Abstract
Fine epitope mapping of EF13D, ahighly potent neutralizing monoclonal antibody specific for the anthrax edema factor (EF), was accomplished through random mutagenesis and yeast surface display. A yeast-displayed library of single point mutants of an EF domain III (DIII), comprising amino acids624-800, was constructed by random mutagenesis and screened for reduced binding to EF13D. With this method, residues Leu 667, Ser 668, Arg 671, and Arg 672 were identified as key residues important for EF13D binding. They form a contiguous patch on a solvent-exposed surface at one end of the four-helixbundle of DIII. Computational protein-protein docking experiments between an EF13D model and a crystal structure of EF indicate that the EF13D heavy chain complementarity-determining region 3 (HCDR3) is deeply buried within a hydrophobic cleft between two helices of DIII and interacts directly with residues Leu 667, Ser 668, Arg 671 and Arg 672, providing an explanation for the high binding affinity. In addition, they show that the HCDR3 binding site overlaps with the binding site of the N-terminal lobe of calmodulin (CaM), an EF enzymatic activator, consistent with a previous finding showing direct competition with CaM that results in neutralization of EF. Identifying the neutralization epitope of EF13D on EF improves our understanding of the neutralization mechanism and has implications for vaccine development.
Keywords: epitope mapping, yeast surface display, neutralization mechanism, antibody modeling, protein-protein docking
Introduction
Edema toxin is one of the two toxins produced by Bacillus anthracis and is composed of edema factor (EF) and protective antigen (PA). While PA functions as a vehicle to mediate translocation of EF into host cells, EF is an adenylatecyclase[1]. Upon translocation into the cells, EF is activated by association with a calcium sensor, calmodulin (CaM) and becomes a highly active adenylatecyclase that raises the intracellular concentration of cyclic AMP (cAMP). The high concentration of cAMP causes sustained and potent activation of host cAMP-dependent signaling pathways, which results in local inflammation, edema and other toxic effects [1,2]. Given its important role in pathogenesis of B. anthracis, EF has been a target for development of anti-anthrax drugs. Several small molecules that can inhibit EF activity have been reported [3,4,5,6,7]. Although they inhibit EF by different mechanisms, the small molecules found so far inhibit the activity of EF in the low micromolar range. In contrast, we have recently reported the isolation of a chimpanzee/human monoclonal antibody (mAb), EF13D that can neutralize EF In vitro in the subnanomolar range [8]. The therapeutic usefulness of the antibody was demonstrated by protection experiments in mice. In addition to binding to a conformational epitope on EF with very high affinity, EF13D also inhibits CaM-mediated activation of EF not only by out competing CaM for binding to EF but also by replacing the pre-bound CaM in the EF/CaM complex. This study attempts to shed light on this neutralization mechanism by fine mapping the epitope of EF13D.
Epitope mapping is the determination of amino acid residues responsible for mediating antibody-antigen interactions. Epitopes can be divided into two categories: continuous, linear epitopes and discontinuous, conformational epitopes. It is relatively easy to map linear epitopes by using synthetic overlapping peptide-based methods. In contrast, it is difficult to map discontinuous, conformational epitopes. Recently, yeast surface display has been used to facilitate mapping conformational epitopes [9,10,11]. Yeast surface display is a method whereby a protein of interest is expressed on the surface of yeast as a fusion with the yeast Aga2 protein[12]. By combination of random mutagenesis, selection of loss-of-binding mutants and localization of the residues critical for antibody-binding based on X-ray crystal structure, the critical contact residues of an epitope can be identified.
EF protein consists of 800 amino acids and is structurally organized into three domains [13]. Domain I is located in the N-terminal portion of EF and is responsible for binding to PA. Domain II lies between domain I and domain III and can be divided structurally into the CA and CB domains. The catalytic site is formed at the interface of CA and CB. Domain III (DIII), known as the “helical domain,” is located in the C-terminal portion of EF. Since mAb EF13D was found to bind to the helical domain of EF, this part of EF was displayed on yeast for epitope mapping. The analysis of the library composed of EF DIII mutants resulted in identification of residues critical for antibody binding.
