Abstract
In vivo tracking of the delivery of therapeutic proteins is a useful tool for preclinical studies. However, many labels are too large to use without disrupting the normal uptake, function, or other properties of the protein. Low-molecular weight fluorescent labels allow in vivo and ex vivo tracking of the distribution of therapeutic proteins, and should not alter the protein’s characteristics. We tested the in vitro properties of fluorescent-labeled recombinant human alpha-L-iduronidase (rhIDU, the enzyme deficient in Hurler syndrome) and compared labeled to unlabeled protein. Labeled rhIDU retained full enzymatic activity and showed similar kinetics to non-labeled rhIDU. Uptake of labeled rhIDU into human Hurler fibroblasts, measured by activity assay, was equivalent to unlabeled rhIDU enzyme and showed an uptake constant of 0.72 nM. Labeled rhIDU was also able to enter cells via the mannose 6-phospate receptor pathway and reduce glycosaminoglycan (GAG) storage in Hurler fibroblasts. Subcellular localization was verified within lysosomes by confocal microscopy. These findings suggest that fluorescent labeling does not significantly interfere with enzymatic activity, stability, or uptake, and validates this method as a way to track exogenously administered enzyme.
Keywords: Fluorescence, lysosomal, enzyme replacement therapy, iduronidase, mucopolysaccharidosis, Hurler
1. Introduction
Protein therapeutics comprise an ever-growing share of drug development. Immunoglobulin therapies are used to treat cancer, autoimmune diseases, immunodeficiencies and chronic viral illnesses [1]. In the field of lysosomal storage diseases, protein therapies in the form of recombinant enzymes are on the U.S. market to treat six different disorders, and many others are in development [2–7]. Preclinical development of protein therapies requires testing their ability to distribute and penetrate target areas. In the case of enzymes, activity assays can be used for this purpose. However, when the therapeutic enzyme is administered to a normal animal, the presence of endogenous activity may make this method impractical. Similarly, the use of specific antibodies to track the protein may be complicated if species cross-reactivity exists. Both of these tracking methods also have the obvious disadvantage of requiring ex vivo tissue analysis. In vivo tracking would require tagging the protein with something that can be detected in an intact animal, but a large tag that alters the protein’s characteristics is not desirable. To accurately track the movement of protein therapeutics through CSF and elsewhere, another method is needed.
Here, we evaluate fluorescent-labeled recombinant alpha-L-iduronidase (rhIDU, E.C. 3.2.1.76), which is currently used as therapy for the lysosomal storage disease mucopolysaccharidosis I (Hurler syndrome). Fluorescent labeling has been used previously to track the distribution of intravenously-administered recombinant N-acetylgalactosamine-6-sulfate sulfatase ex vivo, and intrathecally-administered butylcholinesterase in vivo and ex vivo in mice [8;9]. The properties of labeled protein in each case were not characterized. The tag used here is relatively small (1.15 kDa), compared to green-fluorescent protein, quantum dots, and/or other labels. We show through in vitro testing that low molecular-weight fluorescent labeling does not substantially alter the properties of rhIDU, validating its use for tracking the therapeutic protein.
2. Materials and Methods
2.1. Fluorescent labeling of rhIDU
rhIDU (formulated as laronidase, BioMarin Pharmaceutical, Inc., Novato, CA; 0.58 mg/ml in 100mM sodium phosphate buffer, pH 5.5 containing 0.001% polysorbate 80) was concentrated using an Amicon YM30 concentrator (stirred ultrafiltration cell; Millipore, Billerica, MA) to 2 mg/ml. 1M sodium bicarbonate solution (pH 8.3) was added to the enzyme to final pH 7.5–8.5 for maintaining aliphatic amine groups of protein in the non-protonated state. The protein solution (0.5 ml) was transferred into a vial with Alexa Fluor 680 (Alexa Fluor 680 Protein Labeling Kit, Invitrogen, Carlsbad, CA), covered with foil and stirred for 1 h at 22 °C. Labeled rhIDU was separated from unincorporated dye by size exclusion chromatography (purification resin was supplied with the kit). Protein concentration and degree of labeling (DOL) was determined by absorbance of labeled rhIDU at 280 and 679 nm according to the manufacturer’s instructions.
