Abstract
Traditionally, state-space models are fitted to data where there is uncertainty in the observation or measurement of the system. State-space models are partitioned into an underlying system process describing the transitions of the true states of the system over time and the observation process linking the observations of the system to the true states. Open population capture–recapture–recovery data can be modelled in this framework by regarding the system process as the state of each individual observed within the study in terms of being alive or dead, and the observation process the recapture and/or recovery process. The traditional observation error of a state-space model is incorporated via the recapture/recovery probabilities being less than unity. The models can be fitted using a Bayesian data augmentation approach and in standard BUGS packages. Applying this state-space framework to such data permits additional complexities including individual heterogeneity to be fitted to the data at very little additional programming effort. We consider the efficiency of the state-space model fitting approach by considering a random effects model for capture–recapture data relating to dippers and compare different Bayesian model-fitting algorithms within WinBUGS.
Keywords: Bayesian approach, BUGS, data augmentation, hierarchical model, individual heterogeneity, mixed effects models
1. Introduction
Survival parameter estimation is of particular interest within ecological systems for gaining insight into the underlying biological system and for conservation or management polices [1]. To obtain estimates of survival probabilities, capture–recapture data are often collected. The data collection process involves the repeated (re-)capturing of individuals at a series of capture events, where each individual within the study is uniquely identifiable, either by natural markings or a unique mark applied to the individual at their initial capture, such as a tag or ring. It is then possible to specify a statistical model for the data, including an explicit likelihood function for the data given the model parameters and hence obtain estimates of the survival probabilities and recapture probabilities [2–4]. If individual animals can also be recovered dead within the study period, this is referred to as capture–recapture–recovery data, and the approach extended to obtain estimates for the survival, recapture (of live animals) and recovery (of dead animals) probabilities [5–8]. Survival parameters are often subject to individual heterogeneity and recent interest has included such additional complexities within the statistical framework. For example, specifying the survival parameters as a function of individual covariates [9–16] and/or incorporating random effects [17–20]. Including individual heterogeneity typically increases the complexity of the model specification and fitting process.
State-space (or hierarchical) models have become a very useful modelling tool in the analysis of complex ecological data. For example, state-space models have been used in fisheries [21–23], animal count data [24–27] and telemetry data [28–31]. Traditionally, state-space models are fitted to data where the data obtained are observed with error. We will show how capture–recapture–recovery data can be modelled using a state-space framework, where the observation error component is incorporated as a result of a non-perfect recapture and/or recovery process, i.e. the recapture and recovery probabilities are less than unity. For capture–recapture–recovery models, considering a state-space formulation can simplify the model specification by constructing the model as the composition of a series of simpler models corresponding to each separate process acting on the population and permit additional complexity to be incorporated in a straightforward manner. We initially describe the underlying ideas of the general state-space framework and how these can be fitted within a Bayesian framework before applying this to capture–recapture–recovery data.
1.1. State-space models
State-space models separate an observed process into two components: a system process that models the underlying biological process over time and an observation process that accounts for imperfect detection of the system process, such as measurement error. State-space models have become increasingly popular within ecological problems, often separating the nuisance parameters (observational error) from the biological processes of interest.
In general, the state-space framework takes the following form. Let y1, … , yT denote the (possibly multivariate) observations of the process; x1, … , xT the true underlying states at times t = 1, … ,T and θ the set of model parameters. The state-space model can be described by,
 |
When the underlying states of the system process, x1, … , xT, are discrete-valued the state-space model reduces to a traditional hidden Markov model (although we note that some authors have extended the term hidden Markov model to include continuous state variables, see Cappé et al. [32]). For such models with finitely many states, there is an explicit closed form likelihood function for the observed data, denoted by f(y|θ) (by summing over all possible states in the system process) which can be maximized to obtain parameter estimates. Alternatively, the EM algorithm [33] can be implemented to obtain parameter estimates (see also van Deusen [34] for an application of the EM algorithm to capture–recapture data). For a continuous system process, in general there is no such closed form expression for the likelihood function of the observed data (although it can be expressed in the form of an integral over the x values) with the Kalman filter [35] traditionally used to obtain parameter estimates, assuming a linear Gaussian state-space model. More recently, Bayesian model-fitting approaches have been used that permit a greater degree of flexibility including, for example, nonlinear and non-Gaussian processes. We focus on the Bayesian approach and briefly describe a data augmentation approach for fitting state-space models. See, Newman et al. [36] for further discussion.
1.2. Bayesian model fitting
In order to fit a state-space model, we use a Bayesian data augmentation approach [37], also referred to as an auxiliary or latent variable approach, and, more recently, a complete data likelihood approach [38,39]. We treat the underlying states x as parameters (or auxiliary variables) to be estimated in addition to the model parameters, θ. The joint posterior distribution of the model parameters and auxiliary variables can be written in the form,
where
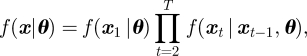 |
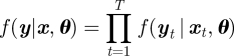 |
and p(θ) denotes the prior specified on the model parameters. In general, the posterior distribution of the model parameters is given by
However, the necessary integration to obtain the posterior distribution is analytically intractable. We will use Markov chain Monte Carlo (MCMC) to obtain a sample from the joint posterior distribution of x and θ. Considering only the sampled values of θ (and ignoring the x values) indirectly performs the necessary integration to obtain a set of sampled values from the marginal distribution π(θ|y) which can be used to obtain summary statistics of interest. We note that the posterior (marginal) distribution of x is often of interest itself, relating to the posterior distribution of the true underlying states of the system under study.
For some studies, some of the underlying states x may be observed without error. In such cases, we let x = {xobs, xmis}, where xobs and xmis denote the observed and unobserved states, respectively. xobs is observed data and so regarded in the same manner as the observations y, while xmis are auxiliary variables and we can write
| 1.1 |
At each iteration of the MCMC algorithm, the auxiliary variables xmis (in addition to the model parameters θ) are updated.
The introduction of auxiliary variables is often used in two particular circumstances: (i) when the likelihood function of the observed data, f(y|θ), is analytically intractable or computationally expensive; and/or (ii) when the inclusion of the additional auxiliary variables simplifies or improves the MCMC algorithm. Thus, we note that an auxiliary variable approach may be used even if an explicit likelihood is available. For example, for hidden Markov models (i.e. discrete system processes), there is an explicit likelihood. However, using an auxiliary variable approach (where xmis corresponds to the discrete underlying states) leads to a complete data likelihood, f(y|x,θ), that can be significantly computationally faster to calculate than the observed data likelihood, f(y|θ). The auxiliary variable approach may also lead to posterior conditional distributions of standard form for the model parameters (and auxiliary variables) and use of the Gibbs sampler compared with the Metropolis–Hastings algorithm when using the observed data likelihood due to non-standard posterior conditional distributions. See for example [27,36,39–42] for further discussion of Bayesian model fitting in the context of a range of ecological state-space models.
Finally, we comment on the issue of parameter redundancy, or local identifiability, within the Bayesian framework. A parameter redundant model has a ridge in the corresponding likelihood surface leading to non-unique maximum-likelihood estimates. Traditionally, to remove this problem, restrictions are specified on the parameter space to allow estimation of all identifiable parameters. In general, parameter redundancy can be difficult to identify, particularly for models with a large number of parameters and/or processes acting, although some advances have been made using symbolic algebraic approaches [43–47]. However, within the Bayesian framework, the likelihood is combined with the prior distribution to form the posterior distribution of the parameters. Thus, parameter redundancy still permits the estimation of the posterior distribution (assuming a proper prior distribution), but due to the identifiability issues, parameters (or functions of parameters) suffer from strong prior sensitivity. For example, suppose that a given parameter is unidentifiable (so that the likelihood is flat with respect to this parameter), then the posterior distribution for this parameter is simply the corresponding prior for the parameter. The idea extends to when functions of parameters are unidentifiable, and the individual parameters within this function will typically exhibit strong prior dependence. Gimenez et al. [48] used this idea to assess potential identifiability issues, by considering the overlap between the posterior and prior distributions. A large overlap may provide an indication that the parameters are not being well estimated and potential parameter identifiability issues. See also King et al. [41] and Kéry & Schaub [42] for further discussion and applications.
1.2.1. Model discrimination
The Bayesian approach can be extended to incorporate model uncertainty. For example, suppose that there are a total of M possible models. We introduce a discrete model indicator parameter, denoted by m ∈ {1, … , M} with corresponding parameters θm (since the set of parameters is model dependent). The joint posterior distribution over the combined parameter and model space is given by
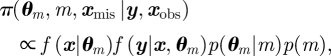 |
where p(m) denotes the prior specified on model m. Marginal posterior model probabilities are often of particular interest, providing a quantitative comparison of competing models, with the posterior probability for model m given by
Bayes factors, defined to be the ratio of posterior model probabilities to prior model probabilities, are also frequently used to compare competing models [49]. Posterior model probabilities (or Bayes factors) are a quantitative comparison of competing models and hence naturally permit posterior model-averaged estimates of parameters of interest. For example, suppose that the parameter θ1 is common to all models. The posterior model averaged distribution for θ1 is given by
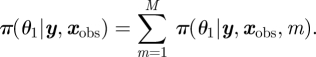 |
MCMC methods can again be used to obtain a sample from the posterior distribution of interest to obtain Monte Carlo estimates of interest. For example the posterior model probabilities are estimated as the proportion of time the chain is in each model. However, an extension to the standard MCMC algorithm is necessary in order for the constructed Markov chain to move between models with a different number of parameters. The most common such algorithm is the reversible jump MCMC algorithm [50] but also see, Godsill [51] for additional discussion. See King et al. [41] and references therein for further discussion of ecological applications.
