig1Cre+/−) mice (Figure 2C). The optic nerve, a well-characterized CNS white matter tract, from Sip1cKO mice was translucent compared to the control (Figure 2B, lower panels), which is a sign of severe deficiency in myelin formation.
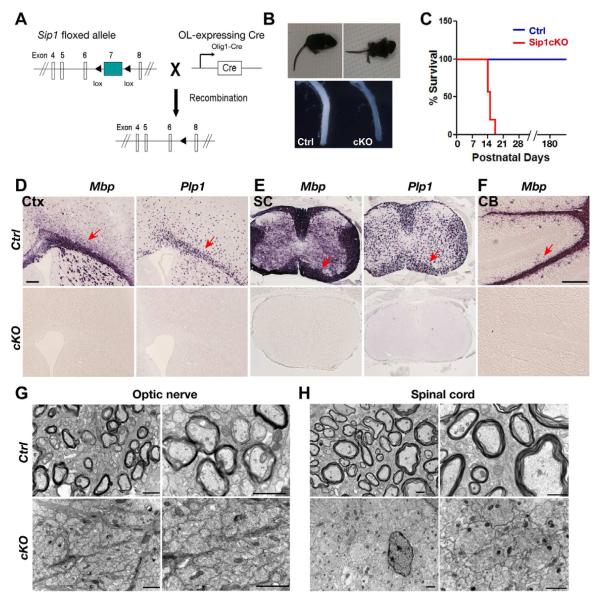
Figure 2. Sip1 is Required for Oligodendrocyte Myelination.
(A) Schematic diagram show that excision of the floxed Sip1 exon 7, which encodes a majority of protein sequence, is mediated by Olig1-Cre.
(B) Sip1cKO (Sip1lox/lox;Olig1Cre+/−) mice developed tremors and hindlimb paralysis starting around P10. Upper panel, a control mouse (Ctrl, Sip1lox/+;Olig1Cre+/−) in comparison to a cKO sibling at P14. Lower panel, optic nerves from control and Sip1cKO littermates at P14.
(C) Survival curve of control and Sip1cKO mice, which died around postnatal week 3 (n ≥ 32).
(D-F) Expression of Mbp and Plp1 in the cortex (Ctx), spinal cord (SC) and cerebellum (CB) from control (Ctrl) and Sip1cKO mice at P14 by in situ hybridization. Arrows indicate MBP+ and Plp1+ cells in the white matter.
(G-H) Electron micrographs of the optic nerve (G) and spinal cord (H) of control and Sip1cKO mice at P14. High power images (right panels in G, H) showing that multilamellar myelin sheaths are apparent around many axons in control mice, whereas all axons in Sip1cKO mice are essentially unmyelinated.
Scale bars in D, F: 100 μm; G, H: left panels, 5 μm; right panels; 1 μm
To confirm the myelin-deficient phenotypes, we examined myelin gene expression in Sip1cKO mice. In contrast to robust expression in control mice, expression of myelin genes such as Mbp (myelin basic protein) and Plp1 (proteolipid protein) is essentially undetectable in the forebrain, spinal cord and cerebellum of mutant mice at P14 (Figure 2D, F). In light of our data demonstrating that expression of mature oligodendrocyte markers was absent in Sip1cKO mice, we further examined myelin sheath assembly in the CNS of these mutants by electron microscopy. In contrast to a large number of myelinated axons that are observed in control mice at P14 (upper panels in Figure 2G,H), they were completely absent in the optic nerve and spinal cord of Sip1cKO mutants (lower panels in Figure 2G,H), indicating that myelin ensheathment has not begun in these animals. These results suggest that Sip1 is required for myelinogenesis in the CNS.
Normal oligodendrocyte precursor development in Sip1 mutant mice
Despite the deficiency in myelin gene expression, the OPC marker PDGFRα was detected in the brain and the spinal cord in the mutant mice (Figure 3A,B). The number of OPCs and their proliferation rate (percentage of Ki67+ proliferating OPCs) in Sip1 mutants were comparable to control mice (Figure 3C,D). We did not detect any significant cell death in the brain and spinal cord of Sip1cKO mice at P7 and P14 based on TUNEL assay and staining for the active form of Caspase-3 (n=3; data not shown). In addition, oligodendrocyte lineage specific Sip1 inactivation did not lead to obvious alterations of astrocytes and neurons marked by GFAP and NeuN, respectively, in the brain of Sip1cKO mice (Figure S2). Our data indicate that OPCs are able to form in the CNS of Sip1cKO mice.
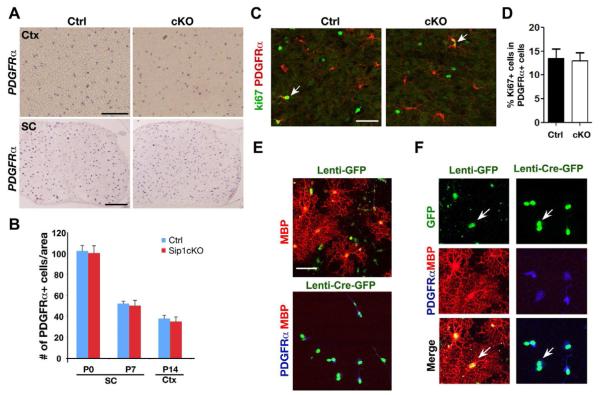
Figure 3. Normal Oligodendrocyte Precursor Development in Sip1cKO Mice.
(A) Expression of PDGFRα was examined by in situ hybridization in the P12 cortex (Ctx) and P7 spinal cord at the cervical level (SC) of control and Sip1cKO mice.
(B) Histogram depicts the number of PDGFRα+ OPCs per area (0.04 mm2) in the spinal cord and cortex at indicated stages (n = 3). Data represent mean ± SEM.
(C) Immunocytochemistry using antibodies to Ki67 and PDGFRα on the cortices of control and Sip1cKO mice at P14. Arrows indicate Ki67+/PDGFRα+ cells.
(D) Histogram depicts the percentage of Ki67-expressing cells among PDGFRα+ OPCs (n = 3). Data represent mean ± SEM.
(E) Primary OPCs isolated from the neonatal cortex of Sip1lox/lox mice at P1 were transduced with lenti-viruses expressing control GFP or Cre-GFP. Expression of MBP and PDGFRα was assessed by immunostaining seven days after switching to the oligodendrocyte differentiation medium.
(F) A higher magnification of E showing mature (MBP+) and immature (PDGFRα+) oligodendrocytes derived from lenti-GFP and lenti-CreGFP infected Sip1lox/lox OPCs. Scale bars in A, 100 μm; in C and E, 25 μm.
To investigate whether the differentiation capacity of OPCs in the absence of Sip1 in vitro is blocked, we carried out Cre-mediated Sip1 excision in cultures of purified OPCs. OPCs from the neonatal cortex of Sip1 flox/flox mice were transduced with lentivirus expressing GFP (lenti-GFP) and lenti-CreGFP. Two days post transduction, OPC cultures were switched to oligodendrocyte differentiation medium to promote oligodendrocyte maturation. In lenti-GFP transduced Sip1flox/flox cells, we observed an increase of mature MBP+ oligodendrocytes typically bearing a complex morphology during differentiation (Figure 3E,F). In contrast, under such differentiation conditions, no MBP+ oligodendrocytes were detected in lenti-CreGFP infected Sip1flox/flox cells (Figure 3E,F). All Sip1flox/flox cells transduced with lenti-CreGFP remained as PDGFRα+ OPCs (Figure 3E,F). As a control, infection of wildtype OPCs with lenti-CreGFP did not affect OPC differentiation (data not shown). These observations indicate that the ablation of Sip1 in the oligodendrocyte lineage in vivo and in vitro even under the differentiation-promoting condition prevents OPCs from further differentiation, suggesting that Sip1 is a key component of the intracellular machinery that is essential for OPC maturation.
