Abstract
Introduction
Over the last 15 – 20 years, targeted anticancer strategies have focused on therapies aimed at abrogating a single malignant protein. Agents that are directed towards the inhibition of a single oncoprotein have resulted in a number of useful drugs in the treatment of cancers (i.e., Gleevec, BCR-ABL; Tarceva and Iressa, EGFR). However, such a strategy relies on the notion that a cancer cell is dependent on a single signaling pathway for its survival. The possibility that a cancer cell may mutate or switch its dependence to another signaling pathway can result in the ineffectiveness of such agents. Recent advances in the biology of heat-shock protein 90 (Hsp90) have revealed intimate details into the complexity of the chaperoning process that Hsp90 is engaged in and, at the same time, have offered those involved in drug discovery several unique ways to interfere in this process.
Areas covered
This review provides the current understanding of the chaperone cycle of Hsp90 and presents the multifaceted approaches used by researchers in the discovery of potential Hsp90 drugs. It discusses the phenotypic outcomes in cancer cells on Hsp90 inhibition by these several approaches and also addresses several distinctions observed among direct Hsp90 ATP-pocket competitors providing commentary on the potential biological outcomes as well as the clinical relevance of such features.
Expert opinion
The significantly different phenotypic outcomes observed from Hsp90 inhibition by the many inhibitors developed suggest that the clinical development of Hsp90 inhibitors would be better served by careful consideration of the pharmacokinetic/pharmacodynamic properties of individual candidates rather than a generic approach directed towards the target.
Keywords: cancer, chaperone, heat-shock protein 90, targeted therapy
1. Introduction
Hsp90 (Heat-shock protein 90) is a chaperone molecule that assists in the correct functioning of several tumor promoting proteins, collectively referred to as ‘client proteins’. Among these are HER2, EGFR, mutant ER, HIF1α, Raf-1, AKT and mutant p53, to list a few [1]. Development of agents that target Hsp90 has, therefore, become a major focus in cancer research because by inhibiting one protein, Hsp90, one may simultaneously inhibit and/or degrade a multitude of oncoproteins [2]. To regulate the complex array of its client proteins that span from kinases, transcription factors and other potential cancer promoting molecules, Hsp90 utilizes an intricate web of associated co-chaperones. The current understanding on Hsp90 presents a scenario in which the chaperone activity is intrinsically linked to conformation, which is in turn dependent on the binding and release of ATP/ADP, co-chaperones and client proteins.
The critical importance of nucleotides and co-chaperones in regulating the Hsp90 cycle offers therapeutic opportunities for modulating Hsp90 by affecting the binding of these molecules. Agents that alter the interaction of these molecules with Hsp90 can be expected to modulate its activity in a non-overlapping fashion. Conceivably, this may be accomplished by targeting binding of ATP/ADP to the Hsp90 regulatory pocket, binding to the co-chaperone directly or targeting sites on Hsp90 (other than the N-terminal nucleotide binding domain) that affect co-chaperone binding to Hsp90. Additionally, molecules that prevent client protein binding to Hsp90 will inhibit their maturation. Therefore, targeting of a specific client protein in the proper context (i.e., HER2 in breast cancer; activated AKT in small cell lung cancer (SCLC); mutant B-Raf in melanoma; Bcr-Abl in chronic myelogenous leukemia (CML); mutant JAK2 in myeloproliferative disorders) may have therapeutic potential in the treatment of specific cancers.
In this review, we describe the Hsp90 catalyzed chaperone cycle and present several strategies for the discovery of molecules that modulate the conformational dynamics of this cycle. We endeavor to describe the numerous ways that are potentially possible to pharmacologically modulate the Hsp90 chaperone machinery and illustrate the current state of affairs in this regard. In doing so, we present available evidence of the therapeutic relevance as well as the differences observed between the alternative modes of modulation. Of the possible modes of affecting Hsp90 activity described in this review, only agents which inhibit the binding of ATP by targeting the nucleotide binding pocket located in the N-terminal domain are currently being evaluated clinically. Even within this class, which have a common binding site and similar tumor retention profile, markedly different properties are observed in preclinical studies. We briefly discuss such distinctions in the mode of interaction of these inhibitors with the chaperone machinery and point out in the expert opinion section the potential important biological activity that may result from these differences.
2. The Hsp90 ATPase cycle and the dynamic nature of Hsp90
Hsp90 is an important chaperone that interacts with and refolds its client proteins in a cycle that is driven by the binding and hydrolysis of ATP [3]. Through the course of its catalytic cycle, Hsp90 undergoes considerable structural changes, and this dynamic nature of Hsp90 is the key in its ability to function as a chaperone [4-7]. Hsp90 is in a state of conformational flux, whose overall structure is constantly altered by the binding of various ligands, including ATP/ADP, and co-chaperones (i.e., HOP, Cdc37, p23, Aha1 and immunophilins) [7]. These ligands bind to specific sites on Hsp90 and alter the conformational equilibrium between the two extreme ‘open’ (apo) and ‘closed’ (ATP-bound) states at any given moment [4].
The ATPase activity of Hsp90 is linked to its conformational state, which for eukaryotic Hsp90 is influenced by > 20 co-chaperones, as well as by the binding of client proteins, which serve to drive it through its catalytic cycle [3,7]. A functional chaperone cycle was first proposed for eukaryotic Hsp90 based on interaction with steroid hormone receptors [8] and is a process that is probably conserved among eukaryotic Hsp90 species [9].
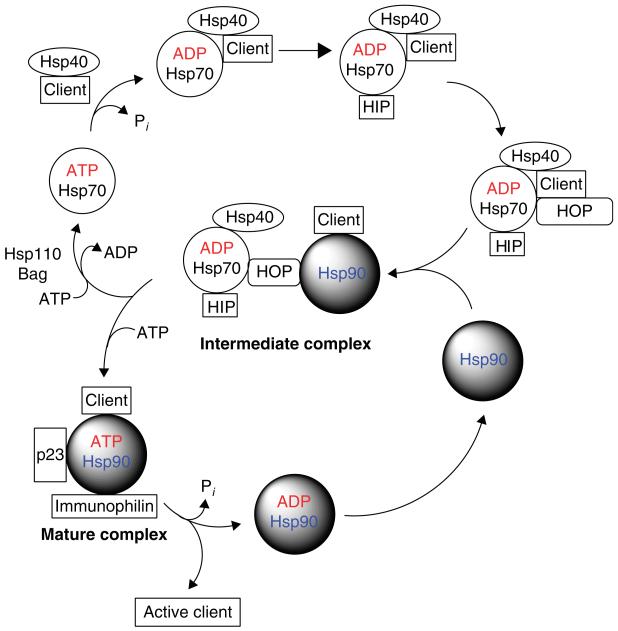
Association of Hsp90 with its client proteins is believed to be initiated by a priori interaction with Hsp70 (Figure 1). The client is presented to Hsp70 by its activator, Hsp40, and binds to it in an ATP-dependent manner. Hsp70 interacting protein then binds to and stabilizes this complex. The dimeric co-chaperone HOP (Sti1 in yeast) binds the Hsp40–Hsp70–client complex to Hsp90, thereby forming an Hsp70–HOP–Hsp90 complex [10]. HOP interacts with the C terminus of Hsp90 through its tetratricopeptide repeat (TPR) domain as well as to additional sites in the middle domain (MD). Co-chaperones and immunophilins bind to the Hsp70–HOP–Hsp90 complex and facilitate the transfer of client from Hsp70 to Hsp90 to form the intermediate complex. On ATP binding, Hsp90 forms a mature complex containing p23 (Sba1 in yeast) and other co-chaperones such as Cdc37 and immunophilins that catalyze the conformational maturation of the client. The co-chaperone p23 as well as the immunophilins FKBP51, FKBP52 and Cyp-40 displace HOP and Hsp70 leading to the mature complex [11]. Large conformational changes that occur to Hsp90 subsequent to ATP binding are probably transduced to the client leading to its activation (described below). Following release of the mature client, presumably, Hsp90 can re-enter the cycle and bind another client protein.
Figure 1. A simplified cartoon describing the ATPase cycle of Hsp90.
The first X-ray crystal structures, along with electron microscopy (EM) and small-angle X-ray scattering (SAXS) data, obtained for full length bacteria (nucleotide free; AMP-PNP-bound; ADP-bound) [12] and yeast (AMP-PNP- and Sba1-bound) [13] Hsp90 as well as mammalian (AMP-PNP; ADP-bound) [14] Grp94 (the endoplasmic reticulum paralog of cytosolic Hsp90) were critical in revealing particular conformations adopted when bound to specific ligand(s). These structures show that the global architecture is conserved across species and that Hsp90 exists as a homodimeric structure in which individual monomers are characterized by three domains; an N-terminal nucleotide binding domain (NBD), site of ATP binding; the MD, site of co-chaperone and client protein binding and involved in ATP hydrolysis; and a C-terminal dimerization domain (CDD), site of dimerization. The NBD is followed by a linker region which connects it to the MD.
Structural and biochemical studies had shown that Hsp90 function was dependent on the binding and hydrolysis of ATP [15,16] and suggested that hydrolysis occurs via a ‘molecular clamp’ mechanism involving dimerization of the NBD in the ATP-bound state [17,18]. The crystal structures of Hsp90, together with EM and SAXS data, confirmed the ATPase-coupled molecular clamp mechanism and provided further insight connecting Hsp90 complex structure and conformation to the ATPase cycle. In the absence of bound nucleotide, Hsp90 exists in an ‘open’ conformation. While the precise details linking the ATPase cycle to conformational state have not been entirely elucidated, it is known that dramatic conformational changes occur subsequent to ATP binding, whereby the N-terminal domains closely associate with one another resulting in a ‘closed’ conformation that is capable of hydrolyzing ATP [17]. EM revealed a distinct ‘compact’ conformation when ADP-bound [12] and in the absence of any bound ligand, the dimer moves to an ‘open’ state. These structures, however, only present a static picture of Hsp90 at its conformational extremes. In order to examine other conformations between these extremes, more dynamic methods must be used.
The solution structure of Escherichia coli Hsp90 (HtpG) determined using SAXS [6] shows some important differences compared to the crystal structure. The apo-conformation in solution is more extended with a wider angle implying that it can accommodate more diverse client proteins. Also, the NBD and the MD are rotated by 40° compared to the crystal structure. This may especially impact the ability of nucleotide binding as Gln122 and Phe123 within the active site lid (residues 100 – 126) are positioned to block nucleotide binding in the apo-conformation. Nucleotide binding requires that the lid region be reorganized and the solution structure is better able to accommodate the necessary structural changes essential for nucleotide binding.
The solution structure of eukaryotic Hsp90 has also been determined using SAXS as well as cryo-EM [19]. Interestingly, these studies showed that Hsp90 can exist in two open conformations, ‘fully-open’ and ‘semi-open’, and revealed an intrinsic flexibility of Hsp90 that is capable of partial closure of the N-terminal domains even in the absence of a nucleotide.
In an attempt to further tease out the conformational cycle of Hsp90 during ATP binding and hydrolysis, Hessling et al. [4] used fluorescence energy resonance transfer (FRET) to propose a model consisting of three distinct conformations between the open and closed conformations. In this model, apo-Hsp90 (open conformation) binds ATP in a rapid manner to yield an ATP-bound conformation, followed by the slow formation of an intermediate (I1) in which the N-terminal domains remain undimerized. While it is not known with certainty, I1 may represent an intermediate in which the ATP lid is closed and the segment on the N-terminal domain required for dimerization is exposed. Subsequent dimerization of the N-terminal domains yields another intermediate (I2). Next, rearrangements allowing for the interaction between the NBD and MD result in the closed conformation which is able to undergo hydrolysis. Following hydrolysis, ADP and Pi are released and Hsp90 returns to the open apo-state. This model does not exclude the possibility of a distinct ADP-bound conformation [12] following hydrolysis as it does not contribute to the rate-limiting step of the hydrolysis reaction, which has been shown for hHsp90 by kinetic and single-turnover experiments to occur after nucleotide binding but before hydrolysis [9].
As was mentioned previously, the binding of co-chaperones to eukaryotic Hsp90 can result in specific conformations that are necessary for driving the chaperone cycle through completion [1,3]. Their role as regulators of the cycle has been enhanced in light of single molecule FRET experiments which have shown that in the absence of co-chaperones or substrate molecules, ATP hydrolysis is not tightly coupled to the conformational cycle [5]. It appears that conformational states of Hsp90 can quickly and reversibly change without committing to hydrolysis and that the co-chaperones function to stabilize a conformation required for progression through the ATPase cycle. Chaperones modulate Hsp90 function by altering ATP turnover or by facilitating client loading and activation. The co-chaperones Cdc37 and HOP are both involved in the recruitment of client proteins and are able to arrest the ATPase cycle of Hsp90 in order to facilitate client protein loading. Cdc37 slows down the ATPase cycle by binding to sites on the lid segment of the N-terminal domain in the open conformation, fixing the ATP lid in an open conformation and preventing interaction of the N-domains [20]. HOP functions by coupling the Hsp70 and Hsp90 chaperones and facilitates client protein transfer between the two. HOP prevents N-terminal dimerization by binding to the open conformation of Hsp90 [21]. p23 slows down the ATPase cycle by binding to and stabilizing the ATP-bound closed conformation which is essential for activation of client proteins [13]. To date, only one activator of the ATPase activity of Hsp90, Aha1 (activator of Hsp90 ATPase1), is known which has been shown to stimulate activity by a factor of 100 or more [9]. Aha1 binds to the open conformation of Hsp90 at both the N terminal and MDs, inducing a conformational change resulting in N-terminal dimerization and stabilization of the ATPase-competent conformation [22]. Interestingly, the binding of only one Aha1 molecule is necessary to fully stimulate ATPase activity and results in an asymmetric complex [22]. Aha1 appears to enhance ATPase activity by reducing the energy barrier accompanying structural rearrangements that occur during the transition between the open and closed states, which have been shown to be rate limiting [4].