1.2.2. Model checking
Model checking (or goodness of fit) is concerned with the absolute fit of a model to the data. Many different models may be fitted to the data, and a model discrimination procedure applied, but none of the models may fit the data well. Within the Bayesian approach, model checking is typically assessed by effectively simulating data from the given model and comparing whether the simulated data appear consistent with the observed data. The most common approach is that of Bayesian p-values [52], a posterior predictive checking approach. The approach takes the form of repeatedly sampling parameter values from the posterior distribution. For each set of sampled values, a dataset is simulated from the model and compared with the observed data using some discrepancy function, g (for example, deviance, Pearson χ2 statistic or Freeman–Tukey statistic). The Bayesian p-value is simply the proportion of times that the discrepancy function of the simulated data is less than that of the observed data. If the data are inconsistent with the model, we would expect the Bayesian p-value to be close to 0 or 1. For examples on the use of Bayesian p-values in the context of capture–recapture–recovery data see earlier studies [12,17,39,41,42,53–55]. Bayesian p-values have also been applied within a model-averaging context [8,12]. Previous applications of Bayesian p-values to capture–recapture–recovery data include discrepancy functions of the observed data xobs given the model parameters θ (i.e. g(xobs|θ)) [53] and discrepancy functions of the observed data xobs given the model parameters and auxiliary variables xmis (i.e. g(xobs|θ, xmis)) [54,55].
2. Capture–recapture–recovery data
Capture–recapture studies involve a series of capture events. At the first capture event, all individuals observed in the study population are uniquely marked and released back into the population. For example, a tag/ring is applied to each individual or more recently photo-identification used when it is possible to use natural physical features, such as individual markings (often used for seals, newts or tigers) and dorsal fin features (of dolphins). At each subsequent capture event, all previously observed individuals are recorded, new individuals are uniquely marked and all released back into the population. A recapture (or initial captures when using photo-identification techniques) of an individual does not necessarily mean a physical capture but often relates to a sighting of the individual; however, for simplicity, we use the term recapture. Typically, for such data a number of assumptions are made, including no mark-loss, migration from the system is permanent (so that apparent survival probabilities are typically estimated), individuals behave independently from each other and are representative of the population. For a review of such data and associated models, see Schwarz & Seber [56]. In some studies, in addition to live resighting, dead recoveries of individuals may also occur, leading to capture–recapture–recovery data.
Capture–recapture–recovery data are typically displayed in the form of the encounter histories of each individual observed within the study. We let n denote the number of individuals observed within the study and T the number of capture events. We let w denote the n × T encounter matrix, where
 |
for i = 1, … , n and t = 1, … , T. The ith row of the matrix w, denoted by wi corresponds to the encounter history of individual i. For example, consider the following encounter histories:
Individual 1 is first observed at time 1, unobserved at times 2, 6 and 7, recaptured at times 3, 4 and 5 and recovered dead at time 8. The second individual is first observed at time 1, recaptured at time 2 but then unseen throughout the remainder of the study—either it survives until the end of the study and remains unobserved, or is unobserved until it dies at some time within the study and is not recovered.
For such data, the parameters of interest are typically demographic parameters (most notably survival probabilities) and/or abundance. The traditional Cormack–Jolly–Seber (CJS) model [2,4] considers only capture–recapture data and the ring-recovery (or tag-recovery) model [57] considers only capture–recovery data. Each of these models, condition on the initial capture of each individual observed within the study with the corresponding likelihood a function of survival and recapture or recovery probabilities (namely the product over each individual of the probability of their corresponding encounter history conditional on their initial capture). These ideas can be extended to integrated capture–recapture–recovery data with an associated likelihood expression [5–8]. Consequently, for CJS-type models (due to the conditioning on the initial capture of individuals), abundance cannot be estimated. The Jolly–Seber (JS) model [3,4] removes this condition on the initial capture of individuals. For both the CJS and JS models, the observed data likelihood are available [58–60] permitting estimation of abundance and demographic parameters.
We initially consider the state-space formulation for the CJS model before extending the approach to allow for additional system processes, including births (leading to JS-type models and abundance estimation) and individual heterogeneity (in the form of discrete and continuous individual covariates and/or individual random effects).
2.1. State-space model formulation
The state-space model formulation of the CJS model has been proposed independently by Royle [18], Gimenez et al. [61] and Schofield & Barker [62] (although this modelling approach dates back to (at least) Dupuis [63]—see §2.3.1). The state-space approach separates the survival process from the observation processes of individuals observed within the study (either alive or dead). We note that (as pointed out by an anonymous referee) this underlying concept can be seen within the original seminal papers, although not formally expressed in a (conditional) state-space representation. For example, both Jolly [3] and Seber [4] describe (in words) the two separate processes acting on each individual member of the population of surviving from one capture event to the next and subsequently being captured (or not).
Formally, within the state-space formulation, capture–recapture–recovery data can be viewed as the combination of two different processes: (i) individuals surviving between successive time points and (ii) individuals being observed (either alive or dead) with some probability. Thus, we can regard each encounter history as a combination of the processes of survival between each capture event and observations of whether they are recaptured (if alive) or recovered (if dead), and thus dissect w into these distinct (conditional) components. Mathematically, to represent the survival process, we define x to be the n × T matrix such that
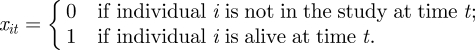 |
We note that xit = 0 corresponds to the cases where an individual is either dead or has not yet entered the study (i.e. before it is first observed). We let f(i) denote the first time individual i is observed in the study and ϕit the probability individual i survives until time t + 1, given they are alive at time t, with ϕ = {ϕit, i = 1, … , n; t = 1, … , T − 1}. Submodels are obtained by setting restrictions on these parameters. For example, a constant survival model is obtained by setting ϕit = ϕ for all i = 1, …, n and t = 1, … , T − 1. However, we retain the more general notation as we will consider a range of models allowing for temporal and/or individual heterogeneity. The underlying system process can be easily described as follows:
| 2.1 |
for t = f(i) + 1, … , T and xif(i) = 1 (since the CJS model conditions on the first time an individual is observed alive). For many encounter histories, the xit values will not always be known. For example, consider encounter history w2 given above, we know that x21 = x22 = 1 but as the individual is unobserved thereafter they may either be alive and not observed or have died and not be recovered. Recall that we let xobs and xmis denote the set of values of xit that are known and unknown, respectively.
For capture–recapture–recovery data, we partition the observation process into two distinct processes corresponding to the (re-)capture (of live animals) and recovery (of dead animals). We initially consider the recapture process and let y denote the n × T matrix such that
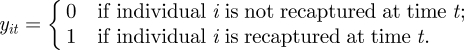 |
We note that for an individual to be recaptured, it is conditional on the individual being alive in the study at that time (i.e. if xit = 0 then yit = 0). Let pit denote the probability individual i is recaptured at time t, given they are alive in the study area at this time, for i = 1, … , n and t = 2, … , T. The observation process can be written as
| 2.2 |
for t = f(i) + 1, … , T and yif(i) = 1.
The recovery process is defined similarly. For simplicity, we assume that the recapture and recovery pro-cesses are observed over the same period of time, though this can be easily relaxed. We let z denote the n × T matrix with elements
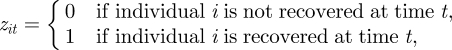 |
for i = 1, … , n and t = 2, … , T. Once more the values of this observation process, z, are conditional on the system process, x. For an individual to be recovered dead at time t, it is usually assumed that the individual must have died in the interval (t − 1,t] (although see below for further discussion of relaxing this assumption). Let λit denote the probability individual i is recovered at time t, given that they die in the interval (t − 1,t], for i = 1, … , n and t = 2, … , T, and set λ = {λit: i = 1, … , n, t = 2, … , T}. The observation process can be written as
| 2.3 |
for i = 1, … , n and t = f(i) + 1, … , T. The product (1 − xit)xit−1 ensures that an individual can only be recovered at time t if they are alive at time t − 1 (i.e. xit−1 = 1) but dead at time t (i.e. xit = 0); in other words, die in the interval (t − 1,t].
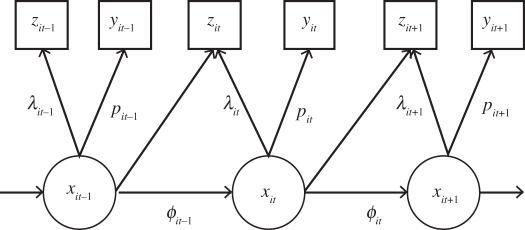
The observed encounter histories w can be separated into the data components xobs, y and z. Note that a glossary of these latent variables, or auxiliary variables (and others introduced later in the manuscript), are listed in table 4 in appendix A. In addition, we note that this state-space formulation is simply a (partial) hidden Markov model, with the system process composed of the two discrete states ‘alive’ and ‘dead’, with some of the states known. The corresponding underlying state-space (or hidden Markov model) representation is provided graphically in figure 1.
Figure 1.
State-space representation of the system process, x, recapture observation process, y, and recovery observation process, z, for capture–recapture–recovery data. Square notes represent observed data, circular nodes unknown states and arrows a stochastic relationship between nodes.
We fit the models using a Bayesian data augmentation approach to form the joint posterior distribution (cf. equation (1.1)),
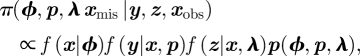 |
where f(x|ϕ), f(y|x,p) and f(z|x,λ) denote the (Bernoulli) probability mass functions associated with the system and observation processes, respectively (such that their product is the complete data likelihood) and p(ϕ, p, λ) the joint prior on the survival, recapture and recovery probabilities.
