Abstract
Aneuploidy refers to karyotypic abnormalities characterized by gain or loss of individual chromosomes. This condition is associated with disease and death in all organisms in which it has been studied. We have characterized the effects of aneuploidy on yeast and primary mouse cells and found aneuploidy to be detrimental at the cellular level. Furthermore, we find that aneuploid cells exhibit phenotypes consistent with increased energy need and proteotoxic stress. These observations together with the finding that the additional chromosomes found in aneuploid cells are active lead us to propose that aneuploidy causes an increased burden on protein synthesis and protein quality control pathways and so induces an aneuploidy stress response.
Alterations in chromosome number that are not a multiple of the haploid complement are called aneuploidies. The effects of such karyotypic changes on human health are profound. Aneuploidy is the leading cause of miscarriages and mental retardation and a key characteristic of cancer. More than 90% of all solid human tumors are aneuploid. Determining how aneuploidy affects cell physiology is therefore critical for understanding the principles underlying many human diseases.
Aneuploidy differs from polyploidy, in which cells harbor a multiple of their haploid karyotype. Polyploidy is well tolerated on both the cellular and organismal level and is part of the normal developmental program in some tissues. In contrast, autosomal aneuploidy is associated with severe abnormalities and death in all organisms analyzed (reviewed in (Torres et al. 2008; Williams and Amon 2009). In budding and fission yeast aneuploidy leads to cell proliferation defects (Niwa et al. 2006; Torres et al. 2007). In flies, with the exception of chromosome 4, all whole-chromosome trisomies and monosomies are lethal (Lindsley et al. 1972). Similar results are observed in worms where all trisomies and monosomies are inviable (Hodgkin 2005). In the mouse all monosomies and all trisomies, except for trisomy 19, are embryonic lethal. In humans, all whole-chromosome aneuploidies except trisomy 13, 18, or 21 result in embryonic lethality. Even these viable trisomies display severe abnormalities. Trisomy 13 or 18 individuals die within the first few months of life and exhibit developmental abnormalities such as cardiovascular and cranio-facial defects, developmental abnormalities of the nervous system as well as growth retardation (Moerman et al. 1988; Lin et al. 2006). These phenotypes are also seen in the only viable human trisomy, trisomy 21 (Antonarakis et al. 2004).
At the organismal level aneuploidy is highly detrimental, yet, at the cellular level, aneuploidy is associated with cancer; a disease characterized by high proliferative potential. These findings raise an interesting conundrum. How is it possible that a single extra chromosome causes developmental defects characterized by growth retardation, yet in the context of cancer, cells with high proliferative potential nevertheless have severe karyotypic abnormalities? It is possible that developmental programs are sensitive to gene copy number changes but cell proliferation is not. For example, copy number imbalances in genes critical for the formation of an essential organ or tissue could lead to malformation thereof and hence death. In contrast, at the cellular level, maintenance of a stable karyotype is perhaps not important as long as each cell has one copy of each chromosome. An alternative hypothesis (that our studies indicate to be the correct one) is that aneuploidy is also detrimental at the cellular level but cancer cells have acquired the ability to overcome the adverse effects of aneuploidy to take advantage of potentially beneficial effects of the condition. To distinguish between these possibilities and to understand the contribution of aneuploidy to tumorigenesis, we thought it was important to determine the effects of aneuploidy on the physiology of normal cells.
A set of shared phenotypes in aneuploid cells
To determine how aneuploidy affects the proliferation and physiology of normal cells, we generated 20 strains of budding yeast, each strain bearing an extra copy of one or more of the yeast chromosomes. These disomic yeast strains display decreased fitness relative to wild type cells. Furthermore, we observed two classes of phenotypes. Phenotypes that are specific for a particular aneuploid strain and traits that are shared among the different aneuploid yeast strains. The most prominent among the shared traits are indicators of proteotoxic stress. Aneuploid yeast strains are temperature sensitive, that is, their proliferation is impaired at elevated temperature (37°C) relative to euploid cells. Most aneuploid yeast cells are also hyper-sensitive to compounds that interfere with protein synthesis, i.e., cycloheximide or hygromycin, and several strains show hyper-sensitivity towards the proteasome inhibitor MG132. In addition, aneuploid yeast strains make less biomass per glucose, share a common gene expression profile and exhibit cell proliferation defects, most prominently a delay at the G1 – S phase transition (Torres et al. 2007).