While it is still unclear precisely how Hsp90 induces client protein conformational changes, it is likely that it is directly linked to the domain movements and conformational changes that occur to Hsp90 as it goes from the ‘closed’ to ‘open’ conformational states. The first structural insight into client protein interaction with Hsp90 was provided by Vaughan et al. [23] who used single-particle EM to determine the structure of Hsp90–Cdc37–CDK4 complex. CDK4 is a protein kinase that is dependent on Hsp90 for activation and on Cdc37 for recruitment [24]. This structure shows that client interactions occur to both the MD and NBD of one Hsp90 subunit while Cdc37 binds to the NBD of the other subunit. While not proven, the fact that this complex contains Cdc37 may suggest that binding of client to Hsp90 occurs before the catalytically competent ATP-bound conformation, which requires that Cdc37 disengage from the complex [23].
3. Agents targeting the Hsp90 chaperone complex
These intricate structural modulations of Hsp90, as presented above, suggest several ways to inhibit its chaperoning activity. To date, most success in Hsp90 modulation has been ascribed to efforts directed towards the development of agents which inhibit the N-terminal nucleotide binding pocket (Figure 2) resulting in the advancement of numerous molecules into clinical trials for the treatment of a variety of cancers [2,25]. Additionally, increasing efforts are being made to develop anticancer agents with alternative modes of inhibition, such as targeting Hsp90 interaction with co-chaperones [26] or client proteins [27], or allosteric binding sites believed to occur on the CDD [28]. Therefore, molecules that abrogate Hsp90 activity may be categorized into agents that cause: i) direct inhibition of ATPase activity by binding at the nucleotide pocket of the NBD (Section 3.1), ii) modulation of Hsp90 activity by binding to the CDD (Section 3.2), iii) disruption of co-chaperone–Hsp90 interactions (Section 3.3), iv) inhibition of client/Hsp90 associations (Section 3.4) and v) interference with post-translational modifications of Hsp90 (Section 3.5). The remainder of this review focuses on the discovery and development of these modulators.
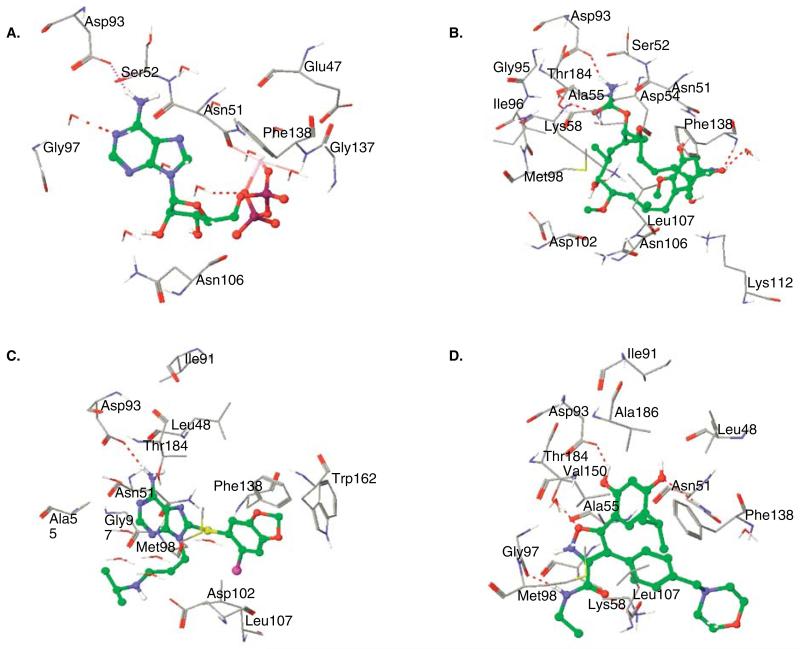
Figure 2. Structure of human Hsp90 bound to: (A) ADP (PDB ID: 1BYQ), (B) geldanamycin (PDB ID: 1YET), (C) PU-H71 (PDB ID: 2FWZ) and (D) NVP-AUY922 (PDB ID: 2VCI).
H-bonds are shown by dotted lines
3.1 NBD interactors
Compounds that modulate Hsp90 chaperone activity by inhibiting the ATP binding site of the NBD were the first compounds identified as Hsp90 inhibitors. Since the serendipitous discovery of geldanamycin (GM) and radicicol (RD) during a phenotypic screen (Section 3.1.1), more targeted approaches such as structure-based drug design (Section 3.1.2), biochemical and cell-based screening (Section 3.1.3), virtual screening (Section 3.1.4), fragment-based drug design (FBDD) (Section 3.1.5) and educated guess (Section 3.1.6) have led to the identification of several novel chemical scaffolds.
3.1.1 Phenotypic screening
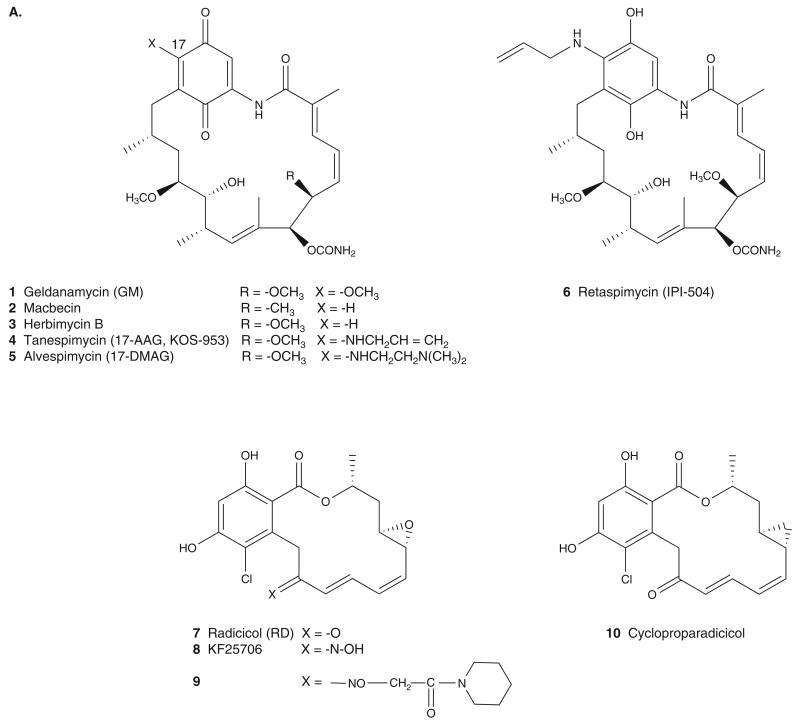
The antitumor properties of GM (1), macbecin (2) and herbimycin B (3) (Figure 3A) were found during a phenotypic screening of compounds to reverse v-src oncogene transformed cells [29]. These compounds belong to the class of benzoquinone ansamycin antibiotics and their anticancer activity was initially thought to be due to direct inhibition of src kinase; however, they were later shown to bind to Hsp90 and interfere with Hsp90–v-src heterocomplex formation [30]. Clinical development of GM has been hampered by a number of limitations including severe hepatotoxicity, metabolic and chemical instability, low solubility and a formulation which was less than ideal. Structural modification to GM led to the discovery of 17-allyl-17-desmethoxy-geldanamycin (17-AAG, 4; also tanespimycin, KOS-953, Figure 3A), which was less hepatotoxic and had an IC50 = 31 nM for inhibition of HER2 in SKBr3 cells [31]. Further development resulted in a water soluble diamine analog 17-(2-dimethylaminoethyl) amino-17-demethoxygeldanamycin (17-DMAG, 5; also alvespimycin, Figure 3A) with an IC50 = 24 nM [32]. 17-DMAG showed promising results in a Phase I clinical trial in acute myelogenous leukemia (AML), but its further development was stopped because of unfavorable toxicity profile [25]. As the toxicity of the ansamycins was associated with the quinone moiety, retaspimycin (6; IPI-504, Figure 3A), the hydroquinone derivative of 17-AAG [31], was synthesized and found to show activity similar to 17-AAG. Retaspimycin has been evaluated in Phase I–II clinical trials in patients with NSCLC, multiple myeloma (MM), breast cancer, castration-resistant prostate cancer, gastrointestinal stromal tumors, metastatic melanoma and metastatic kidney cancer. In the Phase II trial in patients with NSCLC, 28% of the patients achieved stable disease with tumor reduction [33].
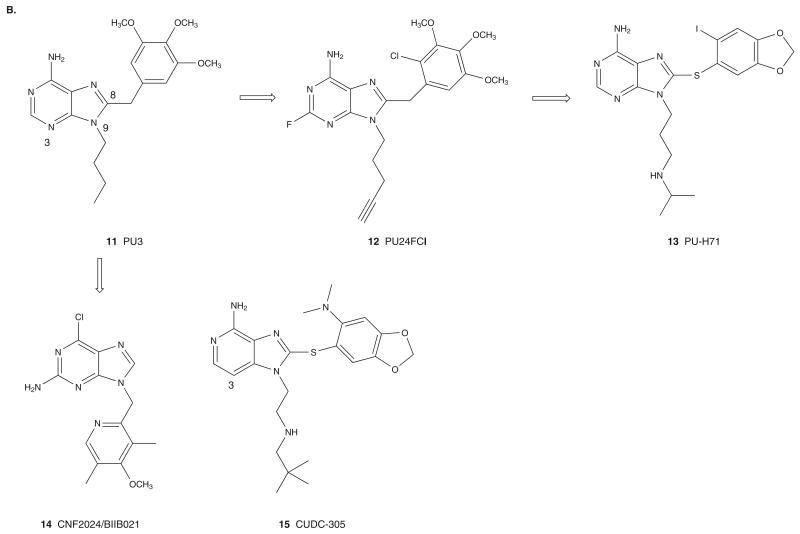
Figure 3. Hsp90 NBD interactors identified by: (A) phenotypic screening and (B) structure-based drug design.
RD (7, Figure 3A), a macrocyclic antibiotic isolated from Monosporium bonorden, was found to reverse the phenotype of v-src transformed cells and cause depletion of Raf-1 and subsequent inhibition of MAPK pathway in K-ras-transformed cells. Hsp90 was determined to be the target of RD through the use of solid-support immobilized analogs [31]. RD competes with GM for binding to the ATP binding site of the NBD, and similar to GM, inhibits the binding of p23 to Hsp90. However, because of its chemical instability, RD failed to show in vivo activity, but oxime (8 and 9) and cyclopropane (10) derivatives showed significant antitumor activity against various human tumor xenograft in animal models [31]. Whereas none of these derivatives has advanced to clinic, the resorcinol core of RD is retained in several agents currently in clinical development (i.e., NVP-AUY922, KW-2478, AT13387, in Figures 4 and 5).
Figure 4. Hsp90 NDB interactors identified by biochemical and cellular screening.
Figure 5. Hsp90 NBD interactors identified by: (A) virtual screening, (B) fragment-based design and (C) educated guess.
The crystal structures of GM and RD with the NBD of yeast Hsp90 (yHsp90) show both molecules inserted into the pocket in a bent conformation, with GM in a C-conformation (in contrast to an extended conformation of unbound GM) and RD in an L-conformation [34]. The binding conformations of GM and RD are similar to bound ADP and, therefore, mimic its critical interactions [34]. The binding interactions of GM with yHsp90 were found to be conserved in human Hsp90 [35]. The macrocyclic ansa-ring and pendant carbamate group of GM are directed toward the bottom of the binding pocket while the benzoquinone ring is oriented towards the top of the pocket with one face solvent-exposed. The orientation of RD is opposite to that of GM, with the resorcinol ring directed toward the bottom of the pocket and the macrocyclic ring toward the top of the pocket. The carbamate and resorcinol moieties of GM and RD, respectively, act as bioisosteres of adenine’s NH2 functionality making direct and indirect (water mediated) H-bond with Leu48 (Leu34, yHsp90), Asp93 (Asp79, yHsp90), Gly97 (Gly83, yHsp90) and Thr184 (Thr171, yHsp90) in the nucleotide-binding site of hHsp90 (Figure 2A, ADP; Figure 2B, GM). GM and RD also make hydrophobic interactions with the pocket formed by Met98 (Met84, yHsp90), Leu103 (Leu89, yHsp90), Leu107 (Leu93, yHsp90), Phe138 (Phe124, yHsp90), Val150 (Val136, yHsp90) and Val186 (Leu173, yHsp90) in hHsp90 (Figure 2B, GM). The inhibitor–protein complex results in the arrest of Hsp90 in its ADP-bound conformation and thereby prevents the ‘clamping’ of Hsp90 around the client protein [36]. This in turn results in premature release of abnormally folded client proteins eventually leading to their ubiquitination and proteasomal degradation [37].