Using a state-space framework, when considering deviations from the above model and/or additional complexities, it is not necessary to explicitly derive a new observed likelihood expression, but simply consider each individual process in turn. For example, the assumption of recovery only in the capture event following the death of an individual is often reasonable, due to the decay of marks used to uniquely identify individuals. However, in some cases, this assumption is not valid, as for abalone (see Catchpole et al. [64], who derive an explicit observed data likelihood for the model where individuals can be recovered at any point following death). The model proposed by Catchpole et al. [64] can be easily fitted within the state-space framework changing only the observation process for recoveries such that
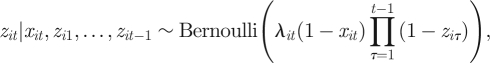 |
for i = 1, … , n and t = f(i) + 1, … , T where 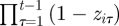 is included since an individual is removed from the study once it is recovered dead and the interpretation of λit now changes to the probability an individual is recovered given they are dead, with no restriction on the time of their death. More generally, models that explicitly allow for a decay of a mark over time can be fitted within the framework, for example, by specifying the recovery probability to be a decreasing function of time since death.
is included since an individual is removed from the study once it is recovered dead and the interpretation of λit now changes to the probability an individual is recovered given they are dead, with no restriction on the time of their death. More generally, models that explicitly allow for a decay of a mark over time can be fitted within the framework, for example, by specifying the recovery probability to be a decreasing function of time since death.
2.2. Estimating abundance
Removing the conditioning of initial capture from the CJS model permits the estimation of birth rates and abundance via the JS model [3,4]. An observed data likelihood can be obtained [58–60] by considering a superpopulation corresponding to all possible individuals who may be observed within the study (i.e. individuals present and available for capture for at least one capture event) and summing over all possible combinations of their entry and exit times (i.e. ‘birth’ and ‘death’ times). Entry to the system could be via birth or immigration and exit from the system, death or permanent emigration. However, for simplicity, we use the terms ‘birth’ and ‘death’ to include such migration. Thus, for the JS model, there is an additional entry process to be included within the system process. We let the number of individuals in the superpopulation be denoted by N, which is a parameter to be estimated. We note that (N − n) individuals are available for capture at least once within the study but are not observed and that x is of dimension N × T, and hence a function of N. We let bit denote the probability that individual i enters the study population in the interval (t − 1,t] and so is available for capture for the first time at capture event t, for t = 1, … , T. This bi1 is interpreted as the probability individual i is in the population and available for capture at the first capture event. Since we condition on all N individuals being available for capture at least once within the study period, we have that ∑t=1Tbit = 1 for all i = 1, … , N.
Within the state-space model, the survival process is replaced by a ‘presence’ process allowing for individuals to both join and leave the study (assuming individuals can only join and leave the study a maximum of one time), corresponding to ‘birth’ and ‘death’. In particular, we have that for t = 1, … , T and i = 1, … , N,
where
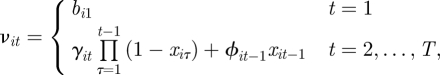 |
2.4 |
such that
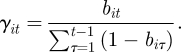 |
The term 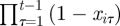 ensures that an individual can only be ‘born’ once within the study and cannot be ‘reborn’ (i.e. re-enter the study once it has died). The parameter γit can be interpreted as the probability individual i enters the population within the interval (t − 1,t], conditional on not entering the study population before or at time t − 1. The recapture and recovery processes essentially remain the same as for the CJS model, given in equations (2.2) and (2.3), but are now specified for t = 1, … , T, since the JS model removes the conditioning on first capture. The posterior distribution for the JS model, assuming no recoveries, is given by
ensures that an individual can only be ‘born’ once within the study and cannot be ‘reborn’ (i.e. re-enter the study once it has died). The parameter γit can be interpreted as the probability individual i enters the population within the interval (t − 1,t], conditional on not entering the study population before or at time t − 1. The recapture and recovery processes essentially remain the same as for the CJS model, given in equations (2.2) and (2.3), but are now specified for t = 1, … , T, since the JS model removes the conditioning on first capture. The posterior distribution for the JS model, assuming no recoveries, is given by
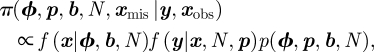 |
where f(x|ϕ, b, N) and f(y|x, N, p) denote the Bernoulli probability mass functions for the system presence process and observation process, respectively. For further discussion, see Schofield & Barker [62].
We note that when specifying the model within the set of BUGS (and associated) computer packages, it is not possible to specify x as a function of a stochastic node (i.e. N). The solution proposed is to specify a fixed upper bound for N, denoted by N*, and let x be of fixed dimension (N* × T) allowing for (N* − N) individuals not available for capture within the study period, or, in other words, do not enter into the study population. See earlier studies [40,42,65–68] for further discussion of this approach, alternative parametrizations and associated computer codes. Finally, we note the link of the JS model to two other common models of occupancy models and closed population models.
Occupancy models relate to the occupancy of a species (rather than individuals) within a given area. Species can colonize an area (enter the population), die out from the area (exit the population), as for capture–recapture data, but also recolonize an area (i.e. be ‘reborn’). The state-space formulation of the JS model can be easily extended to an occupancy model, by adding the term 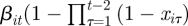 ) (1 − Xit − 1) to νit in (2.4) for capture times t = 3, … , T to allow re-entry into the population (see Royle and Dorazio [40] for further discussion and references therein in relation to occupancy models). Finally, we note that removing the presence process (assuming that all individuals are present for the full duration of the study, so that xit = 1 for all i = 1, … , N and t = 1, … , T) and considering only a recapture process, leads to the special case of a closed population where the estimation of the total population, N, is of particular interest. See earlier research [39–42,67–69] for a range of different closed population models.
) (1 − Xit − 1) to νit in (2.4) for capture times t = 3, … , T to allow re-entry into the population (see Royle and Dorazio [40] for further discussion and references therein in relation to occupancy models). Finally, we note that removing the presence process (assuming that all individuals are present for the full duration of the study, so that xit = 1 for all i = 1, … , N and t = 1, … , T) and considering only a recapture process, leads to the special case of a closed population where the estimation of the total population, N, is of particular interest. See earlier research [39–42,67–69] for a range of different closed population models.
2.3. Individual heterogeneity
In many biological systems, the model parameters may be individual specific, in that they differ between individuals despite identical environmental conditions. For example, individuals typically differ in body condition (which itself may be a function of feeding or breeding behaviour, parasite load, etc.) which in turn has a direct impact on their corresponding survival (animals in better condition are more able to compete for resources and/or survive harsh environmental conditions). Similarly, animals may disperse through an area, with individuals more/less likely to be recaptured/recovered in more remote or dense vegetation areas. To incorporate such heterogeneity individual level factors (or covariates) and/or random effects are often used to model such differences.
For simplicity, we consider capture–recapture data and the CJS model, extensions allowing for further system and/or observation processes follow by including additional separate processes. We consider the model parameters as a function of individual level time-varying covariates. The matrix of observed encounter histories, w, has elements given by
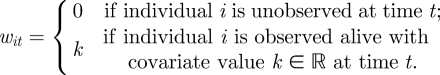 |
The survival and recapture processes for x and y given in equations (2.1) and (2.2) have the same definitions as before but where we now have the survival and recapture probabilities expressed as a function of the covariate values. However, we need an additional system process corresponding to the transition process for the underlying covariates. For the transition process, we let u be an n × T matrix, with elements
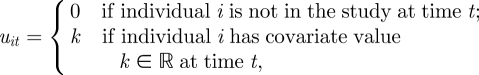 |
for i = 1, … , n and t = 1, … , T. Typically, there are unknown observations within the transition process. For example, for time-varying covariates, if an individual is not observed, generally the corresponding covariate values are also unobserved. We let u = {uobs,umis}, where uobs and umis denote the observed and unobserved states within the transition process, and let ψ denote the parameters relating to the transition process. The joint posterior distribution can be expressed in the form
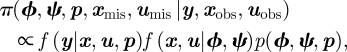 |
2.5 |
where f(x, u|ϕ,ψ) denotes the joint probability function of the survival and transition processes. Within the MCMC algorithm, we update the model parameters and missing states xmis and umis at each iteration of the Markov chain. An estimate of the posterior distribution of the model parameters can be obtained by considering the marginal distribution of these parameters within the algorithm. We consider particular cases for different types of covariate information.
2.3.1. Multi-state data
Discrete-valued time-varying covariates, such as breeding state or discrete location, are most commonly referred to as multi-state (or multi-site) data. These models are an example of a (partially) hidden Markov model, as the system processes have a finite number of states. Notationally, we let the set of possible states for the covariates be denoted by 𝒦 = {1, … , K} (this does not include the state of death). The traditional Arnason–Schwarz (AS) model extends the CJS model by considering a first-order Markov transition process between the different states, with an observed data likelihood available [9,10,70,71], allowing parameter estimation. Dupuis [63] proposed an alternative Bayesian state-space modelling approach, although did not phrase the approach as a state-space model but using an auxiliary variable approach (see also Kéry & Schaub [42, ch. 9]; Marin & Robert [72, ch. 5.5]; Clark et al. [73] and references therein and King & Brooks [54] for an extension to allow for model uncertainty in terms of time and covariate dependence on the model parameters).
We let the transition parameters ψit(j,k) denote the probability that individual i is in state k at time t + 1, given they are alive and in state j at time t for t = f(i), … , T − 1 and set ψit = {ψit(j,k): j,k ∈ 𝒦}. We initially consider a first-order Markovian structure for the transition process, but discuss relaxations to this assumption below. In particular, we can write
where
for i = 1, … ,n, t = f(i) + 1, … , T, ψit(k,·) = {ψit(k, j): j = 1, … , K} and sif(i) = uif(i). In other words, if xit = 1 and uit−1 = sit−1 = j, the probability that individual i is in state k at time t is equal to ψit−1 (j,k); if xit = 0 (individual is dead), then uit = 0 (although sit ∈ 𝒦). The survival and recapture parameters can be extended to be dependent on the additional state parameter, with the analogous Bernoulli processes occurring for the observation and system survival processes.