2.2. Enzyme activity and stability assays
Fluorescent-labeled rhIDU was tested for the retention of enzyme activity. Enzyme activity was determined by fluorogenic assay using 50 μM 4-methylumbelliferyl α-L-iduronide (4-MUI) (Glycosynth, Warrington, UK) at 37 °C for 10 minutes. Fluorescence measurements were read at 365 nm on an LS 45 fluorescence spectrometer (Perkin-Elmer, Waltham, MA). One activity unit is equal to 1 nmole converted substrate per h. For determination of pH optimum, labeled rhIDU activity was tested in various phosphate/citrate (200mM/100mM) buffers ranging from pH 2.6 to 8.0. For stability, the standard activity assay was performed at the indicated pH following storage for various periods of time at 4 °C, 25 °C, and 37 °C. A pH of 4.4 corresponded to lysosomal pH, 5.5 to pH of the storage buffer for rhIDU, and 7.4 to physiologic pH.
2.3. Uptake of fluorescent labeled rhIDU into MPS I fibroblasts
For biochemical (quantitative) measurement of uptake, rhIDU labeled with Alexa Fluor 680 or unlabeled was applied in concentrations ranging from 10 to 160 units/ml to human Hurler fibroblasts (GM 1391) cultured in minimal essential medium (MEM) supplemented with 2 mM glutamine without fetal bovine serum in 6-well plates at 37 °C and 5% CO2. Mannose 6-phosphate was also added to the medium at a final concentration of 5 mM for inhibition testing in a subset of experiments. Following 4 h incubation at 37 °C and 5% CO2, cells were harvested by trypsinization. The pellet was washed in phosphate buffered saline (PBS) and resuspended in 60 μl PAD buffer (10 mM sodium phosphate, pH 5.8, 0.02% sodium azide, 0.1 mM dithiothreitol, 0.1% Triton® X-100). Pellets were sonicated, centrifuged, and cell lysates measured for iduronidase activity as described above. Kinetic data were obtained from Lineweaver–Burk plots. Determination of uptake by direct detection of labeled enzyme in cell lysates was also performed by spectrofluorometry at the excitation/emmission settings of 640/700 nm. Fluorescence intensity was compared to a standard curve generated using dilutions of labeled rhIDU at defined units/ml of activity.
For qualitative determination of uptake by confocal microscopy, Hurler fibroblasts treated with labeled or unlabeled rhIDU were grown on coverslips and stained with LysoTracker Red® DND-99 (Invitrogen, Carlsbad, CA) to identify lysosomes and rabbit polyclonal antibody against rhIDU (BP13, donated by Biomarin Pharmaceutical). Immediately following treatment, cells were incubated with LysoTracker for 5 min and then rinsed in PBS prior to fix. Cells were blocked in 1% goat serum in PBS with 0.3% Triton® X-100, and incubated 30 min with primary antibody (1:100 dilution in blocking solution) followed by three PBS washes and treatment for 1 h at 22° C with FITC goat-anti rabbit IgG secondary antibody (1:5000 dilution in blocking solution) (Invitrogen, Carlsbad, CA). Finally, cells were washed with PBS and mounted onto glass microscope slides using Vectashield (Vector Laboratories, Burlingame, CA). Uptake and colocalization were determined with confocal microscopy on a Leica SP2 (Leica Microsystems, Bannockburn, IL). Untreated Hurler fibroblasts were used as controls.
2.4. Glycosaminoglycan storage reduction
Cultured fibroblasts were labeled with H235SO4 as described elsewhere [10]. Briefly, Hurler fibroblasts were grown to confluence in six-well plates. Fluorescent-labeled or unlabeled rhIDU was applied to the cells (0.0–0.4 units/ml) in serum-free MEM with 25 μM H235SO4 (1050Ci/mM) and incubated for 48 h at 37 °C and 5% CO2. Cells were washed in PBS before and after trypsinization and extracted twice with boiling in 80% ethanol followed by centrifugation in a clinical centrifuge for 15 minutes at the highest speed. Pellets were resuspended in 10% sodium hydroxide and neutralized with 2 M acetic acid, and radiolabeled GAG were measured via scintilliation counting (Tri-Carb 2800TR, Perkin-Elmer, Waltham, MA). Radioactive counts per minute were normalized to protein concentrations as determined using a Bio-Rad protein assay (Bio-Rad, Hercules, CA).