Materials and Methods
Strains, media, vectors and antibodies
The yeast (Saccharomyces cerevisiae) strain EBY100 and pCTCON2 yeast display vector were gifts from Dr. K. D. Wittrup (MIT, Cambridge, MA). Culture medium SDCAA (0.67% Difco yeast nitrogen base, 0.5% Bactocasamino acids, 0.54% Na2HPO4, 0.856% NaH2PO4.H2 and 2% dextrose) was used to select yeast that contained plasmid and SGCAA (same as SDCAA except for 2% galactose instead of 2% dextrose) was used to induce expression as described [14]. Expression in yeast was monitored using anti-c-myc chicken IgY and Alexa Fluor 488 goat anti-chicken IgG (Invitrogen, Carlsbad, CA). EF neutralizing mAb EF13D IgG[8] was directly labeled with Atto633 with a Lightning-Link Atto633 conjugate kit from Innova Biosciences (Cambridge, UK). Bacterial expression vector pET31b was purchased from Novagen (La Jolla, CA) and bacterial strain BL21(DE3)pLysS was purchased from Promega (Madison, WI).
Cloning, expression and detection of DIII
The gene fragment encoding DIII (aa 624–800) of EF was amplified by PCR cloned into pCTCON2 at NheI and SalI sites, resulting in yeast expression vector DIII-pCTCON2.
Yeast EBY100 was transformed with DIII-pCTCON2 and transformants were selected on SDCAA agar plates. The yeast was first cultured in SDCAA with shaking at 30°C overnight and expression was induced by culturing in SGCAA at 22°C for 48 h with shaking. For measuring DIII expression and EF13D mAb binding by flow cytometry, cells were harvested, washed and incubated with anti-c-myc antibody followed by Alexa Fluor488 goat anti-chicken IgG and Atto633-labled EF13D IgG as described below.
Construction and expression of epitope library
A library of DIII mutants was generated by random mutagenesis using a GeneMorph II random mutagenesis kit from Agilent Technologies (Santa Clara, CA) as described [14] under conditions for low error rate. The PCR products were gel-purified and extracted using a gel extraction kit from Qiagen (Valencia, CA). The purified PCR products were mixed with pCTCON2 plasmid linearized by NheI and SalI digestion. The mixture was transformed into yeast EBY100 by electroporation and homologous recombination [14]. The transformation mixture was plated on an SDCAA agar plate and incubated at 30°C for 48 h. The size of the yeast library was calculated from the number of colonies plated with serial dilutions of the transformation mixture. The estimated error rate of the mutagenic library was determined by plasmid recovery using a Zymoprep Yeast Plasmid Miniprep I Kit (Zymo Research) and sequencing of 20 unsorted library clones. Growth and expression of the library using yeast display was performed as described [14].
Flow cytometric sorting of the yeast-display library
To maintain the library diversity, a number of yeast cells at least ten times the library size (2 × 106) were washed with FACS buffer (phosphate buffered saline containing 1 mg/ml of BSA, 2 mM EDTA). The cells were incubated with 4 µg/ml anti-c-myc chicken IgY and 1 µg/ml of Atto633-labled EF13D IgG for 30 min at room temperature. The cells were then washed with FACS buffer and incubated with 8 µg/ml of Alexa Fluor 488 goat anti-chicken IgG for 15 min on ice. The labeled cells were washed and sorted on a BD FACSAria II cell sorter (BD Biosciences) in the NIAID Flow Cytometry Core Facility. Gates were set so that the sorted yeast were positive for Alexa Fluor 488, indicating that they expressed DIII, but negative for Atto633, indicating that they did not bind to EF13D. The cells were grown, labeled and sorted as before for one more round to enrich for DIII-expressing clones that did not bind to EF13D IgG.
Identification and testing of single clones
Following the second round of selection, the clones were pooled and the plasmid DNA was recovered using the Zymoprep kit. The plasmids were then transformed into E. coli JM109 cells. Ninety-six individual clones were picked and grown in a 96-well plate, and plasmids were prepared and sequenced.
For clones with a single amino acid mutation, plasmid DNA was transformed into yeast and the loss-of-binding phenotype was verified by BD FACS Canto II flow cytometry analysis. For clones with 2–3 amino acid changes, single independent mutations were generated by site-directed mutagenesis of DIII-pCTCON2 using mutant oligonucleotides and the Quick Change II Mutagenesis Kit (Agilent Technologies). The same method was used to generate single-alanine mutants. All mutants were confirmed by sequencing and analyzed for EF13D binding on BD FACS Canto II flow cytometer.