The joint probability mass function of the survival and transition processes, f(x, u|ϕ,ψ) given in equation (2.5) can be specified in conditional form
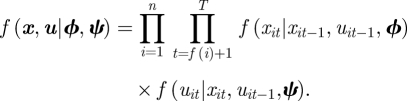 |
The first term corresponds to the Bernoulli survival process and the second to the multinomial transition process. In the standard AS model, if an individual is recaptured (i.e. yit = 1), the corresponding state of the individual, uit, is also known. However, if this is not the case and the state may not be observed when an individual is observed then within the state-space formulation the model specification remains the same. It is simply that the set of umis and uobs values change to reflect the set of missing and observed states, and that any unobserved uit values are auxiliary variables and updated within the MCMC algorithm.The state-space model can be extended to include additional system or observation processes. For example, within the traditional AS model, we assume that there are no recoveries within this process, but these can be easily included by considering an additional observation process corresponding to the recovery process [42,61,74]. Similarly, a birth process can be included (and a parameter for total population size) leading to the multi-state JS model [55,65].
Using a state-space framework, it is straightforward to fit more complex model structures, including a second-order transition process, such that the state of an individual at time t is dependent not only on their state at time t − 1 but also t − 2. An explicit observed data likelihood for a second-order Markov model is available [10,75] but is computationally expensive to calculate compared with the complete data likelihood, conditional on the unobserved states within the transition process. In addition, within the state-space framework the survival and observation processes remain the same, with only the transition system process needing to be modified. Let Ψit(j,k,l) denote the probability that individual i is in state l ∈ 𝒦 at time t + 2, given that they are alive and in states j ∈ 𝒦 and k ∈ 𝒦 at times t and t + 1, respectively, for t = f(i), … , T − 2. We let Ψit = {Ψit(j,k,l): j, k, l ∈ 𝒦}. The transition process for t = f(i) + 1, … , T can be described in the form
where
for t = f(i) + 2, … ,T and Ψit−2(sit−2, sit−1,·) = {Ψit−2 (sit−2, sit−1,l),l ∈ 𝒦}; and for t = f(i) + 1, typically, a first-order process is considered, so that
using the notation as for the first-order Markovian process and where sif(i) = uif(i).
Possible error within the state observation process can also be incorporated, leading to multi-event models proposed by Pradel [76]. In such models, the state of an individual is not observed directly, but an event relating to the state is observed. For example, states may refer to ‘breeding’ and ‘non-breeding’ and events ‘seen near nest’ and ‘not seen near nest’. In this case, there are no observed states (uobs = ∅︀). The state-space framework extends the AS model specification by including an additional event observation process, conditional on the underlying states. We note that the event process can, equivalently, be incorporated into the state process, corresponding to the event an individual is in at each capture event (with the introduction of auxiliary variables for the unobserved events). However, for simplicity, we retain the event process within the observation process (but note that if the model parameters such as the recapture probabilities are dependent on the event process, rather than the true underlying states, the event process needs to be incorporated into the system process and the unobserved events treated as auxiliary variables).
Let ℰ = {1, … , E} denote the set of possible observed events, and let vit denote the event observation process for individual i at time t, conditional on the individual being observed at this time (i.e. yit = 1). For statistical convenience, we let vit = 0 if yit = 0 (i.e. the individual is not observed). Thus, we have
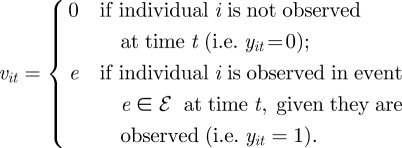 |
We define the additional model parameters βit(kit, eit) to be the probability individual i is in event eit ∈ ℰ at time t, given they are in state kit ∈ 𝒦 and observed at this time and let βit = {βit(k,e): k ∈ 𝒦,e ∈ ℰ}. Then, for i = 1, … , n and t = f(i), … , T, we have
where
and βit(uit,·) = {βit(uit,rit): rit = 1, … , E}, such that ∑e∈ℰ βit(uit, e) = 1 for all uit ∈ 𝒦 (assuming that on initial capture the observed event is independent of the fact that it is their first encounter).
The underlying system process is the same as for the AS model, with the event process incorporated in the observation process. However, we note that within the AS model, the likelihood conditions on the state an individual is in when first observed. Within multi-event models, this conditioning is removed (since we do not observe the true states), and an initial state distribution needs to be specified. Let κi(k) denote the probability individual i is in state k when it is first encountered. The initial state distribution is given by
where κi = {κi(k): k = 1, … , K}. More generally, the multi-event framework allows for both observed known state and partially observed state where an observed event restricts the set of possible true states (see Smout et al. [77] who consider this framework in relation to tag-loss models and King & McCrea [78] who derive an efficient closed form observed data likelihood).
Finally, we consider the special case of mixture models proposed by Pledger et al. [79] to allow for unobservable heterogeneity within the population. Within these models, the population is composed of K homogeneous sub-populations, with each individual belonging to one sub-population and there are no transitions between sub-populations. Typically, the sub-populations are unobservable, so that there is no directly observable covariate information (uobs = ∅︀). The covariate values are time-invariant, so that uit = ui for all, t = 1, … , T, and
where κ = {κ(k): k = 1, … , K} and κ(k) denotes the probability an individual belongs to sub-population k. In general, K may be known or be a parameter to be estimated [79].
2.3.2. Mixed/random effect models
We now consider continuous individual covariates. In these cases, the demographic parameters are typically specified as a parametric function of covariate values [11,12,15–20,41,42,65,80] or recently, more flexible semi-parametric spline models have also been proposed to model the relationship between the demographic parameters and covariates [14,81]. For example, let uit = {uit(j): j = 1, … , J} denote the set J covariate values for individual i at time t. The survival probability of individual i at time t may be (parametrically) logistically regressed on the set of covariates, such that
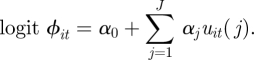 |
We note that if an individual is unobserved at any capture event (or following capture for the CJS model), their corresponding time-varying covariate(s) are also generally unobserved. This typically leads to an analytically intractable observed data likelihood, specified only in integral form, where the integration is over the unknown covariate values. Catchpole et al. [15] proposed an alternative conditional likelihood approach, conditioning on only the observed covariate values while Bonner et al. [16] compared this approach with an alternative Bayesian approach. The comparative study conducted suggested that, in general, the conditional likelihood approach suffers from poorer precision (as a result of discarding a potentially large proportion of the data) and can have boundary value problems for the parameter estimates, but has the potential advantage that no underlying model needs to be specified on the underlying covariate values, as in the Bayesian approach. In particular, the transition process for the covariate values are written in the form of the probability density function associated with the covariates (potentially allowing for dependence between the values). For example, Bonner & Schwarz [13] considered only one time-varying covariate and set
for t = f(i) + 1, … , T, dropping the notation on covariate j and where δt denotes an additive year effect. A probability density function is specified for uif(i) if there are unobserved values at initial capture. The missing covariate values are treated as auxiliary variables and updated within the MCMC algorithm.
Including individual random effects to the model allows for additional unexplained individual heterogeneity, such that
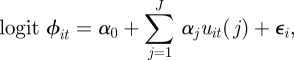 |
where εi ∼ N(0, σε2). Gimenez & Choquet [20] use a numerical integration procedure to approximate the observed data likelihood for random effect models on survival probabilities. Such an approach can be computationally very expensive for more than two individual random effects. Alternatively, within the Bayesian model-fitting approach, the additional random effect terms ε = {εi: i = 1, … , n} are treated as auxiliary variables that are updated within the Markov chain. The joint posterior distribution over the model parameters, unobserved system state values and random effect terms is given by
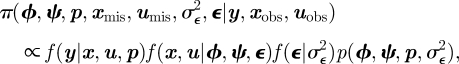 |
where  denotes the probability density function for the random effects (independent Gaussian for the above random effects model). Model selection can be performed on the covariates present in the model as a variable selection problem [12,19,41].
denotes the probability density function for the random effects (independent Gaussian for the above random effects model). Model selection can be performed on the covariates present in the model as a variable selection problem [12,19,41].
2.4. Efficiency
In general, when there are competing model-fitting algorithms that can be used, their relative performance will be both model and data dependent. In order to investigate the computational efficiency of a state-space modelling approach, we consider capture–recapture data relating to dippers (see Lebreton et al. [82] for further details) where there are T = 7 capture occasions and compare different model-fitting algorithms. Brooks et al. [53] compare different time-dependent models for these data, identifying the model with constant recapture probability, p, and two survival probabilities, corresponding a flood effect year (years 2 and 3) with survival probability, ϕf, and non-flood effect year (years 1, 4, 5 and 6) with survival probability, ϕn, as having largest posterior support (assuming uniform priors on the recapture and survival probabilities). Using this temporal model, we specify additional individual random effects, so that the model parameters are of the form
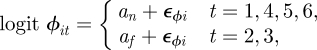 |
| 2.6 |
where 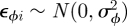 and
and 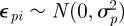 . Royle [18] considers the same data but specifies the fully time-dependent model with individual random effects. We specify the analogous priors to [18], such that logit−1
an ∼ U[0, 1], logit−1af ∼ U[0, 1], logit−1b ∼ U[0, 1] and σp, σϕ ∼ U[0, 10]. Posterior summary statistics of the model parameters are provided in table 1. We note that the posterior distribution of the underlying mean recapture and survival probabilities is similar to the posterior distribution of the recapture and survival probabilities obtained under the analogous temporal model without random effects [53]. For some discussion relating to prior sensitivity analyses on these data, see studies by Brooks et al. [53] and Royle [18], and for further discussion of individual heterogeneity and alternative models, see Choquet & Gimenez [83].