3. Results
3.1. Activity and stability of rhIDU are unchanged following fluorescent labeling
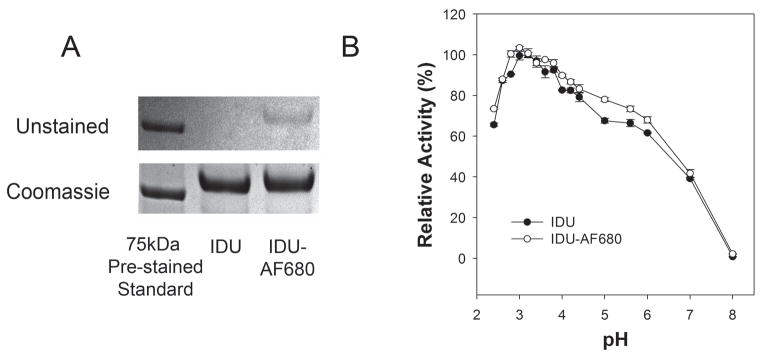
rhIDU was labeled with Alexa Fluor 680 in two separate batches to a DOL of either 1.7 or 3.1 molecules of dye/molecule of protein (Fig. 1A). Specific activity of rhIDU was 108,000 units/mg for DOL of 1.7 and 84,000 units/mg for DOL of 3.1. Following labeling and purification to remove unincorporated dye, 80% or greater of the total activity was recovered in the process. As a lysosomal hydrolase, rhIDU is known to have maximum activity under acidic conditions resembling the lysosome of the cell. We examined the range of pH for enzyme activity pre- and post-labeling towards the artificial substrate 4-MUI. Both labeled and unlabeled rhIDU behaved identically with optimum performance at approximately pH 3.3 (Fig. 1B).
Figure 1.
Labeling and pH optimum of rhIDU. (A) Polyacrylamide gel electrophoresis (4–20%) analysis of rhIDU (IDU) before and after the labeling process. Upper panel shows unstained gel with Alexa Fluor 680 labeled protein (IDU-AF680) clearly visible to the naked eye (DOL of 3.1). Lower panel is the same gel after staining with coomassie blue. (B) Labeled (DOL 3.1) or unlabeled IDU were diluted to 16 μg/ml in 200 mM/100 mM phosphate-citrate buffer at the indicated pHs (2.4–8.0) and immediately assayed for enzyme activity towards the artificial fluorogenic substrate 4-MUI for 15 minutes at 22 °C. Relative activity for each enzyme was plotting from an average of three independent experiments where the maximally observed activity was defined as 100%.
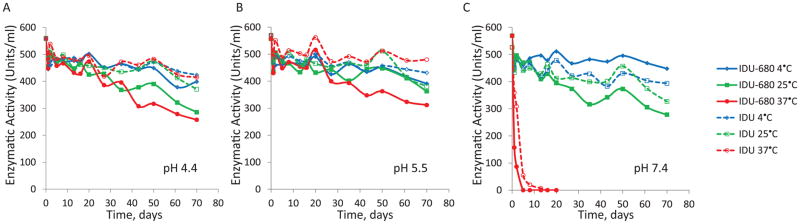
While lysosomal hydrolases are the most active under acidic conditions found in the lysosome, they nonetheless must be structurally stable under the mildly acidic pH of endosomes and neutral pH found in the cytosol and extracellular environments. Purified rhIDU was previously shown to exhibit stability over prolonged periods of storage with very little loss of activity [11]. We compared the impact of labeling on stability at typical storage and physiologically relevant temperatures and pH. Notably, both labeled and unlabeled rhIDU exhibited similar activities for all conditions tested for up to two months (the endpoint of the assay) (Fig. 2A–C). We also observed strict sensitivity of both enzymes to freezing with loss of enzyme activity following a single freeze-thaw cycle (data not shown). We thus routinely stored our rhIDU before and after labeling at 4 °C.
Figure 2.
Stability of fluorescent-labeled rhIDU. Stability of the enzyme as a function of pH and storage temperature for labeled and unlabeled rhIDU are shown over the course of 70 days incubation. Labeled (solid lines) and unlabeled (dashed lines) enzymes were diluted to 16 μg/ml in 100 mM/50 mM phosphate-citrate buffer at pH 4.4 (A), pH 5.5 (B), and pH 7.4 (C) and these stock solutions were incubated at various temperatures, 4°C (blue), 25°C (green), 37°C (red) over a 70-day period. Enzyme aliquots were taken throughout this period of time and activity was estimated under the standard conditions (25 mM 4-MUI for 10 minutes at 22 °C). Units of activity are defined as nmoles of 4-MU product formed per hour.