Cloning and expression of full-length EF and EF mutants in E. coli
To determine if a single mutation identified by yeast surface display for loss-of-binding phenotype on DIII has the same effect on full-length EF, the same single mutation was introduced into the EF-encoding gene in PET31d [8]. The resulting EF mutants as well as wild type EF were expressed in E. coli and purified on a HisTrap FF nickel column. The purity was assessed by SDS-PAGE and the quantity was measured by spectrophotometry and by ELISA for binding intensity to a non-neutralizing anti-EF mAb, EF12A [8] using EF of known concentration as a control.
ELISA
ELISA was performed as described previously [15], with recombinant EF or EF mutants as coating antigens and EF13D, EF12A and EF14H [8] as detecting antibodies.
In vitro cyclic AMP production assay
The enzymatic activity of EF and EF mutants was measured for cyclic AMP (cAMP) production from ATP using the Monoclonal Anti-cAMP EIA Kit from NewEast Biosciences (Malvern, PA) according to the manufacturer’s instruction.
Anti-EF antibody modeling
The sequence for EF13D was uploaded to the RosettaAntibody modeling server [16] and the full protocol with VL-VH refinement was used for modeling. The top ranked solution was taken as input into RosettaDock. For details, please see supplemental methods.
Protein-protein docking
The anti-EF antibody model and DIII taken from the crystal structure (PDB accession code 1K8T) were docked using RosettaDock (v.2.3) [17] on the NIH Biowulf Linux cluster (http://biowulf.nih.gov). For details, please see supplemental methods.
Molecular dynamics
Protein structures were explicitly solvated with TIP3P water molecules and Na+ and Cl-counterions using VMD [18]. Molecular dynamics simulations were performed under isobaric-isothermal conditions using NAMD [19] (v.2.7) on the NIHBiowulf Linux cluster (http://biowulf.nih.gov) and included periodic boundary conditions. Electrostatic interactions were handled using a Particle-Mesh Ewald summation. The CHARMM27 [20] force field was used with CHARMM atom types and charges.