. Royle [18] considers the same data but specifies the fully time-dependent model with individual random effects. We specify the analogous priors to [18], such that logit−1
an ∼ U[0, 1], logit−1af ∼ U[0, 1], logit−1b ∼ U[0, 1] and σp, σϕ ∼ U[0, 10]. Posterior summary statistics of the model parameters are provided in table 1. We note that the posterior distribution of the underlying mean recapture and survival probabilities is similar to the posterior distribution of the recapture and survival probabilities obtained under the analogous temporal model without random effects [53]. For some discussion relating to prior sensitivity analyses on these data, see studies by Brooks et al. [53] and Royle [18], and for further discussion of individual heterogeneity and alternative models, see Choquet & Gimenez [83].
Table 1.
Posterior mean, standard deviation (s.d.) and 95% symmetric credible intervals (CIs) for the parameters in the model with individual random effects on the recapture and survival probabilities. The terms logit−1an and logit−1af correspond to the underlying mean survival probabilities of the individuals in the presence of individual random effects and logit−1b the underlying mean recapture probability.
| parameter | posterior mean (s.d.) | 95% CI |
|---|---|---|
| logit−1an | 0.61 (0.04) | (0.54, 0.69) |
| logit−1af | 0.48 (0.05) | (0.39, 0.58) |
| logit−1b | 0.94 (0.04) | (0.86, 0.99) |
| σϕ | 2.43 (1.28) | (0.33, 5.15) |
| σp | 0.28 (0.19) | (0.04, 0.74) |
We consider three model-fitting algorithms:
— SS1: the underlying state-space formulation described in §2 with individual random effects on the model parameters specified in (2.6);
— SS2: a reparametrized state-space formulation replacing the set of values xil(i)+1, …, xiT, where l(i) corresponds to the final time individual i is recaptured alive, with the parameter d = {di: i = 1, … , n} corresponding to the time of death for individual i (see, appendix B for further details); and
— L1: explicit calculation of the joint probability of the encounter histories given the parameters and auxiliary variables corresponding to the random effect terms, denoted by f(y|xobs, ε) (see appendix B for further details).
Each model-fitting approach was fitted within the computer package WinBUGS [84]. Method SS1 uses an adapted version of the code provided by Royle [18] allowing for the different time-dependence on the model parameters, with the code for SS2 and L1 modified versions of this code. Three different models are considered relating to individual random effects on the recapture probabilities only, survival probabilities only or on both recapture and survival probabilities. These models are denoted by {p(h), ϕ(f)}, {p, ϕ(f + h)} and {p(h), ϕ(f + h)}, respectively, where the h in brackets corresponds to the presence of individual heterogeneity and the f to the flood effect. The length of time to run each model for 10 000 iterations is given in table 2. In general, there is a trade-off between the number of parameters to be updated and the complexity of the posterior distribution. For L1, the probabilities of the encounter histories (following the final time an individual is observed) need to be calculated, whereas for SS1 and SS2 this reduces to a product of Bernoulli terms and imputing the alive/dead status of each individual animal. In this example, the state-space approaches are clearly computationally faster than the L1 approach.
Table 2.
Length of time (in seconds) that each model-fitting approach took to run 10 000 iterations in WinBUGS for three different models, where h in the brackets corresponds individual random effects and f to a flood effect.
| model-fitting approach |
|||
|---|---|---|---|
| Time for 10 000 iterations | SS1 | SS2 | L1 |
| p(h), ϕ(f + h) | 126 | 83 | 180 |
| p(h), ϕ(f) | 65 | 50 | 115 |
| p, ϕ(f + h) | 83 | 61 | 97 |
Of the three models fitted including individual heterogeneity, model {p(h), ϕ(f)}, corresponding to only individual random effects on the recapture probabilities, is preferred in terms of posterior model probabilities. Bayes factors of 3.9 and 32 are obtained for model {p(h), ϕ(f)} versus {p, ϕ(f + h)} and {p(h), ϕ(f + h)}, respectively. (However, we do note that the model excluding any random effects, {p, ϕ(f)}, is preferred with a Bayes factor of 5.2 versus model {p(h), ϕ(f)}.) We use model {p(h), ϕ(f)} to compare the performance of the different model-fitting algorithms including some level of individual heterogeneity. Note that for this model, we set q = logit−1b, corresponding to the underlying mean recapture probabilities of individuals in the presence of individual random effects, and ϕ = {ϕn, ϕf} the survival probabilities for non-flood and flood years, respectively (no individual random effects present).
We consider the convergence of the Markov chains to the stationary distribution, using the Brooks–Gelman–Rubin (BGR) statistic [85]. The BGR statistic for parameters ϕ, b and σp2 consistently tend to one within 10 000 iterations for repeated simulations starting from different initial values, though ϕn and ϕf always tend to one much quicker than the other parameters. This appears to be a result of the poorer mixing of the Markov chain for σp2 (and b). We return to this issue below in relation to autocorrelation. To be conservative, we specify a burn-in of 20 000 iterations for the MCMC simulations, to ensure the chain has sufficiently converged.
To compare the relative performances of the MCMC algorithm for the different model-fitting approaches, accounting for autocorrelation present within the Markov chain, we consider the effective sample size (ESS) [86] of each of the parameters. The ESS is essentially the number of independent samples within the MCMC sampled values, correcting for autocorrelation within the Markov chain and are presented in table 3. There are clear differences in the ESS across the different parameters with the consistent signal that all the MCMC algorithms perform poorly in the updating of the random effect variance term σp, with very high autocorrelation. Generally, the L1 model-fitting approach appears to have the largest ESS, suggesting a better mixing Markov chain; however, we recall that this was also the slowest model-fitting algorithm. Table 3 provides the corresponding ESS per unit time (100 s), which calculates the effective number of independent samples per unit of computational time. Using this ESS per unit time, the state-space approaches SS1 and SS2 each perform better than the L1 approach, for all parameters in this example. However, the SS2 state-space parametrization (auxiliary variables correspond to the time of death of each individual) significantly outperforms the SS1 parametrization (auxiliary variables correspond to alive/dead state of each individual at each capture time following final capture). However, we note the very poor performance of the MCMC algorithm for the random effect variance across all algorithms, and subsequently q (or b), due to the high posterior correlation between b and σp.
Table 3.
(a) Effective sample size (ESS) of each parameter under the different model-fitting algorithms using 100 000 iterations after 20 000 burn-in and (b) ESS per 100 seconds.
| parameter | SS1 | SS2 | L1 |
|---|---|---|---|
| (a) ESS | |||
| ϕn | 4014 | 4529 | 5142 |
| ϕf | 3569 | 3974 | 3979 |
| q | 629 | 943 | 749 |
| σp | 77 | 104 | 121 |
| (b) ESS per 100 seconds | |||
| ϕn | 616 | 900 | 447 |
| ϕf | 547 | 790 | 346 |
| q | 96 | 187 | 65 |
| σp | 12 | 21 | 11 |
We emphasize that, in general, the performance of different model-fitting approaches will be both problem and model dependent. For example, King [87] compares a state-space approach (using the Gibbs sampler) with the observed data likelihood approach (using a Metropolis–Hastings random walk algorithm) for the AS model using data analysed by Dupuis [63] consisting of 96 lizards, six capture events and three sites. In this case (using bespoke written code), the state-space approach was computationally faster, exhibited lower autocorrelation and did not require pilot-tuning compared with the observed data likelihood approach.
3. Discussion
The application of state-space models to capture–recapture–recovery data is very appealing due to its simplicity and ease with which models can be generalized. Partitioning the system and observation processes into distinct components leads to the natural identification of each individual process operating. This allows a complex model to be constructed using a sequence of simpler models, corresponding to each individual process acting on the study population. For example, for the capture–recapture–recovery data, the observation process is partitioned into distinct recapture and recovery processes; for the AS model, the system process is decomposed into the distinct survival process and state transition process. The different processes operating within a system will be problem dependent, but the general modelling approach remains the same. The full model is constructed by considering each process in turn and any conditional dependence between these processes (for example, for capture–recapture data the capture process is conditional on the individual being alive). This type of approach is clearly applicable to integrated analyses, combining together different sources of data within a single analysis. Integrated models simply specify the different processes acting on each individual dataset [42]. To construct and represent the state-space model, a graph is often a useful tool. See Buckland et al. [25,88] for further discussion and examples of different possible processes, including an ageing process (potentially leading to age-dependent parameters) and removals of individuals upon capture. Specifying the complete system process as a combination of simpler distinct system components also leads to the necessary and important consideration of the order in which the system processes occur [88,89]. The state-space approach has the advantage that if the model is modified in some way, the impact on each separate component can be considered, and in many cases, the different components may be unaltered. For example, for the AS model, changing the transition process from a first-order to second-order process only affects the system transition process, with the observation and recapture process being unaltered. This is in contrast to the derivation of the observed data likelihood which typically needs to be recalculated for different underlying models, which itself can be a non-trivial exercise. In addition, standard software including variants of BUGS (such as WinBUGS/OpenBUGS/JAGS) can be used to implement the state-space modelling approach, specifying the underlying distributional form for each individual process, which in general is significantly simpler than providing the explicit likelihood formulation.When fitting capture–recapture–recovery data within a Bayesian state-space approach there is a computational trade-off between the simplicity of the complete data likelihood and the number of parameters and auxiliary variables being updated in the MCMC algorithm. Using the standard state-space approach, imputing the alive/dead status of an individual following their final capture increases the number of parameters to be updated within the MCMC algorithm, but significantly simplifies the form of the posterior distribution. This can also lead to the posterior conditional distribution of the parameters being of standard form so that the Gibbs sampler may be implemented within the MCMC algorithm, for example, within the AS model for the survival/recapture/recovery probabilities (assuming independent beta priors) and transition probabilities (assuming independent Dirichlet priors). When implementing a Bayesian MCMC algorithm to fit such models the performance of the Markov chain (particularly in standard packages) should be examined. For poorly performing chains, alternative updating schemes can be implemented, for example, via the use of blocking of highly correlated parameters, hierarchical centring, parameter expansion and orthogonal reparametrization (see Browne et al. [90] for an overview of such methods and references therein). The use of alternative and efficient updating algorithms is an area of ongoing research.
