SUMMARY
PUF (Pumilio/FBF) RNA-binding proteins and Argonaute (Ago) miRNA-binding proteins regulate mRNAs post-transcriptionally, each acting through similar yet distinct mechanisms. Here, we report that PUF and Ago proteins can also function together in a complex with a core translation elongation factor, eEF1A, to repress translation elongation. Both nematode and mammalian PUF/Ago/eEF1A complexes were identified, using co-immunoprecipitation and recombinant protein assays. Nematode CSR-1 (Ago) promotes repression of FBF (PUF) target mRNAs in in vivo assays, and the FBF-1/CSR-1 heterodimer inhibits EFT-3 (eEF1A) GTPase activity in vitro. Mammalian PUM2/Ago/eEF1A inhibits translation of nonadenylated and polyadenylated reporter mRNAs in vitro. This repression occurs after translation initiation and leads to ribosome accumulation within the open reading frame, roughly at the site where the nascent polypeptide emerges from the ribosomal exit tunnel. Together, these data suggest that a conserved PUF/Ago/eEF1A complex attenuates translation elongation.
Translational regulation impacts metazoan development, learning and human disease1–3. Of the many RNA-binding proteins regulating mRNA stability, localization and translation, the two classes relevant to this work are the PUF and Ago families. PUF proteins regulate a broad spectrum of mRNAs to maintain stem cells, among other functions1,2. Ago proteins bind small RNAs, most notably miRNAs, and act in many biological contexts, including stem cells (reviewed in 3).
PUF proteins regulate mRNA expression in virtually all eukaryotes4. One conserved mechanism, which has been established for Saccharomyces cerevisiae, Caenorhabditis elegans and human PUF proteins, relies on the direct recruitment of the Ccr4-Not deadenylase complex to target mRNAs, resulting in a shorter poly(A)-tail length and either mRNA instability or translational repression5,6. PUF proteins in yeast and flies also use deadenylation-independent mechanisms, although the specifics of these mechanisms remain largely unknown7,8.
Ago proteins function within the miRISC complex to control target mRNA stability and translation. This complex controls deadenylation and stability of its target mRNAs9–11, but its mechanism to control translation has garnered controversy. Best documented is its regulation of translation initiation9–13. However, a mechanism affecting translation elongation also appears to exist since target mRNAs can be bound to polyribosomes14. One model is that miRISC promotes ribosomal drop-off during translation elongation15. Yet the specific mechanism by which miRISC, and hence Ago family members, inhibits translation elongation is not understood.
Here, we report that PUF and Ago proteins form an inhibitory complex with eEF1A, a GTPase required for translation elongation. FBF-1 (a C. elegans PUF protein) binds CSR-1 (a C. elegans Ago family member) in vitro and in vivo, and csr-1 depletion leads to increased expression of FBF target mRNAs. The FBF-1/CSR-1 heterodimer forms a complex with EFT-3 (C. elegans eEF1A), and this FBF-1/CSR-1/EFT-3 ternary complex has inhibited GTPase activity. Importantly, the PUF/Ago/eEF1A complex is conserved: human PUM2 (a PUF protein) associates with human AGO proteins in vivo and with eEF1A. Wild-type human PUM2 inhibits translation of both nonadenylated and polyadenylated mRNAs in rabbit reticulocyte lysate; however, PUM2 mutants that cannot form the PUM2/Ago/eEF1A complex or that cannot bind RNA are severely compromised for translation repression. Mechanistically, PUM2/Ago/eEF1A represses translation during elongation with ribosomes accumulating ~100–140 nts after the AUG within the open-reading frame (ORF). We propose a model in which PUF and AGO proteins complex with eEF1A to inhibit its GTPase activity and attenuate translation elongation.
RESULTS
Association of C. elegans FBF-1 and CSR-1
To explore the molecular mechanisms of PUF protein function, we sought proteins that interact with two nearly identical C. elegans PUF proteins, FBF-1 and FBF-2, known collectively as FBF16. Previous yeast two-hybrid screens identified several FBF partners, but no components of the translation machinery17,18. Here, we used recombinant, full-length GST-FBF-1 and GST-FBF-2 to select proteins from C. elegans lysates and identify interacting proteins by mass spectrometry (Supplementary Fig. 1, Supplementary Table 1). Known FBF-interacting proteins, CPB-1 and NOS-3 (refs. 17,18), were detected in addition to previously unknown putative partners CSR-1, one of 27 C. elegans Ago family members19,20, and EFT-3, the C. elegans homolog of translation elongation factor 1A, eEF1A21.
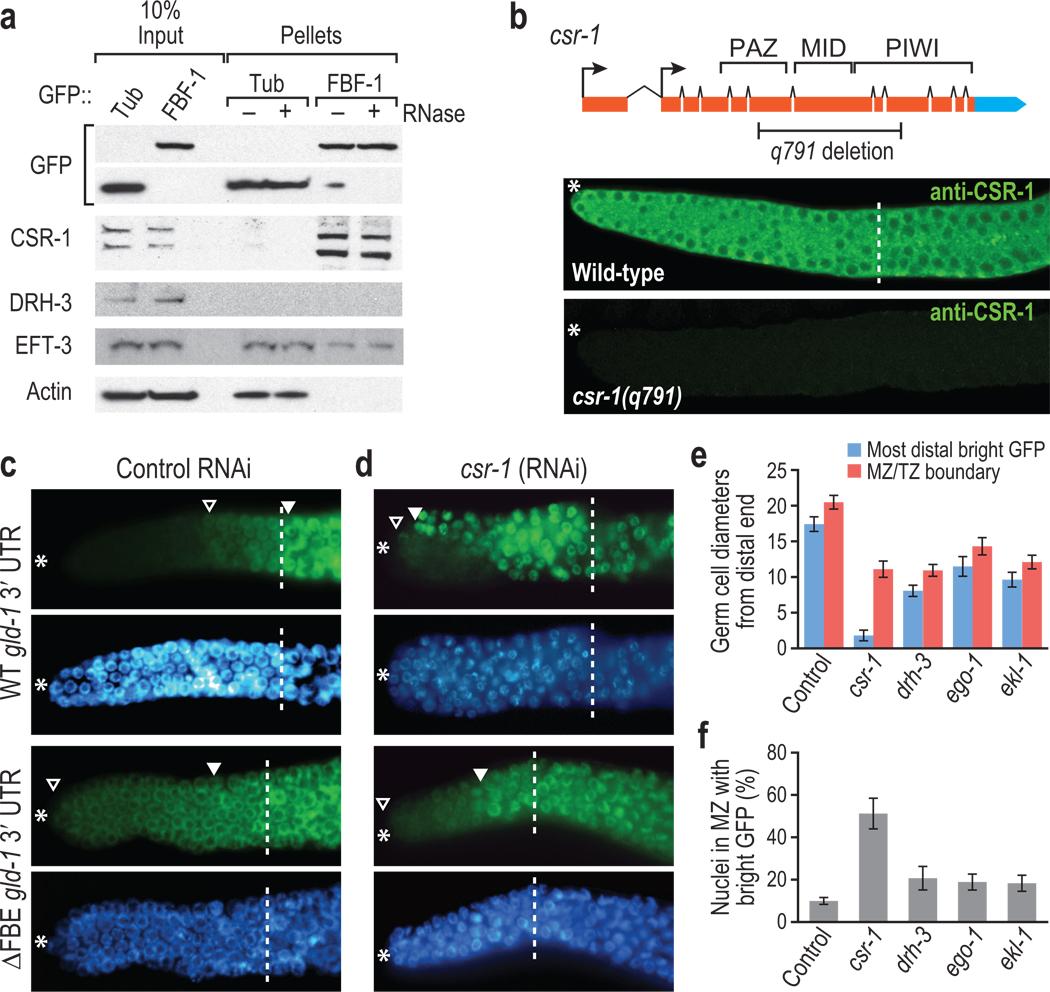