Sip1 promotes OPC differentiation by modulating critical differentiation regulators
Given the essential role of Sip1 in oligodendrocyte maturation in vivo, we then asked whether Sip1 is sufficient to promote OPC differentiation. For this, we isolated OPCs from the neonatal rat brain and cultured these cells in oligodendrocyte growth medium containing the mitogen PDGF-AA, and then transfected these cells with expression vectors carrying a GFP-control and/or Sip1 cDNA, and immunostained for the differentiated oligodendrocyte marker RIP (Friedman et al., 1989) four days after transfection. In the control group, spontaneous OPC differentiation detected as RIP+ cells was less than 3% (Figure 4A left panel, and 4B) in the presence of PDGF-AA mitogen. In contrast, Sip1 overexpression led to a drastic increase of RIP+ mature oligodendrocytes that harbored complex processes (Figure 4A right panel, and 4B), while displaying a concomitant reduction of PDGFRα+ OPCs (Figure 4C). Similarly, there was a significant increase of galactocerebroside O1+ differentiated oligodendrocytes with Sip1 transfection (Figure 4D,E). The extent of process outgrowth measured by average circumference of O1+ oligodendrocytes with transfected Sip1 vector is significantly greater than that of spontaneous differentiated cells with control vector (179 μm ± 42 versus 119 μm ± 18, p < 0.01). These results indicate that high levels of Sip1 promote OPC maturation.
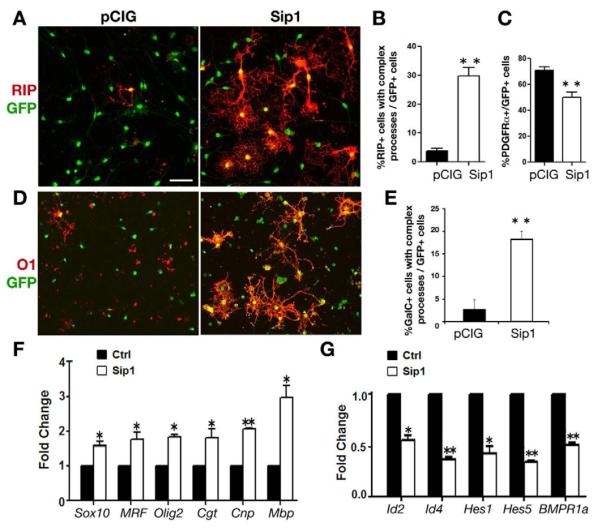
Figure 4. Sip1 Promotes OPC Differentiation by Modulating Oligodendrocyte Differentiation Regulators.
(A,D) Isolated rat OPCs were transfected with GFP-expressing control and Sip1 expressing vector and cultured in the presence of PDGF-AA for four days. Cells are stained for differentiated oligodendrocyte marker RIP (A) and O1 (D). Scale bar, 25 μm.
(B,C,E) Quantification of the percentage of RIP+ (B) and O1+ (E) cells with complex processes and PDGFRα+ cells (C) among transfected cells (n=3).
(F-G) qRT-PCR was carried to analyze myelin-associated genes and positive differentiation regulators (F) and negative regulators (G) from mRNA after 24 h transfection. Error bars indicate the mean ± SEM (n= 3). * p < 0.01, * * p < 0.001; Student’s t test.
To further examine Sip1 as a key regulator for oligodendrocyte differentiation, we performed qRT-PCR analysis of oligodendroglial gene alteration after Sip1 vector transfection. Our data revealed a significant upregulation of myelin genes such as Cnp, Cgt and Mbp, and of the genes encoding crucial differentiation activators such as Sox10, MRF and Olig2 in Sip1-transfected cells compared to the control (Figure 4F). Conversely, we observed significant downregulation of steady-sate levels of transcripts for negative regulators of differentiation, including Id2, Id4, Hes1, Hes5 and BMPR1a (Figure 4G). These results suggest that Sip1 promotes oligodendrocyte differentiation by activating positive regulators while repressing negative regulators of oligodendrocyte differentiation.
Sip1 antagonizes the inhibitory effect of receptor-activated BMP-Smad signaling on the oligodendrocyte differentiation program
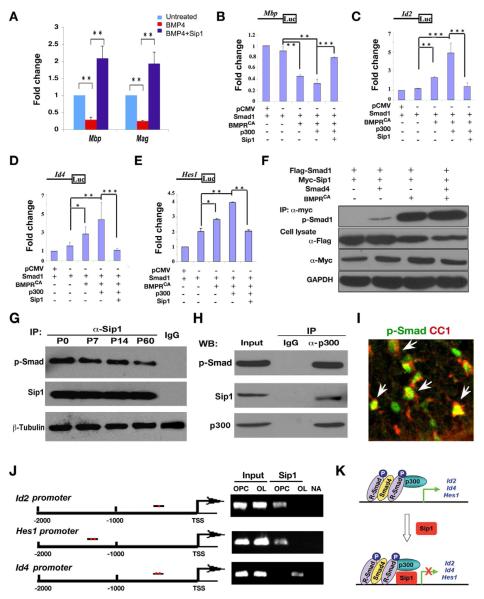
In the presence of BMP4, expression of myelin genes Mbp and Mag in differentiating oligodendrocyte precursors was inhibited (Figure 5A). However, overexpression of Sip1 was able to reverse BMP4-induced suppression of these myelin genes (Figure 5A). To investigate a possible link between the function of Sip1, identified as a Smad-interacting transcriptional repressor (Remacle et al., 1999; Verschueren et al., 1999), and BMP-Smad transcriptional activity in regulating oligodendrocyte differentiation, we examined the promoter activity of myelination-associated genes in the presence of Sip1 and activated BMP receptor signaling, which was shown to inhibit oligodendrocyte differentiation (Cheng et al., 2007; Hall and Miller, 2004). Expression of Smad1, and its subsequent activation by phosphorylation (p-Smad1) was achieved by co-transfection of expression vectors carrying Smad1 and constitutively activated BMP receptor 1b (mutant Q203D) (BMPRCA), the latter obviating the need to stimulate the cells with ligand but recapitulating faithfully receptor-mediated Smad activation (Skillington et al., 2002). This combination was found to significantly repress Mbp reporter activity (Ye et al., 2009), in a BMPRCA-dependent fashion, in the oligodendrocyte cell line Oli-Neu (Kadi et al., 2006). On the other hand, BMPRCA-activated Smad1 significantly enhanced reporter activities directed by the promoter of differentiation inhibitory genes Id2 and Id4, acknowledged downstream target genes of BMPR-Smad signaling (Samanta and Kessler, 2004), as well as of Hes1, an effector of activated Notch signaling (Ogata et al., 2010; Wu et al., 2003). Addition of p300/CBP, a co-activator of p-Smad1 (Nakashima et al., 1999; Pearson et al., 1999), further reduced the Mbp promoter activity and enhanced Hes1, Id2 and Id4 reporter activities (Figure 5B-E). In contrast, overexpression of Sip1 antagonized the inhibitory effects mediated by BMPRCA/Smad1/p300 expression on the Mbp promoter activity while repressing the promoter activity of Id2, Id4 and Hes1 activated by BMP-Smad signaling (Figure 5B-E). These results suggest that Sip1 blocks p-Smad1/p300 complex mediated transcriptional activation of oligodendrocyte differentiation inhibitors.
Figure 5. Sip1 Antagonizes the Inhibition of BMPR-Smad Signaling in Oligodendrocyte Maturation.