The analysis of primary mouse embryonic fibroblasts (MEFs) trisomic for either Chromosome 1, 13, 16 or 19 revealed similar results (Williams et al. 2008). Aneuploidy is deleterious at the cellular level, causing cell proliferation defects. This is in agreement with the finding that primary foreskin fibroblasts of trisomy 21 patients proliferate more slowly than euploid control cells (Segal and McCoy 1974) and the observation that primary mouse cells or human cell lines with decreased chromosome segregation fidelity show impaired growth (Baker et al. 2004; Sotillo et al. 2007; Thompson and Compton 2008). Most important, the aneuploid MEFs have similar properties as the disomic yeast strains; aneuploid mouse cells too exhibit signs of increased energy need and of proteotoxic stress. Trisomic MEFs show elevated uptake of glutamine, the major carbon source of the TCA cycle in tissue culture cells (DeBerardinis et al. 2007; Williams et al. 2008). Levels of the chaperone Hsp70 and of autophagy, which are indicators of proteotoxic stress, are also increased in trisomic MEFs (Y.-C. Tang, unpubl.).
In summary, the characterization of normal cells with a defined aneuploid karyotype led to two key findings. First, aneuploidy is detrimental at the cellular level. Second, aneuploid cells exhibit two types of phenotypes: (1) Traits that are unique to a specific karyotypic abnormality and (2) traits that are shared by most aneuploid yeast and mouse cells. Based on the observation that many of the shared traits are reminiscent of cellular stresses, we collectively call the shared phenotypes the “aneuploidy stress response”.
Our findings have important implications for how one thinks about cancer. Cancer cells must overcome the adverse effects of aneuploidy in order to outgrow euploid cells and take advantage of potential benefits that arise from the aneuploid condition. Furthermore, our results raise the interesting possibility that cancer cells are under energy and proteotoxic stress caused by, at least in part, the aneuploid state of cancer cells.
Origins of the phenotypes shared by aneuploid cells
Which aspects of the aneuploid state are responsible for the phenotypes shared by disomic yeast and trisomic mouse cells? Is it the additional DNA, additional transcripts and/or the additional proteins? In budding yeast, it is clear that the phenotypes are due to the additional transcripts and/or proteins. Introduction of artificial mammalian chromosomes similar in size to yeast chromosomes, but not producing any yeast proteins and most likely producing few if any mammalian transcripts and even fewer peptides/proteins in yeast, do not lead to the phenotypes observed in strains with additional yeast chromosomes (Torres et al. 2007). Furthermore, the severity of the shared phenotypes increases with the amount of additional yeast DNA present in cells (Torres et al. 2007). These data together with the observation that the genes located on the additional chromosomes are transcribed and translated indicate that it is the proteins produced from the additional chromosomes and/or their production that leads to the phenotypes seen in aneuploid cells.
The same is likely to be true in trisomic MEFs. The additional chromosomes are transcribed and the degree of fitness loss in trisomic MEFs correlates with the size of the additional chromosome and the number of transcripts that are generated (Williams et al. 2008). So how do the active additional chromosomes cause the phenotypes we observe? In what follows, we will consider the origins of the general aneuploidy associated phenotypes.