To directly test whether FBF-1 interacts with CSR-1 and EFT-3 in vivo, we immunoprecipitated tagged FBF-1 (ref. 22) from C. elegans lysate and probed for associated proteins. CSR-1 was specifically enriched, and this interaction was RNA-independent (Fig. 1a), indicating that FBF-1 and CSR-1 form a protein complex. By contrast, Dicer-related helicase (DRH-3), which is required to produce the 22-nt RNAs bound by CSR-1 (refs. 23,24), did not co-immunoprecipitate (IP) with FBF-1 (Fig. 1a), demonstrating that FBF-1 specifically binds CSR-1. EFT-3 was present in both FBF-1 and control IPs (Fig. 1a), so we performed further assays to confirm specificity of binding (see below). Our co-IP experiments suggested that CSR-1 should be expressed in the same germline region as FBF. FBF is enriched in the germ cell cytoplasm at the distal end of the gonad25, and we found CSR-1 in the same location (Fig. 1b), in addition to its known location in the proximal germline20. Since FBF-1 and CSR-1 co-IP and are expressed in the same region, FBF-1 and CSR-1 likely interact in vivo.
Figure 1.
FBF-1 binds CSR-1 to repress target mRNA. (a) FBF-1 co-IPs with CSR-1. Transgenic GFP::FBF-1 (FBF-1) was immunoprecipitated from adult hermaphrodites with or without RNase. Bound proteins (Pellets) were probed on Western blots for indicated proteins. CSR-1 and EFT-3 were both detected in FBF-1 IPs, whereas DRH-3 and actin were not. Two CSR-1 isoforms exist in vivo20,23, and both co-IP with FBF-1. CSR-1 did not co-IP with GFP::Tubulin (Tub). (b) CSR-1 is expressed in the mitotic zone. Top, diagram of csr-1 locus. The q791 deletion removes key domains resulting in a frameshift. Arrows, alternative promoter elements. Bottom: CSR-1 antibody23 stains the wild-type mitotic zone cytoplasm (*, distal end of the gonad; dashed line, mitotic zone/transition zone boundary), but not in a csr-1 mutant. (c, d) Adult germlines expressing GFP::H2B (green) under control of wild-type (WT) or FBE-lacking (ΔFBE) gld-1 3′ UTR; nuclei seen with DAPI (blue). Marked include: *, distal end; open triangle, distal-most GFP-positive cell; closed triangle, distal-most bright GFP; dashed line, mitotic zone/transition zone boundary. (c) Control RNAi, empty vector. GFP::H2B normally repressed in distal region (top panels), but expands into distal germ cells without FBEs (lower panels). (d) csr-1(RNAi). GFP::H2B expanded more distally than control in panel c when reporter harbors an FBE. (e) GFP::H2B extent after RNAi against genes indicated, scoring only sterile animals to ensure effective RNAi. Blue columns, most distal germ cell row with GFP::H2B-positive cell; red columns, mitotic zone/transition zone boundary. (f) Percent cells in mitotic zone with bright GFP::H2B.
CSR-1 promotes repression of FBF target mRNAs
PUF and Ago family members both repress translation4,10, and models have suggested that miRNA binding sites proximal to PUF mRNA target sites may indicate an interaction26. To ask if FBF and CSR-1 might function together, we examined expression of an FBF reporter mRNA after CSR-1 depletion. In the reporter mRNA, the 3′ UTR of gld-1, an FBF target mRNA25, was linked to an ORF encoding a GFP histone H2B fusion protein (GFP::H2B), which labels nuclei. We used RNAi to deplete csr-1 from animals transgenic for reporters with either the wild-type gld-1 3′ UTR (WT gld-1 3′ UTR) or a mutant gld-1 3′ UTR (ΔFBE gld-1 3′ UTR) without FBF binding elements (FBEs)27. Briefly, fourth-stage larvae were subjected to feeding RNAi, and the GFP::H2B reporter was scored one generation later in adult progeny. In control animals not depleted for CSR-1, GFP::H2B was absent from the distal-most germ cells with the WT gld-1 3′ UTR reporter (Fig. 1c, top panels), but GFP::H2B extended to the distal end in animals with the ΔFBE gld-1 3′ UTR reporter (Fig. 1c, bottom panels), as seen previously27. After CSR-1 depletion in animals carrying the WT gld-1 3′ UTR reporter, bright GFP::H2B expression was always shifted distally compared to controls (n = 31/31) (Fig. 1d, top panels) and was often in distal-most germ cells (n = 12/31). By contrast, csr-1 RNAi had little effect on expression of the ΔFBE gld-1 3′ UTR reporter (Fig. 1d, bottom panels). As a control, the same reporter assay was used after depleting proteins thought to form a complex with CSR-1 (ref. 24), but these depletions did not affect the WT gld-1 3′ UTR reporter (Figs. 1e, f). Because total loss of FBE-mediated repression always leads to bright GFP::H2B in the distal-most germ cells28, csr-1 depletion does not completely eliminate FBE-mediated repression but instead severely compromises that repression. We conclude that CSR-1 reduces expression of FBF reporter mRNA and that this effect requires FBEs.
To validate our findings with the gld-1 reporter mRNAs, we assayed two FBF target mRNAs, gld-1 (ref. 25) and cye-1 (ref. 29) mRNAs, in a csr-1 mutant. GLD-1 protein is normally expressed at a low but graded level in the mitotic zone (Supplementary Fig. 2a, top panel), while CYE-1 is more uniform in the mitotic zone (Supplementary Fig. 2a, middle panel). In a csr-1 deletion mutant, GLD-1 and CSR-1 both increased dramatically in the mitotic zone, though GLD-1 was still graded (Supplementary Fig. 2b, top and middle panels). For controls, we also analyzed GLD-1 and CYE-1 levels in mutants of drh-3, ego-1, and ekl-1, which are thought to function jointly with csr-1 (refs. 20,24,30). In these mutants, we observed more GLD-1 and CYE-1 than wild-type, but not to the high level typical of csr-1 mutants (Supplementary Figs. 2c–e). Therefore, wild-type CSR-1 lowers expression of at least two FBF target mRNAs.