(A) qRT-PCR analysis of Mbp and Mag in OPC culture treated with BMP4 and/or transfected with pCIG-Sip1 for 48 hr as indicated.
(B-E) Oli-Neu cells were transfected with luciferase reporters driven by Mbp, Id2, Id4 or Hes1 promoter together with expression vectors carrying Smad1 and BMPRCA, p300 with/without Sip1 as indicated. Values represent the average of three independent experiments (n=3). Error bars indicate the mean ± SEM (* p < 0.05, * * p < 0.01, * * * p < 0.001, ANOVA).
(F) Expression vector carrying Smad1 was co-transfected with Smad4, BMPRCA and Sip1, respectively, as indicated. Co-immunoprecipitation with an antibody to Myc tagged Sip1 was carried out from cell lysates after 48 h transfection and Western-blotted with antibodies to p-Smad1, Flag and Myc-tags as indicated. IP, immunoprecipitation; GAPDH, the loading control.
(G) Co-immunoprecipitation was carried out with anti-Sip1 using cortices at P0, P7, P14 and P60 or with IgG using P7 cortex. Immunoprecipitated complexes were assayed for p-Smad and Sip1. β-tubulin was used as the loading control.
(H) Oligodendrocytes derived from purified rat OPCs cultured in differentiation medium for three days were immunoprecipitated with an antibody against p300. Immunoprecipitated complexes were Western-blotted with antibodies against p-Smad and Sip1.
(I) Immunostaining of p-Smad with an oligodendrocyte marker CC1 in P7 wildtype spinal cord. Arrows indicate the co-labeling cells.
(J) Left; a diagram shows the promoter of Id2, Hes1 and Id4 carrying the consensus Sip1 binding sites [CACCT(G)] (red dot). Right; Sip1-ChIP assay for the promoter region with consensus binding site(s) from proliferating OPCs and differentiating oligodendrocytes (OL) after 3 days exposure to the differentiation medium. Input DNA was used as positive control. NA; no antibody IP control.
(K) A schematic diagram shows that receptor-activated Smad1 forms a complex with Smad4/p300 to activate expression of differentiation inhibitors Id2/4 and Hes1 (upper panel). Sip1 is upregulated during oligodendrocyte differentiation, interacts directly with Smad1/p300 to blocks BMP-Smad activation of differentiation inhibitor expression (lower panel).
To determine whether Sip1 would interfere with p-Smad/p300 complexes and physically interact with p-Smad1, we introduced Smad4, the co-Smad of Smad1, and BMPRCA individually or in combination with Sip1, and performed co-immunoprecipitation assays. In the absence of BMPRCA, Sip1 interacted weakly with p-Smad1 as long as Smad4 was present (Figure 5F). This interaction increased dramatically when BMPRCA was introduced (Figure 5F), suggesting that Sip1 is associated with receptor-activated Smad1 likely in a complex with Smad4. To verify the Sip1-pSmad interaction at the endogenous protein level, we carried out co-immunoprecipitation assays using mouse brain tissues at different stages. Sip1 was found to interact with p-Smad in cortical tissues at P0, P7, P14 and P60 (Figure 5G). The decrease of p-Smad pulled-down by Sip1 with ages might reflect a reduction of activated BMPR-Smads when OPCs differentiate into mature oligodendrocytes (Cheng et al., 2007). To further demonstrate this interaction during oligodendrocyte differentiation, we performed a co-immunoprecipitation assay in differentiating oligodendrocytes using an antibody against p300, which was previously shown to interact with p-Smad and bridge the p-Smad transcriptional activity (Nakashima et al., 1999). Sip1 was detected in the complex of p-Smad together with p300 (Figure 5H). Given that p-Smad is observed in CC1+ differentiating oligodendrocytes in the developing spinal cord at P7 (Figure 5I), the physical interaction Sip1 with p-Smad suggests that Sip1 inhibits the p-Smad/p300-mediated negative regulatory activity during oligodendrocyte maturation.
Furthermore, endogenous Sip1 was found to bind to the Sip1-consensus binding sites of promoter regions of Id2 and Hes1 in OPCs and Id4 in differentiating oligodendrocytes (Figure 5J) by ChIP assays, suggesting that Sip1 targets directly the promoter of the genes for these differentiation inhibitors. Together, these observations suggest that Sip1 interacts with activated p-Smad and directly regulates the expression of a set of genes encoding differentiation inhibitors, thereby blocking the inhibitory effects of BMPR-Smad-p300 signaling on oligodendrocyte differentiation (Figure 5K).
Smad7 is a downstream target gene induced by Sip1 in oligodendrocyte lineage cells
As an unbiased approach to determine the downstream genes of Sip1 that regulate oligodendrocyte differentiation, we also carried out mRNA microarray profiling analysis in the spinal cord of control and Sip1cKO mice at P14. Consistent with our in situ hybridization analysis (Figure 2), myelination-associated genes including myelin genes for mature oligodendrocytes and critical differentiation regulatory genes (such as MRF and Sox10) were found remarkably downregulated in the spinal cord of Sip1 mutants (Table S2) (Figure S3).
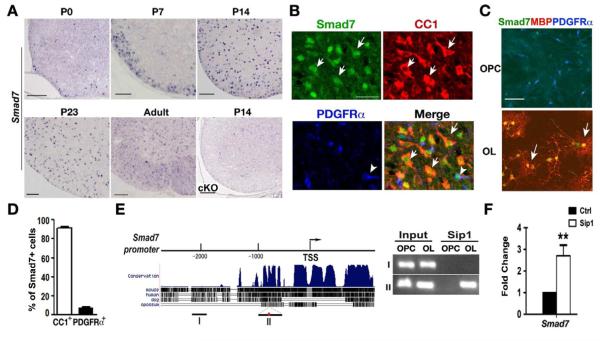
Besides previously known transcriptional regulators for myelination, the clustering analysis of the transcriptome for myelin genes revealed that Smad7 was drastically downregulated in Sip1 mutants (Figure 6A; Table S2). Smad7, a member of I-Smads, is a negative feedback regulator of signaling by liganded TGFβ and BMP receptor complexes (Massague et al., 2005). Smad7 expression appeared in the ventral spinal cord at P0, increased strongly in the spinal white matter at perinatal stages, and persisted into adulthood (Figure 6A). Consistent with the Sip1 expression pattern, intense Smad7 protein staining (Figure 6B,C) and RNA levels (Figure 6A) were predominantly detected in differentiated oligodendrocytes but not in OPCs in developing spinal cord and primary oligodendrocyte culture (Figure 6D).
Figure 6. Smad7 is an Oligodendrocyte-specific Downstream Target of Sip1.
(A) In situ hybridization using a probe to Smad7 in wildtype spinal cord at P0, P7, P14, P23 and P60 (adult), and Sip1cKO spinal cord at P14 as indicated.
(B) Immunostaining of Smad7 with CC1 and PDGFRα in the wildtype spinal cord at P14. Arrows and arrowhead indicate the CC1/Smad7 and PDGFRα/Smad7 co-labeling cells, respectively.
(C) Primary rat OPCs and differentiated oligodendrocytes were immunostained with Sip1 or Smad7 together with MBP and PDGFRα. Arrows indicate the co-labeling cells.
(D) Quantification of the percentage of CC1+ and PDGFRα+ cells among Smad7+ cells in P14 wildtype spinal cord (n=3).
(E) Left; the conserved map of the endogenous Smad7 promoter containing consensus Sip1 binding sites (red dot) among vertebrates. Right; ChIP assay for Sip1 recruitment to the Smad7 promoter with and without Sip1 binding sites in OPCs and differentiating oligodendrocytes (OL), respectively. Input DNA was used as positive control.