Origins of the proteotoxic stress
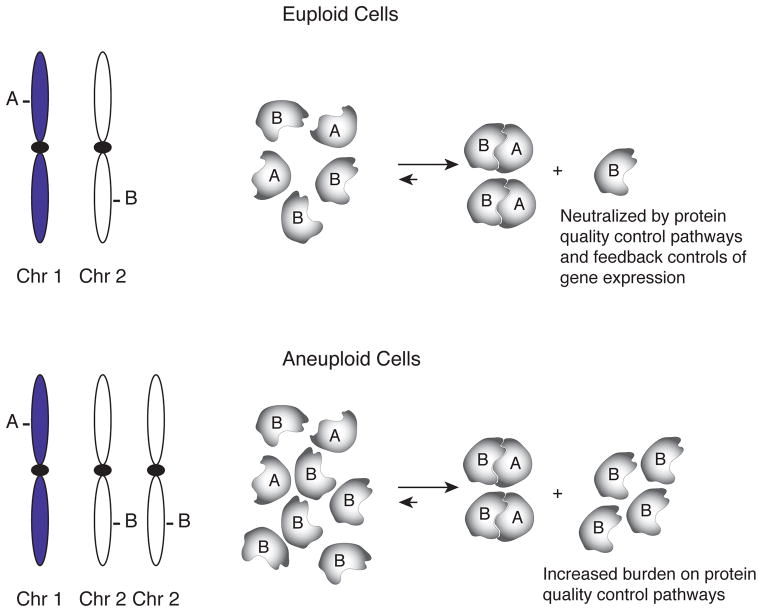
The fact that the additional chromosomes in aneuploid yeast and mouse cells are active must mean that aneuploid cells face protein stoichiometry imbalances. The finding that aneuploid cells exhibit phenotypes indicative of proteotoxic stress suggests that these imbalances cause an increased burden on the protein quality control pathways. This is best illustrated when thinking about two proteins, A and B, that function in a complex and whose genes are located on two different chromosomes (Figure 1A). Producing and maintaining the exact number of each subunit is regulated by protein quality control pathways, such as protein degradation or chaperone-mediated sequestration and sometimes feedback controls that regulate gene expression to maintain protein A and B in stoichiometric amounts. Several such protein stoichiometry control pathways have been described. Unassembled ribosomal subunits are rapidly degraded (elBaradi et al. 1986; Agrawal and Bowman 1987; Maicas et al. 1988). Gene expression feedback mechanisms ensure the stoichiometry of histones and α- and β-tubulin (Katz et al. 1990; Theodorakis and Cleveland 1992; Bachurski et al. 1994; Gunjan and Verreault 2003; Libuda and Winston 2006). Translational control mechanisms determine the stoichiometry of the ATP synthase subunits in the chloroplast (Drapier et al. 2007). Similar mechanisms must exist for many other protein complexes, because it is well established that experimental inactivation of one subunit of a protein complex leads to destabilization of the other subunits.
Figure 1. Protein stoichiometry imbalances associated with aneuploidy.
See text for details.
In aneuploid cells protein stoichiometry imbalances are greatly exaggerated (Figure 1B). This leads to an increased burden on the protein quality control pathways. Importantly, although the identity of the proteins whose stoichiometries are imbalanced will be different for each aneuploid chromosome, the mechanisms whereby cells attempt to correct these imbalances – protein degradation and chaperone mediated sequestration – are the same.
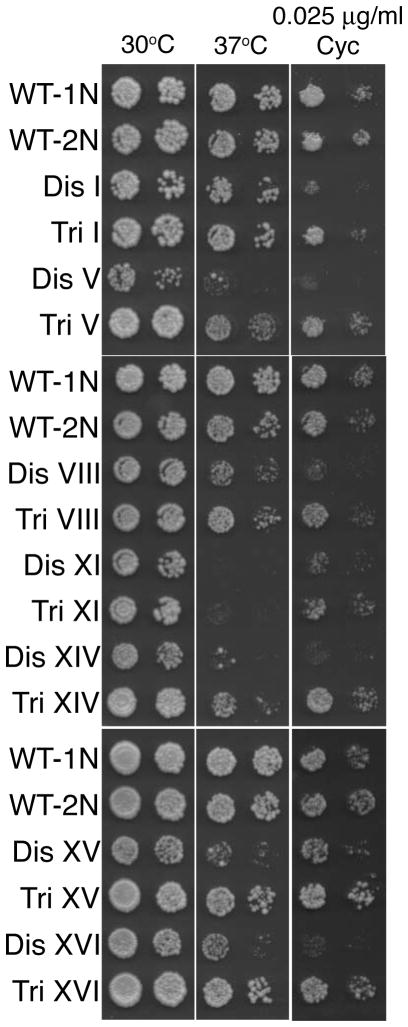
A prediction of the protein stoichiometry imbalance hypothesis is that haploid yeast cells carrying an additional chromosome (disomes) should be more sensitive to proteotoxic stress than diploid cells carrying an extra chromosome (trisomes) because the degree of stoichiometric imbalance is lower in diploid cells carrying an extra chromosome than in haploid cells harboring an additional chromosome. To test this we compared the ability of strains disomic for chromosome I, V, VIII, XI, XIV, XV or XVI with that of diploid cells trisomic for the same chromosome to grow at high temperature (37°C), which causes protein mis-folding or in the presence of the translation inhibitor cycloheximide, which causes proliferation defects in yeast owing to ubiquitin depletion (Hanna et al. 2003). Figure 2 shows that all disomic strains that we analyzed are more sensitive to high temperature and cycloheximide than trisomic cells. This observation is consistent with the idea that protein stoichiometry imbalances are at least in part responsible for the sensitivity of cells to conditions that interfere with protein turn over and folding.