3.1.2 Structure-based drug design
The availability of X-ray crystallographic data for Hsp90 bound to ATP/ADP was critical for the design of novel chemotypes as Hsp90 inhibitors. Taking advantage of the C-shaped conformation adopted by GM (PDB ID: 1YET) and RD when bound to Hsp90, Chiosis et al. [38] designed the first reported synthetic Hsp90 inhibitor, the purinescaffold inhibitor, PU3 (11; Figure 3B) [38]. Optimization of this compound led to the more active PU24FCl (12; Figure 3B) having a fluorine at C2 and a pentynyl chain at N9 of the purine, in addition to a chlorine at C2′ of the trimethoxyphenyl ring. PU3 and PU24FCl while maintaining important H-bond interactions similar to ADP induce a conformational change between helix 3 and 4 of Hsp90 to accommodate the 8-aryl ring [39]. Further optimization by means of structure–activity relationship (SAR) studies led to the 8-arylsulfanyl adenine class with PU-H71 (13) being one of the most active compounds (IC50 = 50 nM). The X-ray crystal structure of PU-H71 bound to the NBD of human Hsp90α (PDB ID: 2FWZ) revealed that the adenine moiety bound Hsp90 in a manner similar to ATP (Figure 2C). Key interactions of the adenine ring include the direct hydrogen bond between N6 with Asp93, and the water mediated hydrogen bonds to Leu48, Asn51, Ile91, Asp93, Gly97, Asp102 and Thr184, as well as hydrophobic interactions with Met98 and Ala55 [39]. Similar to PU3 and PU24FCl, PU-H71 induced a conformational change between helix 3 and 4 of Hp90 in order to accommodate the 8-aryl ring. However, the binding of PU-H71 is significantly different from that of PU24FCl in that the 2′-iodo substituent is oriented nearly 180° relative to the 2′-chloro of PU24FCl. As a result, the 8-aryl ring of PU-H71 makes additional π–π interactions with Trp162 and Phe138, and with a water mediated hydrogen bond from the iodine to the carbonyl oxygen of Leu107. One reason for this drastic change in conformation is the necessity to avoid a steric clash with Tyr139. Whereas the methoxy groups of PU24FCl can freely rotate away from the tyrosine ring, the fixed orientation of the 4′,5′-methylenedioxy group prevents simple rotation as a means of alleviating steric strain. In order to avoid steric clash, the whole 8-aryl ring system in PU-H71 must rotate and, as a result, the 2′-halogen in these molecules are oriented in completely opposite directions. PU-H71 has shown potent activity in preclinical models of SCLC [40], hepatocellular carcinoma [41], triple-negative breast cancer [42], diffuse large B-cell lymphomas [43] and myeloproliferative disorders [44] and is scheduled for clinical translation in cancer.
Kasibhatla et al. [45] modified the purine series by rotating the C8-attached aryl ring to the N9 position [45]. This resulted in the compound CNF2024/BIIB021 (14; Figure 3B), the first synthetic Hsp90 inhibitor to enter Phase I clinical trials [39].
Curis replaced the N3 amine in the purine series with a carbon to result in CUDC-305 (15; Figure 3B) [46]. CUDC-305 is brain permeable and can potentially be useful in primary and metastatic brain cancers. CUDC-305 has been licensed to Debiopharm SA and is currently undergoing Phase I clinical evaluation under the name Debio 0932.
3.1.3 Biochemical and cell-based screening
A better understanding of Hsp90 biochemistry and tumor biology has led to the development of several biochemical and cellular assays that were used to identify novel Hsp90 inhibitors. Among these assays are those measuring Hsp90 ATPase activity (Section 3.1.3.1), competitive binding to Hsp90 (Section 3.1.3.2), competitive binding to a purine affinity column (Section 3.1.3.3) and selective mutant p53 degradation (Section 3.1.3.4).
3.1.3.1 Hsp90 ATPase activity inhibition
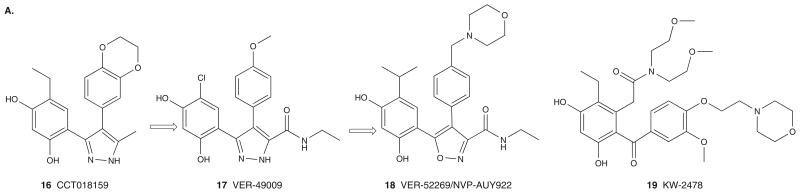
A library of 56,000 compounds was screened for inhibition of yHsp90 ATPase activity using a colorimetric readout for detection of inorganic phosphate [47]. This effort resulted in the identification of a resorcinolic pyrazole derivative, CCT018159 (16; Figure 4A), as an Hsp90 inhibitor. The X-ray structure of yHsp90 bound CCT018159 revealed that the resorcinol hydroxyls and the pyrazole nitrogen atoms make significant direct and water mediated interactions with the Asp79, Gly83 and Thr171 side chains, and that CCT018159 mimics the binding interactions made by RD [31].
Co-crystal structures of resorcinol-type inhibitors with Hsp90 resulted in a better understanding of their binding mode and assisted in the further development of these compounds. Along these lines, structure-based optimization of pyrazole CCT018159 led to the more potent inhibitor VER-49009 (17; Figure 4A). Further optimizations led to VER-52296/NVP-AUY922 (18; Figure 4A) whereby the pyrazole was replaced by its bioisostere, isoxazole, for the purpose of keeping a more defined hydrogen bonding network with Hsp90 (Figure 2D) [31]. VER-52296/NVP-AUY922 shows improved cellular uptake and retention in cancer cells compared to the corresponding pyrazole derivative, which may explain the enhanced cellular activity of this compound. Exposure of cancer cells to VER-52296/NVP-AUY922 resulted in concentration- and time-dependent Hsp90 client modulation and induction of Hsp70 expression, and the agent was reported to have antitumor activity in colon and breast cancer xenograft models [31,48]. VER-52296/NVP-AUY922 is currently undergoing clinical evaluation in cancers [1,2].
Another novel resorcinol analog, KW-2478 (19; Figure 4A), was reported by Kyowa Hakko Kirin Co [49]. KW-2478 showed significant reduction in tumor growth in a mouse model bearing NCI-H929 human tumor xenonograft following intravenous administration once daily for 5 days at doses of 25 – 100 mg/kg. These effects were associated with a decrease in several Hsp90-chaperoned onco-client proteins. Currently, KW-2478 is under Phase I clinical investigation in MM and in Phase II in combination with bortezomib in relapsed (or refractory) MM patients.
3.1.3.2 Competitive binding inhibition
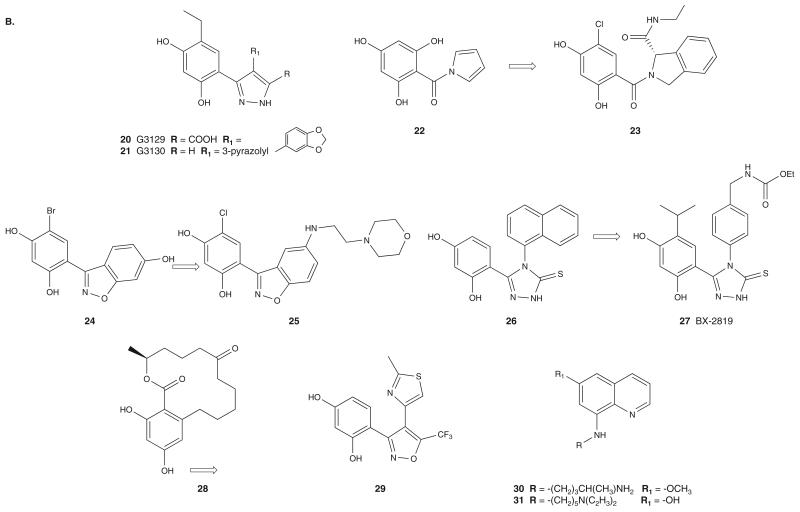
Resorcinolic pyrazoles G3129 (20) and G3130 (21) (Figure 4B) were also identified as Hsp90 inhibitors using a timeresolved FRET-based high-throughput screening (HTS) assay [50] that measures the binding of biotinylated GM to the His-tagged hHsp90 NBD.
Scientists at Pfizer developed a HTS based on the compounds ability to displace tritium-labeled 17-propylamino-benzoquinone ansamycin (17-PGA) from Hsp90 bound to copper on yttrium-silicate scintillant beads. This effort led to the discovery of the tri-hydroxy containing compound 22 (Hsp90 binding affinity Ki = 200 nM and cellular activity IC50 > 20 μM) [51]. X-ray crystallography driven structure modification led to the discovery of 23 (Hsp90 binding affinity Ki = < 1 nM and cellular activity IC50 = 300 nM) [52] (Figure 4B). Similar to other resorcinol containing inhibitors, 23 binds to the NBD of Hsp90.
HTS using a fluorescence polarization (FP) competition assay using BODIPY-GM identified the benzisoxazole derivative 24 as an Hsp90 inhibitor (IC50 = 0.19 μM) with poor cellular activity (IC50 > 20 μM) [53]. Further optimization led to compound 25 (IC50 = 30 nM) (Figure 4B), which exhibited antiproliferative activity against a panel of cancer cell lines at submicromolar concentrations. The co-crystal structure of this compound with the NBD of hHsp90 revealed that it binds similar to ADP and other resorcinol-containing compounds such as RD. In addition, binding of 25 induces the rearrangement of a flexible loop to accommodate the water solubilizing morpholine group, which was closed in case of hit compound 24.
The resorcinol analog 26 (Figure 4B), containing a triazolothione ring, was also identified as an Hsp90 inhibitor by HTS of molecules that compete with the binding of GM-BOD-IPY [54]. Optimization resulted in BX-2819 (27) that binds potently to Hsp90, displaying an IC50 = 41 nM for inhibition of GM-BODIPY binding. BX-2819 blocked the expression of HER2 in SKBr3 breast or SKOV3 ovarian cancer cells and also stimulated the expression of Hsp70. The X-ray crystal structure of 27 with the NBD of Hsp90 indicates that it binds in the nucleotide-binding pocket of Hsp90 in a manner similar to ADP, GM and the resorcinol-containing molecules.
A HTS effort using a FP assay that measured the interaction of a red-shifted fluorescently labeled geldanamycin (GM-cy3B) with Hsp90 in tumor cell lysates identified compounds 28 and 29 as Hsp90 inhibitors [55] (Figure 4B). Use of cancer cell derived lysates instead of recombinant Hsp90 is advantageous as lysate protein contains the therapeutically relevant form of Hsp90, which is a high affinity, co-chaperone-bound state. Compounds 28 and 29 are derivatives of the resorcinol and pyrazole scaffolds, respectively. This effort also identified aminoquinoline 30 as a novel inhibitor [56]. Quinocide dihydrochloride (30) inhibits Hsp90 in the FP assay with an IC50 = 5.8 μM and has cellular activity at similar concentrations. Further optimization efforts yielded compound 31 with an IC50 of 1 μM in the Hsp90 FP assay.
3.1.3.3 Purine-column affinity purification
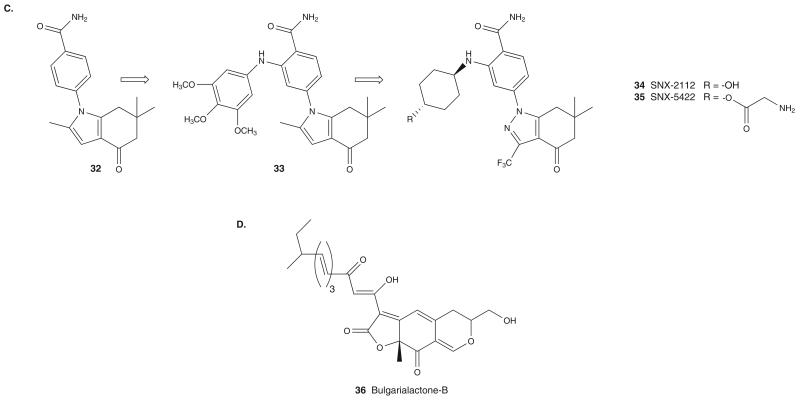
A chemoproteomics-based drug design approach was used by Serenex to identify a new Hsp90 inhibitor chemotype [57]. In this approach, purine-binding proteins from porcine lung or liver were loaded onto an affinity column and were subsequently challenged with a library of structurally diverse 8000 compounds. Mass spectrum analysis of proteins eluted by compound 32 (Figure 4C) resulted in the identification of Hsp90 as a potential binder of 32 (Kd = 3.7 μM) [57]. Initial optimization of 32 provided compound 33 (Figure 4C) (Hsp90 binding affinity of Kd = 0.22 μM) that was optimized to result in the pyrazole SNX-2112 (34; Figure 4C), a compound of improved Hsp90 binding affinity and better in vivo properties [58].
The binding mode of this class of compounds was deduced from the co-crystal structure of 33 with the NBD (residues 1 – 232) of hHsp90α. The amide oxygen and the NH2 group of the benzamide moiety mimic adenine N1 and (C6)-NH2 of ATP, respectively, and interact by forming both direct and water mediated hydrogen bonds to Thr184 and Asp93. As seen with the purine-based inhibitors, conformational rearrangement of Hsp90 on 33 binding results in displacement of Leu107 from its natural position and creates a hydrophobic binding pocket for the indolone moiety. Currently in development by Pfizer, SNX-5422 (35), the glycine prodrug of SNX-2112, is undergoing Phase I and II clinical trials in cancers [2].
3.1.3.4 Cell-based assay
Bulgarialactone B (36; Figure 4D), an azaphilone derived from ascomycetes, was identified in a cellular screen looking for compounds that selectively degrade mutant but not WT p53 protein [59]. Based on surface plasmon resonance binding analysis and limited proteolysis-mass spectrometry techniques, bulgarialactone B is believed to bind to the NBD (90 – 280 region) of Hsp90 [60]. Interestingly, while bulgarialactone B and other natural azaphilones downregulate several Hsp90 client proteins, such as Raf-1, survivin, CDK4, AKT and EGFR, they fail to induce a feedback heat-shock response, as indicated by absence of Hsp70 upregulation.
3.1.4 Virtual screening (in silico)
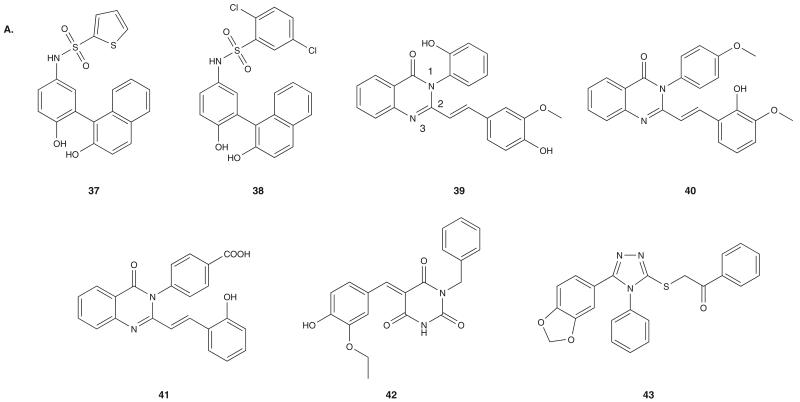
Virtual screening has also led to the identification of novel chemical scaffolds as initial structural leads targeting Hsp90. For example, scientists at Vernalis identified 1-(2-phenol)-2-naphthol [61] as a new Hsp90 inhibitor class by docking a library of 700,000 commercial compounds against the GM-bound and PU3-bound conformations of hHsp90 NBD. These efforts led to the identification of 37 and 38 (Figure 5A) with binding affinities for Hsp90 determined to be 600 and 700 nM, respectively, in an FP assay. 37 and 38 showed moderate cancer cell growth inhibition activity associated with reduction of an Hsp90 client, CDK4 and with induction of Hsp70. The crystal structure of 38 with Hsp90 suggests a binding similar to the resorcinol class, with the naphthol hydroxyl making H-bonding interactions with Asp93 [61].