(F) qRT-PCR was performed to measure expression of Smad7 in OPCs transfected with control and Sip1 expressing vectors. Data represent mean ± SEM from at least three independent experiments. **p < 0.01 (Student’s t test).
Scale bars in A, 50 μm; B, 25 μm; C; 30 μm.
Analysis of the expression pattern of Sip1 and Smad7 in the spinal cord at early developmental stages indicates that Sip1 mRNA was detected as early as E16.5 while Smad7 was initially detected at P0 in the developing white matter (Figure S4), suggesting that expression of Sip1 precedes that of Smad7 in the oligodendrocyte lineage. In addition, we identified Sip1 consensus binding sites (Remacle et al., 1999) in the highly conserved Smad7 promoter (Figure 6E). To determine whether Smad7 is a direct target gene of Sip1, we performed ChIP on the chromatin isolated from OPCs and differentiated oligodendrocytes. Sip1 was recruited to the Smad7 promoter region that carries Sip1 consensus binding sites in differentiating oligodendrocytes but this enrichment was barely detectable in proliferating OPCs (Figure 6E). In addition, overexpression of Sip1 in OPCs significantly promoted Smad7 mRNA expression assayed by qRT-PCR (Figure 6F). Collectively, these data suggest that Smad7 is a direct Sip1-induced target gene in the oligodendrocyte lineage.
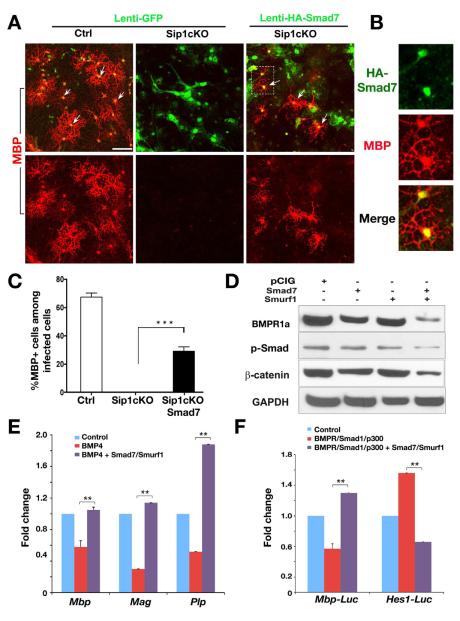
Smad7 overexpression rescues the differentiation defect of Sip1-deficient OPCs and targets inhibitory signaling pathways
If Smad7 is a critical target gene of Sip1 in myelination, introducing and overexpressing Smad7 should rescue the defect caused by Sip1 deletion. OPCs were isolated from cortices of control and Sip1cKO pups at P1 and transduced with GFP control or HA-tagged Smad7 encoding lentivirus. Under differentiation condition, robust MBP expression was detected in the culture derived from control OPCs; in contrast, no MBP+ oligodendrocytes were observed in Sip1 mutant OPCs (Figure 7A). When Sip1 mutant OPCs were transduced with Smad7 expressing lentivirus, a significant increase in MBP+ oligodendrocyte formation was detected (Figure 7A-C). Mature oligodendrocytes formed after Smad7 transduction of Sip1cKO cells were confirmed by the detection of the HA-epitope tag on Smad7 (Figure 7B). These observations suggest that Smad7 rescues, at least partially, the differentiation defect of OPCs in the absence of Sip1. In addition, Smad7 transduction in developing chick neural tube was able to promote ectopic expression of the OPC marker PDGFRα and a differentiated oligodendrocyte marker Sox10 (Figure S5), indicating that Smad7 is capable of inducing oligodendrocyte differentiation in vivo.
Figure 7. Smad7 Overexpression Rescues OPC Differentiation Defects Caused by Sip1 loss.
(A) Primary OPCs isolated from the cortices of control and Sip1cKO pups at P1 were transduced with control (lenti-GFP) and Smad7 expressing (lenti-HA-Smad7) lenti-viruses and then cultured in the differentiation medium for five days. Immunostaining was performed with antibodies to MBP and HA-tag.
(B) The white box region of the Smad7 transduced cells in panel A was shown at a high magnification. Arrows indicate MBP+/HA+ co-labeled cells.
(C) Histogram depicts the number of MBP positive or MBP and HA double positive cells after lentiviral infection. Data from three independent experiments represent mean ± SEM. *** p < 0.001 (Student’s t test).
(D) Expression vectors encoding Smad7 and/or Smurf1 were cotransfected in OPCs under the growth condition containing PDGF-AA. Western blotting with indicated antibodies was carried out from cell lysates after 48 h transfection. GAPDH was included as a loading control.
(E) Rat OPC cultures were treated with BMP4 and transfected with control vector or vectors expressing Smad7 and Smurf1. Myelin-associated genes Mbp, Mag and Plp from cell lysates were analyzed by qRT-PCR 48h after transfection. Fold change over control vector transfected cells was present. Error bars indicate the mean ± SEM (n= 3). ** p < 0.01 (Student’s t test).
(F) Expression vectors carrying BMPRCA, Smad1 and p300 with or without expression vectors or Smad7 and Smurf1 were transfected into Oli-Neu cells with the luciferase reporter driven by the Mbp or Hes1 promoter. Fold-changes over control-transfected cells were presented. Error bars indicate the mean ± SEM (n=3), ** p < 0.01 (Student’s t test).
Scale bar in A, 30 μm.
Smad7 can negatively regulate TGFβ/BMP signaling in various ways, including via forming a complex with Smurf proteins or other E3 ubiquitin ligases. The Smad7-Smurf complex was shown to target and degrade TGFβ/BMP receptors by ubiquitination, thereby attenuating TGFβ/BMP signaling at the receptor level (Kavsak et al., 2000; Suzuki et al., 2002). Smad7 was also reported to negatively regulate Wnt/β-catenin signaling (Han et al., 2006; Millar, 2006), while β-catenin stabilization inhibits oligodendrocyte myelination (Fancy et al., 2009; Ye et al., 2009). To investigate the effects of Smad7 and its cofactor E3 ubiquitin ligase Smurf1 on BMPR/Smad and Wnt/β-catenin signaling, we expressed Smad7 and Smurf1 individually or in combination in rat OPCs. Smad7 alone could slightly decrease BMPR1a and β-catenin protein levels. When cotransfected with Smurf1, Smad7 substantially downregulated BMPR1a and β-catenin steady-state protein levels (Figure 7D). Similarly, the level of p-Smad is also reduced (Figure 7D), indicating that a decrease of BMP-Smad signaling parallels with downregulation of the BMPR1a level, possibly underlying a reduced sensitivity to BMPs (Figure 7D).
Consistently, expression of Smad7 together with Smurf1 was found to reverse the inhibition of expression of myelin genes Mbp, Mag and Plp in rat OPC culture exposed to BMP4 (Figure 7E). In addition, Smad7/Smurf1 expression antagonized the inhibitory effects mediated by BMPRCA-Smad1/p300 expression on the Mbp promoter activity while repressing the Hes1 promoter activity (Figure 7F). These data agree fully with other biochemical studies in the TGFβ field that inhibitory Smads negatively regulate receptor-activated Smad signaling in BMP-stimulated cells (Massague et al., 2005). Collectively, our observations suggest that Smad7 is a critical downstream target of Sip1 and promotes oligodendrocyte differentiation indirectly by inhibiting BMP-Smad signaling and perhaps β-catenin-mediated negative regulatory pathways.