Figure 2. Sensitivity of trisomic and disomic yeast cells to high temperature and cycloheximide.
Proliferative capability of disomes and trisomes in the presence of 37°C and Cycloheximide. 10-fold dilutions are shown. Strains utilized (from top): Wild type-1N (A11311), wild type-2N (A19550), disome I (A12683), trisome I (A18345), disome V (A14479), trisome V (A18346), disome VIII (A13628), trisome VIII (A18347), disome XI (A13771), trisome XI (A18348), disome XIV (A13979), trisome XIV (A18349), disome XV (A12697), trisome XV (A18350), disome XVI (A12700) and trisome XVI (A18351).
How many yeast proteins engage the cell’s protein quality control pathways and gene copy number feedback mechanisms on gene expression? Gene expression analysis in disomic yeast cells showed that the genes located on the additional chromosome are on average transcribed according to gene copy number. We observe an average 1.8 – 2.0 fold increase in gene expression of genes located on the duplicated chromosome (Torres et al. 2007). Thus, in yeast the number of genes that are regulated by feedback mechanisms that down-regulate gene expression in response to increased gene copy-number is likely to be low. A notable exception is the histone locus (Libuda and Winston 2006). In other organisms this form of feedback regulation may be more prominent. Many transcriptional feedback mechanisms have been described in plants, worms and flies (Straub and Becker 2007).
Whether aneuploid cells engage protein degradation pathways to attenuate protein stoichiometry imbalances caused by aneuploidy is an important and eminently answerable question. In a small-scale study of 16 proteins, we previously showed that proteins that function in large macromolecular complexes, i.e. ribosomal subunits, are not up-regulated according to gene copy number in disomic yeast strains (Torres et al. 2007). SILAC based proteomic analyses of disomic yeast strains confirmed these results and suggest that between 20 – 30 % of the yeast proteome is attenuated at a posttranscriptional level in disomic yeast strains (N. Dephoure and S. Gygi, pers. comm.). Identifying the pathways that bring about this attenuation and determining their importance in aneuploid cells is an important question that we must address.
Identifying the proteins which, when present in excess, are sequestered by chaperones will be more challenging. Proteins that require chaperones for their folding in wild-type cells, such as WD40-repeat proteins, protein kinases, membrane proteins and disulfide-containing proteins are likely among them (Bukau et al. 2006; Caplan et al. 2007). But whether cells use chaperone-mediated sequestration as a general mechanism to neutralize protein stoichiometry imbalances and whether such mechanisms are important for the survival of aneuploid cells remains to be determined.
Origins of the increased need for energy
The amount of biomass produced per glucose molecule was lower in disomic yeast cells than in wild-type cells (Torres et al. 2007). Trisomic mouse cells take up more glutamine (Williams et al. 2008), which represents the nitrogen and major carbon source of the TCA cycle in tissue culture cells (DeBerardinis et al. 2007; Williams et al. 2008). Yet, mouse cells produce less biomass suggesting that trisomic mouse cells also exhibit an increased need for energy. From these observations, it follows that aneuploid cells use energy for things other than producing biomass. Energy could be spent making the proteins from the additional chromosomes rather than the proteins that are needed to increase biomass and then neutralizing them by protein quality control mechanisms such as protein degradation. It is worth noting that protein degradation is a costly process. It has been estimated that the degradation of a single protein by the proteasome requires between 300–400 ATPs (Benaroudj et al. 2003).