Results
Construction and expression of epitope library
The DIII of EF was expressed as a fusion protein on the yeast cell surface. The expression of DIII and binding to the mAb were monitored by flow cytometry for the intensity of Alexa Fluor 488 that is directed to the c-myc tag at the c-terminus of DIII (display fluorescence) and Atto633 that is coupled to the mAb (binding fluorescence) (Figure S1a). Detection of the dual fluorescence by flow cytometry revealed that DIII was expressed on the yeast surface and was reactive with EF13D (Figure. S1b)
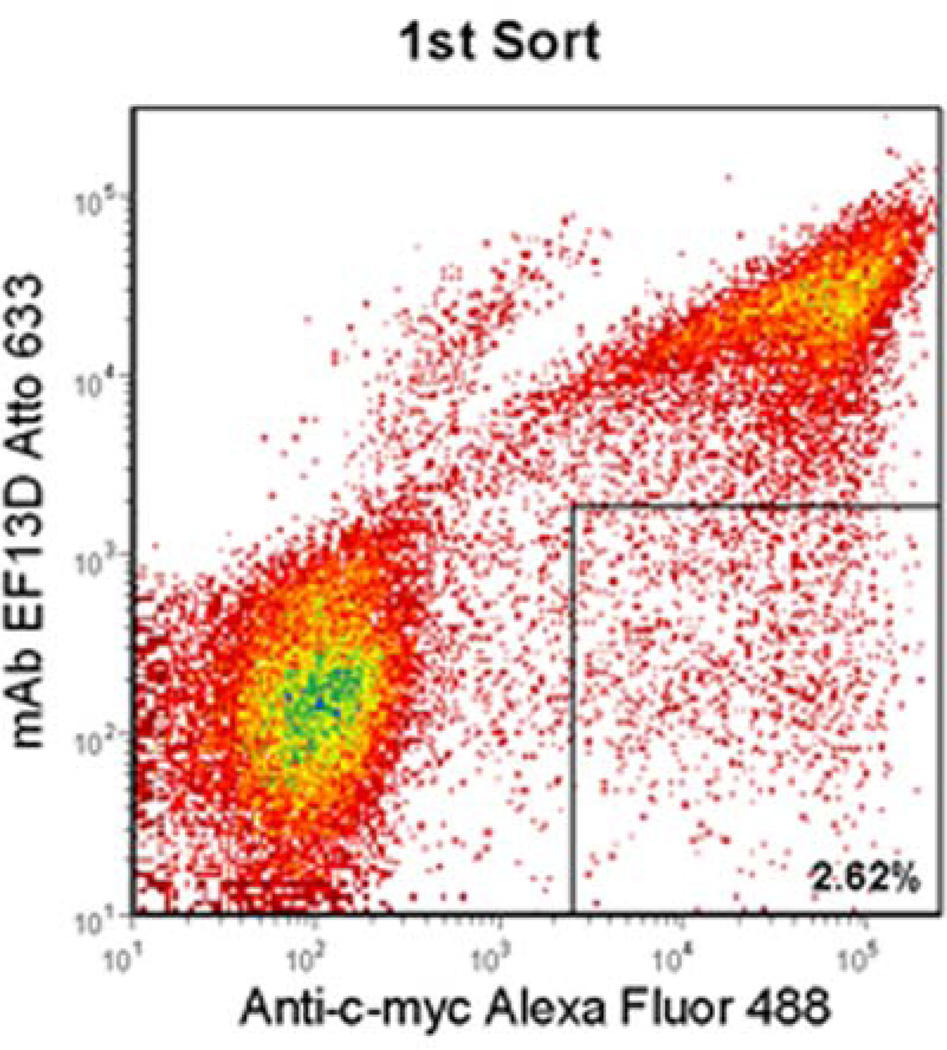
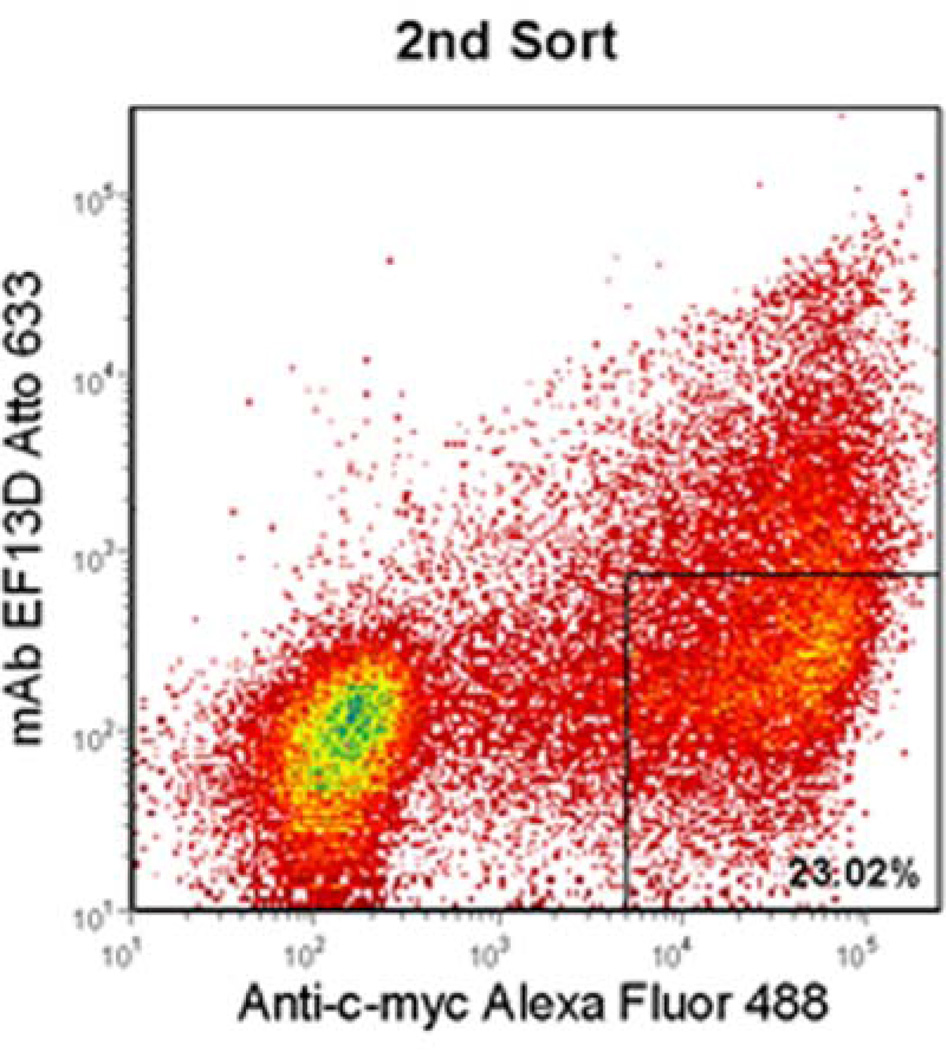
A library of DIII mutants was constructed by error-prone PCR under conditions resulting in a low mutation rate. The initial library size was 3 × 104, and sequencing of 20 clones from the unsorted library indicated that 21% were wild-type DIII, 32% contained single amino acid mutations, 32% contained two or three amino acid mutations, and 15% contained more than three mutations. This corresponds to a size of 9.6 × 103 for the library containing single amino acid mutations, which is 3-fold higher than the largest theoretical diversity of single amino acid mutations for this 176-residue DIII (3.3 × 103). In principle, every mutant of each amino acid is well-represented in the library. Mutants that resulted in a loss of binding to mAb presumably contain a change of residues critical for binding to the mAb. DIII mutants that were displayed on the surface of yeast, but exhibited a loss of binding to mAb were isolated for analysis. Two successive rounds of flow cytometric sorting were performed based on positive display fluorescence and negative mAb binding fluorescence; the desired mutants were enriched by 2.6% after 1st sort and by 23% after 2nd sort (Figure 1a and b).