Park et al. used virtual screening to identify 3-phenyl-2-styryl-3H-quinazolin-4-one derivatives as Hsp90 inhibitors [62]. A library of 85,000 commercially available compounds was screened against a receptor model created by using the coordinates in the crystal structure of Hsp90 in complex with a benzenesulfonamide inhibitor [61] and with water molecules found within 3.5Å of ligand. Compounds 39 – 41 (Figure 5A) were identified as inhibitors in this screen and subsequently validated by an in vitro colorimetric ATPase assay using yHsp90. They showed modest activity in inhibiting Hsp90 ATPase activity and in inhibiting the proliferation of cancer cells (GI50 = 25.4, 27.5 and 35.6 μM, respectively). Docking analysis of these compounds shows the N-3 of each inhibitor making direct H-bond contacts with the side chain amide of Asn51, while the carbonyl oxygen makes indirect H-bond contacts with Asp93 via a structural water molecule, exemplifying the importance of water mediated binding interactions in the ATP-binding site. In addition to quinazoline compounds, the same group used structure-based virtual screening to identify pyrimidine-2,4,6-trione (42) and 4H-1,2,4-triazole-3-thiol (43) as novel Hsp90 binding scaffolds (Figure 5A) [63]. Each of these compounds resulted in modest cellular activity.
3.1.5 Fragment-based drug discovery
FBDD is another approach used to identify Hsp90 binders. The basic concept of this method is to identify by NMR (Section 3.1.5.1) or biochemical (Section 3.1.5.2) techniques small molecular mass fragments that form quality interactions with different amino acids in the therapeutic target. Although individual fragments may have weak potency, they may be combined to provide an attractive starting point for medicinal chemistry efforts.
3.1.5.1 NMR
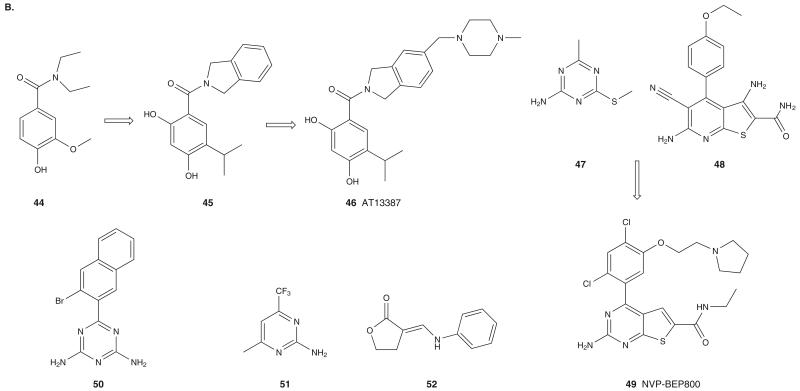
Scientists at Astex Therapeutics used a FBDD approach to identify fragments with Hsp90 binding affinity. Initially, 1600 fragment library compounds were screened by NMR to compete with ADP for binding to the NBD of Hsp90. This effort led to identification of fragment 44 (Figure 5B) with an affinity for Hsp90 of 790 μM, as determined by isothermal titration calorimetry [64]. Optimization of this hit fragment provided lead compound 45 with Hsp90 binding affinity of 0.54 nM. Further optimization resulted in the clinical compound AT13387 (46; Figure 5B) that binds at the NBD of Hsp90 (Ki = 0.6 nM) [65]. The co-crystal structure of AT13387 with the NBD of human Hsp90 suggests that it interacts with the Hsp90 pocket in a fashion resembling RD, where the resorcinol moiety makes critical H-bond contacts with Asp93. In addition, binding of the inhibitor causes the side chain of Lys58 to move, exposing a hydrophobic pocket that is occupied by the phenyl ring of the isoindoline moiety, which forms hydrophobic contacts with Ala55, Lys58 and Ile96. AT13387 resulted in significant tumor growth delay in xenograft models of melanoma, NSCLC and AML [66]. A Phase I study to assess the safety of escalating doses of AT13387 in patients with metastatic solid tumors is currently ongoing.
A novel series of 2-aminothieno[2,3-d]pyrimidine compounds were developed by combining hits identified from FBDD and in silico screening approaches [67]. Screening of 1351 fragments for binding affinity to the NBD of human Hsp90 in the presence of PU3 led to identification of the hit fragment 47 (FP IC50 = 365 μM) (Figure 5B). In a parallel approach, virtual screening of a library of 700,000 compounds against GM-bound and PU3-bound forms of hHsp90 NBD led to the identification of compound 48 (Figure 5B) (FP IC50 = 0.9 μM). Optimization of these hit fragments using X-ray crystallographic data and SAR led to NVP-BEP800 (49; Figure 5B) as a new Hsp90 inhibitor (IC50 = 58 nM). 49 showed potent activity in mice bearing either A375 human melanoma or BT-474 human breast cancer xenograft [67].
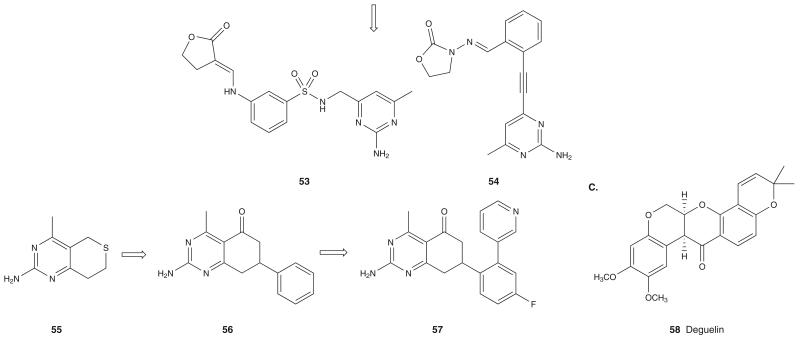
NMR-based screening of 11,520 compounds for Hsp90 binding affinity by scientists at Abbott resulted in identification of two fragments, aminotriazine 50 and aminopyrimidine 51 (Figure 5B) (Ki = 0.32 and 18 μM, respectively, by FRET assay) [68]. Binding affinities were determined as a measure of the ability of the compounds to cause chemical shifts in the NMR spectra of Leu, Val and Ile methyl groups found in the NBD of hHsp90. Co-crystal structures of both 50 and 51 with the NBD of hHsp90 suggest that these compounds bind to Hsp90 in a manner similar to ADP. Interestingly, the naphthalene moiety of 50 induces a conformational change that results in opening of a larger binding site that can be further exploited to increase potency. A second NMR screening of a 3360 compound library testing for Hsp90 binding in the presence of saturating levels of 51 led to the identification of a furanone moiety containing derivative 52 (Figure 5B) (Kd = 150 μM in the presence and > 5000 μM in the absence of 51 suggesting co-operative binding). Linking fragments 51 and 52 led to compounds 53 (sulfonamide linker, Ki = 1.9 μM) and 54 (acetylene linker, Ki = 4 μM) (Figure 5B) that bind to the closed and open conformation of Hsp90, respectively. The methylene sulfonamide linker in 53 induces a 180° bend between the aminopyrimidine and the furanone groups resulting in a stacked orientation that prefers the closed conformation of Hsp90. On the other hand, in compound 54, the acetylene linker causes a 90° angle between the linking groups, resulting in compound preference for the open conformation of Hsp90 [68].
3.1.5.2 Biochemical assay
A fragment library of 20,000 compounds was screened for Hsp90 binding using a high concentration confocal fluorescence-based biochemical assay whereby fragments were identified that displaced a Tamra-labeled analog of GM [69]. This process led to identification of the nonplanar bicylic aminopyrimidine 55 as an Hsp90 binder (IC50 = 15 μM) (Figure 5B). Additional screening of compounds for substructural features of 55 and subsequent docking of hits in two Hsp90 crystal structures containing either of the open or the closed helical pocket led to the discovery of tetrahydrobenzopyrimidine 56 (IC50 = 0.8 μM) (Figure 5B). This compound accesses the helical pocket adjacent to the ATP binding site of Hsp90. Further structure-guided optimization approach led to the discovery of 57 (Figure 5B) (IC50 = 30 nM) with submicromolar cellular activity in NSCLC and colon cancer cells. This compound caused the degradation of Raf-1 and induced Hsp70 in select cancer cells. The crystal structure of 57 with the NBD of hHsp90 shows that it makes direct and indirect H-bond interactions with Hsp90 and that the phenyl ring triggers an opening of the helical binding pocket for the ortho pyridyl ring to make π-stacking interactions with Phe138 of the binding pocket [69].
3.1.6 Educated guess
The rotenoid derivative deguelin (58; Figure 5C), known to have anticancer activity against a variety of cancers, was found to disrupt the binding of Hsp90 to one of its client proteins, HIF-1 [70]. Follow-up investigation into the mechanism of action of deguelin demonstrated that it binds to the Hsp90 ATP-binding pocket. Similar to other Hsp90 inhibitors, addition of deguelin to cancer cells led to ubiquitin mediated degradation of Hsp90 client proteins such as CDK4, AKT, eNOS, MEK1/2 and mutant p53.
3.2 C-terminal inhibitors
The CDD of Hsp90 is believed to allosterically modulate the NBD ATPase activity through a second nucleotide binding site, thus providing another strategy towards altering Hsp90 chaperone activity [71]. The putative binding site is believed to be buried in the Hsp90 dimer; however, it can be unveiled after transient separation of the CDD caused by inter-domain communication following ATP binding to the NBD [71]. Though the specific site(s) is unknown, binding of compounds to the CDD causes conformational changes to the chaperone structure that disrupt the interaction between Hsp90 and co-chaperones, eventually leading to the destabilization of client proteins.
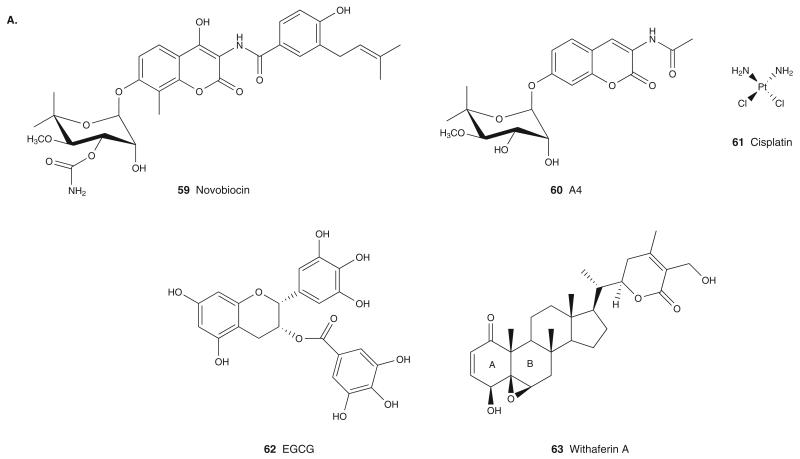
Novobiocin (59; Figure 6A) was the first molecule found to inhibit Hsp90 by binding to the CDD. Novobiocin is a coumeromycin antibiotic and inhibitor of DNA gyrase. Like Hsp90, DNA gyrase is a member of the GHKL family, and due to high structural similarities between DNA gyrase and Hsp90, novobiocin was initially investigated for its binding to the NBD of Hsp90 [72]. Though novobiocin weakly inhibits Hsp90 (IC50 = 700 μM in SKBr3 cells) and depletes several Hsp90 client proteins, such as HER2, v-src, Raf-1 and mutated p53, it fails to compete with GM or RD for binding to the NBD. In fact, it was determined via truncation studies that novobiocin binds to the CDD of Hsp90 resulting in destabilization of the chaperone complex, release of co-chaperones and substrates, with the subsequent degradation of Hsp90 client proteins. Removal of the hydroxyl group on the coumarin scaffold and of the carbamate moiety on the noviose sugar resulted in A4 (60) as a selective Hsp90 inhibitor (IC50 = 1 μM) with only weak gyrase activity [72]. In vivo activity for this class of compounds is yet to be reported.
Figure 6. Hsp90 inhibitors that act by: (A) binding to the C-terminal of Hsp90, (B) targeting co-chaperone–Hsp90 interaction and (C) targeting Hsp90–client protein binding.
Cisplatin (61; Figure 6A), a platinum containing chemotherapeutic agent, was also shown to have high affinity for the CDD of Hsp90 [73]. In neuoroblastoma cells, cisplatin specifically inhibited the steroid receptor–Hsp90 complex and caused the selective degradation of androgen and glucocorticoid steroid receptors without affecting other Hsp90 client proteins.
Epigallocatechin-3-gallate (EGCG; 62; Figure 6A), a polyphenol found in green tea, inhibits the activity of telomerase, multiple kinases and the aryl hydrocarbon receptor by binding to Hsp90. Based on affinity chromatography, EGCG binds to amino acids 538 – 728 of Hsp90, which encompass the putative ATP binding site in the CDD [74].