Origins of the cell proliferation defect
All aneuploid yeast and primary mouse cells that we examined show cell proliferation defects. The basis for the decreased ability of trisomic mouse cells to accumulate in culture is not yet known but specific cell cycle defects have been identified in aneuploid yeast strains. Most of the aneuploid yeast strains that we analyzed exhibit a G1 delay (Torres et al. 2007). A smaller number of aneuploid cells have additional cell cycle defects.
Are the cell cycle delays observed in the aneuploid cells due to a response to the aneuploid state of the cell (i.e. elicited by the proteotoxic or energy stress) or due to the mis-regulation of individual cell cycle proteins or both? Heat shock, for example, causes cells to delay in G1 (Nover 1991). The proteotoxic stress or energy stress could also cause a slowing of the cell cycle in G1 as part of an aneuploidy response mechanism. Adapting to or compensating for various stresses may be more effective or lead to a higher survival rate when cells are in G1. It is also possible that the excess proteins produced in the aneuploid cells engage the protein quality control pathways of the cell thereby interfering with the timely degradation and/or folding of key cell cycle regulators. Finally, the two -fold increase of G1 regulators in disomic cells could interfere with cell proliferation especially the G1 – S phase transition. The number of such copy-number sensitive cell cycle regulators genes is however likely to be small. Only a few proteins are known that, when present in two copies, hamper cell proliferation. The best characterized example and only gene that causes lethality when present in two copies in the cell is the β-tubulin-encoding TUB2 gene (Schatz et al. 1988). Other genes are known to subtly impair proliferation when present in two copies, for example, the actin- encoding ACT1 gene or the protein phosphatase encoding CDC14 gene (Kaizu et al. ; Liu et al. 1992). It is also formally possible that excess proteins interfere with the production or regulation of essential cell cycle regulators. Why this would preferentially lead to a cell cycle delay in G1 is however not obvious. Clearly, it will be necessary to characterize the cell cycle defects of disomic yeast strains in depth as well as to design strategies to identify all genes that impair cell proliferation when present in two copies.
Relationship between aneuploidy and cancer
Theodor Boveri was the first to raise the possibility of a link between aneuploidy and tumor formation. Boveri speculated that some aneuploid cells could proliferate better than wild-type cells or in ways that wild-type cells would not. Our results indicate that aneuploidy causes a proliferative disadvantage in primary yeast and mouse cells. Is aneuploidy a byproduct of cancer or does it promote tumorigenesis despite its adverse effect on cell proliferation? To get at this question it is important to review the evidence for and against a tumor-promoting role of aneuploidy.
Evidence against a role for aneuploidy in inducing tumorigenesis
Individuals carrying an extra copy of Chromosome 21 have a 50% lower probability of developing solid tumors than individuals with the correct chromosome number (Hasle et al. 2000; Satge et al. 2003). Mice carrying segmental trisomies also exhibit a reduced incidence of intestinal neoplasia in the sensitized APCMin genetic background (Sussan et al. 2008). Furthermore, a mouse model in which low-level aneuploidy was induced by interfering with the chromosome segregation machinery prevented tumor formation in most tissues and caused tumor formation only very late in the others (Weaver et al. 2007). Perhaps the strongest argument against a role for aneuploidy in the selection for cancer cell fitness is the cytogenetic characterization of primary tumors. Many structural and numerical aberrations have been identified in solid tumors, yet relatively few are shared among specific types of solid tumors and even between cells within a tumor (reviewed in (Albertson et al. 2003)). Even fewer such aberrations have been shown to contribute to tumor formation (reviewed in (Albertson et al. 2003)).
If aneuploidy does not contribute to tumorigenesis, why are solid tumors aneuploid? Aneuploidy could be a consequence of loss of p53. Loss of function of the tumor suppressor gene p53 leads to aneuploidy in cultured cells. The basis for p53’s role in preventing karyotype destabilization is not fully understood but inactivation of p53 results in tetraploidization (Bunz et al. 2002), a state that might facilitate aneuploidy. In general, inactivation of p53 is thought to be a late event during tumorigenesis. Thus, aneuploidy could simply be a consequence of loss of p53 that occurs late during tumorigenesis and does not contribute to tumor development.