These experiments suggest that FBF functions with CSR-1 to repress target mRNAs. Yet, the documented germline defects in fbf and csr-1 mutants differ in many respects. The fbf-1 fbf-2 double mutant germlines are defective for germline self-renewal and hence have no mitotic zone25 while csr-1 is defective for chromosome segregation and embryonic viability19. We examined csr-1 deletion mutants for mitotic zone defects and found the csr-1 mitotic zone to be present but smaller than normal (Supplementary Fig. 2b). The reduced csr-1 mitotic zone is consistent with a functional overlap with FBF, but its presence demonstrates that CSR-1 is not required for all FBF functions. We suggest that CSR-1 and FBF can work together to repress mRNA expression via FBEs, but that they can also work independently. The independent functions likely involve interactions with other paralogs (e.g. FBF with other Agos, CSR-1 with other PUFs) as well as interactions with other complexes (e.g. FBF with the Ccr4/Not deadenylase and CSR-1 with RISC).
FBF-1 interacts with the CSR-1 MID-PIWI domains
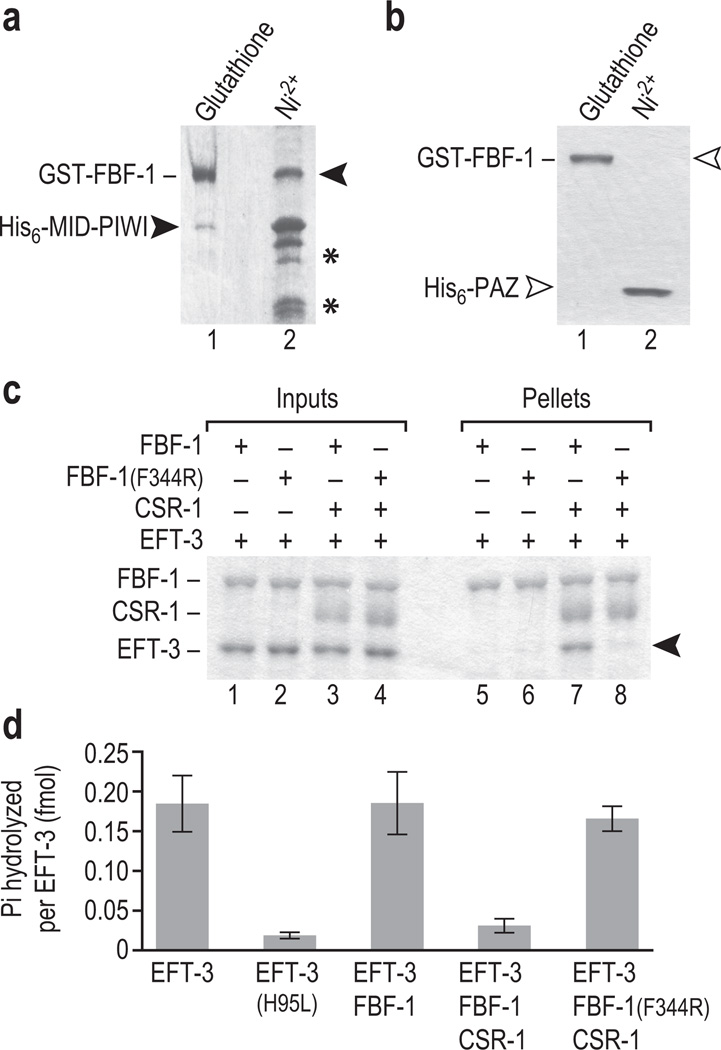
To directly assay FBF-1 interactions with CSR-1, we performed biochemical analyses with purified recombinant proteins. Initially, we explored the interaction, focusing on conserved domains – the PUF repeats of FBF-1 (hereafter called FBF-1) and the MID-PIWI or PAZ domains of CSR-1. GST-FBF-1 (a.a. 121–612) was co-expressed with either His6-MID-PIWI (a.a. 557–993) or His6-PAZ (a.a. 378–505) in the pETDuet system, which allows simultaneous expression of multiple proteins. Affinity selection for GST-FBF-1 enriched His6-MID-PIWI (Fig. 2a, lane 1) but not His6-PAZ (Fig. 2b, lane 1); the reciprocal selection for His6-MID-PIWI enriched GST-FBF-1 (Fig. 2a, lane 2), but selection for His6-PAZ did not enrich GST-FBF-1 (Fig. 2b, lane 2). Therefore, purified FBF-1 interacts specifically with the CSR-1 MID-PIWI domains but not with its PAZ domain. Our next experiments employed the CSR-1 MID-PIWI region, referred to as CSR-1 for brevity.
Figure 2.
FBF-1/CSR-1/EFT-3 ternary complex formation reduces EFT-3 GTPase activity. (a) GST-FBF-1 and His6-CSR-1(MID-PIWI) were co-expressed in the pETDuet system. Affinity selection was performed with either glutathione to select GST-FBF-1 or Ni2+ to select His6-MID-PIWI respectively. GST-FBF-1 and His6-MID-PIWI associate. His6-MID-PIWI degradation products are indicated (*). (b) As in panel a, but GST-FBF-1 was co-expressed with CSR-1 PAZ (His6-PAZ). FBF-1 and His6-PAZ do not associate. (c, d) CSR-1 refers to His6-CSR-1(MID-PIWI); FBF-1 refers to GST-FBF-1. (c) EFT-3 binds the FBF-1/CSR-1 complex. Recombinant EFT-3 was added to wild-type FBF-1, FBF-1(F344R), FBF-1/CSR-1 or FBF-1(F344R)/CSR-1 prepared from bacteria (Inputs, 25% loaded). Glutathione was used to select GST-FBF-1 (wild-type and mutant) after incubation (Pellets, 50% loaded). EFT-3 (arrowhead) specifically associated with wild-type FBF-1/CSR-1, but not with FBF-1 alone or with mutant FBF-1(F344R)/CSR-1. We note that the FBF-1/CSR-1/EFT-3 complex forms with apparent 1:1:1 stoichiometry. (d) FBF-1/CSR-1 specifically inhibits EFT-3 GTPase activity. [32P]-γ-GTP was incubated with EFT-3, catalytically-dead EFT-3(H95L), EFT-3 and FBF-1, EFT-3/FBF-1/CSR-1 or EFT-3 and FBF-1(F344R)/CSR-1. Liberated Pi was scintillation counted after phase extraction.