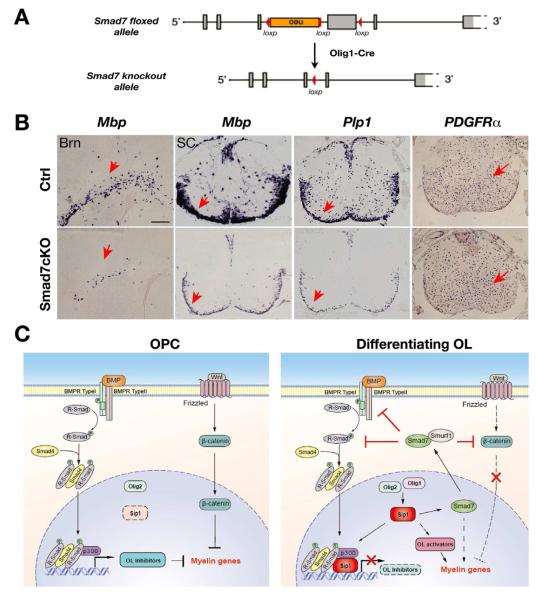
Smad7 is required for oligodendrocyte differentiation
To further determine whether Smad7 is required for oligodendrocyte development, we generated and analyzed conditional Smad7 knockout mice, with the Smad7 allele deleted in the oligodendrocyte lineage by Olig1-Cre (Chen et al., 2009a) (Figure 8A). Conventional Smad7 null embryos die in utero due to multiple defects in cardiovascular development (Chen et al., 2009a). Although Smad7cKO (Smad7flox/flox;Olig1Cre+/−) mice are viable, they developed tremors at postnatal week two. To determine the role of Smad7 in oligodendrocyte development, we examined expression of the markers for mature oligodendrocytes and their precursors in the CNS of Smad7cKO animals at P7. In the brain and spinal cord of Smad7cKO mice, the expression of the myelin genes Mbp and Plp1 was diminished in the white matter in contrast to robust expression in control mice (Figure 8B). In contrast, the OPC marker PDGFRα was detected throughout the spinal cord and the number of positive cells was comparable to that of control littermates (Figure 8B). We did not detect any significant alteration of astrocytic GFAP expression in the spinal cord of Smad7 mutant mice (data not shown). The severe downregulation of myelin gene expression in Smad7cKO mice suggests that Smad7 is critically required for oligodendrocyte differentiation.
Figure 8. Smad7 is Required for Oligodendrocyte Differentiation.
(A) Schematic diagram shows excision of the floxed exon encoding MH2 domain of Smad7 in the oligodendrocte lineage by Olig1-Cre.
(B) In situ hybridization using probes to Mbp and PDGFRα in P8 brain (Brn) and spinal cord (SC) from control (Ctrl: Olig1Cre+/−;Smad7lox/+) and Smad7cKO (Olig1Cre+/−;Smad7lox/lox) mice. Arrows indicate the labeled cells. Scale bar: 50 μm.
(C) Diagram illustrating Sip1 as a nexus that integrates extrinsic signals and intrinsic regulators to control CNS myelination. In OPCs with low Sip1 expression, activation of negative regulatory pathways such as BMP and β-catenin signaling inhibits OPC differentiation. During oligodendrocyte (OL) differentiation, elevated expression of Olig1 coordinates with Olig2 to activate Sip1 production. Sip1 promotes myelination at least in part by physically antagonizing BMP-Smad activity, and by activating critical OL activators and the I-Smad effector Smad7. Smad7 promotes oligodendrocyte differentiation directly or indirectly by blocking BMP and β-catenin signaling pathways. Thus, Sip1 may represent a novel molecular node of the regulatory network that coordinates extrinsic signaling pathways and transcriptional regulators to promote myelination in the CNS.
Discussion
BMP, Wnt and Notch signaling activation is a major obstacle for remyelination by oligodendrocytes in acute and subacute demyelinating lesions, as these pathways inhibit oligodendrocyte precursor differentiation (Fancy et al., 2010; Franklin and Ffrench-Constant, 2008; Kotter et al., 2011). However, how these myelination-inhibitory pathways are regulated at the level of receptor activation, their intracellular signal transduction and feedback components during oligodendrocyte myelination is not fully understood. In this study, we identify Sip1 as a common downstream target gene of Olig1 and Olig2. Overexpression or upregulation of Olig1 and Olig2 can activate Sip1 expression. Sip1 appears to bridge Olig activities to balance the signaling pathways mediated by BMP/Smad and Wnt/β-catenin to control the timing of oligodendrocyte myelination. Our findings point to Sip1 as a master regulator that coordinates opposing signaling pathways to promote myelination and a nexus that connects extracellular signaling pathways to intracellular transcriptional programs for myelination in the CNS.
Sip1 controls the transition from OPCs to myelinating oligodendrocytes by regulating the BMP-Smad pathway
The severe myelination defect but preservation of OPCs in the CNS of Sip1 mutants suggests that Sip1 is a key regulator for the transition from immature to mature myelinating oligodendrocytes. Sip1 is robustly upregulated during OPC differentiation in vitro and in the postnatal CNS, consistent with the requirement for Sip1 in oligodendrocyte maturation. However, low levels of Sip1 in OPCs may still regulate early steps of differentiation e.g. by targeting negative regulatory genes Id2 and Hes1 in OPCs (Figure 5), which may in turn lead to more Sip1 accumulation in a positive feedback loop.
Interaction of Sip1 with Smad1/Smad4/p300 complexes was found here to block BMP-Smad activated expression of differentiation inhibitors, leading to de-repression of myelin gene expression. Besides Smad1, we expect similar outcome of Sip1 action on the activity of other closely related Smads (i.e. the other BMP-Smads Smad5 and Smad8) by blocking the activity of p-Smads. In addition to interacting physically with p-Smads, Sip1 also antagonizes BMP signaling by activating at the transcriptional level an I-Smad, Smad7, which in turn downregulates BMP receptor signaling. Recently, Sip1 was also found to inhibit expression of BMP ligands, like the BMP4 gene (van Grunsven et al., 2007). Given that inhibition of BMP signaling e.g. by ablating BMPR1a or by adding BMP antagonists was shown to increase the number of mature oligodendrocytes and promote remyelination (Sabo et al., 2011; Samanta et al., 2007), the findings from our present studies and others suggest that Sip1 inhibits the BMP signaling pathway at multiple levels including the BMP ligand, its receptor and intracellular effectors to promote oligodendrocyte myelination.
The modulation of various differentiation regulators by Sip1 appears to be stage-dependent. For instance, Sip1 binds the Smad7 promoter when OPCs begin to differentiate, while being recruited to Id2 and Hes1 promoters in OPCs and the Id4 promoter in differentiating oligodendrocytes, respectively, suggesting that stage-specific co-factors may direct the binding of Sip1 to different targets to modulate their expression. This temporal specificity and the dual effect of Sip1-mediated promotion and repression of positive and negative regulators of myelination, respectively, may eventually promote and reinforce the process of myelination. Sip1 is a multi-domain zinc-finger E-box-binding homeobox transcription factor that may also interact with many distinct protein complexes other than p-Smads, like CtBP (Postigo et al., 2003) and the NuRD chromatin remodeling complex (Verstappen et al., 2008), to regulate the oligodendrocyte differentiation program. Whether these effects converge or exist in parallel at different stages during oligodendrocyte development, and whether Sip1 also regulates other signaling pathways as seen in different contexts (Goossens et al., 2011; Miquelajauregui et al., 2007; Seuntjens et al., 2009), are compelling new questions for future investigation. We show here that during oligodendrocyte differentiation, Sip1 inhibits BMP-Smad signaling activity by interacting directly with the receptor-activated Smad complex while activating expression of Smad7, encoding a negative feedback regulator of TGF-b/BMP signaling. These two action modes via Sip1 work in concert to inhibit negative BMP-Smad signaling activity on expression of myelin genes and therefore indirectly promote myelination (Figure 8C). Other potential Sip1 downstream components such as these encoded by MRF and Sox10 may coordinate with Smad7 to regulate myelin gene expression. Thus, Sip1 may act, even within the same cell, both as repressor and activator in a context-dependent manner, probably depending on the transcriptional co-regulators with which it cooperates at a specific time during oligodendrocyte differentiation. In either case, our findings suggest that Sip1 exerts a dualistic function via controlling the activity of distinct Smad effectors and functionally coordinate the positive and negative regulatory cues to establish the program that promotes myelination (Figure 8C).