Evidence for a role for aneuploidy in promoting tumorigenesis
Several findings argue for aneuploidy being an early and causative event during tumorigenesis. The observation that low grade, small adenomas and pre-malignant atypical ductal hyperplastic cells show a low degree of aneuploidy (Bomme et al. 1998; Bomme et al. 2001; Shih et al. 2001; Larson et al. 2006) suggests that discrete chromosome gains play a causative role early during tumorigenesis. Furthermore, even though tumors form late in mice carrying mutations that induce chromosome instability, they do arise with a statistically significant increased frequency in some tissues (Michel et al. 2001; Iwanaga et al. 2007; Jeganathan et al. 2007; Sotillo et al. 2007; Weaver et al. 2007; Baker et al. 2009). These findings indicate that under specific circumstances, i.e. in specific tissues or during certain developmental stages an additional copy of specific chromosomes accelerates some aspects of tumorigenesis. For example, gaining an additional copy of an oncogene or losing a copy of a tumor suppressor gene could promote inappropriate cell proliferation in differentiated non-dividing tissues.
Is single chromosome aneuploidy a good model for aneuploidy in cancer?
The role of aneuploidy in tumorigenesis continues to be controversial. Single chromosomal gains rarely occur in cancer. Instead, severe karyotypical abnormalities, involving many chromosomes and often multiple copies of individual chromosomes are common. These observations beg the question whether the study of disomic and trisomic cells represents a good model for studying the role of aneuploidy in tumorigenesis. We believe that it is for the following reasons. First, unlike genome-instability inducing mutations, the karyotypes of the disomic and trisomic strains do not change and all cells harbor the same karyotype. Second, important features and traits of the aneuploid state can be deduced from the analysis of multiple single chromosomal abnormalities because phenotypes shared by cells carrying disomies or trisomies of different identity will speak to the aneuploid condition in general. Finally, it is important to note that in several cancers aneuploidy involving many numerical abnormalities is seen only late during tumorigenesis; premalignant lesions or low grade tumors show limited chromosomal gains or losses. For example, small adenomas and atypical ductal hyperplastic cells show a low degree of loss of heterozygosity (Bomme et al. 1998; Bomme et al. 2001; Shih et al. 2001; Larson et al. 2006). Low-grade aneuploidy may therefore exist among premalignant lesions and in early stages of tumorigenesis. Thus, we believe that our studies of single chromosomal abnormalities contribute to our understanding of the role of aneuploidy during tumorigenesis and complement the analyses of aneuploidy models that rely on chromosome mis-segregation inducing mutations to generate aneuploid cells (Baker et al. 2004; Sotillo et al. 2007; Weaver et al. 2007; Thompson and Compton 2008; Baker et al. 2009).
A model for how aneuploidy promotes tumorigenesis
While high-grade aneuploidy may be a late stage event in the evolution of tumors and precipitated by the loss of p53, it is quite clear that low-grade aneuploidies exist in premalignant lesions and early stages of the disease (Bomme et al. 1998; Bomme et al. 2001; Shih et al. 2001; Larson et al. 2006). Furthermore, several mouse studies have shown that aneuploidy-inducing mutations predispose animals for cancer (Sotillo et al. 2007; Weaver et al. 2007; Baker et al. 2009). How can this be, given that the aneuploid state per se impairs proliferation rather than promotes it?
We propose two, not mutually exclusive possible explanations for these observations. Aneuploidy could cause a proliferative advantage under conditions when normal euploid cells in the tissue do not divide (i.e. through loss of growth or G1 – S phase transition control). In such a situation even slowly proliferating aneuploid cells will have an advantage over euploid cells. A similar logic could apply to the ability of cells to adapt to new environments. Aneuploidy may allow cells to migrate to distant sites and/or colonize new niches. This “selective edge” is best summarized by the proverb: “In the land of the blind, the one-eyed man is king.” Even if aneuploid cells proliferate slowly, when their euploid neighbors cannot, they will eventually outcompete their euploid parents.