Figure 1.
Flow cytometric enrichment for DIII-expressing yeast variants that did not bind to EF13D. Yeast cells that expressed DIII, but lost binding to the antibody (boxed region) were sorted by two successive rounds (a and b). After each round, an increased percentage of DIII-expressing yeast did not recognize EF13D. After the final round, the desired variants were harvested for further analysis.
Identification and testing of single clones for loss of mAb binding
Individual mutant clones recovered after the second sort were sequenced. Of 96 clones sequenced, 6 distinct single mutant clones, 16 distinct double mutant clones and 3 distinct triple mutant clones were found. To establish that all binding phenotypes could be unambiguously attributed to a single mutation, all the double and triple mutants were converted to single mutants through site-directed mutagenesis. This resulted in 41 distinct single mutants. The analysis of binding phenotypes of these mutants by flow cytometry identified 12 loss-of-binding clones and 9 partial loss-of-binding clones (Table 1). A large number of point mutations leading to loss of binding by EF13D implies that the overall conformation of the EF DIII is very sensitive to perturbation. This is in agreement with the previous finding that EF13D recognized a conformational epitope on DIII of EF [8].
Table 1.
MAb EF13D binding mutations identified by random or site-directed mutagenesis
| EF DIII MUTANT | BINDING | PARTIAL BINDING | LOSS OF BINDING |
|---|---|---|---|
| E662V | x | ||
| L667F | x | ||
| L667S | x | ||
| S668P | x | ||
| S669C | x | ||
| I670F | x | ||
| R671I | x | ||
| R672G | x | ||
| V676A | x | ||
| V678D | x | ||
| K680N | x | ||
| K690I | x | ||
| V694A | x | ||
| V694G | x | ||
| S693R | x | ||
| K695E | x | ||
| K696T | x | ||
| I697N | x | ||
| L701S | x | ||
| Y704C | x | ||
| Y705C | x | ||
| N709D | x | ||
| H710R | x | ||
| F712L | x | ||
| Q714R | x | ||
| S721P | x | ||
| Q727L | x | ||
| A728D | x | ||
| I732T | x | ||
| L736P | x | ||
| K737N | x | ||
| Y748N | x | ||
| Y748C | x | ||
| L762S | x | ||
| L763P | x | ||
| I771T | x | ||
| Q779L | x | ||
| N785K | x | ||
| V792D | x | ||
| I797T | x | ||
| E799G | x |
Locating the mAb epitope on EF and mechanism of neutralization
Molecular modeling of the locations of mutations on the X-ray crystal structure of EF [13] enabled the selection of individual amino acid residues that could potentially form a functional epitope for EF13D. Location of loss-of-binding residues (in red) and partial loss-of-binding residues (in yellow) on DIII are shown in Figure 2a. To determine which DIII solvent-accessible residues are likely to participate in the formation of a functional epitope based on this data, a crystal structure of EF (PDB accession code 1K8T) was read into VMD and a Connolly surface calculated using a probe radius of 1.4Å. DIII residues were chosen visually based on their contact with the Connolly surface. Mutations at buried DIII positions that likely participate in maintaining the core of the 4 helix bundle, may be disruptive, leading either to a conformational change or misfolding and subsequent loss of mAb EF13D binding. Therefore, non-surface residues were eliminated from the analysis, and the location of the remaining mutations is shown in Figure 2b. The loss of binding residues Leu 667, Ser 668, Arg 671 and Arg 672; as well as the partial loss of binding residues Val 678 and Val 694; are located on adjacent helices [13] and form a contiguous patch on a solvent-exposed surface that interfaces with the DII of EF. These residues most likely form the functional epitope for mAb EF13D.