FBF-1 and CSR-1 form a complex with EFT-3 (eEF1A)
This work identifies EFT-3 as a potential FBF-1 interactor (Supplementary Table 1), and work by others identified human eEF1A as a potential AGO1 and AGO2 interactor31, although the interaction was not confirmed for either nematode or human components. We hypothesized that the FBF-1/CSR-1 heterodimer might form a ternary complex with the translation elongation factor EFT-3. To explore that idea, we purified FBF-1 alone, the FBF-1/CSR-1 heterodimer and EFT-3 alone (Fig. 2c, lanes 1–4); attempts to purify CSR-1 by itself were unsuccessful. FBF-1 alone did not bind EFT-3 (Fig. 2c, lane 5), but the FBF-1/CSR-1 heterodimer did bind EFT-3 (Fig. 2c, lane 7), suggesting existence of a ternary complex. This ternary complex was observed in the absence of GTP (Fig. 2c, lane 7), but FBF-1/CSR-1 also bound EFT-3 when GTP was included (Supplementary Fig. 3a). We confirmed the identity of CSR-1 and EFT-3 proteins in these pull-down assays by western blotting (Supplementary Fig. 3b). We examined FBF-1 for conserved residues that are not involved in RNA-binding and found a broadly-conserved Phe residue (Phe344) (Supplementary Fig. 3c). We hypothesized that mutant FBF-1(F344R) might be defective for ternary complex formation. Although FBF-1(F344R) was able to bind CSR-1 (Supplementary Fig. 3d), the FBF-1(F344R)/CSR-1 heterodimer failed to bind EFT-3 (Fig. 2c, lane 8). We propose that FBF-1/CSR-1/EFT-3 exists as a complex.
The FBF-1/CSR-1 heterodimer inhibits EFT-3 GTPase activity
How might the complex affect translation? eEF1A family members possess GTPase activity, which is required during translation elongation to release aminoacyl-tRNAs upon delivery to the ribosome. Purified eEF1A has intrinsic GTPase activity, which is stimulated by ribosomes32. We hypothesized that the PUF/Ago complex might inhibit eEF1A activity since PUF and Ago proteins can inhibit translation4,9–11. To test this idea, we first confirmed that wild-type EFT-3 possesses GTPase activity compared to a catalytically-dead mutant EFT-3(H95L)33 (Fig. 2d; Supplementary Fig. 3e). We then assayed FBF-1 alone and the FBF-1/CSR-1 heterodimer for an effect on EFT-3 GTPase activity. FBF-1 alone did not reduce GTPase activity, but the FBF-1/CSR-1 heterodimer reduced GTPase activity to a level similar to that of catalytically-dead EFT-3(H95L) (Fig. 2d). Importantly, the mutant FBF-1(F344R)/CSR-1 heterodimer, which did not form a complex with EFT-3, also did not inhibit EFT-3 GTPase activity (Fig. 2d). Inhibition of EFT-3 GTPase activity in the FBF-1/CSR-1/EFT-3 complex suggests that the complex may repress translational elongation.
A human PUM2/AGO/eEF1A complex
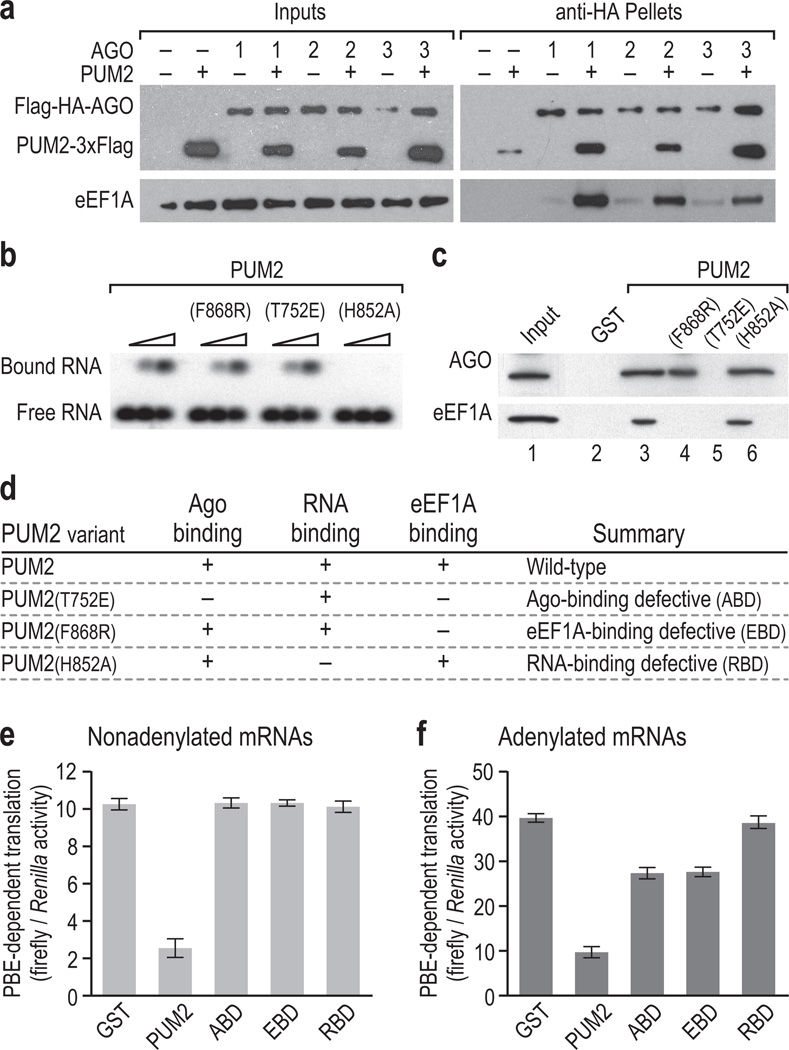
We predicted that a PUM2/AGO/eEF1A complex might also form in humans. Initially, we tested whether PUM2 binds AGO family members in vivo. HEK293T cells were transiently transfected with constructs encoding the Flag-tagged PUF repeats of human PUM2 (PUM2-3xFlag) and Flag-HA-tagged, full-length human AGOs1-3 (ref. 31). Using anti-HA antibodies, AGOs1-3 were immunoprecipitated from formaldehyde-crosslinked cells under denaturing conditions with RNase to isolate endogenous complexes34. Anti-Flag western blots revealed that PUM2 and AGOs1-3 co-IP (Supplementary Fig. 4a). We confirmed that PUM2 co-IPs with AGOs1-3 without crosslinking (Supplementary Fig. 4b). To test whether PUM2 is critical for the association of Ago and eEF1A, we performed the same transfection experiments as above in the presence or absence of PUM2. When PUM2 was co-expressed, AGO1-3 IPs pulled down dramatically more eEF1A than when PUM2 was omitted, arguing that PUM2/AGO binds eEF1A in vivo (Fig. 3a). We conclude that the PUM2/AGO/eEF1A complex is conserved.
Figure 3.