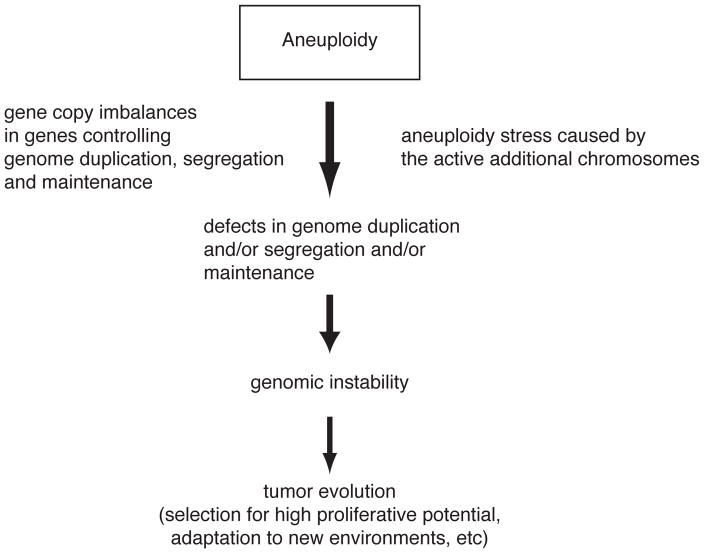
Aneuploidy could also promote tumorigenesis by promoting genomic instability, hence increasing evolvability of tumors (Torres et al. 2007; Torres et al. 2008). Aneuploidy could cause genomic instability by multiple mechanisms (Figure 3). Gene copy imbalances in factors essential for genome duplication and segregation as well as genome maintenance exist in aneuploid cells thereby promoting genome instability. It is also possible that the stresses associated with the aneuploid state, the increased burden on protein quality control mechanisms and metabolic alterations could lead to increased genomic instability by interfering with pathways controlling genome maintenance and propagation. Precedent for stress induced genomic instability exists in B. subtilis, E. coli, Candida albicans and mouse fibroblasts (Edlund and Normark 1981; Mihaylova et al. 2003; Sung et al. 2003; Ponder et al. 2005; Selmecki et al. 2006). Increased genomic instability in turn could promote the evolution of tumors with high proliferative potential and ability to adapt to different environmental conditions. Thus, we propose that in a rather counterintuitive manner, the adverse properties of aneuploidy are the reasons why aneuploidy promotes tumor growth and development. We are currently testing the hypothesis that aneuploidy promotes genomic instability in yeast.
Figure 3. A model for how aneuploidy promotes tumorigenesis.
See text for details.
What’s next?
Our analysis of primary aneuploid yeast cells and trisomic MEFs led to several hypotheses as to how the aneuploid state affects cellular physiology. Clearly, it will be necessary to continue to characterize the effects of aneuploidy on the cell in molecular detail. Perhaps most exciting is the prospect of identifying genetic alterations that enhance or suppress the adverse effects of the aneuploid state. Genes that when inactivated improve the proliferative ability of single or multiple different aneuploidies will not only provide us with an understanding of the defects underlying the aneuploid condition but may also shed light on the evolution of tumors. Genetic alterations that exhibit synthetic lethality with the aneuploid state either by exaggerating the adverse effects of aneuploidy and/or interfering with pathways essential for the survival of aneuploid cells could provide the basis for the discovery of new tumor treatments.
Acknowledgments
We are grateful to Frank Solomon for his critical reading of the manuscript and suggestions. Work in the Amon lab was supported by a grant from the National Institutes of Health grant GM56800. E.T. was supported by a Charles King Trust postdoctoral Fellowship. Y.C. Tang is the recipient of a Human Frontiers Postdoctoral Fellowship. A.A. is also an Investigator of the Howard Hughes Medical Institute.