Figure 2.
Mapping of EF-neutralizing mAb EF13D on the surface of DIII. (a) Ribbon cartoon of the X-ray crystal structure of DIII (PDB accession code 1K8T) showing all the mutated amino acid residues that led to partial loss (yellow) or complete loss (red) of binding by EF13D. (b) A molecular surface representation, based on the X-ray crystal structure of DIII (PDB accession code 1K8T). The indicated amino acid residues associated with abolishing and reducing binding by mAb EF13D are shown in red and yellow, respectively. These residues are predicted to form the epitope for mAb EF13D.
Computer modeling including antibody modeling, protein-protein docking and molecular dynamics was used to better understand the interactions between antibody, EF and CaM. Modeling reveals that it is the antibody heavy chain that interacts with the proposed epitope on DIII with HCDR3 deeply buried within a hydrophobic cleft between two helices, directly interacting with Leu 667, Ser 668, Arg 671, Arg 672, Val 678, Val 694, K691 and K695 through salt-bridges, hydrogen-bonds and hydrophobic interactions (Figure 3a). The model agrees well with the first six residues which were found through mutagenesis and identified as crucial for antibody binding.. Two additional contact residues K691 and K695 were predicted only by the model and when mutated, were shown to have a very small effect on binding. Examination of EF/CaM crystal structure (PDB accession code 1K93 and 1XFV)[13,21] indicated that residues Ser 668, Arg 671 and Val 694 identified as forming the EF13D epitope on DIII are within 3Å and capable of contacting CaM, suggesting that the HCDR3 competes with the N-terminal lobe of CaM for binding to EF.
Figure 3.
Interaction between EF13D and DIII and the relative positions of mAb EF13D and CaM on EF. (a) Aspredicted by a low energy RosettaDock solution, all potential contact residues between the heavy chain complementarity-determining region 3 loop of EF13D (green) and DIII (light gray) are shown. Basic, acidic, hydrophobic and hydrophilic residues are shown in dark blue, red, dark gray and pink, respectively. Salt-bridges and hydrogen-bonds are indicated as dashed lines. Amino acid residues derived from mAb EF13D are labeled in green whereas those from DIII are labeled in light gray. (b) Amino acid sequences of variable domains of heavy (I) and light (II) chains of EF13D. Complementarity-determining regions (CDR1-3) and framework regions (FWR1-4) are assigned according to Kabat nomenclature.
Correlation between the enzymatic activity of full-length EF and its binding to EF13D
To confirm if the finding from the yeast-displayed DIII is truly representative of the full-length EF, we performed site-directed mutagenesis on four key positions for EF13D binding, Leu 667, Ser 668, Arg 671 or Arg 672, resulting in the single mutations EFL667F, EFS668P, EFR671I and EFR672G, respectively. A single mutation at any one of the four residues abolished binding to the mAb whereas EFwild reacted well with EF13D (Figure. 4). The two unique non-neutralizing EF-binding mAbs, EF12A and EF14H, reacted equally well with wild-type EF and mutant EFs (data not shown), suggesting that the binding to EF, but not to EF mutants by mAb EF13D was the result of the specific mutation on the epitope.
Figure 4.
The binding of EF13D at 1 µg/ml to solid phase-immobilized wild type EF, and to mutants EFL667F, EFS668P, EFR671I and EFR672G at indicated concentrations. The assay was performed in triplicate.
To understand the impact of these loss-of-binding mutations on enzymatic activity, the enzymatic activity of EFwild as well as of EF mutants was measured for their cAMP production from ATP using a cAMP ELISA. Mutations at any of the four residues resulted in reduced enzymatic activity, but the degree of reduction was not uniform: mutants EFL667F, EFS668P, EFR671I and EFR672G had 0.1–0.8%, 4–8%, 5–8% and 25–40% of activity of EFwild, respectively.
Discussion
Analysis of results obtained by yeast surface display led us to identify residues Leu 667, Ser 668, Arg 671, Arg 672, Val 678 and Val 694 as critical residues for binding of EF13D to EF, thus providing insight into the molecular basis for the antibody’s potent neutralization activity, high affinity binding and extraordinary ability to replace CaM bound to EF.