PUM2/Ago/eEF1A complex inhibits protein production. (a) Human PUM2 promotes eEF1A association with Ago. Western blots using Flag antibody to detect 3xFlag-PUM2 and Flag-HA-AGOs1-3 (top) oreEF1A antibody (bottom). PUM2 co-expression dramatically enhanced eEF1A co-IP. (b) RNA binding by PUM2 variants. Recombinant PUM2, PUM2(F868R), PUM2(T752E) and PUM2(H852A) at increasing concentrations (triangle) were assayed for binding the hunchback PBE sequence. (c) Complex formation with PUM2 variants. AGO co-precipitates with all PUM2 mutants except PUM2(T752E). eEF1A co-precipitates with wild-type and PUM2(H852A) but not with PUM2(F868R) or PUM2(T752E). Equivalent amounts of recombinant PUM2 proteins were added to reticulocyte lysate (Input) in the presence of RNase. (d) PUM2 variant summary. (e) PBE-dependent translation repression of nonadenylated reporter mRNA. Recombinant GST or GST-tagged PUM2 variants, nonadenylated firefly luciferase mRNA with 3xPBEs in its 3′ UTR and control nonadenylated Renilla luciferase reporter were added to reticulocyte lysate. To monitor PBE-dependent translation, firefly luciferase production was normalized to Renilla luciferase production. Only wild-type PUM2 inhibited translation of nonadenylated firefly luciferase mRNA. Note that protein output was not corrected for mRNA level since all reactions contained same initial quantity of reporter mRNAs, and wild-type PUM2 reactions had more final mRNA than others. (f) As in panel e, but with polyadenylated mRNAs.
PUM2 mutants abolish complex formation
We turned to rabbit reticulocyte lysate to characterize PUM2/Ago/eEF1A effects on translation. For these experiments, we first identified PUM2 mutants that abolish complex formation or RNA binding. Each PUM2 variant tested was comprised of the PUF repeats of human PUM2 (a.a. 706–1062) and was generated as a GST-PUM2 fusion protein. PUM2 refers to the wild-type version; PUM2(F868R) alters the conserved Phe corresponding to FBF-1(F344); PUM2(T752E) changes a conserved Thr residue (Supplementary Figs. 5a–c); and PUM2(H852A) was predicted to abrogate RNA binding35. Of these four proteins (Supplementary Fig. 5d), only PUM2(H852A) failed to bind an RNA carrying PUF-binding elements (PBEs) (Fig. 3b). When assayed for complex formation, wild-type PUM2 and PUM2(H852A) co-precipitated Ago and eEF1A from rabbit reticulocyte lysate (Fig. 3c, lanes 3 and 6). PUM2(F868R) bound Ago, but not eEF1A (Fig. 3c, lane 4), and PUM2(T752E) failed to bind either Ago or eEF1A (Fig. 3c, lane 5). In summary, PUM2(T752E) is Ago-binding defective (ABD); PUM2(F868R) is eEF1A-binding defective (EBD); and PUM2(H852A) is RNA-binding defective (RBD) but binds both Ago and eEF1A (Fig. 3d). The PUM2/Ago heterodimer recruits eEF1A, and PUM2 RNA-binding activity is not required for complex formation.
PUM2/Ago/eEF1A represses mRNA translation
To assay effects of the PUM2/Ago/eEF1A complex on translation, we used reporter mRNAs in reticulocyte lysate. We first quantitated Ago levels in the lysate (Supplementary Fig. 6a) and added an equivalent amount of PUM2. To ensure that reticulocyte lysate was not limiting for translation, we titrated mRNA encoding firefly luciferase with 3xPBEs in its 3′ UTR [PBEs bind human PUM1 and PUM2 (refs. 36,37)] against constant control Renilla luciferase reporter lacking PBEs (Supplementary Fig. 6b). PUM2 did not destabilize reporter mRNAs; in fact, PUM2 stabilized them (Supplementary Fig. 6c). Therefore, the ratio of firefly to Renilla luciferase enzyme activities monitors PBE-dependent translation. We first assayed nonadenylated mRNA (Fig. 3e) to exclude from our analysis PUF-mediated deadenylation effects5. Wild-type PUM2 inhibited mRNA translation, but the three mutant PUM2 proteins failed to repress reporter expression (Fig. 3e). Most importantly, the ABD and EBD mutants, which disrupt PUF/Ago/eEF1A complex formation (Fig. 3b), did not repress the reporter.
Most natural mRNAs are polyadenylated, so we also assayed polyadenylated reporter mRNAs (Fig. 3f). Similar to the effects observed on the nonadenylated reporter mRNA, wild-type PUM2 repressed translation of polyadenylated mRNA while PUM2(RBD) did not. Notably, the PUM2 ABD and EBD mutants remained capable of limited repression. One possibility might have been that the ABD and EBD mutants had this minor effect by promoting mRNA deadenylation. However, mRNA adenylation was unaffected in our assays (Supplementary Fig. 6d). An open question is how PUM2(ABD and EBD) repress the polyadenylated reporter in the absence of both deadenylation and PUM2/Ago/eEF1A complex formation. Indeed, these mutants suggest the existence of yet another mechanism of PUF repression. Since PUF proteins associate with Nanos38, we considered the possibility that the region of PUM2 important for its Nanos association might be important for PUM2 repression. However, PUM2(G987D) repressed both nonadenylated and polyadenylated reporter mRNAs as well as wild-type PUM2 (Supplementary Fig. 7). Together, experiments using nonadenylated and polyadenylated mRNA reporters argue that the PUF/Ago/eEF1A complex represses translation and that it does so independent of mRNA deadenylation.
PUM2/Ago/eEF1A represses translation after initiation
We next investigated the mechanism of PUM2 translation inhibition, again in rabbit reticulocyte lysates. Initially, we performed a standard assay for inhibition of translation initiation: we fractionated polyribosomes to ask if PUM2 inhibits the association of its target mRNA with ribosomes. In vitro translation reactions were performed as above with firefly mRNA (3xPBEs) and Renilla mRNA (Control) in the presence of PUM2 or PUM2(RBD). Reactions were quenched with cycloheximide (maintains ribosomes on mRNA) or puromycin (removes ribosomes from mRNA) and separated over a sucrose gradient. After centrifugation, polyribosome profiles were indistinguishable when PUM2 or PUM2(RBD) was added, although puromycin did disrupt polyribosomes (Supplementary Figs. 8a, b). Moreover, by Northern blots, the 3xPBE-containing mRNA associated with polyribosomes to the same level as the control mRNA if either PUM2 or PUM2(RBD) was added (Supplementary Fig. 8c). Since the 3xPBE-containing mRNA migrated in lighter fractions after puromycin addition, it is bound to ribosomes (Supplementary Fig. 8d). Although these data contrast observations that miRNA-mediated translation repression occurs at initiation in rabbit reticulocyte lysate39, we note that our experiments query PUF/Ago/eEF1A, not miRNAs. Therefore, ribosomal association with PUM2-bound mRNA appears unaffected by this assay, leading us to ask further questions about how translation is inhibited.