Inhibitory Smad signaling promotes oligodendrocyte differentiation
Although BMP-Smad signaling has been reported to block oligodendrocyte maturation (Cheng et al., 2007; Miller et al., 2004; See et al., 2004), the function of negative feedback Smad effectors in the regulation of oligodendrocyte differentiation is not known. The identification of the Smad7 gene as a direct target of Sip1 suggests that Sip1 exerts its function in oligodendrocyte myelination at least in part by activating I-Smad gene expression. Of particular interest, Smad7 is found uniquely and highly elevated in oligodendrocytes both in vivo and in vitro, in contrast to the second I-Smad gene, Smad6, whose mRNA is hardly detectable in oligodendrocytes by in situ hybridization, although Smad6 overexpression in OPCs downregulates BMP signaling (data not shown). This suggests that Sip1 regulation of oligodendrocyte-specific Smad7 is a unique and novel aspect of Smad feedback regulation in the blocking of BMP/TGFβR signaling during oligodendrocyte differentiation, and which may also apply to other cell types where Sip1 regulates differentiation. Our unprecedented observations of Sip1-Smad7 regulatory cascade for OPC differentiation by antagonizing BMP signaling fit in a general picture where the BMP pathway is regulated by various mechanisms including synexpression and negative feedback mechanisms. In our case, forced expression of Smad7 not only inhibits BMP signaling in OPCs, but also leads to downregulation of β-catenin levels through a mechanism involving Smad7 and its cognate E3 ubiquitin ligase Smurf1, suggesting that Smad7 can block both BMP and β-catenin negative regulatory pathways for oligodendrocyte differentiation.
Importantly, we show that Smad7 overexpression is able to promote differentiation of OPCs even in the absence of Sip1 and induce oligodendrocyte formation from their precursors in ovo (Figure S5), suggesting that Smad7 acts downstream of Sip1 as a potent positive regulator for oligodendrocyte differentiation. These findings reveal a previously unrecognized pivotal role of Smad7 in promoting myelination at least in part through antagonizing BMP and β-catenin negative regulatory pathways.
SIP1/ZFHX1B in human diseases
Human mutations in SIP1/ZFHX1B cause Mowat-Wilson Syndrome (MWS), which is characterized by the combination of defects with variable penetrance, including severe mental retardation, white matter defects such as corpus callosum agenesis, Hirschsprung disease and variable congenital malformations like heart and craniofacial defects (Dastot-Le Moal et al., 2007; Garavelli and Mainardi, 2007; Zweier et al., 2005). This single-gene disorder also leads to delayed motor development, seizures and epilepsy in many MWS patients. Although Sip1 is critical for neurogenesis in the embryonic cortex (Seuntjens et al., 2009), the critical role of Sip1 in CNS myelination discovered here through oligodendrocyte lineage-specific mutagenesis suggests that mutations in SIP1/ZFHX1B may contribute to delayed myelination and white matter defects seen in patients with MWS (Figure S6) (Schell-Apacik et al., 2008; Sztriha et al., 2003). In addition, by using a lysolecithin-induced demyelination/remyelination animal model, we found that Sip1 was substantially upregulated in oligodendrocyte lineage cells in the lesion during remyelination (Figure S7), pointing to a potential important role of Sip1 in the remyelination process. As an integral component of the Smad regulatory circuitry, Sip1 may represent a novel molecular node of the regulatory network that integrates and balances negative signaling pathways and transcriptional signals to govern CNS myelinogenesis, and perhaps other neurological aspects. Modulation of the Smad signaling pathway may provide a future effective means to promote brain repair in patients with devastating demyelinating diseases and other neurological disorders of the CNS.
Supplementary Material
Acknowledgements
The authors would like to thank Y. Yu, A. Nishiyama, B. Kim, W. Liu, L. Liu, C. Shen, Melinda K. Duncan and A. Francis and A. Conidi for technical support. We thank C. Stiles, J. Svaren, S. Yoon, E. Olson, J. Johnson, J. Li, E. Hurlock, N. Ma and O. Barca-Mayo for critical comments and suggestions. This study was funded in part by grants from the US National Institutes of Health (R01NS072427) and the National Multiple Sclerosis Society (RG3978) to QRL, and the Research Council of KULeuven (OT-09/053 and GOA-11/012), FWO-V (G.0954.11N to DH and ES), the Queen Elisabeth Medical Foundation (GSKE 1113) and Interuniversity Attraction Poles (IUAP 6/20), and the type 3 large-infrastructure support InfraMouse by the Hercules Foundation to DH.
APPENDIX
Material and Methods
Animals
Sip1lox/lox mice were crossed with Olig1-Cre heterozygous mice to generate Olig1Cre+/−; Sip1lox/+ mice, which were then bred with Sip1lox/lox mice to produce control (Sip1lox/+;Olig1Cre+/−) and Sip1cKO offspring (Sip1lox/lox;Olig1Cre+/−). Sip1lox/+;Olig1Cre+/− mice were used as control mice since they developed and behaved the same as WT. A similar mating strategy was used for generating Smad7 control (Smad7lox/+;Olig1Cre+/−) and conditional knockout (Smad7lox/lox;Olig1Cre+/−) mice. All animal use and studies were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center at Dallas, USA.
Human Participants
Patients with Mowat Wilson syndrome were enrolled in a clinical, imaging and genetics study of individuals with callosal disorders approved by the Committee on Human Research at the University of California, San Francisco.
ChIP-sequencing, gene-chip microarray and quantitative RT-PCR
Differentiating oligodendrocytes (1×107 cells) were harvested from purified rat OPC cultured in the oligodendrocyte differentiation medium for three days. Chromatin preparation, ChIP, DNA purification, and library preparation for Illumina sequencing were performed using ChIP-sequencing DNA Prep kit (Illumina) according to manufacturer instruction. ChIP-seq was performed using a rabbit Olig2 antibody (Abcam) and control IgG on differentiating oligodendrocytes. Sequencing was done on an Illumina high throughput sequencer. For gene-chip microarray, RNAs from the myelinating optic nerve or spinal cord of control and Olig1 or Sip1 mutant mice at P14 were labeled for microarray analysis (Affymetrix gene-chip, ST1.0). qRT-PCR was carried out using the ABI Prism 7700 Sequence Detector System. The PCR primer sequences are available upon request.
Tissue and histology
The brain, spinal cord and optic nerve of mice at defined ages were dissected and fixed overnight in 4% paraformaldehyde and processed for vibratome- and cryo-sections. Sections with lysolecithin-induced demyelinating/remyelinating lesion in the adult rat spinal cord were kindly provided by Dr. Akiko Nishiyama. For immunostaining, we used antibodies to Olig2 (gift of C. Stiles), Sip1 (gift of D. Huylebroeck and Santa Cruz), PDGFRα (BD Bioscience, 558774), CC1 (Oncogene Research, OP80), O1 (gift of A. Gow), MBP (Covance, SMI-94R), p-Smad (Cell Signaling), Smad7 (Santa Cruz SC-11392). Monoclonal antibody to RIP was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa. RNA in situ hybridization was performed using digoxigenin-labeled riboprobes as described previously (Lu et al., 2002). The probes used were: murine PDGFRα, Plp1/Dm-20, Mbp, Smad7 and chick Pdgfrα and Sox10. Detailed protocols are available upon request. Electron microscopy was performed as previously described (Xin et al., 2005).
