Abstract
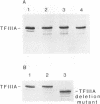
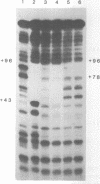
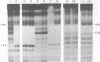
Zinc binding domains and the conserved Thr-Gly-Glu-Lys (TGEK) tetrapeptide in the N-terminal half of transcription factor IIIA (TFIIIA) were subjected to in vitro mutagenesis to biochemically assess their role in factor interaction with the 5S gene internal control region (ICR). TFIIIA containing a Leu in place of His33 in the Cys2His2 zinc binding site of finger I lost the ability to protect the entire 5S RNA gene ICR (nucleotides +96 to +43) from DNase I digestion. Thus, mutation of one potential zinc ligand in the N-terminal finger inhibited specific DNA binding by the N-terminal as well as downstream fingers. Cooperativity apparently exists among TFIIIA zinc fingers in metal binding/finger folding and DNA binding. Substituting a Ser for Gly69 or a Glu for Lys 71 in the conserved TGEK tetrapeptide in finger II of TFIIIA resulted in the loss of DNA binding. A Gly-dependent bend structure and a terminal positive charge in this tetrapeptide are important for TFIIIA interaction with DNA. Whereas TFIIIA with a Ser substituted for Cys20 in finger I (proposed zinc ligand) did not protect the ICR from DNase I digestion, TFIIIA containing a Ser substituted for Cys35 (not a proposed zinc ligand) retained the ability to bind the ICR. When Cys112 or Cys 164 (proposed zinc ligands in fingers IV and VI) were replaced by Ser, the DNase I footprint patterns afforded by the respective mutant proteins were similar, protection on the ICR from about nucleotides +96 up to +78. A similar pattern was obtained with a TFIIIA mutant in which fingers V, VI, VII, and a portion of VIII were deleted. Maintenance of zinc coordination spheres in necessary for DNA binding by downstream fingers. The six fingers comprising the N-terminal half of TFIIIA appear to act in two groups of three with binding of the second group dependent upon initial binding of the N-terminal group to the +90 to +80 region of the 5S gene ICR.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A. 1988 Jan;85(1):99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985 Jul 8;186(2):271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Christy B. A., Lau L. F., Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with "zinc finger" sequences. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J. L., Lipscomb W. N., Schellman C. G. The reverse turn as a polypeptide conformation in globular proteins. Proc Natl Acad Sci U S A. 1973 Feb;70(2):538–542. doi: 10.1073/pnas.70.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fiser-Littell R. M., Duke A. L., Yanchick J. S., Hanas J. S. Deletion of the N-terminal region of Xenopus transcription factor IIIA inhibits specific binding to the 5 S RNA gene. J Biol Chem. 1988 Feb 5;263(4):1607–1610. [PubMed] [Google Scholar]
- Gaskins C. J., Fiser-Littell R. M., Duke A. L., Hanas J. S. Species variation in transcription factor IIIA. Nucleic Acids Res. 1989 Jan 25;17(2):781–794. doi: 10.1093/nar/17.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins C. J., Hanas J. S. Sequence variation in transcription factor IIIA. Nucleic Acids Res. 1990 Apr 25;18(8):2117–2123. doi: 10.1093/nar/18.8.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. Cooperative model for the binding of Xenopus transcription factor A to the 5S RNA gene. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2142–2145. doi: 10.1073/pnas.80.8.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas J. S., Duke A. L., Gaskins C. J. Conformation states of Xenopus transcription factor IIIA. Biochemistry. 1989 May 2;28(9):4083–4088. doi: 10.1021/bi00435a068. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Hazuda D. J., Bogenhagen D. F., Wu F. Y., Wu C. W. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J Biol Chem. 1983 Dec 10;258(23):14120–14125. [PubMed] [Google Scholar]
- Hanas J. S., Littell R. M., Gaskins C. J., Zebrowski R. Internal deletion mutants of Xenopus transcription factor IIIA. Nucleic Acids Res. 1989 Dec 11;17(23):9861–9870. doi: 10.1093/nar/17.23.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kuwahara J., Coleman J. E. Role of the zinc(II) ions in the structure of the three-finger DNA binding domain of the Sp1 transcription factor. Biochemistry. 1990 Sep 18;29(37):8627–8631. doi: 10.1021/bi00489a019. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Russel M., Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984 Sep;159(3):1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Eckstein F. 5'-3' exonucleases in phosphorothioate-based oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Feb 11;16(3):791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana K. E., Churchill M. E., Tullius T. D., Brown D. D. Mapping functional regions of transcription factor TFIIIA. Mol Cell Biol. 1988 Apr;8(4):1684–1696. doi: 10.1128/mcb.8.4.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]