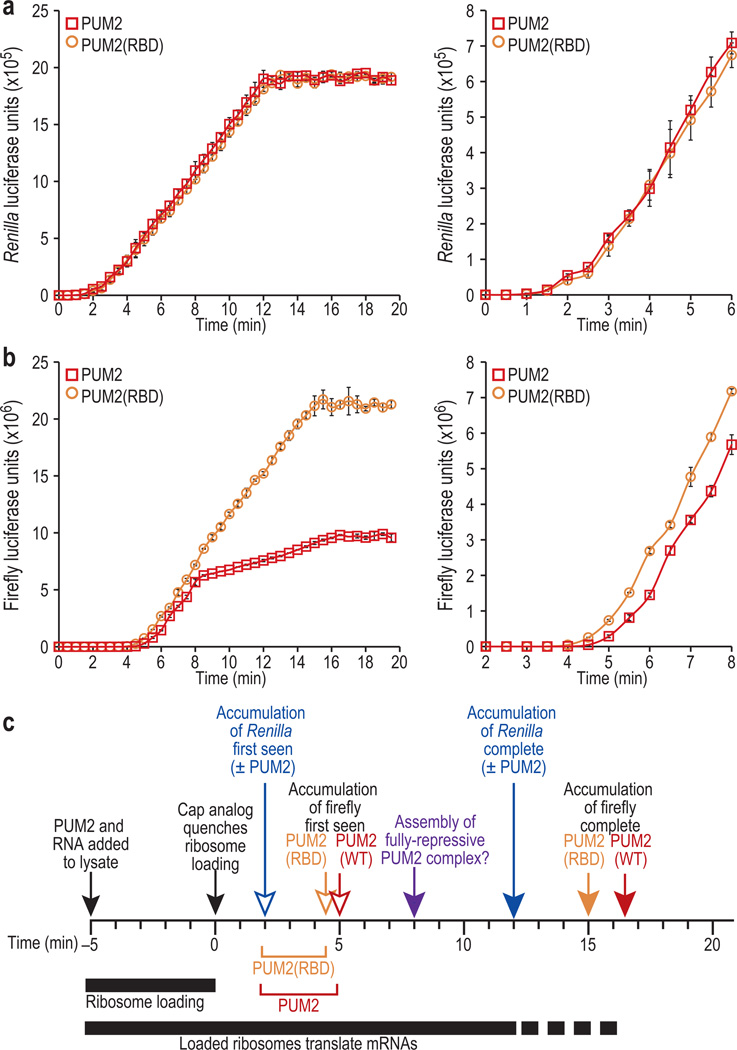
We next varied the translation assays to ask if PUM2-mediated repression occurs after translation initiation. Reporter RNAs plus purified recombinant PUM2 protein were incubated in lysate for 5 minutes to begin translation; then excess mRNA cap analog was added to quench additional ribosomal loading, and thus to block translation initiation. Production of firefly (subject to PUM2 regulation) and Renilla luciferase was monitored in the presence of either PUM2 or PUM2(RBD) over time. In the absence of cap analog, reporters produced luciferase linearly for about 45 minutes; however, in the presence of cap analog, luciferase production stopped much earlier. The control Renilla luciferase RNA plateaued after only ~13 minutes in the presence of either PUM2 or PUM2(RBD) (Fig. 4a). This early plateau indicates that the excess cap analog had abolished de novo 40S ribosomal subunit loading. With the RNA-binding defective PUM2 variant, expression from the firefly luciferase mRNA plateaued after ~15 minutes, a bit after the Renilla control, presumably because its protein product is larger. However, wild-type PUM2 had a dramatic effect on firefly luciferase expression, decreasing sharply after only 8 minutes (Fig. 4b, left panel), an effect that can account for the difference in synthesized protein observed in Figures 3e, f. Wild-type PUM2 also modestly, but reproducibly, delayed the initial production of firefly luciferase relative to PUM2(RBD), an effect that likely reflects slower translation elongation (Fig. 4b, right panel; summarized in Fig. 4c). That slowed translation elongation is consistent with inhibited eEF1A GTPase activity in the PUM2/Ago/eEF1A complex, but the effect was not large enough to account for the four-fold reduction in synthesized protein seen in Figures 3e, f. We conclude that PUM2 slows translation elongation and blocks protein production from its target mRNA.
Figure 4.
Kinetics of PUM2 translational repression. (a,b) PUM2 proteins were incubated with reporter mRNAs in reticulocyte lysate for 5 minutes to ensure ribosome loading; at time = 0, excess mRNA cap analog was added to inhibit de novo 40S ribosomal subunit loading. Luciferase production was monitored every 30-seconds. (a) Renilla luciferase production. Left, kinetics of Renilla luciferase production were equivalent for PUM2 and PUM2(RBD); right, enlargement of early time points. (b) Firefly luciferase production. Left, firefly luciferase production was delayed with PUM2 compared to PUM2(RBD) and dramatically inhibited midway through the reaction. The PUM2 curve was similar to that of PUM2(RBD) from the ~5 to 8 min time points (albeit with a 30 second delay), suggesting that a fully-functional repressive complex forms slowly. Right, enlargement of data points when firefly luciferase is first produced. (c) To estimate translational elongation rate, we compared the time required to first produce either firefly or Renilla luciferase (firefly luciferase is 319 residues longer than Renilla luciferase). In reactions with PUM2(RBD), the first firefly luciferase protein was detected 2.5 minutes after the first Renilla luciferase (4.5 minutes for firefly vs. 2 minutes for Renilla). From this difference, we infer an estimated translational elongation rate (eTER) of 2.12 a.a./sec (319 residues/150 second). In reactions with wild-type PUM2, the same calculation yields an eTER of 1.77 a.a/sec. This modest decrease was seen at a time well before PUM2 becomes fully repressive (8 minutes after cap analog addition). Therefore, PUM2 may affect translation elongation using multiple mechanisms.
PUM2/Ago/eEF1A attenuates translation elongation
We considered possible mechanisms by which PUM2 might inhibit protein production: it might attenuate translation elongation, consistent with the ribosome drop-off model proposed for miRNA-mediated translation repression15; it might inhibit translation termination; or it might promote destruction of nascent polypeptides during translation, as observed for miRNA-mediated repression40. The first two mechanisms make predictions about effects on ribosome position. If ribosomes drop off the mRNA during elongation, ribosomal density should be higher at the 5′ end of the ORF and taper off towards the 3′ end. If translation termination is inhibited, ribosomes should accumulate at the 3′ end of the ORF. High-resolution ribosomal footprinting was previously described at a genomic level41, and we modified this approach for our single mRNA reporter (Fig. 5a). Briefly, in vitro translation reactions were performed with radiolabeled firefly luciferase mRNA (with 3xPBEs). Reactions contained PUM2, PUM2(ABD), PUM2(EBD) or PUM2(RBD). Translation reactions were quenched with cycloheximide and treated with RNase One, then fractionated over a sucrose gradient to isolate ribosome-bound RNA fragments. These RNA fragments were extracted from the monoribosome peak and hybridized to a spotted array containing overlapping DNA oligonucleotides complementary to the firefly luciferase mRNA (Supplementary Fig. 9). RNA signals from monoribosome fractions were normalized to signal from unfractionated samples to control for hybridization efficiency, uridine content and RNase sensitivity and then plotted across the mRNA (Fig. 5b). Strikingly, in the presence of PUM2, a large peak occurs ~100–140 nts into the ORF, and then ribosomal footprints are sharply reduced across the remainder of the ORF. The peak coincides with the position at which nascent polypeptide should emerge from the ribosomal exit tunnel (~30–40 amino acids42). Importantly, the ribosomal footprints observed in samples with PUM2(ABD and EBD) were similar to that with PUM2(RBD). Therefore, the ribosomal footprinting profile is only altered when PUM2 can form a ternary complex on its target mRNA. To ensure that the observed ribosomal profile was independent of nucleotide sequence, we repeated our ribosomal footprinting experiment with firefly luciferase mRNA lacking ~500 nts after the initiation codon (including the sequence where ribosomes accumulated above). Again, we observed decreased ribosomal density after the first ~140 nts into the ORF only when wild-type PUM2 was added (Fig. 5c). We conclude that PUM2/Ago/eEF1A complex attenuates translation elongation, causing ribosomes to accumulate within the ORF.