Primary oligodendroglial cell culture
Primary rat OPCs were isolated from cortices of pups at P2 using a differential detachment procedure as previously described (Chen et al., 2007). Isolated rat OPCs were grown in the OPC Growth Medium (Sato medium supplemented mitogens 10 ng/ml PDGF-AA and 20 ng/ml bFGF), and differentiated in Oligodendrocyte Differentiation Medium (Sato medium supplemented with 15 nM thyroid hormone T3 and 10 ng/ml ciliary neurotrophic factor). A similar differential detachment method was used for mouse OPC isolation using P1 neocortices. Briefly, the cortices of mouse pups were dissociated in a Dulbecco’s Modified Eagle Media (DMEM) medium containing 10% FBS and 1% penicillin-streptomycin by gently triturating through G18, G21, 23G needles for 3 times each. Cells collected through a sterile 70 μm filter were plated onto poly-D-lysine coated 75 cm2 flasks in the above medium. The medium was changed every other day till cells became 50%-60% confluent. The medium was then switched to a serum-free B104 conditional medium (DMEM/F12 medium containing 15% B104 CM, 1× N2 and 50 μm/ml insulin) to enrich OPC production (Chen et al., 2007). After removing microglia and astrocytes through shaking the mixed glia-culture and differential attachment, the isolated mouse OPCs (approximately 80% pure) were cultured in the OPC Proliferation Medium plus B27, 1 ng/ml NT3 and 5 μM forskolin (Emery et al., 2009). OPCs were transduced with lentivirus or transfected with expressing vectors using Amaxa electroporator according to manufacturer’s protocol and assayed for immunocytochemistry and qRT-PCR analysis. The extent of oligodendrocyte process outgrowth was measured by the area surrounding the nuclei including the outermost tips occupied by processes using Image J.
Chromatin immunoprecipitation assays
Chip assay was performed as previously described (Chen et al., 2009b) using genomic DNAs from OPCs and differentiating oligodendrocytes (after T3/CNTF treatment of OPCs) were immunoprecipitated with anti-Sip1 antibody. Briefly, primary OPCs isolated from rat neonatal pups or oligodendrocytes were fixed in 1% formaldehyde for 10 mins and stop fixing by 2.5M glycine for 5 min at room temperature. Cells were washed in PBS, resuspended in a cell lysis solution containing 150mM NaCl, 10% glycerol, 50mM Hepes, 1mM EDTA, 0.5% NP40, 0.25% Triton X-100 and homogenized. Lysates were sonicated with a Bioruptor sonicator (Diagenode) into fragmented DNAs around 300 bp in a sonication buffer containing 1mM EDTA, 0.5mM EGTA, 10mM Tris and 0.1% SDS. Sonicated chromatin (100 μg DNA) was used for immunoprecipitation by incubated with 2 μg of anti-Sip1 antibody. Primers used for ChIP analysis on promoters are Smad7(I)-F: gtcacctgtagcctggtttagc, Smad7(I)-R: gcatcggcactgtattctcac; Smad7(II)-F: gtcacctgtagcctggtttagc, R: gcatcggcactgtattctcac. Id4 F: cgcagcagtatttgtagagcc, Id4-R: gcgttgacggaatggagtgt; Id2-F:acagacccgcttggagttgc; R: gtcacgggcggaatggacac; Hes1-F: tacctttagccacatcttcatcag; R: gactcagcatatttcaaccacctc.
Lentivirus generation
Sip1 and HA-tagged Smad7 were cloned into a lentiviral expressing vector (lenti-CSCsp-pw-ires-GFP, a gift from Dr. Jenny Hsieh). Lentiviruses were prepared by cotransfected lentiviral expressing vectors with packaging vectors pMD2.G and psPAX2 (Addgene) into 293T cells using Polyjet transfection reagents (SignaGen labs) or CaCl2 transfection methods. 48 hours after transfection, viral supernatants were collected and filtered through a 0.45 μm filter, then concentrated by ultracentrifuging at 19,400 rpm for 2 hours at 4 degree. OPCs were infected at multiplicity of infection (MOI) of 50 (MOI was determined in human 293T cells). The infection rate was > 90% in these cultures.
Transient transfection, luciferase assays and co-immunoprecipitation
The mouse oligodendrocyte precursor cell line Oli-neu (Jung et al., 1995) was a kind gift from Dr. P. Wright (University of Arkansas, USA). The Oli-neu cells were maintained in growth medium consisting of DMEM supplemented with N2 supplemented and 1% horse serum. Oli-neu cells were transfected with luciferase reporters and assayed 24 h post-transfection for luciferase activities by using a Promega luciferase assay kit according to the manufacturer’s instruction. For immunoprecipitation, whole-cell lysates were prepared from brain tissues and cells using 1x Passive lysis buffer (Promega) supplemented with a protease inhibitor cocktail (1:200, Sigma). 300 μg of cell lysate proteins were incubated with 2 μg antibody. Phosphatase inhibitors used for IP are 5 μM microcystin, 2mM imidazole, 1.15mM sodium molybdate, and 0.184mg/ml sodium orthovahedate. Western blotting was performed using chemiluminescence with the ECL kit (Pierce) according to the manufacturer’s instruction. Chick embryo in ovo electroporation in developing neural tube was conducted as previously described (Ye et al., 2009).