Withaferin A (WA; 63; Figure 6A), a steroidal lactone extracted from Withania somnifera, shows potent antiproliferative activity in several cancer cells [75]. WA binds to the CDD of Hsp90 and causes the proteasomal degradation of several Hsp90 clients, such as AKT, CDK4 and glucocorticoid receptor. WA also disrupts the Hsp90–Cdc37 complex either by binding to the CDD of Hsp90 and causing a change in Hsp90 conformation that prevents Cdc37 binding or by directly labeling cysteine residues of Cdc37 or Hsp90. The ketone containing unsaturated A ring, the epoxide within B ring and the unsaturated lactone ring E are three moieties crucial for the interaction between WA and Hsp90. As these groups are reactive Michael acceptors, they probably react with thiol-nucleophiles in Hsp90, leading to covalent protein–WA adducts. In accord with this mechanism of action, preincubation of cancer cells with N-acetylcysteine, a thiol antioxidant, reversed the Hsp90-induced effects of WA, such as onco-client protein degradation and induction of Hsp70. These data suggest that WA may inhibit Hsp90 function through covalent modification of cysteines located in the C-terminal of Hsp90, whose identity remains to be further elucidated.
Though significant work has been carried out on the C-terminal Hsp90 inhibitors, besides the unspecific protein modifier, cisplatin, none has advanced to clinical trials. The lack of a reported co-crystal structure between any such potential interactor, their modest reported biological activity and potential pleiotropic mechanisms of action may be the major reasons for their lack of advancement in spite of exponential interest over the last few years in the development of Hsp90 inhibitors for cancers.
3.3 Targeting co-chaperone–Hsp90 interactions
In eukaryotic Hsp90, co-chaperones play an important role in driving the chaperone cycle through to completion. Therefore, affecting co-chaperone function by specifically targeting their interaction with Hsp90 offers an alternative way to modulate Hsp90 activity. While this strategy has proven difficult, some progress has been made in identifying molecules that affect the interaction of Hsp90 with Cdc37 (Section 3.3.1), HOP (Section 3.3.2) and Aha1 (Section 3.3.3).
3.3.1 Cdc37/Hsp90
As was previously discussed, the co-chaperone Cdc37 functions in the recruitment of client proteins, predominantly kinases such as EGFR, Src, Lck, Raf-1 and CDK4, to Hsp90 [26]. It inhibits Hsp90 ATPase activity by binding to sites on the lid segment of the NBD in the open conformation and by preventing N-terminal domain dimerization [20,76]. Therefore, molecules that disrupt Cdc37-Hsp90 interaction would offer an additional opportunity to affect the chaperone cycle. Cdc37 is overexpressed in cancer cells, potentially offering additional tumor-targeting specificity and an improved side effect profile to that of direct targeting of Hsp90. Because most of the Hsp90 clients regulated by Cdc37 are kinases, it is hypothesized that Cdc37 targeting may offer a therapeutic benefit in cancers that are kinase driven [26]. In human colon cancer cells, silencing of Cdc37 using siRNA resulted in proteasome mediated degradation of Hsp90 client kinases, including HER2, Raf-1, CDK4, CDK6 and AKT and in inhibition of cell proliferation [77]. Interestingly, depletion of Cdc37 failed to result in a heat-shock response that is typical with Hsp90 NBD ATPase inhibitors. Combination of Cdc37 silencing with 17-AAG or pyrazole VER-49009 Hsp90 inhibition induced more extensive and sustained depletion of kinase clients than either approach alone, suggesting a potential therapeutic benefit for combining Hsp90 and Cdc37 inhibitors [77].
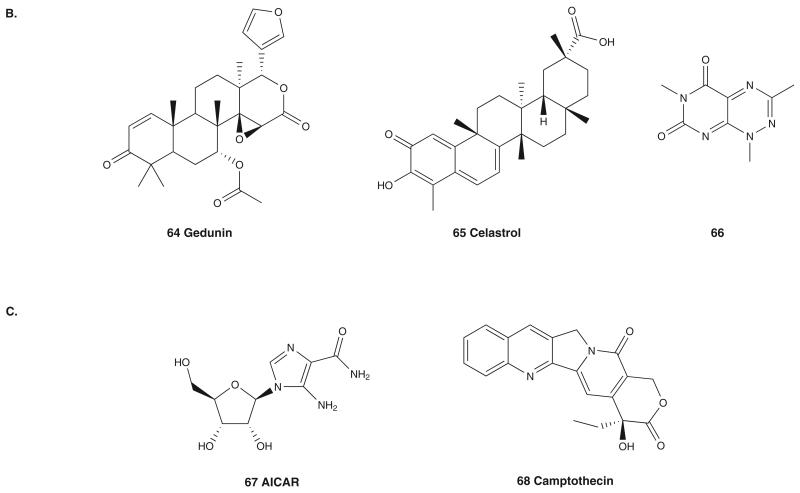
Gedunin (64; Figure 6B), a tetranortriterpenoid isolated from Azadirachta indica, and celastrol (65; Figure 6B), a quinone methide triterpene isolated from Tripterygium wilfordii, were initially reported to inhibit the Hsp90 pathway through the use of gene expression signature-based screening (GE-HTS) [78], specifically by the use of the gene expression Connectivity Map. In this map, the biological activity of a compound is connected to the biological activities of drugs with known mode of action by comparing gene expression in cells treated with compounds to the compendium of gene expression profile of drugs. Based on this Connectivity Map, and because gedunin and celastrol invoked gene expression signatures highly similar to those of GM, 17-DMAG and 17-AAG, it was determined that they exhibited their biological activity through Hsp90 pathway modulation. Gedunin decreased the level of androgen receptor (AR) in LNCaP prostate cancer cells in a concentration-dependent manner and also caused the depletion of other Hsp90 client proteins such as EGFR, Bcr-Abl and FLT3 in various cancer cells [78]. Precisely, how gedunin modulates AR-Hsp90 interaction is not known. Gedunin inhibited ATP binding to Hsp90 in LNCaP cells but failed to compete with the binding of Cy3B-GM to purified Hsp90, suggesting a mechanism other than that of the NBD ATPase inhibitors.
Follow-up work on celastrol found that it disrupted Cdc37-Hsp90 association, resulting in degradation of AKT and CDK4 and induction of apoptosis in the pancreatic cell line Panc-1 [79]. Celestrol does not inhibit ATP binding, and molecular docking suggests the binding site to be in the NBD of Hsp90 [79]. However, recent NMR studies suggest that celastrol binds to Cdc37 and not the Hsp90 NBD [80]. Furthermore, MS analysis indicates that celastrol forms a covalent adduct with a cysteine residue of Cdc37, presumably through Michael addition to an electrophilic site on celastrol. Based on this result, the observed effects described above are potentially due to pharmacological targeting of Cdc37. However, it cannot be ignored that celastrol is also a known proteasome inhibitor [81] and similar to withaferin A, it probably acts on cancer cells through a multitude of mechanisms.
Because of the potential importance of Cdc37 as an anticancer target, and because its inhibition fails to activate a heat-shock response, efforts should direct towards the discovery of more drug-like chemical scaffolds that can selectively disrupt Cdc37–Hsp90 interaction.
3.3.2 HOP/Hsp90
As mentioned earlier, HOP mediates the formation of a complex among Hsp90, Hsp70 and a client protein. The TPR domains of HOP, TPR1 and TPR2A bind the EEVD motif found on the C-terminal domain tails of Hsp70 (GPTIEEVD) and Hsp90 (MEEVD), respectively, thereby joining Hsp70 and Hsp90 for client protein transfer [82]. Inhibiting these protein–protein interactions may prevent formation of the intermediate complex and thus modulate Hsp90 chaperone activity. In this regard, a TPR mimic, CTPR390+, was designed. CTPR390 is a peptide consisting of three TPR motif repeats, where each TPR motif consists of 34 amino-acid residues forming two antiparallel α-helices that are stacked together to produce a superhelical structure. Amino acids with high global propensity to occur at each of the 34 positions of the TPR motif in different proteins were determined by statistical analysis to yield consensus TPR with 3 repeats (CTPR3). Grafting of Hsp90-binding residues from TPR2A onto the CTPR3 scaffold resulted in CTPR390. This peptide mimic binds with high affinity to the Hsp90 C-terminal domain (Kd = < 1 μM) [83] and prevents the formation of a functional Hsp70–HOP–Hsp90 complex. In breast cancer cells, addition of CTPR390+ resulted in the degradation of HER2 and inhibition of proliferation [83]. More importantly, unlike N-terminal domain binders, CTPR390+ did not induce Hsp70 when added to breast cancer cells.
Novel small molecules that hinder the Hsp90–TPR2A interaction were also identified from an AlphaScreen technology-based HTS effort. In all, 76,314 compounds from the NIH Chemical Genomics Center and 20,000 compounds from the Maybridge diversity library were screened for their ability to disrupt the interaction of TPR2A with a C-terminal Hsp90 peptide (TEEMPPLEGDDDTSRMEEVD) [84] and resulted in three hits, each having in common a core 7-azapteridine ring system [85]. 66, a representative of this class (Figure 6B), inhibited proliferation of BT474 and SKBr3 (IC50 = 0.946 and 1.25 μM, respectively) breast cancer cells, and resulted in a short-lived decrease in HER2 levels. Interestingly, these molecules failed to induce Hsp70 levels in cells.
Clearly, more work is required to fully appreciate the implications of modulating the HOP–Hsp90 interaction. Initial work with molecules such as 66, while encouraging, suggest that 66, like WA and celastrol, is a potential cysteine modifier. Discovery of novel inhibitors with increased potency/selectivity would be crucial to translate this strategy into a clinically useful agent.
3.3.3 Aha1/Hsp90
As was previously discussed, Aha1 is a co-chaperone that enhances the ATPase activity of Hsp90 and helps to drive the chaperone cycle forward in the maturation of client proteins [86,87]. siRNA silencing of Aha1 failed to affect the expression of Hsp90 clients such as Raf-1, HER2 and CDK4, but resulted instead in decreased kinase activity for Raf-1 and in reduced levels of phosphorylated MEK1/2 and ERK1/2 in HCT116 colon cancer cells. Based on these findings, it is proposed that Aha1 may play a role in activation rather than stabilization of Hsp90 client proteins [88].
3.4 Targeting client–Hsp90 interactions
The maturation of client proteins requires extensive physical contact with Hsp90. Therefore, affecting these protein–protein interactions by targeting sites on Hsp90 or client that are required for their interaction may offer an additional way of modulating Hsp90 activity.
3.4.1 Hsp90-survivin
Survivin is a member of the inhibitor of apoptosis protein family whose function is governed by Hsp90 in cancer cells [89]. Survivin binds to the NBD of Hsp90 and disruption of this interaction destabilizes survivin, initiates mitochondrial apoptosis and suppresses cell proliferation [89]. A peptide sequence of survivin (sequence Lys79-Leu87, KHSSGCAFL), called shepherdin [27,89], inhibited survivin–Hsp90 interaction. Because of the extensive contacts it makes with the NBD of Hsp90, it is believed that shepherdin blocks binding of ATP and Cdc37 to Hsp90. When added to cancer cells, shepherdin results in apoptosis, degradation of survivin and other Hsp90 client proteins such as AKT, CDK4 and CDK6. In preclinical mouse models of cancer, shepherdin exhibited anticancer activity against various tumor types.
A pharmacophore model was generated by in silico docking of shepherdin into the crystal structure of the GM-bound hHsp90 NBD, which led to the discovery of 5-aminoimidazole-4-carboxamide-1-α-d-ribofuranoside (AICAR; 67; Figure 6C) as an Hsp90 inhibitor [90]. Docking studies suggest that AICAR interacts with the NBD of Hsp90, with binding and functional properties mimicking those of shepherdin. However, probably due to poor cell-permeability properties, AICAR only exhibited moderate antiproliferative activity in cancer cells, although it spared WI38 human lung fibroblasts at similar concentrations.
3.4.2 Hsp90–AR complex
Prostate cancer is dependent on AR-mediated signaling. In the cytoplasm, Hsp90 is responsible for stabilization of unliganded AR and participates in the activation process by maintaining apoAR in a high-affinity ligand binding conformation [1]. Hsp90 is also required for AR to acquire active conformation following agonist binding and plays a role in nuclear transfer [91] and disruption of Hsp90-AR association leads to cytoplasmic aggregates of AR. In LNCaP prostate cancer cells, camptothecin (68; Figure 6C), a topoisomerase inhibitor, causes Hsp90 to dissociate from AR, thereby blocking its nuclear translocation [92].
3.5 Inactivators of Hsp90 function by posttranslational modifications
Post-translational modifications such as acetylation [93] (Section 3.5.1), nitrosylation [94,95] (Section 3.5.2) and phosphorylation [96] (Section 3.5.3) are also thought to regulate Hsp90 chaperone activity by allosterically affecting binding of co-chaperones and/or nucleotides. Thus, targeting these modifications offers an alternative way to inhibit Hsp90 chaperone activity.
3.5.1 Acetylation
HDACs such as HDAC6 [93,97] and histone acetyltransferases such as p300 [98] regulate Hsp90 by controlling the reversible acetylation of Hsp90. Mutational analysis showed acetylation of Hsp90 at Lys294 in the MD weakens interaction with a variety of client proteins as well as with co-chaperones, but does not affect ATP binding [99]. Hyperacetylation of Hsp90 either by HDAC6 pharmacological inhibition or by knockdown using siRNA [100] leads to dissociation of co-chaperone p23 from Hsp90, thus preventing Hsp90-dependent maturation of client proteins such as glucocorticosteroid receptor [93] and Bcr-Abl [97]. HDAC inhibitors LAQ824 and LBH589 induce Hsp90 hyperacetylation, which results in inhibition of chaperone functions and subsequent polyubiquitylation, proteasomal degradation and depletion of several client proteins, such as Bcr-Abl, AKT and Raf-1 in CML cells [97]. In AML cells expressing mutant oncoprotein FLT3, MC-275, a synthetic inhibitor of HDAC1, induced hyperacetylation of Hsp90, leading to inhibition of Hsp90–FLT3 interaction and proteasomal degradation of FLT3 [101]. Interestingly, HDAC6 inhibition by siRNA increased the affinity of 17-AAG for Hsp90 [100]. Co-treatment of 17-AAG and the HDAC inhibitor LBH589 produced synergistic effects in attenuation of Bcr-Abl activity and in induction of apoptosis in CML and AML cells [102].