3.2. Uptake of labeled rhIDU into Hurler fibroblasts
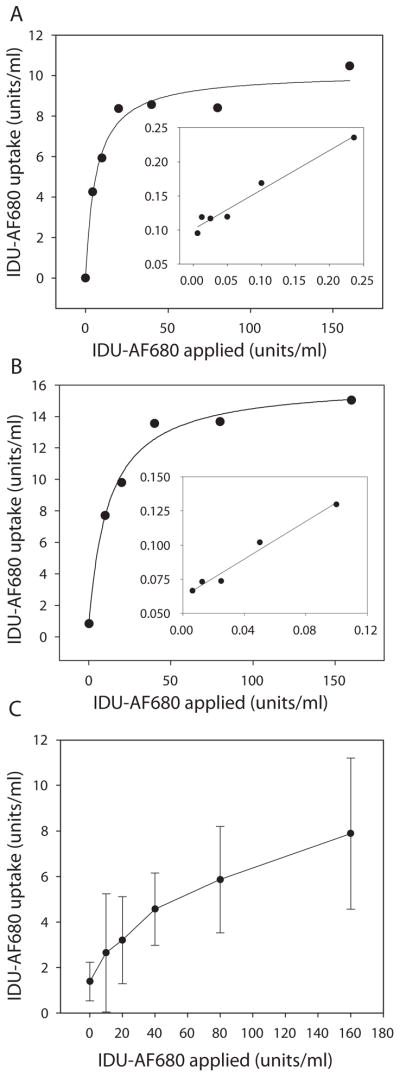
Labeled rhIDU (rhIDU-AF680) was applied to Hurler fibroblasts in 6-well plates and incubated for 4 h to determine cellular uptake. The uptake constant (Kuptake) for rhIDU-AF680 (DOL of 1.7) was 0.72 nM (Fig. 3A), equal to that previously published for unlabeled rhIDU (0.7 nM) [11]. Only a slight increase in the uptake constant was observed with the higher degree of labeling (DOL 3.1) to a Kuptake of 1.4 nM, well within the physiological range for normal performance (Fig. 3B). We also tested the ability to evaluate cellular uptake by directly measuring the appearance of fluorescently labeled protein inside treated Hurler cell lysates and observed strong dose-dependent signal increases by spectrofluorometry (Fig. 3C).
Figure 3.
Kinetics of cellular uptake of rhIDU labeled with Alexa Fluor 680. Labeled rhIDU (IDU-AF680) at either DOL of 1.7 (A) or DOL 3.1 (B) was applied to confluent Hurler fibroblasts (GM 1391) in 6 well dishes at the indicated doses in MEM medium without serum and incubated for 4 hours. Double reciprocal (Lineweaver-Burk) plots of the kinetic data are shown as insets. (C) Qualitative direct detection of Alexa Fluor 680 taken up by Hurler cells. Fluorescence was monitored in cell lysates at 640/700 nm excitation/emission wavelengths, and uptake was determined by comparison to a standard curve of fluorescent intensity emitted at 700 nm vs. input amount of Idu-AF680 (units/ml). All experiments were performed in triplicate.
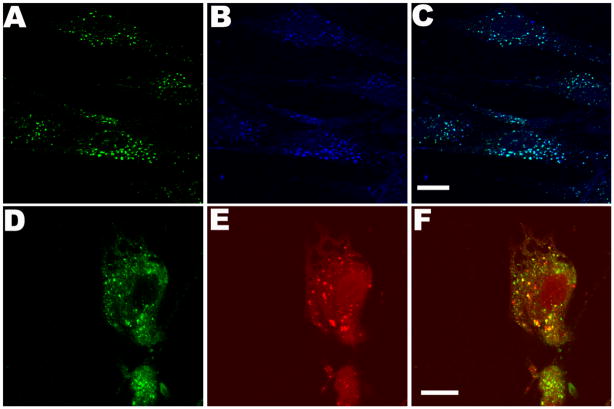
Intracellular uptake of rhIDU-AF680 into Hurler skin fibroblasts was further verified qualitatively by confocal microscopy (Fig. 4). Fluorescence clustered within Hurler fibroblasts (Fig. 4A) and was confirmed to be associated with labeled rhIDU-AF680 (vs. free dye) by co-localization with polyclonal anti-iduronidase antibody (Fig. 4B–C). Co-localization of anti-iduronidase antibody and LysoTracker demonstrated lysosomal targeting of rhIDU-AF680 (Fig. 4D–F).