Figure 5.
Human PUM2 attenuates translation elongation. (a) Ribosomal footprinting protocol. In vitro translation reactions were performed with radiolabeled firefly luciferase mRNA harboring 3xPBEs, reactions quenched with cycloheximide and treated with RNase. RNA from monoribosome fractions was collected and hybridized to a spotted array with oligonucleotides complementary to firefly mRNA. Signals from monoribosome-bound RNA fragments were normalized with input RNA fragments. (b) PUM2 affects ribosome position. PUM2, PUM2(ABD), PUM2(EBD) or PUM2(RBD) were added to reticulocyte lysate, and ribosomal footprinting was performed as diagrammed in panel a. At the 5′ end of the ORF, ribosomal density is equivalent for all samples, a level that continues across the ORF when PUM2(ABD, EBD and RBD) mutants are added. With wild-type PUM2, ribosomal density peaks at ~100–140 nts and decreases to a level lower than that seen with PUM2 mutants. Therefore translation is attenuated during elongation. Note: samples with wild-type PUM2 had more input RNA signal due to stabilized mRNA (Supplementary Fig. 6b), which dampens the signal observed for ribosomal footprints once normalized. (c) As in panel b, but footprinting was performed with a mutant firefly luciferase mRNA deleted for ~500 nts after the start codon (red). As above, wild-type PUM2 caused ribosomes to accumulate over the first ~40–140 nts. Ribosomal density again dropped after this site of accumulation to a level lower than that seen with the PUM2 mutants, which all had equivalent ribosomal density across the ORF. (d) Model for how the PUF/Ago/eEF1A complex attenuates translation elongation. See text for explanation.
DISCUSSION
Our data support three main conclusions that advance our understanding of how PUF and Ago proteins regulate translation. First, PUF and Ago proteins can form a conserved complex containing the core translation elongation factor, eEF1A. Second, the nematode complex inhibits eEF1A GTPase activity, and the human complex represses translation. Third, the complex blocks elongation at a site ~100–140 nts into the open reading frame, where the nascent peptide is expected to emerge from the ribosomal exit tunnel. Below we discuss the implications of these findings and propose a model to describe our observations: that PUF/Ago inhibits eEF1A GTPase activity and blocks translation elongation.
A conserved PUF/Ago/eEF1A complex
Studies prior to this work focused on either PUF or Ago proteins4,9–11. One hint that the two might work together was a bioinformatics analysis of human 3′ UTRs, revealing a nonrandom proximity of PUF protein binding elements and miRNA binding sites26. Here, we demonstrate not only that PUF and Ago proteins interact, but also that the PUF/Ago heterodimer associates with eEF1A and inhibits its GTPase activity. Our focus in nematodes was the FBF-1 PUF protein and the CSR-1 Ago protein, but we suspect that other PUF/Ago combinations may occur, as the nematode genome encodes 11 PUFs and 27 Agos. FBF-1 and CSR-1 are divergent members of their respective families, yet both proteins possess signature domains. Moreover, CSR-1 has slicing activity23 despite its non-canonical function in chromosome segregation19. One possibility might have been that the FBF-1/CSR-1 association was atypical. However in mammals, human PUM2 can associate with three AGOs (AGO1, AGO2 and AGO3), suggesting conservation. We suggest that multiple PUF/Ago combinations exist both in nematodes and mammals. A question for the future is whether individual PUFs and Agos behave differently with respect to their participation in the PUF/Ago/eEF1A complex.
Small RNAs cannot be essential for formation of the PUF/Ago/eEF1A complex. The MID-PIWI domain of Ago is sufficient to form the complex, and together with its PUF partner, the MID-PIWI domain inhibits eEF1A GTPase activity. Yet the MID-PIWI domain cannot bind small RNAs on its own. By contrast, the PAZ domain of Ago is not required to assemble the complex (Fig. 2c), but this PAZ domain drives small RNA loading43. Nonetheless, PUF/Ago/eEF1A complexes formed in vivo contain full-length Agos and likely associate with small RNAs. An attractive idea is that the PUF/Ago/eEF1A complex coordinates PUF and miRNA regulation.
The interaction between the PUF/Ago heterodimer and eEF1A complex is reminiscent of the original identification of Ago in a complex with eIF2 (ref. 44), which is structurally similar to eEF1A and also possesses GTPase activity. Moreover, two Drosophila RNA-binding proteins, dFXR and VIG, coordinate with Ago2 in the context of miRISC to repress target mRNAs45. An exciting possibility is that Ago proteins can work with other RNA-binding proteins and other translation factors to modulate multiple steps of protein synthesis.
PUF/Ago/eEF1A regulates translation elongation
Figure 5d presents a model for how the PUF/Ago/eEF1A complex may control translation elongation. Central to this model is its anchor via the PUF/Ago heterodimer to regulatory elements in the 3′ UTR and its association with an inhibited core translation elongation factor, eEF1A. Also central to this model is a block within the ORF at a position corresponding roughly to where the nascent polypeptide emerges from the ribosomal exit tunnel. Our kinetic analysis of protein production from a PUM2-repressed reporter mRNA (Fig. 4) suggested two phases of translational inhibition. An initial modest inhibition was inferred from a slower elongation rate, consistent with inhibition of eEF1A GTPase activity; a subsequent full repression was inferred from a complete block in protein synthesis seen later. Full repression may require a conformational change in the PUF/Ago/eEF1A complex, its modification, or recruitment of additional factors. If ribosomes do indeed accumulate where the nascent polypeptide leaves the ribosomal exit tunnel, an additional factor that recognizes the nascent polypeptide seems likely.