Acknowledgements
I thank Roland Langrock, Byron Morgan, Diana Cole and Ian Goudie for their very helpful comments and discussions on earlier versions of this manuscript. I also thank three anonymous reviewers for their insightful comments on the initial manuscript which has led to the current improved version.
Appendix A
Table 4.
Glossary of auxiliary variables.
| auxiliary variable | value | definition | |
|---|---|---|---|
| (a) system processes | |||
| xit | 0 | individual i is not in the study at time t; | |
| 1 | individual i is in the study at time t. | ||
| uit | 0 | individual i is not in the study at time t; | |
| k | individual i has covariate value k at time t. | ||
| (b) observation processes | |||
| yit | 0 | individual i is not recaptured alive at time t; | |
| 1 | individual i is recaptured alive at time t. | ||
| zit | 0 | individual i is not recovered dead at time t; | |
| 1 | individual i is recovered dead at time t. | ||
| vit | 0 | individual i is not observed at time t; | |
| e | individual i has event value e at time t, | ||
| given individual is observed. |
Appendix B. Model-fitting algorithms
Here, we describe in greater detail the model-fitting approaches of SS2 and L1 described in §2.4. Method SS2 uses a very similar state-space formulation as to method SS1, but reparametrizes the model replacing the set of values xil(i)+1, …, xiT, where l(i) corresponds to the final time individual i is recaptured alive, with the parameter di corresponding to the time of death for individual i. Clearly, the parametrizations are equivalent, but the updating of such parameters in generic packages (including WinBUGS) will be different with only a single parameter needing to be updated for each individual using the parameters d = {d1, … , dn}. Conversely, if using the x parametrization each parameter following the last time an individual is observed alive will be updated (though many of the probability mass functions of these terms may correspond to a point mass on a single value). It is straightforward to show that
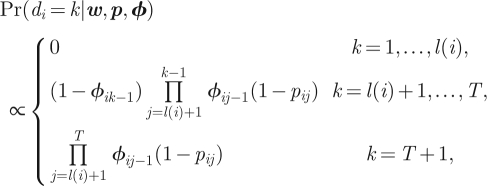 |
where di = T + 1 corresponds to an individual surviving to the end of the study and 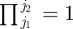 for j2 < j1.
for j2 < j1.
The third approach for fitting the MCMC algorithm, denoted by L1, explicitly calculates the joint probability of the encounter histories given the parameters and auxiliary variables corresponding to the random effects present in the study, denoted by ε. Let χit denote the probability individual i is not observed again in the study following time t, given they are alive at time t. The term χit can be calculated using the recursive formulae
with χiT = 1 for all i = 1, … , n [7]. The joint posterior distribution is given by
where σ2 denotes the random effect variances present in the model and f(w|p,ϕ,ε) is the joint probability of the observed data, given the model and random effect parameters. Thus, this approach does not involve any updating of the x (or d) values, but uses an explicit expression for the probability of not being observed again, taking into account all possible times an individual may have died.
The WinBUGS code for model fitting approach SS1 is available in Royle [18], with the adapted codes for approaches SS2 and L1 available from the author on request. Finally, we note that these approaches can be extended to other models, such as multi-state models (including extensions), but that approach L1 becomes more complicated to code, owing to the complexity of the observed data likelihood.
References
- 1.Williams B. K., Nichols J. D., Conroy M. J. 2002. Analysis and management of animal populations. London, UK: Academic Press. [Google Scholar]
- 2.Cormack R. M. 1964. Estimates of survival from the sighting of marked animals. Biometrika 51, 429–438. [Google Scholar]
- 3.Jolly G. M. 1965. Explicit estimates from capture–recapture data with both death and immigration stochastic model. Biometrika 52, 225–247. [PubMed] [Google Scholar]
- 4.Seber G. A. F. 1965. A note on the multiple-recapture census. Biometrika 52, 249–259. [PubMed] [Google Scholar]
- 5.Burnham K. P. 1993. A theory of combined analysis of ring recovery and recapture data. In Marked individuals in bird population studies (eds Lebreton J.-D., North P.), pp. 199–213. Basel, Switzerland: Birkhauser Verlag. [Google Scholar]
- 6.Barker R. J. 1997. Joint modelling of live recapture, tag-resight and tag-recovery data. Biometrics 53, 666–677. 10.2307/2533966 ( 10.2307/2533966) [DOI] [Google Scholar]
- 7.Catchpole E. A., Freeman S. N., Morgan B. J. T., Harris M. P. 1998. Integrated recovery/recapture data analysis. Biometrics 54, 33–46. 10.2307/2533993 ( 10.2307/2533993) [DOI] [Google Scholar]
- 8.King R., Brooks S. P. 2002. Model selection for integrated recovery/recapture data. Biometrics 58, 841–851. 10.1111/j.0006-341X.2002.00841.x ( 10.1111/j.0006-341X.2002.00841.x) [DOI] [PubMed] [Google Scholar]
- 9.Schwarz C. G., Schweigert J. F., Arnason A. N. 1993. Estimating migration rates using tag-recovery data. Biometrics 49, 177–193. 10.2307/2532612 ( 10.2307/2532612) [DOI] [Google Scholar]
- 10.Brownie C., Hines J. E., Nichols J. D., Pollock K. H., Hestbeck J. B. 1993. Capture–recapture studies for multiple strata including non-Markovian transitions. Biometrics 49, 1173–1187. 10.2307/2532259 ( 10.2307/2532259) [DOI] [Google Scholar]
- 11.Catchpole E. A., Morgan B. J. T., Coulson T. N., Freeman S. N., Albon M. P. 2000. Factors influencing Soay sheep survival. J. R. Stat. Soc. Ser. C 49, 453–472. 10.1111/1467-9876.00205 ( 10.1111/1467-9876.00205) [DOI] [Google Scholar]
- 12.King R., Brooks S. P., Morgan B. J. T., Coulson T. 2006. Bayesian analysis of Soay sheep survival. Biometrics 62, 211–220. 10.1111/j.1541-0420.2005.00404.x ( 10.1111/j.1541-0420.2005.00404.x) [DOI] [PubMed] [Google Scholar]
- 13.Bonner S. J., Schwarz C. J. 2006. An extension of the Cormack–Jolly–Seber model for continuous covariates with application to Microtus Pennsylvanicus. Biometrics 62, 142–149. 10.1111/j.1541-0420.2005.00399.x ( 10.1111/j.1541-0420.2005.00399.x) [DOI] [PubMed] [Google Scholar]
- 14.Gimenez O., Crainiceanu C., Barbraud C., Jenouvrier S., Morgan B. J. T. 2006. Semiparametric regression in capture–recapture modelling. Biometrics 62, 691–698. 10.1111/j.1541-0420.2005.00514.x ( 10.1111/j.1541-0420.2005.00514.x) [DOI] [PubMed] [Google Scholar]
- 15.Catchpole E. A., Morgan B. J. T., Tavecchia G. 2008. A new method for analysing discrete life history data with missing covariate values. J. R. Stat. Soc. Ser. B 70, 445–460. 10.1111/j.1467-9868.2007.00644.x ( 10.1111/j.1467-9868.2007.00644.x) [DOI] [Google Scholar]
- 16.Bonner S. J., Morgan B. J. T., King R. 2010. Continuous covariates in mark–recapture–recovery analysis: a comparison of methods. Biometrics 66, 1256–1265. 10.1111/j.1541-0420.2010.01390.x ( 10.1111/j.1541-0420.2010.01390.x) [DOI] [PubMed] [Google Scholar]
- 17.Barry S. C., Brooks S. P., Catchpole E. A., Morgan B. J. T. 2003. The analysis of ring-recovery data using random effects. Biometrics 59, 54–65. 10.1111/1541-0420.00007 ( 10.1111/1541-0420.00007) [DOI] [PubMed] [Google Scholar]
- 18.Royle J. A. 2008. Modeling individual effects in the Cormack–Jolly–Seber model: a state-space formulation. Biometrics 64, 364–370. 10.1111/j.1541-0420.2007.00891.x ( 10.1111/j.1541-0420.2007.00891.x) [DOI] [PubMed] [Google Scholar]
- 19.King R., Brooks S. P., Coulson T. 2008. Analysing complex capture–recapture data in the presence of individual and temporal covariates and model uncertainty. Biometrics 64, 1187–1195. 10.1111/j.1541-0420.2008.00991.x ( 10.1111/j.1541-0420.2008.00991.x) [DOI] [PubMed] [Google Scholar]
- 20.Gimenez O., Choquet R. 2010. Incorporating individual heterogeneity is studies on marked animals using numerical integration: capture–recapture mixed models. Ecology 91, 951–957. 10.1890/09-1903.1 ( 10.1890/09-1903.1) [DOI] [PubMed] [Google Scholar]