Figure 4.
Uptake and lysosomal targeting of fluorescent-labeled rhIDU into human Hurler patient fibroblast cultures. AF680 labeled enzyme (160 units) was directly applied to Hurler fibroblasts grown on coverslips in serum-free MEM and incubated for 4 hours at 37 °C and 5% CO2. (A–C) Detection of AF680 label (blue) anti-rhIDU antibodies (green) and co-localization (aqua) in Hurler cells. (D–F) Lysosomal targeting of rhIDU-AF680. Detection with anti-iduronidase antibody (green), LysoTracker Red DND-99 (red), and merge. All colors are generated by Leica LCS imaging software. Scale bars are 15 μm.
3.3. Labeled rhIDU enters cells via the mannose 6-phosphate receptor and reduces lysosomal storage in vitro
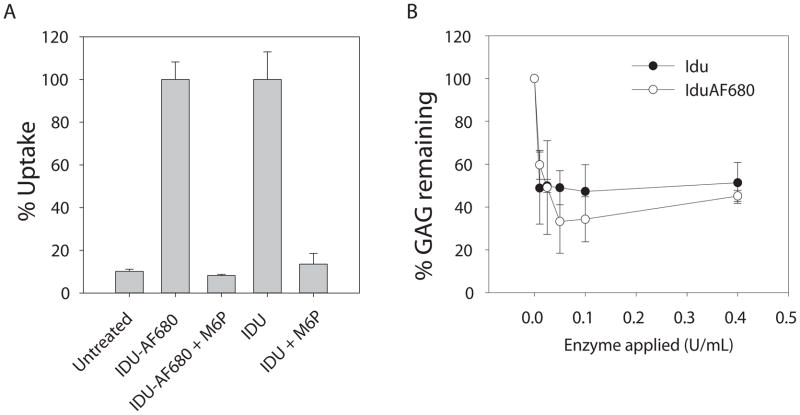
While labeled rhIDU appeared to be able to enter cells by activity assay and co-localize with lysosomal compartments, it was important to verify that the presence of the AF680 molecules attached to the surface of the enzyme did not alter the mechanism of delivery. The uptake of labeled rhIDU into Hurler fibroblasts was completely inhibited by the addition of free mannose 6-phosphate into the cell culture medium illustrating normal delivery via the mannose 6-phospate/insulin like growth factor II receptor pathway (Fig. 5A).
Figure 5.
Mannose 6-phosphate receptor mediated delivery and GAG storage reduction. (A) 80 activity units per ml rhIDU with Alexa Fluor 680 label (IDU-AF680) or unlabeled (IDU) were applied to confluent Hurler fibroblasts in six-well plates containing 1 ml of serum free MEM with or without competing 5 mM mannose 6-phosphate (M6P) for 4 hours. Enzymatic activity assays using 4-MUI substrate were performed on cell lysates and percent uptake was plotted relative to maximal uptake observed in the absence of inhibitor. (B) IDU or IDU-AF680 was applied (0.0–0.4 units/ml) to cultured Hurler fibroblasts in the presence of H235SO4 and incubated for 48 h at 37 °C and 5% CO2 as described in the methods and reported in detail previously [11]. Radioactive labeled GAG were isolated from harvested cell pellets by two rounds of ethanol extraction, acid solubilized, and quantified by scintillation counting. Radioactive counts per minute were normalized to total protein concentration in the extracted pellet and plotted as relative percent labeled GAG compared to the untreated samples at 0 units of enzyme applied.
The ability of labeled rhIDU (DOL 1.7) to reduce pathological GAG accumulation in Hurler fibroblasts was tested in vitro using a 35S assay (Fig. 5B). A concentration of as little as 0.01 units/ml labeled rhIDU reduced GAG storage in Hurler cells by 40.0% following 48 hour incubation. Unlabeled rhIDU reduced GAG by 54.0%. Both labeled and unlabeled rhIDU reached a plateau of detectable labeled storage reduction (around 60% reduction) as reported previously [11].
4. Discussion
In this study, a small molecular weight label (1.15 kDa) with in vivo and ex vivo applications was applied to a therapeutic protein without substantially altering its properties. rhIDU labeled with a near-infrared fluorophore shows enzymatic activity, stability, receptor-mediated cellular uptake, and storage reduction that is equivalent to unlabeled enzyme. At a degree of labeling of 3.1 moles dye per mole protein, labeled rhIDU retained 80% of its activity and showed similar kinetics to unlabeled rhIDU. Labeled rhIDU was able to enter Hurler fibroblasts with similar, high-affinity uptake and reduced glycosaminoglycan storage in Hurler fibroblasts. Lysosomal localization was verified by confocal microscopy.