Statistic analysis
Quantifications were performed from at least three independent experimental groups. Data are presented as mean ± SEM in the graphs. p values are from Student’s two-tailed t test to compare two sets of data. For multiple comparisons, which were done using one-way analysis of variance analysis (ANOVA). p value < 0.05 is considered to be statistically significant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–2115. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen H, Zheng D, Kuang C, Fang H, Zou B, Zhu W, Bu G, Jin T, Wang Z, et al. Smad7 is required for the development and function of the heart. J Biol Chem. 2009a;284:292–300. doi: 10.1074/jbc.M807233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Balasubramaniyan V, Peng J, Hurlock EC, Tallquist M, Li J, Lu QR. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc. 2007;2:1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wu H, Wang S, Koito H, Li J, Ye F, Hoang J, Escobar SS, Gow A, Arnett HA, et al. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci. 2009b;12:1398–1406. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang Y, He Q, Qiu M, Whittemore SR, Cao Q. Bone morphogenetic protein signaling and olig1/2 interact to regulate the differentiation and maturation of adult oligodendrocyte precursor cells. Stem Cells. 2007;25:3204–3214. doi: 10.1634/stemcells.2007-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastot-Le Moal F, Wilson M, Mowat D, Collot N, Niel F, Goossens M. ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat. 2007;28:313–321. doi: 10.1002/humu.20452. [DOI] [PubMed] [Google Scholar]
- Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, Sanai N, Franklin RJ, Rowitch DH. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–1585. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Kotter MR, Harrington EP, Huang JK, Zhao C, Rowitch DH, Franklin RJ. Overcoming remyelination failure in multiple sclerosis and other myelin disorders. Exp Neurol. 2010;225:18–23. doi: 10.1016/j.expneurol.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Franklin RJ. Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- Friedman B, Hockfield S, Black JA, Woodruff KA, Waxman SG. In situ demonstration of mature oligodendrocytes and their processes: an immunocytochemical study with a new monoclonal antibody, rip. Glia. 1989;2:380–390. doi: 10.1002/glia.440020510. [DOI] [PubMed] [Google Scholar]
- Garavelli L, Mainardi PC. Mowat-Wilson syndrome. Orphanet J Rare Dis. 2007;2:42. doi: 10.1186/1750-1172-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AK, Miller RH. Emerging roles for bone morphogenetic proteins in central nervous system glial biology. J Neurosci Res. 2004;76:1–8. doi: 10.1002/jnr.20019. [DOI] [PubMed] [Google Scholar]
- Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, Ten Dijke P, Lin X, et al. Smad7-induced beta-catenin degradation alters epidermal appendage development. Dev Cell. 2006;11:301–312. doi: 10.1016/j.devcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Maruhashi M, Nelles L, Van de Putte T, Verschueren K, Miyoshi T, Yoshimoto A, Kondoh H, Huylebroeck D. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis. 2002;32:82–84. doi: 10.1002/gene.10048. [DOI] [PubMed] [Google Scholar]
- Howng SY, Avila RL, Emery B, Traka M, Lin W, Watkins T, Cook S, Bronson R, Davisson M, Barres BA, et al. ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev. 2010;24:301–311. doi: 10.1101/gad.1864510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Aimone JB, Kaspar BK, Kuwabara T, Nakashima K, Gage FH. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Kramer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, Khazaie K, Chlichlia K, von Blankenfeld G, Kettenmann H, et al. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci. 1995;7:1245–1265. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- Kadi L, Selvaraju R, de Lys P, Proudfoot AE, Wells TN, Boschert U. Differential effects of chemokines on oligodendrocyte precursor proliferation and myelin formation in vitro. J Neuroimmunol. 2006;174:133–146. doi: 10.1016/j.jneuroim.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Kotter MR, Stadelmann C, Hartung HP. Enhancing remyelination in disease--can we wrap it up? Brain. 2011 doi: 10.1093/brain/awr014. [DOI] [PubMed] [Google Scholar]
- Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009 doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Mar S, Noetzel M. Axonal damage in leukodystrophies. Pediatr Neurol. 2010;42:239–242. doi: 10.1016/j.pediatrneurol.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Millar SE. Smad7: licensed to kill beta-catenin. Dev Cell. 2006;11:274–276. doi: 10.1016/j.devcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Miller RH, Dinsio K, Wang R, Geertman R, Maier CE, Hall AK. Patterning of spinal cord oligodendrocyte development by dorsally derived BMP4. J Neurosci Res. 2004;76:9–19. doi: 10.1002/jnr.20047. [DOI] [PubMed] [Google Scholar]
- Miquelajauregui A, Van de Putte T, Polyakov A, Nityanandam A, Boppana S, Seuntjens E, Karabinos A, Higashi Y, Huylebroeck D, Tarabykin V. Smad-interacting protein-1 (Zfhx1b) acts upstream of Wnt signaling in the mouse hippocampus and controls its formation. Proc Natl Acad Sci U S A. 2007;104:12919–12924. doi: 10.1073/pnas.0609863104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Nave KA. Neurological mouse mutants and the genes of myelin. J Neurosci Res. 1994;38:607–612. doi: 10.1002/jnr.490380602. [DOI] [PubMed] [Google Scholar]
- Ogata T, Ueno T, Hoshikawa S, Ito J, Okazaki R, Hayakawa K, Morioka K, Yamamoto S, Nakamura K, Tanaka S, et al. Hes1 functions downstream of growth factors to maintain oligodendrocyte lineage cells in the early progenitor stage. Neuroscience. 2010 doi: 10.1016/j.neuroscience.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Pearson KL, Hunter T, Janknecht R. Activation of Smad1-mediated transcription by p300/CBP. Biochim Biophys Acta. 1999;1489:354–364. doi: 10.1016/s0167-4781(99)00166-9. [DOI] [PubMed] [Google Scholar]
- Postigo AA, Depp JL, Taylor JJ, Kroll KL. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 2003;22:2453–2462. doi: 10.1093/emboj/cdg226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, Smith JC, Huylebroeck D. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 1999;18:5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo JK, Aumann TD, Merlo D, Kilpatrick TJ, Cate HS. Remyelination is altered by bone morphogenic protein signaling in demyelinated lesions. J Neurosci. 2011;31:4504–4510. doi: 10.1523/JNEUROSCI.5859-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Burke GM, McGuire T, Pisarek AJ, Mukhopadhyay A, Mishina Y, Kessler JA. BMPR1a signaling determines numbers of oligodendrocytes and calbindin-expressing interneurons in the cortex. J Neurosci. 2007;27:7397–7407. doi: 10.1523/JNEUROSCI.1434-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Schell-Apacik CC, Wagner K, Bihler M, Ertl-Wagner B, Heinrich U, Klopocki E, Kalscheuer VM, Muenke M, von Voss H. Agenesis and dysgenesis of the corpus callosum: clinical, genetic and neuroimaging findings in a series of 41 patients. Am J Med Genet A. 2008;146A:2501–2511. doi: 10.1002/ajmg.a.32476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See J, Zhang X, Eraydin N, Mun SB, Mamontov P, Golden JA, Grinspan JB. Oligodendrocyte maturation is inhibited by bone morphogenetic protein. Mol Cell Neurosci. 2004;26:481–492. doi: 10.1016/j.mcn.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Skillington J, Choy L, Derynck R. Bone morphogenetic protein and retinoic acid signaling cooperate to induce osteoblast differentiation of preadipocytes. J Cell Biol. 2002;159:135–146. doi: 10.1083/jcb.200204060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikauchi Y, Imamura T, Miyazono K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J Biol Chem. 2002;277:39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- Sztriha L, Espinosa-Parrilla Y, Gururaj A, Amiel J, Lyonnet S, Gerami S, Johansen JG. Frameshift mutation of the zinc finger homeo box 1 B gene in syndromic corpus callosum agenesis (Mowat-Wilson syndrome) Neuropediatrics. 2003;34:322–325. doi: 10.1055/s-2003-44671. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Peterson J, Ransohoff RM, Rudick R, Mork S, Bo L. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338:278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- van Grunsven LA, Taelman V, Michiels C, Verstappen G, Souopgui J, Nichane M, Moens E, Opdecamp K, Vanhomwegen J, Kricha S, et al. XSip1 neuralizing activity involves the co-repressor CtBP and occurs through BMP dependent and independent mechanisms. Dev Biol. 2007;306:34–49. doi: 10.1016/j.ydbio.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Verstappen G, van Grunsven LA, Michiels C, Van de Putte T, Souopgui J, Van Damme J, Bellefroid E, Vandekerckhove J, Huylebroeck D. Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Hum Mol Genet. 2008;17:1175–1183. doi: 10.1093/hmg/ddn007. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu Y, Levine EM, Rao MS. Hes1 but not Hes5 regulates an astrocyte versus oligodendrocyte fate choice in glial restricted precursors. Dev Dyn. 2003;226:675–689. doi: 10.1002/dvdy.10278. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T, Xian K, Hurlock E, Xin M, Kernie SG, Parada LF, Lu QR. A critical role for dorsal progenitors in cortical myelination. J Neurosci. 2006;26:1275–1280. doi: 10.1523/JNEUROSCI.4717-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalc B, Colman DR. Origins of vertebrate success. Science. 2000;288:271–272. doi: 10.1126/science.288.5464.271c. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Argaw AT, Gurfein BT, Zameer A, Snyder BJ, Ge C, Lu QR, Rowitch DH, Raine CS, Brosnan CF, et al. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc Natl Acad Sci U S A. 2009;106:19162–19167. doi: 10.1073/pnas.0902834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zweier C, Thiel CT, Dufke A, Crow YJ, Meinecke P, Suri M, Ala-Mello S, Beemer F, Bernasconi S, Bianchi P, et al. Clinical and mutational spectrum of Mowat-Wilson syndrome. Eur J Med Genet. 2005;48:97–111. doi: 10.1016/j.ejmg.2005.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.