- 21.Millar R. B., Meyer R. 2000. Non-linear state space modelling of fisheries biomass dynamics by using Metropolis–Hastings within-Gibbs sampling. J. R. Stat. Soc. Ser. C 49, 327–342. 10.1111/1467-9876.00195 ( 10.1111/1467-9876.00195) [DOI] [Google Scholar]
- 22.Rivot E., Prévost E., Parent E., Bagliniére J. L. 2004. A Bayesian state-space modelling framework for fitting a salmon stage-structured population dynamics model to multiple time series of field data. Ecol. Model. 179, 463–485. 10.1016/j.ecolmodel.2004.05.011 ( 10.1016/j.ecolmodel.2004.05.011) [DOI] [Google Scholar]
- 23.Schnute J. T., Kronlund A. R. 2002. Estimating salmon stock-recruitment from catch and escapement data. Can. J. Fish. Aquat. Sci. 59, 433–449. 10.1139/f02-016 ( 10.1139/f02-016) [DOI] [Google Scholar]
- 24.Besbeas P., Freeman S. N., Morgan B. J. T., Catchpole E. A. 2002. Integrating mark–recapture–recovery and census data to estimate animal abundance and demographic parameters. Biometrics 58, 540–547. 10.1111/j.0006-341X.2002.00540.x ( 10.1111/j.0006-341X.2002.00540.x) [DOI] [PubMed] [Google Scholar]
- 25.Buckland S. T., Newman K. N., Thomas L., Koesters N. B. 2004. State-space models for the dynamics of wild animal populations. Ecol. Model. 171, 157–175. 10.1016/j.ecolmodel.2003.08.002 ( 10.1016/j.ecolmodel.2003.08.002) [DOI] [Google Scholar]
- 26.King R., Brooks S. P., Mazzetta C., Freeman S. N., Morgan B. J. T. 2008. Identifying and diagnosing population declines: a Bayesian assessment of lapwings in the UK. J. R. Stat. Soc. Ser. C 57, 609–632. 10.1111/j.1467-9876.2008.00633.x ( 10.1111/j.1467-9876.2008.00633.x) [DOI] [Google Scholar]
- 27.Newman K. N., Fernández C., Thomas L., Buckland S. T. 2009. Monte Carlo inference for state-space models of wild animal populations. Biometrics 65, 572–583. 10.1111/j.1541-0420.2008.01073.x ( 10.1111/j.1541-0420.2008.01073.x) [DOI] [PubMed] [Google Scholar]
- 28.Jonsen D. S., Flemming J., Myers R. 2005. Robust state-space modeling of animal movement data. Ecology 86, 2874–2880. 10.1890/04-1852 ( 10.1890/04-1852) [DOI] [Google Scholar]
- 29.Jonsen D. S., Myers R., Flemming J. 2003. Meta-analysis of animal movement using state-space models. Ecology 84, 3005–3063. 10.1890/02-0670 ( 10.1890/02-0670) [DOI] [Google Scholar]
- 30.Patterson T. A., Thomas L., Wilcox C., Ovaskeinen O., Matthiopoulos J. 2008. State-space models of individual animal movement. Trends Ecol. Evol. 23, 87–94. 10.1016/j.tree.2007.10.009 ( 10.1016/j.tree.2007.10.009) [DOI] [PubMed] [Google Scholar]
- 31.McClintock B. T., King R., Thomas L., Matthiopoulos J., McConnell B. J., Morales J. M. 2011. A general modeling framework for animal movement and migration using multi-state random walks. Technical Report. St Andrews, UK: University of St Andrews. [Google Scholar]
- 32.Cappé O., Moulines E., Ryden T. 2005. Inference in hidden Markov Models. New York, NY: Springer. [Google Scholar]
- 33.Dempster A. P., Laird N. M., Rubin D. B. 1977. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B 39, 1–38. [Google Scholar]
- 34.van Deusen P. C. 2002. An EM algorithm for capture–recapture estimation. Environ. Ecol. Stat. 9, 151–165. 10.1023/A:1015118120406 ( 10.1023/A:1015118120406) [DOI] [Google Scholar]
- 35.Kalman R. R. 1960. A new approach to linear filtering and prediction problems. Trans. ASME J. Basic Eng. 82, 35–45. 10.1115/1.3662552 ( 10.1115/1.3662552) [DOI] [Google Scholar]
- 36.Newman K. B., Borchers D. L., Buckland S. T., King R., Thomas L., Besbeas P. T., Gimenez O., Morgan B. J. T. In preparation. Estimating animal abundance: open populations. [Google Scholar]
- 37.Tanner M. A., Wong W. H. 1987. The calculation of posterior distribution by data augmentation. J. Am. Stat. Assoc. 82, 528–540. 10.2307/2289457 ( 10.2307/2289457) [DOI] [Google Scholar]
- 38.Schofield M. R., Barker R. J., Mackenzie D. I. 2009. Flexible hierarchical mark–recapture modeling for open populations using WinBUGS. Environ. Ecol. Stat. 16, 369–387. 10.1007/s10651-007-0069-1 ( 10.1007/s10651-007-0069-1) [DOI] [Google Scholar]
- 39.Link W. A., Barker R. J. 2010. Bayesian inference with ecological applications. London, UK: Academic Press. [Google Scholar]
- 40.Royle J. A., Dorazio R. M. 2008. Hierarchical modeling and inference in ecology. London, UK: Academic Press. [Google Scholar]
- 41.King R., Morgan B. J. T., Gimenez O., Brooks S. P. 2009. Bayesian analysis for population ecology. Boca Raton, FL: CRC Press. [Google Scholar]
- 42.Kéry M., Schaub M. 2011. Bayesian population analysis using WinBUGS: a hierarchical perspective. London, UK: Academic Press. [Google Scholar]
- 43.Catchpole E. A., Morgan B. J. T. 1997. Detecting parameter redundancy. Biometrika 84, 187–196. 10.1093/biomet/84.1.187 ( 10.1093/biomet/84.1.187) [DOI] [Google Scholar]
- 44.Cole D. J., Morgan B. J. T., Titterington D. M. 2010. Determining the parametric structure of non-linear models. Math. Biosci. 228, 16–30. 10.1016/j.mbs.2010.08.004 ( 10.1016/j.mbs.2010.08.004) [DOI] [PubMed] [Google Scholar]
- 45.Cole D. J., Morgan B. J. T. 2010. A note on determining parameter redundancy in age-dependent tag return models for estimating fishing mortality, natural mortality and selectivity. J. Agric. Biol. Environ. Stat. 15, 431–434. 10.1007/s13253-010-0026-6 ( 10.1007/s13253-010-0026-6) [DOI] [Google Scholar]
- 46.Cole D. J., Morgan B. J. T. 2010. Parameter redundancy with covariates. Biometrika 97, 1002–1005. 10.1093/biomet/asq041 ( 10.1093/biomet/asq041) [DOI] [Google Scholar]
- 47.Cole D. J. In press. Determining parameter redundancy of multi-state mark–recapture models for sea birds. J. Ornithol. [Google Scholar]
- 48.Gimenez O., Brooks S. P., Morgan B. J. T. 2009. Weak identifiability in models for mark–recapture–recovery data. In Modeling demographic processes in marked populations, vol. 3 (eds Thomson D. L., Cooch E. G., Conroy M. J.), pp. 1055–1067. Environmental and Ecological Statistics Series New York, NY: Springer. [Google Scholar]
- 49.Kass R. E., Raftery A. E. 1995. Bayes factors. J. Am. Stat. Soc. 90, 773–793. [Google Scholar]
- 50.Green P. J. 1995. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82, 711–732. 10.1093/biomet/82.4.711 ( 10.1093/biomet/82.4.711) [DOI] [Google Scholar]
- 51.Godsill S. J. 2001. On the relationship between Markov chain Monte Carlo methods for model uncertainty. J. Comput. Graph. Stat. 10, 230–248. 10.1198/10618600152627924 ( 10.1198/10618600152627924) [DOI] [Google Scholar]
- 52.Gelman A., Meng X. 1996. Model checking and model improvement. In Markov chain Monte Carlo in practice (eds Gilks W. R., Richardson R., Spiegelhalter D. J.), pp. 189–210. London, UK: Chapman and Hall. [Google Scholar]
- 53.Brooks S. P., Catchpole E. A., Morgan B. J. T. 2000. Bayesian animal survival estimation. Stat. Sci. 15, 357–376. 10.1214/ss/1009213003 ( 10.1214/ss/1009213003) [DOI] [Google Scholar]
- 54.King R., Brooks S. P. 2002. Bayesian model discrimination for multiple strata capture–recapture data. Biometrika 89, 785–806. 10.1093/biomet/89.4.785 ( 10.1093/biomet/89.4.785) [DOI] [Google Scholar]
- 55.Dupuis J. A., Schwarz C. J. 2007. A Bayesian approach to multistate Jolly–Seber capture–recapture model. Biometrics 63, 1015–1022. 10.1111/j.1541-0420.2007.00815.x ( 10.1111/j.1541-0420.2007.00815.x) [DOI] [PubMed] [Google Scholar]
- 56.Schwarz C. J., Seber G. A. F. 1999. A review of estimating animal abundance III. Stat. Sci. 14, 427–456. 10.1214/ss/1009212521 ( 10.1214/ss/1009212521) [DOI] [Google Scholar]
- 57.Seber G. A. F. 1970. Estimating time-specific survival and reporting rates for adult birds from band returns. Biometrika 57, 313–318. 10.1093/biomet/57.2.313 ( 10.1093/biomet/57.2.313) [DOI] [Google Scholar]