There is a concern that the loss of binding associated with some of the mutants, such as R672G and S667P, may result from main-chain conformation changes since Gly and Pro can cause changes in helix conformation. To address this concern, an alanine-scanning mutagenesis approach [22] was used. One single-alanine mutant was generated for each of residues Leu 667, Ser 668, Arg 671 and Arg 672 and analyzed for their binding to EF13D in yeast display. Both S668A and R672A mutants displayed complete loss of binding whereas L667A and R671A had reduced binding to EF13D. The small change in binding affinity can be explained for the substitution of Ala for Leu at position 667 as some hydrophobic interactions with W104 of the antibody are maintained (Fig. 3a). To understand why R672A, but not R672I mutant can bind with EF13D with relatively lower affinity, we performed rigorous molecular dynamics simulations and found a plausible explanation. For R671A, it was observed that W104 is able to position itself deeper into the EF DIII hydrophobic cleft than wildtype EF, thereby reducing solvent accessibility and increasing van der Waals contacts (Fig. 3a). However, this decrease in solvent accessibility does not fully compensate for the absence of salt-bridging between R671 and the side-chain and backbone atoms of DIII residue D110 observed in simulations of wildtype protein. More similar to wildtype DIII, but again missing the R671 salt-bridge contact, R671I retains a bulky, aliphatic group at position 671, preventing W104 from burying itself as completely as seen in R671A (Fig S2). Therefore, molecular dynamics simulations suggest that the reduction in EF13D binding affinity for both R671A and R671I compared to wildtype can be explained by the absence of R671 being available to form a salt bridge with D110 and the further reduction of binding affinity for R671I is compounded by the relative lack of W104 burial.
Previous crystallographic analysis showed that residues Leu 667, Ser 668, Arg 671, Arg 672 and Val 694 are involved in binding of CaM to EF [13] (insert citation for Shen et al, EMBO 2005). A NMR study indicated that the binding of CaM to EF was a sequential process starting with the initial binding of the N-terminal domain of CaM to the EF helical domain [23]. Once the N-terminal CaM is anchored to the helical domain, C-terminal CaM is allowed to insert between the catalytic core and helical domains of EF. The binding of CaM leads to a conformational change of the switch C region to stabilize the critical catalytic loop of EF for a high rate of enzymatic activity. Therefore, antibody binding at the residues Leu 667, Ser 668, Arg 671 and Arg 672 disrupts the initial binding of the N-terminal domain of CaM to the EF helical domain and thus inhibits subsequent C-terminal CaM insertion and activation of EF.
In conclusion, the binding epitope of EF-neutralizing mAb EF13D was identified by using random mutagenesis and yeast surface display, and substantiated by computational modeling. The identification of the epitope may improve our understanding of EF13D mechanisms of action and may have important implications for immunoprophylaxis.
Highlights.
We determined the epitope for EF13D binding of EF and modeled the interaction
The proposed binding site explains the high affinity of EF13D for EF
Neutralization of EF occurs by direct competition between CaM and EF13D
DIII of EF is necessary and sufficient to induce therapeutic antibodies
This study has an important implications for future vaccine design
Supplementary Material
Figure S1: Mapping epitope of EF13D to EF domain III by yeast surface display. (a) A schematic showing surface display of EF domain III on yeast. A fusion protein consisting of EF domain III and yeast Aga2 protein is attached to the Aga1 protein through disulfide bonds on the yeast cell wall. The level of expression of DIII and the binding by the mAb were monitored by fluorescence flow cytometry (Alexa Fluor 488 directed to the c-myc tag at the C-terminus of DIII and the Atto633 attached to the mAb). (b) Fifty four percent of yeast cells displayed properly folded DIII that was reactive with EF13D, as judged by dual fluorescence.