The model in Figure 5d does not include any small RNA associated with Ago, because our work focused on Ago, PUF and eEF1A proteins, not small RNAs. For example, a miRNA binding site was not engineered into the reporter 3′ UTRs, and miRNAs were not added to the reactions. One intriguing possibility is that the PUF/Ago/eEF1A mechanism reveals a role for Ago that does not rely on small RNAs. Perhaps more likely is the idea that small RNAs influence recruitment, either in an essential or facilitating manner. Addressing the role of small RNAs in this mechanism is an obvious next step for future analyses.
Repression at the level of translation initiation is emerging as a favored mechanism of translational control by miRNAs9–11. We provide two lines of evidence that the PUF/Ago/eEF1A complex does not have a major effect on translation initiation. First, polyribosomal profiles were the same for a PUM2-repressed mRNA and a control mRNA (Supplementary Fig. 8). Second, the same initial slope was observed in our kinetic analysis of protein production by PUM2-repressed and control mRNAs (Fig. 4). In addition, PUM2 repression occurred in the presence of excess mRNA cap analog, suggesting that its repression is exerted after loading 40S ribosomal subunits. Although we cannot exclude the possibility of a minor effect on translation initiation, our results strongly point to translational elongation as the major level at which the PUF/Ago/eEF1A complex exerts its repressive effects.
How does the proposed PUF/Ago/eEF1A mechanism compare to established mechanisms of regulation at the level of translation elongation? Perhaps the best-known mechanism is that adopted by the signal recognition particle (SRP) during synthesis of membrane proteins46, where translation elongation is inhibited47 until SRP docks with the endoplasmic reticulum46. The PUF/Ago/eEF1A mechanism resembles this case in that ribosomes accumulate at a similar position within the ORF. However, no role for 3′ UTR binding proteins is known for the SRP block. Recently discovered is the hnRNP E1 association with eEF1A to inhibit translation elongation; however, hnRNP E1 does not inhibit eEF1A GTPase activity but instead inhibits eEF1A dissociation from the ribosome48. Another case is miRISC, which is thought to promote ribosomal drop-off from a targeted mRNA but not at a given position within the ORF15. Therefore no established mechanism fits the PUF/Ago/eEF1A model completely.
Why multiple PUF and Ago mechanisms?
The PUF/Ago/eEF1A mechanism controlling translation elongation (Fig. 5d) is not the only mechanism used by PUF and Ago proteins, and perhaps not even the primary mechanism. Its relative importance to other well-established mechanisms (e.g. deadenylation and instability, see Introduction) remains unknown. Why employ multiple mechanisms? We do not know but suggest three ideas. One is security. If the PUF protein or Ago fails to silence a target mRNA with one mechanism, the regulator may invoke a different mechanism to repress the escaped mRNA. A second idea is reversibility. Destruction is a dead end for an mRNA, but blocked elongation might ensure robust activation at a later step. A third is subcellular compartmentalization. As an mRNA emerges from the nucleus, it meets one subcellular environment, but upon localization, for example to stress granules, the mRNA enters a different environment. Translational repression in (or localization to) distinct subcellular locales may differentially employ one mechanism over another. Key issues for the future include unraveling the biological functions of the various PUF and Ago mechanisms, their relationships and their regulation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank H. Tabara (Kyoto University) for providing CSR-1 and DRH-3 antibodies and E. Kipreos (University of Georgia) for CYE-1 antibody. We thank Kimble and Wickens lab members for discussion; we also thank Elsebet Lund and Scott Kennedy for critical reading of the manuscript and Anne Helsley-Marchbanks and Laura Vanderploeg for help with the manuscript and figure preparation. Mass spectrometry was performed with support from the Human Proteomics Program at the University of Wisconsin-Madison.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
K.F. performed the experiments with the exception of phylogenetic analysis (Z.C.), luciferase reporter mRNA production (A.C.) and csr-1 mutant generation (P.K.-C.). K.F., M.W. and J.K. prepared the manuscript. K.F. is supported by PF-10-127-01-DDC from the American Cancer Society, Z.C. by National Institutes of Health postdoctoral fellowship F32 GM095169, A.C. by N.I.H. training grant T32 GM07215 and an Advanced Opportunity Fellowship from UW-Madison, M.W. by N.I.H. grants GM031892 and GM050942 and J.K. by N.I.H. grant GM069454. J.K. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Thompson B, Wickens M, Kimble J. Translational control in development. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 507–544. [Google Scholar]
- 2.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 4.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 5.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger mRNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 6.Suh N, et al. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chagnovich D, Lehmann R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 2001;98:11359–11364. doi: 10.1073/pnas.201284398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hook BA, Goldstrohm AC, Seay DJ, Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 2007;282:15430–15438. doi: 10.1074/jbc.M611253200. [DOI] [PubMed] [Google Scholar]
- 9.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 11.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 12.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 13.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 15.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 17.Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14:2596–2609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraemer B, et al. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- 19.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 20.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhoads RE, Dinkova TD, Korneeva NL. Mechanism and regulation of translation in C. elegans. In: Community TCeR., editor. WormBook. WormBook; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M-H, et al. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crittenden SL, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 26.Galgano A, et al. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merritt C, Rasoloson D, Ko D, Seydoux G. 3'UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merritt C, Seydoux G. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development. 2010;137:1787–1798. doi: 10.1242/dev.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. USA. 2010;107:3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000624. e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meister G, et al. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 32.Parmeggiani A, Sander G. Properties and regulation of the GTPase activities of elongation factors Tu and G, of initiation factor 2. Mol. Cell. Biochem. 1981;35:129–158. doi: 10.1007/BF02357085. [DOI] [PubMed] [Google Scholar]
- 33.Cool RH, Parmeggiani A. Substitution of histidine-84 and the GTPase mechanism of elongation factor Tu. Biochemistry. 1991;30:362–366. doi: 10.1021/bi00216a008. [DOI] [PubMed] [Google Scholar]
- 34.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, McLachlan J, Zamore PD, Hall TMT. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 36.Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 37.Fox MS, Reijo Pera RA. Male infertility, genetic analysis of the DAZ genes on the human Y chromosome and genetic analysis of DNA repair. Mol.Cell. Endocrinol. 2001;184:41–49. doi: 10.1016/s0303-7207(01)00646-3. [DOI] [PubMed] [Google Scholar]
- 38.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricci EP, et al. Activation of a microRNA response in trans reveals a new role for poly(A) in translational repression. Nucleic Acids Res. 2011;39:5215–5231. doi: 10.1093/nar/gkr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 41.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beckmann R, et al. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 43.Miyoshi K, Okada TN, Siomi H, Siomi MC. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009;15:1282–1291. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakravarty I, Bagchi MK, Roy R, Banerjee AC, Gupta NK. Protein synthesis in rabbit reticulocytes. Purification and properties of an Mr 80,000 polypeptide (Co-eIF-2A80) with Co-eIF-2A activity. J. Biol. Chem. 1985;260:6945–6949. [PubMed] [Google Scholar]
- 45.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolin SL, Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 1989;109:2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussey GS, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.