Other, larger labels have been used to track proteins in vivo. Quantum dots can be conjugated to antibodies or other proteins for tumor imaging in mice [12]. Green-fluorescent protein can be fused to a protein, allowing very sensitive detection of the protein in vivo and ex vivo [13]. However, these labels are large, and may interfere with the kinetics, distribution, or uptake of an enzyme into tissue and cells. This is especially critical for optimization of protein therapy formulations for hard to treat tissues. Fusions also preclude the use of formulated protein. Low molecular-weight fluorescent labels can be applied post-translationally, allowing direct labeling of the product as formulated and avoiding additional time and costs associated with producing and purifying the protein from DNA fusion constructs. Clinically used dyes that do not bind the protein are also problematic. Gadolinium was co-administered with adeno-associated virus (AAV) intraparenchymally and tracked in vivo using magnetic resonance imaging [14]. However, the tissue distribution of AAV and gadolinium was not precisely equivalent. 18I-labeled and fluorescent-labeled substrate analogs have recently been developed for in vivo imaging of glucocerebrosidase [15;16]. These molecules bind to the enzyme’s active site, and are therefore enzyme-specific. No similar labeled substrate analogs exist for rhIDU.
The fluorescent label used in this study was a near-infrared label with high stability for in vivo imaging. Succinimidyl esters are preferred over other amine-reactive fluorescent dyes due to their ability to form very stable amide bonds between the dye and non-protonated aliphatic amine groups of the protein of interest. These dyes show higher signal intensity, enhanced resistance to photobleaching, pH insensitivity and a high degree of water solubility. The pH insensitivity is a key feature to enable studies on enzymes that may be rapidly internalized into the highly acidic lysosomal environment. Microgram amounts of protein can be detected in live mice using a small animal imager [17]. This method can be applied to preclinical testing of a therapeutic enzyme for which a large affected animal model is too costly or not readily available. For example, Johnson et al. labeled the enzyme butylcholinesterase, injected it into the spinal fluid of mice, and were able to successfully track the protein in vivo, showing delivery to brain up to 25 h following a lumbar injection [9].
Currently, in vivo imaging of fluorescent-labeled proteins is limited to small animals, and optimum quality images require that the mice be nude or depilated. However, ex vivo imaging can be performed on sections from any tissue using fluorescence microscopy, potentially enabling determination of the distribution of a therapeutic protein in even larger animal models. Of particular use would be the ability to track the movement of a therapeutic protein across the blood-brain barrier, and other hard to treat tissues (such as the heart valve or cartilage) in a canine MPS I model. This method may also prove useful for development of other therapeutic proteins for CNS delivery.
We have demonstrated that a human recombinant lysosomal protein used in enzyme replacement therapy can be easily labeled for in vivo imaging and ex vivo analysis without any significant alteration to enzyme kinetics, receptor binding affinity, sub-cellular targeting, or therapeutic function. The maintenance of normal behavior should allow reliable tracking of tissue distribution without major concern over altered affinity because of the tag used. The use of a labeled protein allows ready determination of the administered protein without background from a normal animal often associated with immunohistochemistry.
Acknowledgments
rhIDU (as laronidase) and BP13 antibody were donated by BioMarin Pharmaceutical Inc. (Novato, CA). The Los Angeles Biomedical Research Institute and the Harbor-UCLA Department of Pediatrics have financial interest in laronidase. Support was provided by the National Institutes of Health [NS054242 to PID].
Footnotes
Conflicts of interest: The Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center has a financial interest in recombinant human alpha-L-iduronidase (laronidase). Dr. Dickson receives research support from Biomarin Pharmaceuticals, Inc. and Genzyme Corporation.