3.5.2 S-nitrosylation
S-Nitrosylation of Hsp90 by its client protein, eNOS, represents another level of Hsp90 regulation. In a feedback mechanism, S-nitrosylation of Cys597 by eNOS results in decreased ATPase activity, which results in decreased binding and activation of eNOS by Hsp90 [94]. Cys597 is located in the middle of a conformational switch region in Hsp90 CDD, and in silico-based analysis suggests that this residue is involved in regulating the conformation in Hsp90 [103]. In mutants of both human and yHsp90, S-nitrosylation at this position results in a change in Hsp90 conformational equilibrium leading to decreased affinity for Aha1, resulting in decreased Aha1 stimulated Hsp90 ATPase activity [95].
3.5.3 Phosphorylation
The phosphorylation state of several serine, threonine and tyrosine residues on Hsp90 is reported to uniquely modulate its chaperone function. Ppt1 dephosphorylates Hsp90 in vitro and genomic deletion of the ppt1 gene in yeast results in hyperphosphorylation of Hsp90 and in an apparent decrease in the efficacy of the Hsp90 chaperone system [96]. In the Hsp90 mediated activation of the reovirus attachment protein ó1, it was suggested that phosphorylation was linked to release of client protein, as only unphosphorylated Hsp90 was associated withó1 [104]. Phosphorylation of Hsp90 and/or v-src also reduced their association [105]. Hsp90 is also a substrate of casein kinase 2 (CK2), where CK2 mediated phosphorylation is required for Hsp90 to chaperone several kinases including CK2 itself [106]. A positive feedback relationship between Hsp90 and its kinases is further supported by tyrosine kinase WEE1 [107]. Although WEE1 is an Hsp90 client protein, it also modulates Hsp90 activity by directly phosphorylating Tyr38 in the NBD of Hsp90. Phosphorylation of Tyr38 positively influences Hsp90 chaperone activity for certain cancer kinases such as HER2, Raf-1, CDK4 and WEE1, but negatively affects the binding of GM to Hsp90. The apoptotic agent IC101 induces Hsp90 tyrosine dephosphorylation which causes inhibition of AKT-Hsp90 binding, leading to release of AKT from the chaperone complex and causing dephosphorylation of AKT [108]. Dephosphorylation of Hsp90 by ICI101 led to inhibition of ATP binding in a non-competitive manner with subsequent degradation of Raf-1. A better understanding of the role of kinases and phosphatases in regulating Hsp90 function could potentially lead to novel drug targets.
4. Conclusion
Molecules that inhibit the binding of ATP to the N-terminal nucleotide binding pocket, such as GM and RD, have offered the most insight into the effects of pharmacological modulation of Hsp90. Early studies using these inhibitors have underscored the importance of Hsp90 function to the transformed cell and resulted in the general acceptance of it as a viable therapeutic target in the treatment of cancer and potentially other diseases.
The goal of this review is to present the reader with the many potential ways to interfere with Hsp90 function that result in biological activity. We assemble the manuscript by starting with a section to point the reader to the complex structural regulation of Hsp90 activity. Here, we describe the ATPase cycle of Hsp90 and show that throughout this cycle Hsp90 undergoes considerable structural changes caused by the binding of ligands including ATP/ADP as well as co-chaperones such as HOP, Cdc37, p23 and Aha1. Each of these seemingly plays a critical role by binding to Hsp90 and altering its conformation in a highly coordinated process with the purpose of binding and activating misfolded proteins. From this, it becomes abundantly clear that there is ample opportunity in modulating Hsp90 activity. The most evident and one that much of the drug discovery efforts have focused thus far is inhibition of Hsp90 by molecules that bind to its N-terminal nucleotidebinding pocket. Our review first focuses on describing these inhibitors. In these sections, we pay special attention to the mode of interaction of these pharmacophores with the Hsp90 pocket and to the specific ways they were discovered. Our goal is to indicate that while these molecules interact with the same pocket, they bind to it in a dissimilar fashion and some induce pocket rearrangements. We also show that the discovery tool used to identify each pharmacophore appears to influence the interaction mode each inhibitor has with Hsp90. We move on to demonstrate the potential for discovering novel chemical agents that affect Hsp90 function by alternative modes. These include molecules that: i) bind to the CDD of Hsp90, ii) cause disruption of co-chaperone–Hsp90 interactions, iii) inhibit client–Hsp90 associations and iv) interfere with post-translational modifications of Hsp90.
5. Expert opinion
As discussed above, most in vivo data to date are available on the NBD ATP-competitive Hsp90 binders. Surprisingly in light of a common binding pocket, clear biological differences have been observed among the several identified chemotypes. Some of the most significant differences observed involve the spectrum of Hsp90–client proteins modulated by specific inhibitors and the kinetics of client protein modulation, which both can have a significant effect on the clinical efficacy and the therapeutic window of the Hsp90-agents. While the reason(s) for these differences are yet unknown, the complex array of conformational changes and co-chaperones that regulate Hsp90 activity, as detailed above, may shed light on this apparent paradox. In addition, as detailed above, it is evident that several ATP-competitive inhibitors induce on binding pocket rearrangements which may also affect binding affinity as well as the Koff of these agents. These pocket rearrangements may also account for a binding preference to any of the several conformational states of Hsp90.
An interesting difference among such inhibitors was noted in their effect on kinases. A body of literature exists stating that Hsp90 is an essential factor in restraining kinase clients, and it has been shown that Hsp90 inhibition by first generation Hsp90 inhibitors GM and RD releases and transiently activates kinases such as ERK and AKT, MAPK and src, among others [109-111]. In prostate cancer, Hsp90 inhibition by 17-AAG leads to transient src activation, an event with the consequence of enhanced bone remodeling and prostate tumor growth in bone [111]. Breast cancers similarly metastasize to bone, and it would be expected that this similar problem may arise, which may pose a significant clinical challenge. Indeed, 17-AAG and RD enhance the incidence of bone metastasis and osteolytic lesions following intracardiac inoculation of MDA-MB-231 cells in the nude mouse [112]. In contrast, in breast cancer cells, PU-H71 treatment immediately and potently inactivated ERK and AKT, without detectable shortterm activation [42]. Such difference may be explained by a preference of GM for the ADP-conformation, in which client protein is released from the complex on ligand binding, whereas PU-H71 may prefer an ATP-conformation with client protein trapping. It will be of interest to assess the Hsp90 clinical candidates for such preferential binding, as it clearly will impact their activity and implementation in metastatic disease.
Such distinct mode of interaction with the Hsp90 conformational states may also help explain the dissimilar pharmacodynamic (PD) profiles reported for the several Hsp90 inhibitors in preclinical and clinical evaluation. Many exhibit an analogous tumor-retention profile, which in the context of comparable Hsp90 affinity would predict identical tumor pharmacodynamics for these agents. The tumor PD profile of these agents is far from uniform, however, with Hsp90 onco-client protein recovery ranging from 16 – 24 h to beyond 48 h post-administration (Table 1). These findings may be partly explained by a distinct onco-client trapping potency of these inhibitors that may result from distinct selectivity of these agents for the many conformational states taken by Hsp90 during its activity cycle.
Table 1. PD effect of several Hsp90 inhibitors in breast cancer tumors.
| Drug/dose (mg/kg)* |
Tumor | Markers, degradation |
Markers, induction |
Drug tumor retention |
Comments |
|---|---|---|---|---|---|
| 17-DMAG | |||||
| 75, i.v. | MDA-MB-231 | Raf-1 | Hsp70 | 2.5 μg/g at 24 h p.a 2 μg/g at 48 h p.a. |
Minimal Raf-1 degradation (< 20%) at 24 h p.a. [114] |
| IPI-504 | |||||
| 75, i.v. or 100, p.o. |
BT-474 | HER2 | NA | NA | HER2 degraded at 7 h p.a. but starts to recover at 24 h and is back to baseline at 48 h p.a. [115] |
| PU-H71 | |||||
| 75, i.p. | MDA-MB-468 MDA-MB-231 |
Raf-1 p-AKT AKT |
cPARP Hsp70 |
2.5 μg/g at 24 h p.a. 1.8 μg/g at 48 h p.a. |
PD markers not recovered at 48 h p.a. [42] |
| 75, i.p. | BT-474 | HER2 | cPARP Hsp70 |
NA | HER2 degraded and returns to baseline at 72 h p.a. cPARP induction up to 72 h p.a. [116] |
| BIIB021 | |||||
| 150, p.o. | BT-474 | HER2 Raf-1 |
Hsp70 | 2 μg/g at 24 h p.a. 1 μg/g at 48 h p.a. |
HER2 degraded at 7 h p.a. but starts to recover at 24 h and is back to baseline at 48 h p.a. [117] |
| NVP-AUY922 | |||||
| 50, i.v. | BT-474 | HER2 | Hsp70 | 5 μg/g at 24 h p.a. 2 μg/g at 48 h p.a. |
HER2 degraded at 6 – 18 h p.a. is back to baseline between 24 and 48 h p.a. [118] |
| SNX-5422 | |||||
| 50, p.o. | BT-474 | HER2 p-AKT |
cPARP | 5 μg/g at 24 h p.a. 1.8 μg/g at 48 h p.a. |
HER2 degraded at 6 – 10 h p.a. recovers at 24 h and is back to baseline at 48 h p.a. cPARP induction up to 10 h p.a. [119] |
Single dose administration.
BT-474: HER2-overexpressing breast cancer; i.p.: Intraperitoneal; i.v.: Intravenous; MDA-MB-231 and MDA-MB-468: Triple-negative breast cancer; NA: Data not available; PD: Pharmacodynamic; p.o.: By mouth.
Another important distinction that we would like to point out is the lack of Hsp70 induction on Hsp90 inhibition observed for certain agents that act on Hsp90 by mechanisms other than direct occupancy of the NBD ATP-pocket. HSF-1 activation and consequent Hsp70 induction by Hsp90 inhibitors is one of the mechanisms theorized to be behind some of the disappointing results in clinical trials with the N-terminal Hsp90 inhibitors [113]. While this may potentially be overcome by the development and concurrent use of inhibitors of HSF-1 or Hsp70, alternative strategies such as inhibition of Hsp90/Cdc37, Hsp90/HOP and Hsp90–client protein interaction may represent promising avenues to alleviate the above mentioned problem with NBD inhibitors.
Altogether, clear biochemical and phenotypic differences observed among the several Hsp90 inhibitors are noted, and these should be taken in consideration in the clinical development of these agents. Nonetheless, the clinical development of all Hsp90 inhibitors to date follows a cookie cutter mimicry of the path designed for 17-AAG. With distinct pharmacokinetic profiles and probably non-overlapping toxicity profiles, a better understanding in preclinical settings should be given to the cancer types more likely to be responsive to each agent. Further, it remains unclear whether in clinic therapeutic doses are indeed delivered to tumors, and whether the limited responses seen in clinic are simply due to an insufficient drug delivery and not a feedback heat-shock response or the potential P-glycoprotein liability of certain inhibitors, such as 17-AAG. More effort, therefore, should be spent on ways to analyze the tumor delivered Hsp90 inhibitor concentration, as it is clear that these agents tend to have a tumor retention profile, and on non-invasive ways to monitor the PD response on Hsp90 administration.
To conclude, Hsp90, the complex and multifaceted protein, remains an attractive but yet not easily understood molecular target. While the initial excitement on the potential of the target has led to the clinical introduction of several inhibitors, we now see that this rush for the first-in-class has left much biology unanswered. A better understanding of the target in the context of each inhibitor will probably move the field forward and lead to better clinical effects. While it is easy to blame the target, fingers should also be pointed at the inhibitors we put in clinic and the strategies we develop for their clinical implementation.
Article highlights.
Heat-shock protein 90 (Hsp90) is currently an attractive target in anticancer drug research because it plays a prominent role in the maturation of many tumor promoting client proteins and enables for the targeting of multiple aberrant signaling pathways. Hsp90 is an ATPase whose function is intimately linked to its conformational state, which in turn is regulated by the binding of nucleotides (ATP/ADP) and co-chaperones (HOP, Cdc37, p23, Aha1 and immunophilins).
Drug discovery efforts to date have resulted in the development of several Hsp90 inhibitors in various states of clinical trials for the treatment of a wide variety of cancers. All molecules which have advanced to clinical trials inhibit the ATPase activity of Hsp90 by binding to the N-terminal nucleotide binding pocket. Efforts to discover these inhibitors have included phenotypic screening, structure-based drug design, biochemical and cell-based screening, virtual screening, fragment-based drug design and educated guess.
Despite an apparently common binding site, the ATP-pocket of Hsp90, it is becoming increasingly clear that the Hsp90 ATP-competitive inhibitors exhibit remarkably different pharmacodynamic (PD) profiles, which can have significant effects in the outcome of clinical trials. The differences in PD profiles cannot be explained by pharmacokinetics alone and may result from drug binding to different conformational states of Hsp90.
An increased understanding into the complex nature of Hsp90 has inspired research groups to look at alternative ways in modulating its activity other than binding to the nucleotide pocket of the nucleotide binding domain. These include agents that bind to the C-terminal dimerization domain, cause disruption of co-chaperone–Hsp90 interactions, inhibit client–Hsp90 associations and interfere with post-translational modifications of Hsp90.
This box summarizes key points contained in the article.