Figure S2: Molecular dynamics simulations of interaction between EF13D and wild type EF, EFR671A or EFR671I. It shows that the reduction in EF13D binding affinity for both R671A and R671I compared to wild-type can be explained by the absence of R671 being available to form a salt bridge with D110 and the further reduction of binding affinity for R671I is compounded by the relative lack of W104 burial.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, NIAID. We thank Dr. Stephen Leppla for providing the recombinant EF and plasmid containing EF-coding gene and Dr. Dane Wittrup for providing yeast strain EBY100 and yeast display vector pCTCON2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Firoved AM, Miller GF, Moayeri M, Kakkar R, Shen Y, Wiggins JF, McNally EM, Tang WJ, Leppla SH. Bacillus anthracis edema toxin causes extensive tissue lesions and rapid lethality in mice. Am J Pathol. 2005;167:1309–1320. doi: 10.1016/S0002-9440(10)61218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Misra M, Sower L, Peterson JW, Kellogg GE, Schein CH. Novel inhibitors of anthrax edema factor. Bioorg Med Chem. 2008;16:7225–7233. doi: 10.1016/j.bmc.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laine E, Goncalves C, Karst JC, Lesnard A, Rault S, Tang WJ, Malliavin TE, Ladant D, Blondel A. Use of allostery to identify inhibitors of calmodulin-induced activation of Bacillus anthracis edema factor. Proc Natl Acad Sci U S A. 2010;107:11277–11282. doi: 10.1073/pnas.0914611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YS, Bergson P, He WS, Mrksich M, Tang WJ. Discovery of a small molecule that inhibits the interaction of anthrax edema factor with its cellular activator, calmodulin. Chem Biol. 2004;11:1139–1146. doi: 10.1016/j.chembiol.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Shen Y, Zhukovskaya NL, Zimmer MI, Soelaiman S, Bergson P, Wang CR, Gibbs CS, Tang WJ. Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection. Proc Natl Acad Sci U S A. 2004;101:3242–3247. doi: 10.1073/pnas.0306552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soelaiman S, Wei BQ, Bergson P, Lee YS, Shen Y, Mrksich M, Shoichet BK, Tang WJ. Structure-based inhibitor discovery against adenylyl cyclase toxins from pathogenic bacteria that cause anthrax and whooping cough. J Biol Chem. 2003;278:25990–25997. doi: 10.1074/jbc.M301232200. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Moayeri M, Zhao H, Crown D, Leppla SH, Purcell RH. Potent neutralization of anthrax edema toxin by a humanized monoclonal antibody that competes with calmodulin for edema factor binding. Proc Natl Acad Sci U S A. 2009;106:13487–13492. doi: 10.1073/pnas.0906581106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao G, Cochran JR, Wittrup KD. Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J Mol Biol. 2004;342:539–550. doi: 10.1016/j.jmb.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 10.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy R, Forsyth CM, LaPorte SL, Geren IN, Smith LA, Marks JD. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J Mol Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 13.Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, Grabarek Z, Bohm A, Tang WJ. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 14.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Moayeri M, Zhou YH, Leppla S, Emerson S, Sebrell A, Yu F, Svitel J, Schuck P, St Claire M, Purcell R. Efficient neutralization of anthrax toxin by chimpanzee monoclonal antibodies against protective antigen. Journal of Infectious Diseases. 2006;193:625–633. doi: 10.1086/500148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sircar A, Kim ET, Gray JJ. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 2009;37:W474–W479. doi: 10.1093/nar/gkp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 19.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Zhukovskaya NL, Guo Q, Florian J, Tang WJ. Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. EMBO Journal. 2005;24:929–941. doi: 10.1038/sj.emboj.7600574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 23.Ulmer TS, Soelaiman S, Li S, Klee CB, Tang WJ, Bax A. Calcium dependence of the interaction between calmodulin and anthrax edema factor. J Biol Chem. 2003;278:29261–29266. doi: 10.1074/jbc.M302837200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Mapping epitope of EF13D to EF domain III by yeast surface display. (a) A schematic showing surface display of EF domain III on yeast. A fusion protein consisting of EF domain III and yeast Aga2 protein is attached to the Aga1 protein through disulfide bonds on the yeast cell wall. The level of expression of DIII and the binding by the mAb were monitored by fluorescence flow cytometry (Alexa Fluor 488 directed to the c-myc tag at the C-terminus of DIII and the Atto633 attached to the mAb). (b) Fifty four percent of yeast cells displayed properly folded DIII that was reactive with EF13D, as judged by dual fluorescence.
Figure S2: Molecular dynamics simulations of interaction between EF13D and wild type EF, EFR671A or EFR671I. It shows that the reduction in EF13D binding affinity for both R671A and R671I compared to wild-type can be explained by the absence of R671 being available to form a salt bridge with D110 and the further reduction of binding affinity for R671I is compounded by the relative lack of W104 burial.