- 58.Crosbie S. F., Manly B. F. J. 1985. Parsimonious modelling of capture–mark–recapture studies. Biometrics 41, 385–398. 10.2307/2530864 ( 10.2307/2530864) [DOI] [Google Scholar]
- 59.Schwarz C. J., Arnason A. N. 1996. A general methodology for the analysis of capture–recapture experiments in open populations. Biometrics 52, 860–873. 10.2307/2533048 ( 10.2307/2533048) [DOI] [Google Scholar]
- 60.Pledger S., Pollock K. H., Norris J. L. 2010. Open capture–recapture models with heterogeneity: II. Jolly–Seber model. Biometrics 66, 883–890. 10.1111/j.1541-0420.2009.01361.x ( 10.1111/j.1541-0420.2009.01361.x) [DOI] [PubMed] [Google Scholar]
- 61.Gimenez O., Rossi V., Choquet R., Dehais C., Doris B., Varella H., Vila J.-P., Pradel R. 2007. State-space modelling of data on marked individuals. Ecol. Model. 206, 431–438. 10.1016/j.ecolmodel.2007.03.040 ( 10.1016/j.ecolmodel.2007.03.040) [DOI] [Google Scholar]
- 62.Schofield M. R., Barker R. J. 2008. A unified capture–recapture framework. J. Agric. Biol. Environ. Stat. 13, 459–477. 10.1198/108571108X383465 ( 10.1198/108571108X383465) [DOI] [Google Scholar]
- 63.Dupuis J. A. 1995. Bayesian estimation of movement and survival probabilities from capture–recapture data. Biometrika 82, 761–772. 10.2307/2337343 ( 10.2307/2337343) [DOI] [Google Scholar]
- 64.Catchpole E. A., Morgan B. J. T., Freeman S. N., Nash W. J. 2001. Abalone I: analysing mark–recapture–recovery data, incorporating growth and delayed recovery. Biometrics 57, 136–144. 10.1111/j.0006-341X.2001.00469.x ( 10.1111/j.0006-341X.2001.00469.x) [DOI] [PubMed] [Google Scholar]
- 65.Schofield M. R., Barker R. J. 2011. Full open capture–recapture models with individual covariates. J. Agric. Biol. Environ. Stat. 16, 253–268. 10.1007/s13253-010-0052-4 ( 10.1007/s13253-010-0052-4) [DOI] [Google Scholar]
- 66.Gardner B., Reppucci J., Lucherini M., Royle J. A. 2010. Spatially explicit inference for open populations: estimating demographic paramters from camera-trap studies. Ecology 91, 3376–3383. 10.1890/09-0804.1 ( 10.1890/09-0804.1) [DOI] [PubMed] [Google Scholar]
- 67.Durban J. W., Elston D. A. 2005. Mark–recapture with occasion and individual effects: abundance estimation through Bayesian model selection in a fixed dimensional parameter space. J. Agric. Biol. Environ. Stat. 10, 291–305. 10.1198/108571105X58630 ( 10.1198/108571105X58630) [DOI] [Google Scholar]
- 68.Royle J. A., Dorazio R. M., Link W. A. 2007. Analysis of multinomial models with unknown index using data augmentation. J. Comput. Graph. Stat. 16, 67–85. 10.1198/106186007X181425 ( 10.1198/106186007X181425) [DOI] [Google Scholar]
- 69.King R., Brooks S. P. 2008. On the Bayesian estimation of a closed population size in the presence of heterogeneity and model uncertainty. Biometrics 64, 816–824. 10.1111/j.1541-0420.2007.00938.x ( 10.1111/j.1541-0420.2007.00938.x) [DOI] [PubMed] [Google Scholar]
- 70.King R., Brooks S. P. 2003. Closed form likelihoods for Arnason–Schwarz models. Biometrika 90, 435–444. 10.1093/biomet/90.2.435 ( 10.1093/biomet/90.2.435) [DOI] [Google Scholar]
- 71.McCrea R. S. 2012. Sufficient statistic likelihood construction for age- and time-dependent multi-state joint recapture and recovery data. Stat. Prob. Lett. 82 357–359. 10.1016/j.spl.2011.10.020 ( 10.1016/j.spl.2011.10.020) [DOI] [Google Scholar]
- 72.Marin J., Robert C. P. 2007. Bayesian core: a practical approach to computational Bayesian statistics. New York, NY: Springer. [Google Scholar]
- 73.Clark J. S., Ferraz G., Oguge N., Hays H., DiCostanzo J. 2005. Hierarchical Bayes for structured variable populations: from recapture data to life-history prediction. Ecology 86, 2232–2244. 10.1890/04-1348 ( 10.1890/04-1348) [DOI] [Google Scholar]
- 74.Dupuis J. A., Badia J., Maublanc M. L., Bon R. 2002. Spatial fidelity of mouflon: a Bayesian analysis of an age-dependent capture–recapture model. J. Agric. Biol. Environ. Stat. 7, 277–298. 10.1198/10857110260141292 ( 10.1198/10857110260141292) [DOI] [Google Scholar]
- 75.Rouan L., Choquet R., Pradel R. 2009. A general framework for modeling memory in capture–recapture data. J. Agric. Biol. Environ. Stat. 14, 338–355. 10.1198/jabes.2009.06108 ( 10.1198/jabes.2009.06108) [DOI] [Google Scholar]
- 76.Pradel R. 2005. Multievent: an extension of multistate capture–recapture models to uncertain states. Biometrics 61, 442–447. 10.1111/j.1541-0420.2005.00318.x ( 10.1111/j.1541-0420.2005.00318.x) [DOI] [PubMed] [Google Scholar]
- 77.Smout S. C., King R., Pomeroy P. 2011. Integrating heterogeneity of detection and mark loss to estimate survival and transience in UK grey seal colonies. J. Appl. Ecol. 48, 364–372. 10.1111/j.1365-2664.2010.01913.x ( 10.1111/j.1365-2664.2010.01913.x) [DOI] [Google Scholar]
- 78.King R., McCrea R. S. 2011. A generalised matrix form likelihood for mark–recapture–recovery models. Technical Report. St Andrews, UK: University of St Andrews. [Google Scholar]
- 79.Pledger S., Pollock K. H., Norris J. L. 2003. Open capture–recapture models with heterogeneity: I. Cormack–Jolly–Seber model. Biometrics 59, 786–794. 10.1111/j.0006-341X.2003.00092.x ( 10.1111/j.0006-341X.2003.00092.x) [DOI] [PubMed] [Google Scholar]
- 80.North P. M., Morgan B. J. T. 1979. Modeling heron survival using weather data. Biometrics 35, 667–681. 10.2307/2530260 ( 10.2307/2530260) [DOI] [Google Scholar]
- 81.Bonner S. J., Thomson D. L., Schwarz C. J. 2009. Time-varying covariates and semi-parametric regression in capture–recapture: an adaptive spline approach. In Modeling demographic processes in marked populations, vol. 3 (eds Thomson D. L., Cooch E. G., Conroy M. J.), pp. 657–675. Environmental and Ecological Statistics Series. New York, NY: Springer. [Google Scholar]
- 82.Lebreton J. D., Burnham K. P., Clobert J., Anderson D. R. 1992. Modelling survival and testing biological hypotheses using marked animals: a unified approach with case studies. Ecol. Monogr. 62, 67–118. 10.2307/2937171 ( 10.2307/2937171) [DOI] [Google Scholar]
- 83.Choquet R., Gimenez O. In press. Towards built-in capture–recapture mixed models in program E-SURGE. J. Ornithol. ( 10.1007/s10336-010-0613-x) [DOI] [Google Scholar]
- 84.Lunn D. J., Thomas A., Best N., Spiegelhalter D. 2000. WinBUGS: a Bayesian modelling framework: concepts, structure, and extensibility. Stat. Comput. 10, 325–337 ( 10.1023/A:1008929526011) [DOI] [Google Scholar]
- 85.Brooks S. P., Gelman A. 1998. Alternative methods for monitoring convergence of iterative simulations. J. Comput. Graph. Stat. 7, 434–455. 10.2307/1390675 ( 10.2307/1390675) [DOI] [Google Scholar]
- 86.Liu J. S., Chen R. 1995. Blind deconvolution via sequential imputations. J. Am. Stat. Assoc. 90, 567–576. 10.2307/2291068 ( 10.2307/2291068) [DOI] [Google Scholar]
- 87.King R. 2001. Bayesian model discrimination in the analysis of capture–recapture and related data. PhD thesis, University of Bristol, UK. [Google Scholar]
- 88.Buckland S. T., Newman K. N., Fernández C., Thomas L., Harwood J. 2007. Embedding population dynamics models in inference. Stat. Sci. 22, 44–58. 10.1214/088342306000000673 ( 10.1214/088342306000000673) [DOI] [Google Scholar]
- 89.Duriez O., Sæther S. A., Ens B. J., Choquet R., Pradel R., Lamback R. H. D., Klaassen M. 2009. Estimating survival and movements using both live and dead recoveries: a case study of oystercatchers confronted with habitat change. J. Appl. Ecol. 46, 144–153. 10.1111/j.1365-2664.2008.01592.x ( 10.1111/j.1365-2664.2008.01592.x) [DOI] [Google Scholar]
- 90.Browne W. J., Steele F., Golalizadeh M., Green M. J. 2009. The use of simple reparameterizations to improve the efficiency of Markov chain Monte Carlo estimation for multilevel models with applications to discrete time survival models. J. R. Stat. Soc. Ser. A 172, 579–598. 10.1111/j.1467-985X.2009.00586.x ( 10.1111/j.1467-985X.2009.00586.x) [DOI] [PMC free article] [PubMed] [Google Scholar]