Acknowledgments
Declaration of interest
G Chiosis discloses grant supports from the Geoffrey Beene Cancer Research Center of MSKCC, Leukemia and Lymphoma Society, Breast Cancer Research Fund, the SPORE Pilot Award and Research &Therapeutics Program in Prostate Cancer, the Hirshberg Foundation for Pancreatic Cancer, the Byrne Fund, National Institutes of Health (1U01 AG032969-01A1, 1R01 CA155226-01, 1R21AI090501-01, 3P30CA008748), Department of Defense (R03-BC085588), Susan G Komen for the Cure, the Institute for the Study of Aging and The Association for Frontotemporal Dementias (Grant #281207 AFTD), and the WH Goodwin and A Good-win and the Commonwealth Cancer Foundation for Research and the Experimental Therapeutics Center of MSKCC. T Taldone discloses a grant support from the Department of Defense (PDF-BC093421) and S Modi from ASCO Cancer Foundation/Breast Cancer Research Foundation: Advanced Clinical Research Award.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Trepel J, Mollapour M, Giaccone G, et al. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–49. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter JR, Fritz CC, Depew KM. Discovery and development of Hsp90 inhibitors: a promising pathway for cancer therapy. Curr Opin Chem Biol. 2010;14:412–20. doi: 10.1016/j.cbpa.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–28. doi: 10.1038/nrm2918. •• References [1-3] are comprehensive and up-to-date reviews on Hsp90 covering a broad range of topics.
- 4.Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–93. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- 5.Mickler M, Hessling M, Ratzke C, et al. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–6. doi: 10.1038/nsmb.1557. •• References [4,5] dissect the conformational intermediates of the Hsp90 chaperone cycle using FRET.
- 6.Krukenberg KA, Forster F, Rice LM, et al. Multiple conformations of E. coli Hsp90 in solution: Insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–65. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer MP. Gymnastics of molecular chaperones. Mol Cell. 2010;39:321–31. doi: 10.1016/j.molcel.2010.07.012. • A review article which briefly discusses the conformational dynamics of the Hsp90 chaperone cycle as well as other ATP-dependent chaperones such as Hsp60, Hsp70 and Hsp100.
- 8.Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17:229–35. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Richter K, Soroka J, Skalniak L, et al. Conserved conformational changes in the ATPase cycle of human hsp90. J Biol Chem. 2008;283:17757–65. doi: 10.1074/jbc.M800540200. [DOI] [PubMed] [Google Scholar]
- 10.Murphy PJ, Kanelakis KC, Galigniana MD, et al. Stoichiometry, abundance, and functional significance of the Hsp90/Hsp70-based multiprotein chaperone machinery in reticulocyte lysate. J Biol Chem. 2001;276:30092–8. doi: 10.1074/jbc.M103773200. [DOI] [PubMed] [Google Scholar]
- 11.Kosano H, Stensgard B, Charlesworth MC, et al. The assembly of progesterone receptor-Hsp90 complexes using purified proteins. J Biol Chem. 1998;273:32973–9. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- 12.Shiau AK, Harris SF, Southworth DR, et al. Structural analysis of E. coli Hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–40. doi: 10.1016/j.cell.2006.09.027. •• This paper describes full-length structures of Escherichia coli Hsp90 with and without nucleotide as determined by EM and X-ray crystallography.
- 13.Ali MMU, Roe SM, Vaughan CK, et al. Crystal structure of an Hsp90–nucleotide–p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–17. doi: 10.1038/nature04716. •• This paper describes the first crystal structure of a full-length yeast Hsp90–co-chaperone complex.
- 14.Dollins DE, Warren JJ, Immormino RM, et al. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the Hsp90 chaperones. Mol Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. •• This paper reports the crystal structure of full-length mammalian GRP94 in complex with AMPPNP or ADP.
- 15.Panaretou B, Prodromou C, Roe SM, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO. 1998;17:4829–36. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obermann WMJ, Sondermann H, Russo AA, et al. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–10. doi: 10.1083/jcb.143.4.901. •• References [15,16] show that Hsp90 is dependent on its ATPase activity.
- 17.Prodromou C, Panaretou B, Chohan S, et al. The ATPase cycle of Hsp90 drives a molecular ‘clamp” via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–92. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter K, Muschler P, Hainzl O, et al. Coordinated ATP Hydrolysis by the Hsp90 Dimer. J Biol Chem. 2001;276:33689–96. doi: 10.1074/jbc.M103832200. •• References [17,18] show that ATP binding induces N-terminal dimerization of Hsp90.
- 19.Bron P, Giudice E, Rolland J-P, et al. Apo-Hsp90 coexists in two open conformational states in solution. Biol Cell. 2008;100:413–25. doi: 10.1042/BC20070149. [DOI] [PubMed] [Google Scholar]
- 20.Roe SM, Ali MM, Meyer P, et al. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50 (cdc37) Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- 21.Onuoha SC, Coulstock ET, Grossmann JG, et al. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J Mol Biol. 2008;379:732–44. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Retzlaff M, Hagn F, Mitschke L, et al. Asymmetric activation of the Hsp90 dimer by its cochaperone aha1. Mol Cell. 2010;37:344–54. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Vaughan CK, Gohlke U, Sobott F, et al. Structure of an Hsp90–Cdc37–Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. •• This paper describes the first structure of an Hsp90–co-chaperone–client protein complex.
- 24.Pearl LH. Hsp90 and Cdc37—a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Kim YS, Alarcon SV, Lee S, et al. Update on Hsp90 inhibitors in clinical trial. Curr Top Med Chem. 2009;9:1479–92. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith JR, Workman P. Targeting CDC37. Cell Cycle. 2009;8:362–72. doi: 10.4161/cc.8.3.7531. • A review article describing the possibility of targeting CDC37 as a strategy for treating cancer.
- 27.Plescia J, Salz W, Xia F, et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7:457–68. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 28.Donnelly A, Blagg BSJ. Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr Med Chem. 2008;15:2702–17. doi: 10.2174/092986708786242895. • A review article on novobiocin and analogs as well as other molecules targeting the CDD of Hsp90.
- 29.Uehara Y, Hori M, Takeuchi T, et al. Phenotypic change from transformed to normal induced by benzoquinonoid ansamycins accompanies inactivation of p60src in rat kidney cells infected with Rous sarcoma virus. Mol Cell Biol. 1986;6:2198–206. doi: 10.1128/mcb.6.6.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitesell L, Mimnaugh EG, De Costa B, et al. Inhibition of heat shock protein Hsp90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8. doi: 10.1073/pnas.91.18.8324. •• This paper describes v-src as an oncoprotein that requires Hsp90 for its malignant function.
- 31.Taldone T, Sun W, Chiosis G. Discovery and development of heat shock protein 90 inhibitors. Bioorg Med Chem. 2009;17:2225–35. doi: 10.1016/j.bmc.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollingshead M, Alley M, Burger AM, et al. In vivo antitumor efficacy of 17-DMAG (17-dimethylaminoethylamino-17-demethoxygeldanamycin hydrochloride), a water-soluble geldanamycin derivative. Cancer Chemother Pharmacol. 2005;56:115–25. doi: 10.1007/s00280-004-0939-2. [DOI] [PubMed] [Google Scholar]
- 33.Gao Z, Garcia-Echeverria C, Jensen MR. Hsp90 inhibitors: clinical development and future opportunities in oncology therapy. Curr Opin Drug Discovery Dev. 2010;13:193–202. [PubMed] [Google Scholar]
- 34.Roe SM, Prodromou C, O’Brien R, et al. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42:260–6. doi: 10.1021/jm980403y. • This paper provides crystal structures of GM and RD bound to the N-terminal domain of yeast Hsp90.
- 35.Stebbins CE, Russo AA, Schneider C, et al. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–50. doi: 10.1016/s0092-8674(00)80203-2. • This paper provides the first crystal structure of GM bound to the N-terminal domain of human Hsp90.
- 36.Neckers L. Chaperoning oncogenes: Hsp90 as a target of geldanamycin. Handb Exp Pharmacol. 2006;172:259–77. doi: 10.1007/3-540-29717-0_11. [DOI] [PubMed] [Google Scholar]
- 37.Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271:22796–801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 38.Chiosis G, Timaul MN, Lucas B, et al. A small molecule designed to bind to the adenine nucleotide pocket of Hsp90 causes Her2 degradation and the growth arrest and differentiation of breast cancer cells. Chem Biol. 2001;8:289–99. doi: 10.1016/s1074-5521(01)00015-1. • Describes the design and validation of the first reported synthetic Hsp90 inhibitor, PU3.
- 39.Taldone T, Chiosis G. Purine-scaffold Hsp90 inhibitors. Curr Top Med Chem. 2009;9:1436–46. doi: 10.2174/156802609789895737. • A review article which describes the discovery and development of purine-scaffold Hsp90 inhibitors.
- 40.Rodina A, Vilenchik M, Moulick K, et al. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat Chem Biol. 2007;3:498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 41.Breinig M, Caldas-Lopes E, Goeppert B, et al. Targeting heat shock protein 90 with non-quinone inhibitors: a novel chemotherapeutic approach in human hepatocellular carcinoma. Hepatology. 2009;50:102–12. doi: 10.1002/hep.22912. [DOI] [PubMed] [Google Scholar]
- 42.Caldas-Lopes E, Cerchietti L, Ahn JH, et al. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci USA. 2009;106:8368–73. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerchietti LC, Lopes EC, Yang SN, et al. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat Med. 2009;15:1369–76. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marubayashi S, Koppikar P, Taldone T, et al. HSP90 is a therapeutic target in JAK2-dependent myeloproliferative neoplasms in mice and humans. J Clin Invest. 2010;120:3578–93. doi: 10.1172/JCI42442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasibhatla SR, Hong K, Biamonte MA, et al. Rationally designed high-affinity 2-amino-6-halopurine heat shock protein 90 inhibitors that exhibit potent antitumor activity. J Med Chem. 2007;50:2767–78. doi: 10.1021/jm050752+. [DOI] [PubMed] [Google Scholar]
- 46.Bao R, Lai C-J, Qu H, et al. CUDC-305, a novel synthetic HSP90 inhibitor with unique pharmacologic properties for cancer therapy. Clin Cancer Res. 2009;15:4046–57. doi: 10.1158/1078-0432.CCR-09-0152. [DOI] [PubMed] [Google Scholar]
- 47.Rowlands MG, Newbatt YM, Prodromou C, et al. High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal Biochem. 2004;327:176. doi: 10.1016/j.ab.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 48.Drysdale MJ, Brough PA. Medicinal chemistry of Hsp90 inhibitors. Curr Top Med Chem. 2008;8:859–68. doi: 10.2174/156802608784911644. [DOI] [PubMed] [Google Scholar]
- 49.Nakashima T, Ishii T, Tagaya H, et al. New molecular and biological mechanism of antitumor activities of KW-2478, a novel nonansamycin heat shock protein 90 inhibitor, in multiple myeloma cells. Clin Cancer Res. 2010;16:2792–802. doi: 10.1158/1078-0432.CCR-09-3112. [DOI] [PubMed] [Google Scholar]
- 50.Zhou V, Han S, Brinker A, et al. A time-resolved fluorescence resonance energy transfer-based HTS assay and a surface plasmon resonance-based binding assay for heat shock protein 90 inhibitors. Anal Biochem. 2004;331:349–57. doi: 10.1016/j.ab.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Kung P-P, Funk L, Meng J, et al. Dihydroxylphenyl amides as inhibitors of the Hsp90 molecular chaperone. Bioorg Med Chem Lett. 2008;18:6273–8. doi: 10.1016/j.bmcl.2008.09.081. [DOI] [PubMed] [Google Scholar]
- 52.Kung P-P, Huang B, Zhang G, et al. Dihydroxyphenylisoindoline amides as orally bioavailable inhibitors of the heat shock protein 90 (Hsp90) molecular chaperone. J Med Chem. 2010;53:499–503. doi: 10.1021/jm901209q. [DOI] [PubMed] [Google Scholar]
- 53.Gopalsamy A, Shi M, Golas J, et al. Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90. J Med Chem. 2008;51:373–5. doi: 10.1021/jm701385c. [DOI] [PubMed] [Google Scholar]
- 54.Feldman RI, Mintzer B, Zhu D, et al. Potent triazolothione inhibitor of heat-shock protein-90. Chem Biol Drug Des. 2009;74:43–50. doi: 10.1111/j.1747-0285.2009.00833.x. [DOI] [PubMed] [Google Scholar]
- 55.Du Y, Moulick K, Rodina A, et al. High-throughput screening fluorescence polarization assay for tumor-specific Hsp90. J Biomol Screen. 2007;12:915–24. doi: 10.1177/1087057107306067. [DOI] [PubMed] [Google Scholar]
- 56.Ganesh T, Min J, Thepchatri P, et al. Discovery of aminoquinolines as a new class of potent inhibitors of heat shock protein 90 (Hsp90): synthesis, biology, and molecular modeling. Bioorg Med Chem. 2008;16:6903–10. doi: 10.1016/j.bmc.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fadden P, Huang KH, Veal JM, et al. Application of chemoproteomics to drug discovery: identification of a clinical candidate targeting Hsp90. Chem Biol. 2010;17:686–94. doi: 10.1016/j.chembiol.2010.04.015. • This paper describes an interesting approach in the discovery of SNX-5422, whereby both biological target and chemical species are simultaneously screened.