References
- Agrawal MG, Bowman LH. Transcriptional and translational regulation of ribosomal protein formation during mouse myoblast differentiation. J Biol Chem. 1987;262:4868–4875. [PubMed] [Google Scholar]
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- Bachurski CJ, Theodorakis NG, Coulson RM, Cleveland DW. An amino-terminal tetrapeptide specifies cotranslational degradation of beta-tubulin but not alpha-tubulin mRNAs. Mol Cell Biol. 1994;14:4076–4086. doi: 10.1128/mcb.14.6.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Bomme L, Bardi G, Pandis N, Fenger C, Kronborg O, Heim S. Cytogenetic analysis of colorectal adenomas: karyotypic comparisons of synchronous tumors. Cancer Genet Cytogenet. 1998;106:66–71. doi: 10.1016/s0165-4608(98)00047-8. [DOI] [PubMed] [Google Scholar]
- Bomme L, Lothe RA, Bardi G, Fenger C, Kronborg O, Heim S. Assessments of clonal composition of colorectal adenomas by FISH analysis of chromosomes 1, 7, 13 and 20. Int J Cancer. 2001;92:816–823. doi: 10.1002/ijc.1275. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Bunz F, Fauth C, Speicher MR, Dutriaux A, Sedivy JM, Kinzler KW, Vogelstein B, Lengauer C. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 2002;62:1129–1133. [PubMed] [Google Scholar]
- Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D, Rimbault B, Vallon O, Wollman FA, Choquet Y. Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J. 2007;26:3581–3591. doi: 10.1038/sj.emboj.7601802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund T, Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981;292:269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- elBaradi TT, van der Sande CA, Mager WH, Raue HA, Planta RJ. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr Genet. 1986;10:733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]
- Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. WormBook. 2005. Karyotype, ploidy, and gene dosage; pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizu K, Moriya H, Kitano H. Fragilities caused by dosage imbalance in regulation of the budding yeast cell cycle. PLoS Genet. 6:e1000919. doi: 10.1371/journal.pgen.1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson PS, de las Morenas A, Cerda SR, Bennett SR, Cupples LA, Rosenberg CL. Quantitative analysis of allele imbalance supports atypical ductal hyperplasia lesions as direct breast cancer precursors. J Pathol. 2006;209:307–316. doi: 10.1002/path.1973. [DOI] [PubMed] [Google Scholar]
- Libuda DE, Winston F. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature. 2006;443:1003–1007. doi: 10.1038/nature05205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HY, Lin SP, Chen YJ, Hung HY, Kao HA, Hsu CH, Chen MR, Chang JH, Ho CS, Huang FY, Shyur SD, Lin DS, Lee HC. Clinical characteristics and survival of trisomy 18 in a medical center in Taipei, 1988–2004. Am J Med Genet A. 2006;140:945–951. doi: 10.1002/ajmg.a.31173. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Sandler L, Baker BS, Carpenter AT, Denell RE, Hall JC, Jacobs PA, Miklos GL, Davis BK, Gethmann RC, Hardy RW, Steven AH, Miller M, Nozawa H, Parry DM, Gould-Somero M, Gould-Somero M. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicas E, Pluthero FG, Friesen JD. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol Cell Biol. 1988;8:169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman P, Fryns JP, van der Steen K, Kleczkowska A, Lauweryns J. The pathology of trisomy 13 syndrome. A study of 12 cases. Hum Genet. 1988;80:349–356. doi: 10.1007/BF00273650. [DOI] [PubMed] [Google Scholar]
- Niwa O, Tange Y, Kurabayashi A. Growth arrest and chromosome instability in aneuploid yeast. Yeast. 2006;23:937–950. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- Nover L. Heat shock response. CRC Press; Boca Raton: 1991. [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Satge D, Sasco AJ, Lacour B. Are solid tumours different in children with Down's syndrome? Int J Cancer. 2003;106:297–298. doi: 10.1002/ijc.11212. [DOI] [PubMed] [Google Scholar]
- Schatz PJ, Solomon F, Botstein D. Isolation and characterization of conditional-lethal mutations in the TUB1 alpha-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics. 1988;120:681–695. doi: 10.1093/genetics/120.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, McCoy EE. Studies on Down's syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. J Cell Physiol. 1974;83:85–90. doi: 10.1002/jcp.1040830112. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih IM, Wang TL, Traverso G, Romans K, Hamilton SR, Ben-Sasson S, Kinzler KW, Vogelstein B. Top-down morphogenesis of colorectal tumors. Proc Natl Acad Sci U S A. 2001;98:2640–2645. doi: 10.1073/pnas.051629398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- Sung HM, Yeamans G, Ross CA, Yasbin RE. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J Bacteriol. 2003;185:2153–2160. doi: 10.1128/JB.185.7.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- Theodorakis NG, Cleveland DW. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol. 1992;12:791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Williams BR, Amon A. Aneuploidy: cancer's fatal flaw? Cancer Res. 2009;69:5289–5291. doi: 10.1158/0008-5472.CAN-09-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]