References
- 1.Leader B, Baca QJ, Golan DE. Nat Rev Drug Discovery. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 2.Barton NW, Brady RO, Dambrosia JM, Di Bisceglie AM, Doppelt SH, Hill SC, Mankin HJ, Murray GJ, Parker RI, Argoff CE, Grewal RP, Yu KT. N Engl J Med. 1991;324:1464–1470. doi: 10.1056/NEJM199105233242104. [DOI] [PubMed] [Google Scholar]
- 3.Eng CM, Guffon N, Wilcox WR, Germain DP, Lee P, Waldek S, Caplan L, Linthorst GE, Desnick RJ the International Fabry Disease Study Group. N Engl J Med. 2001;345:9–16. doi: 10.1056/NEJM200107053450102. [DOI] [PubMed] [Google Scholar]
- 4.Kakkis ED, Muenzer J, Tiller GE, Waber L, Belmont J, Passage M, Izykowski B, Phillips J, Doroshow R, Walot I, Hoft R, Neufeld E. N Engl J Med. 2001;344:182–188. doi: 10.1056/NEJM200101183440304. [DOI] [PubMed] [Google Scholar]
- 5.Harmatz P, Giugliani R, Schwartz I, Guffon N, Teles EL, Miranda MCS, Wraith JE, Beck M, Arash L, Scarpa M, Yu ZF, Wittes J, Berger KI, Newman MS, Lowe AM, Kakkis E, Swiedler SJ. J Pediatrics. 2006;148:533–539. doi: 10.1016/j.jpeds.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Muenzer J, Wraith JE, Beck M, Giugliani R, Harmatz P, Eng CM, Vellodi A, Martin R, Ramaswami U, Gucsavas-Calikoglu M, Vijayaraghavan S, Wendt S, Puga AC, Ulbrich B, Shinawi M, Cleary M, Piper D, Conway AM, Kimura A. Genet Med. 2006;8:465–473. doi: 10.1097/01.gim.0000232477.37660.fb. [DOI] [PubMed] [Google Scholar]
- 7.Amalfitano A, Bengur AR, Morse RP, Majure JM, Case LE, Veerling DL, Mackey J, Kishnani P, Smith W, McVie-Wylie A, Sullivan JA, Hoganson GE, Phillips JA, Schaefer GB, Charrow J, Ware RE, Bossen EH, Chen YT. Genet Med. 2001;3:132–138. [PubMed] [Google Scholar]
- 8.Tomatsu S, Montano AM, Gutierrez M, Grubb JH, Oikawa H, Dung VC, Ohashi A, Nishioka T, Yamada M, Yamada M, Tosaka Y, Trandafirescu GG, Orii T. Mol Genet Metab. 2007;91:69–78. doi: 10.1016/j.ymgme.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Johnson ND, Duysen EG, Lockridge O. Neuro Toxicol. 2009;30:386–392. doi: 10.1016/j.neuro.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao KW, Neufeld EF. Protein Expression Purif. 2000;19:202–211. doi: 10.1006/prep.2000.1230. [DOI] [PubMed] [Google Scholar]
- 11.Kakkis ED, Matynia A, Jonas AJ, Neufeld EF. Protein Expression Purif. 1994;5:225–232. doi: 10.1006/prep.1994.1035. [DOI] [PubMed] [Google Scholar]
- 12.Bentolila LA, Ebenstein Y, Weiss S. J Nucl Med. 2009;50:493–496. doi: 10.2967/jnumed.108.053561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arun KHS, Kaul CL, Romarao PJ. Pharmacol Toxicol Methods. 2005;51:1–23. doi: 10.1016/j.vascn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Fiandaca MS, Varenika V, Eberling J, McKnight T, Bringas J, Pivirotto P, Beyer J, Hadaczek P, Bowers W, Park J, Federoff H, Forsayeth J, Bankiewicz KS. Neuro Image. 2009;47:T27–T35. doi: 10.1016/j.neuroimage.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phenix CP, Rempel BP, Colobong K, Doudet DJ, Adam MJ, Clarke LA, Withers SG. Proc Natl Acad Sci USA. 2010;107:10842–10847. doi: 10.1073/pnas.1003247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witte MD, Kallemeijn WW, Aten J, Li KY, Strijland A, Donker-Koopman WE, van den Nieuwendijk AMCH, Bleijlevens B, Kramer G, Florea BI, Hooibrink B, Hollak CEM, Ottenhoff R, Boot RG, van der Marel GA, Overkleeft HS, Aerts JMFG. Nat Chem Biol. 2010;6:907–913. doi: 10.1038/nchembio.466. [DOI] [PubMed] [Google Scholar]
- 17.Levenson RM, Mansfield JR. Cytometry Part A. 2006;69A:748–758. doi: 10.1002/cyto.a.20319. [DOI] [PubMed] [Google Scholar]