- 58.Huang KH, Veal JM, Fadden RP, et al. Discovery of novel 2-aminobenzamide inhibitors of heat shock protein 90 as potent, selective and orally active antitumor agents. J Med Chem. 2009;52:4288–305. doi: 10.1021/jm900230j. [DOI] [PubMed] [Google Scholar]
- 59.Stadler M, Anke H, Dekermendjian K, et al. Novel bioactive azaphilones from fruit bodies and mycelial cultures of the ascomycete bulgaria inquinans. Nat Prod Lett. 1995;7:7–14. [Google Scholar]
- 60.Musso L, Dallavalle S, Merlini L, et al. Natural and semisynthetic azaphilones as a new scaffold for Hsp90 inhibitors. Bioorg Med Chem. 2010;18:6031–43. doi: 10.1016/j.bmc.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 61.Barril X, Brough P, Drysdale M, et al. Structure-based discovery of a new class of Hsp90 inhibitors. Bioorg Med Chem Lett. 2005;15:5187–91. doi: 10.1016/j.bmcl.2005.08.092. [DOI] [PubMed] [Google Scholar]
- 62.Park H, Kim Y-J, Hahn J-S. A novel class of Hsp90 inhibitors isolated by structure-based virtual screening. Biorg Med Chem Lett. 2007;17:6345–9. doi: 10.1016/j.bmcl.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 63.Hong T-J, Park H, Kim Y-J, et al. Identification of new Hsp90 inhibitors by structure-based virtual screening. Bioorg Med Chem Lett. 2009;19:4839–42. doi: 10.1016/j.bmcl.2009.06.040. [DOI] [PubMed] [Google Scholar]
- 64.Murray CW, Carr MG, Callaghan O, et al. Fragment-based drug discovery applied to Hsp90. Discovery of two lead series with high ligand efficiency. J Med Chem. 2010;53:5942–55. doi: 10.1021/jm100059d. [DOI] [PubMed] [Google Scholar]
- 65.Woodhead AJ, Angove H, Carr MG, et al. Discovery of (2,4-dihydroxy-5-isopropylphenyl)-[5-(4-methylpiperazin-1-ylmethyl)-1,3-dihydroisoindol-2-yl] methanone (AT13387), a novel inhibitor of the molecular chaperone Hsp90 by fragment based drug design. J Med Chem. 2010;53:5956–69. doi: 10.1021/jm100060b. [DOI] [PubMed] [Google Scholar]
- 66.Curry J, Angove H, Graham B, et al. Significance of the long term pharmacodynamic actions of Hsp90 inhibitor AT13387. AACR abstract. 2009 [Google Scholar]
- 67.Brough PA, Barril X, Borgognoni J, et al. Combining hit identification strategies: Fragment-based and in silico approaches to orally active 2-aminothieno [2,3-d]pyrimidine inhibitors of the Hsp90 molecular chaperone. J Med Chem. 2009;52:4794–809. doi: 10.1021/jm900357y. [DOI] [PubMed] [Google Scholar]
- 68.Huth JR, Park C, Petros AM, et al. Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies. Chem Biol Drug Des. 2007;70:1–12. doi: 10.1111/j.1747-0285.2007.00535.x. [DOI] [PubMed] [Google Scholar]
- 69.Barker JJ, Barker O, Boggio R, et al. Fragment-based identification of Hsp90 inhibitors. ChemMedChem. 2009;4:963–6. doi: 10.1002/cmdc.200900011. • References [67,69] provide interesting approaches to the discovery of Hsp90 inhibitors using fragment-based approaches.
- 70.Oh SH, Woo JK, Yazici YD, et al. Structural basis for depletion of heat shock protein 90 client proteins by deguelin. J Natl Cancer Inst. 2007;99:949–61. doi: 10.1093/jnci/djm007. [DOI] [PubMed] [Google Scholar]
- 71.Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure. 2004;12:1087–97. doi: 10.1016/j.str.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 72.Brandt GEL, Blagg BSJ. Alternate strategies of Hsp90 modulation for the treatment of cancer and other diseases. Curr Top Med Chem. 2009;9:1447–61. doi: 10.2174/156802609789895683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenhagen MC, Soti C, Schmidt U, et al. The heat shock protein 90-targeting drug cisplatin selectively inhibits steroid receptor activation. Mol Endocrinol. 2003;17:1991–2001. doi: 10.1210/me.2003-0141. [DOI] [PubMed] [Google Scholar]
- 74.Palermo CM, Westlake CA, Gasiewicz TA. Epigallocatechin gallate inhibits aryl hydrocarbon receptor gene transcription through an indirect mechanism involving binding to a 90 KDa heat shock protein. Biochemistry. 2005;44:5041–52. doi: 10.1021/bi047433p. [DOI] [PubMed] [Google Scholar]
- 75.Yu Y, Hamza A, Zhang T, et al. Withaferin A targets heat shock protein 90 in pancreatic cancer cells. Biochem Pharmacol. 2010;79:542–51. doi: 10.1016/j.bcp.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Siligardi G, Panaretou B, Meyer P, et al. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J Biol Chem. 2002;277:20151–9. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 77.Smith JR, Clarke PA, de Billy E, et al. Silencing the cochaperone CDC37 destabilizes kinase clients and sensitizes cancer cells to HSP90 inhibitors. Oncogene. 2009;28:157–69. doi: 10.1038/onc.2008.380. • This paper provides a scientific basis for targeting Cdc37 as an anticancer strategy.
- 78.Hieronymus H, Lamb J, Ross KN, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of Hsp90 pathway modulators. Cancer Cell. 2006;10:321–30. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 79.Zhang T, Hamza A, Cao X, et al. A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Mol Cancer Ther. 2008;7:162–70. doi: 10.1158/1535-7163.MCT-07-0484. [DOI] [PubMed] [Google Scholar]
- 80.Sreeramulu S, Gande SL, Gobel M, et al. Molecular mechanism of inhibition of the human protein complex Hsp90–Cdc37, a kinome chaperone–cochaperone, by triterpene celastrol. Angew Chem Int Ed. 2009;48:5853–5. doi: 10.1002/anie.200900929. [DOI] [PubMed] [Google Scholar]
- 81.Yang H, Chen D, Cui QC, et al. Celastrol, a triterpene extracted from the Chinese “Thunder of God Vine,” is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66:4758–65. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 82.Scheufler C, Brinker A, Bourenkov G, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 83.Cortajarena AL, Yi F, Regan L. Designed TPR modules as novel anticancer agents. ACS Chem Biol. 2008;3:161–6. doi: 10.1021/cb700260z. [DOI] [PubMed] [Google Scholar]
- 84.Yi F, Zhu P, Southall N, et al. An AlphaScreen™-based high-throughput screen to identify inhibitors of Hsp90-cochaperone interaction. J Biomol Screen. 2009;14:273–81. doi: 10.1177/1087057108330114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yi F, Regan L. A novel class of small molecule inhibitors of Hsp90. ACS Chem Biol. 2008;3:645–54. doi: 10.1021/cb800162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Panaretou B, Siligardi G, Meyer P, et al. Activation of the ATPase activity of Hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–18. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 87.Meyer P, Prodromou C, Liao C, et al. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:1402–10. doi: 10.1038/sj.emboj.7600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holmes JL, Sharp SY, Hobbs S, et al. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008;68:1187–96. doi: 10.1158/0008-5472.CAN-07-3268. • This paper shows that targeting Aha1 can result in increased sensitivity to Hsp90 inhibitors.
- 89.Fortugno P, Beltrami E, Plescia J, et al. Regulation of survivin function by Hsp90. Proc Natl Acad Sci USA. 2003;100:13791–6. doi: 10.1073/pnas.2434345100. • This paper shows that targeting the Hsp90–survivin complex may provide an alternative anticancer strategy.
- 90.Meli M, Pennati M, Curto M, et al. Small-molecule targeting of heat shock protein 90 chaperone function: rational identification of a new anticancer lead. J Med Chem. 2006;49:7721–30. doi: 10.1021/jm060836y. [DOI] [PubMed] [Google Scholar]
- 91.Georget V, Terouanne B, Nicolas J-C, et al. Mechanism of antiandrogen action: Key role of Hsp90 in conformational change and transcriptional activity of the androgen receptor. Biochemistry. 2002;41:11824–31. doi: 10.1021/bi0259150. [DOI] [PubMed] [Google Scholar]
- 92.Liu S, Yuan Y, Okumura Y, et al. Camptothecin disrupts androgen receptor signaling and suppresses prostate cancer cell growth. Biochem Biophys Res Commun. 2010;394:297–302. doi: 10.1016/j.bbrc.2010.02.164. [DOI] [PubMed] [Google Scholar]
- 93.Kovacs JJ, Murphy PJ, Gaillard S, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–7. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 94.Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, et al. S-Nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci USA. 2005;102:8525–30. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Retzlaff M, Stahl M, Eberl HC, et al. Hsp90 is regulated by a switch point in the C-terminal domain. EMBO Rep. 2009;10:1147–53. doi: 10.1038/embor.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wandinger SK, Suhre MH, Wegele H, et al. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 2006;25:367–76. doi: 10.1038/sj.emboj.7600930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–34. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 98.Yang Y, Rao R, Shen J, et al. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68:4833–42. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scroggins BT, Robzyk K, Wang D, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–9. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rao R, Fiskus W, Yang Y, et al. HDAC6 inhibition enhances 17-AAG-mediated abrogation of Hsp90 chaperone function in human leukemia cells. Blood. 2008;112:1886–93. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 101.Nishioka C, Ikezoe T, Yang J, et al. MS-275, a novel histone deacetylase inhibitor with selectivity against HDAC1, induces degradation of FLT3 via inhibition of chaperone function of heat shock protein 90 in AML cells. Leuk Res. 2008;32:1382–92. doi: 10.1016/j.leukres.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 102.George P, Bali P, Annavarapu S, et al. Combination of the histone deacetylase inhibitor LBH589 and the Hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;105:1768–76. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- 103.Morra G, Verkhivker G, Colombo G. Modeling signal propagation mechanisms and ligand-based conformational dynamics of the Hsp90 molecular chaperone full-length dimer. PLoS Comput Biol. 2009;5:e1000323. doi: 10.1371/journal.pcbi.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao YG, Gilmore R, Leone G, et al. Hsp90 phosphorylation is linked to its chaperoning function. Assembly of the reovirus cell attachment protein. J Biol Chem. 2001;276:32822–7. doi: 10.1074/jbc.M105562200. [DOI] [PubMed] [Google Scholar]
- 105.Mimnaugh EG, Worland PJ, Whitesell L, et al. Possible role for serine/threonine phosphorylation in the regulation of the heteroprotein complex between the Hsp90 stress protein and the pp60v-src tyrosine kinase. J Biol Chem. 1995;270:28654–9. doi: 10.1074/jbc.270.48.28654. [DOI] [PubMed] [Google Scholar]
- 106.Miyata Y. CK2: the kinase controlling the Hsp90 chaperone machinery. Cell Mol Life Sci. 2009;66:1840–9. doi: 10.1007/s00018-009-9152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mollapour M, Tsutsumi S, Donnelly AC, et al. Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol Cell. 2010;37:333–43. doi: 10.1016/j.molcel.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fujiwara H, Yamakuni T, Ueno M, et al. IC101 induces apoptosis by Akt dephosphorylation via an inhibition of heat shock protein 90-ATP binding activity accompanied by preventing the interaction with Akt in L1210 cells. J Pharmacol Exp Ther. 2004;310:1288–95. doi: 10.1124/jpet.104.065979. [DOI] [PubMed] [Google Scholar]
- 109.Koga F, Xu W, Karpova TS, et al. Hsp90 inhibition transiently activates Src kinase and promotes Src-dependent Akt and Erk activation. Proc Natl Acad Sci USA. 2006;103:11318–22. doi: 10.1073/pnas.0604705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Donze O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–80. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yano A, Tsutsumi S, Soga S, et al. Inhibition of Hsp90 activates osteoclast c-Src signaling and promotes growth of prostate carcinoma cells in bone. Proc Natl Acad Sci USA. 2008;105:15541–6. doi: 10.1073/pnas.0805354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Price JT, Quinn JM, Sims NA, et al. The heat shock protein 90 inhibitor, 17-allylamino-17-demethoxygeldanamycin, enhances osteoclast formation and potentiates bone metastasis of a human breast cancer cell line. Cancer Res. 2005;65:4929–38. doi: 10.1158/0008-5472.CAN-04-4458. [DOI] [PubMed] [Google Scholar]
- 113.McCollum AK, TenEyck CJ, Stensgard B, et al. P-Glycoprotein–mediated resistance to Hsp90-directed therapy is eclipsed by the heat shock response. Cancer Res. 2008;68:7419–27. doi: 10.1158/0008-5472.CAN-07-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eiseman JL, Lan J, Lagattuta TF, et al. Pharmacokinetics and pharmacodynamics of 17-demethoxy-17-[[(2-dimethylamino) ethyl]amino]geldanamycin (17DMAG, NSC 707545) in C.B-17 SCID mice bearing MDA-MB-231 human breast cancer xenografts. Cancer Chemother Pharmacol. 2005;55:21–32. doi: 10.1007/s00280-004-0865-3. [DOI] [PubMed] [Google Scholar]
- 115.Leow CC, Chesebrough J, Coffman KT, et al. Antitumor efficacy of IPI-504, a selective heat shock protein 90 inhibitor against human epidermal growth factor receptor 2-positive human xenograft models as a single agent and in combination with trastuzumab or lapatinib. Mol Cancer Ther. 2009;8:2131–41. doi: 10.1158/1535-7163.MCT-08-1038. [DOI] [PubMed] [Google Scholar]
- 116.Holland JP, Caldas-Lopes E, Divilov V, et al. Measuring the pharmacodynamic effects of a novel Hsp90 inhibitor on HER2/neu expression in mice using Zr-DFO-trastuzumab. PLoS One. 2010;5:e8859. doi: 10.1371/journal.pone.0008859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lundgren K, Zhang H, Brekken J, et al. BIIB021, an orally available, fully synthetic small-molecule inhibitor of the heat shock protein Hsp90. Mol Cancer Ther. 2009;8:921–9. doi: 10.1158/1535-7163.MCT-08-0758. [DOI] [PubMed] [Google Scholar]
- 118.Jensen MR, Schoepfer J, Radimerski T, et al. NVP-AUY922: a small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models. Breast Cancer Res. 2008;10:R33. doi: 10.1186/bcr1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chandarlapaty S, Sawai A, Ye Q, et al. SNX2112, a synthetic heat shock protein 90 inhibitor, has potent antitumor activity against HER kinase-dependent cancers. Clin Cancer Res. 2008;14:240–8. doi: 10.1158/1078-0432.CCR-07-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]