Abstract
Most physiological and biological processes are regulated by endogenous circadian rhythms under the control of both a master clock, which acts systemically and individual cellular clocks, which act at the single cell level. The cellular clock is based on a network of core clock genes, which drive the circadian expression of non-clock genes involved in many cellular processes. Circadian deregulation of gene expression has emerged to be as important as deregulation of estrogen signaling in breast tumorigenesis. Whether there is a mutual deregulation of circadian and hormone signaling is the question that we address in this study. Here we show that, upon entrainment by serum shock, cultured human mammary epithelial cells maintain an inner circadian oscillator, with key clock genes oscillating in a circadian fashion. In the same cells, the expression of the estrogen receptor α (ERA) gene also oscillates in a circadian fashion. In contrast, ERA-positive and -negative breast cancer epithelial cells show disruption of the inner clock. Further, ERA-positive breast cancer cells do not display circadian oscillation of ERA expression. Our findings suggest that estrogen signaling could be affected not only in ERA-negative breast cancer, but also in ERA-positive breast cancer due to lack of circadian availability of ERA. Entrainment of the inner clock of breast epithelial cells, by taking into consideration the biological time component, provides a novel tool to test mechanistically whether defective circadian mechanisms can affect hormone signaling relevant to breast cancer.
Key words: circadian rhythm, clock genes, estrogen receptor alpha (ERA), breast cancer cells, entrainment, serum shock
Introduction
As a consequence of the Earth's rotation about its axis, most organisms on this planet have evolved a clock that allows them to tailor their physiology and behavior rhythmically, with a period length of about 24 h. These endogenous, circadian (from the Latin “ca. diem”) rhythms can sustain themselves even in the absence of external cues. Disruption of endogenous circadian rhythms seem to have a major impact on overall health and has been correlated with aging and diseases, such as coronary heart disease, various neurodegenerative disorders and cancer.1–9
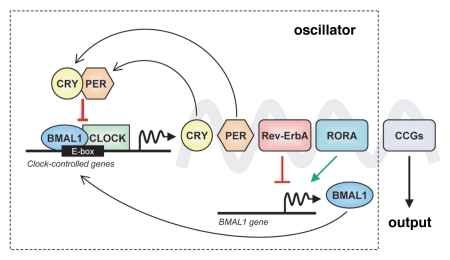
The generation of endogenous circadian rhythms is now known to be an attribute of each individual cell's own molecular time piece, hereafter referred to as circadian oscillator or clock. This clock is based on a genetic network of transcriptional and post-translational negative feedback loops that generate rhythmic 24 h expression of core clock components.1,2,10,11 In mammals, the molecular clocks of all cells are centrally controlled by a main coordinator in the brain, the suprachiasmatic nucleus, which, upon perceiving light from the environment, synchronizes all peripheral clocks to sustain coordinated rhythms for the organism.1,2,10,11 Interestingly, even when removed from the organism, isolated cells, such as fibroblasts, maintain their autonomous timing mechanism (ref. 12 and the references within). The core clock components are represented by the genes that encode the proteins necessary for the generation and maintenance of normal circadian rhythms in individual cells1,13 (Fig. 1). The primary feedback loop comprises eight core clock genes: CLOCK, Casein kinase Iε (CKIE), Cryptochrome 1 (CRY1), Cryptochrome 2 (CRY2), Period 1 (PER1), Period 2 (PER2), Period 3 (PER3) and BMAL1. Another regulatory loop includes two retinoic acid-related orphan nuclear receptors, Rev-Erbα (RevErbA) and RORα (RORA). CLOCK and BMAL1 genes encode for basic helix-loop-helix transcriptional activators that can heterodimerize and regulate the transcription of the PER and CRY genes by binding to E-box elements (CACGTG) in their promoters. The PER proteins build up in the cytoplasm and are degraded by CKIE. The CRY proteins, once they accumulate in the cytoplasm up to a critical threshold, bind the PER proteins, and form a stable complex that can reenter the nucleus and interfere with the heterodimerization of CLOCK and BMAL1. The CLOCK-BMAL1 heterodimer can also drive the transcription of Rev-ErbA and RORA, which repress and drive, respectively, the transcription of BMAL1 itself. The end result of these regulatory pathways is a 24-h rhythmic oscillation of genes of the inner clock. CLOCK and CKIE are the only components of the circadian clock whose expression does not oscillate. Importantly, the CLOCK/BMAL1 complex is able to output to other signaling pathways via clock-controlled genes (CCGs) (Fig. 1), which can influence a wide range of functions outside the clock.2,14,15 It is thought that > 10% of all mammalian transcripts undergo circadian oscillations.16–18
Figure 1.
The mammalian circadian clock. Simplified representation of the cell molecular clock, consisting of an oscillator in which clock gene transcripts and proteins accumulate in a circadian fashion due to a complex feedback loop. The output of this inner oscillator is the circadian transcriptional regulation of many clock-controlled genes (CCGs) directly bound by BMAL1-CLOCK.
Remarkably, nuclear receptors are functionally intertwined with the circadian oscillator.19 In the mouse, the mRNAs of more than half the nuclear receptors accumulates periodically to enable the rhythmic control of energy, glucose and lipid metabolism.20 Moreover, estrogen receptor α (ERA) function was found to be linked to the circadian oscillator in breast cancer cells.21,22 Apparently, the periodic accumulation of PER2 and ERA transcripts/proteins is reciprocally controlled. On one hand, PER2 is an estrogen-inducible gene that contains an estrogen response element (ERE) in its promoter region that can be directly bound by ERA in the presence of estrogen. On the other hand, the PER2 protein seems capable of interacting directly with the ERA protein, thus affecting its stability. This suggests that PER2 circadian oscillation may influence ERA oscillation as well.
ERA-mediated estrogen signaling plays a key role in mammary gland morphogenesis and development, as shown by the inability of proper mammary gland ductal elongation in ERA-knockout mice.23 Aberrant estrogen-mediated signaling through ERA is intimately linked to a large majority of breast cancer.24 Several studies over the years have shown a correlation between loss of circadian rhythm and breast cancer in women (reviewed in refs. 25–30).
To understand the reciprocal influence of circadian oscillator and estrogen signaling in breast tumorigenesis, we established a cell system that enables us to couple the analysis of both circadian oscillator and estrogen signaling in human mammary epithelial cells in a circadian fashion. To this end, we took advantage of a serum shock strategy, first described by Balsalobre et al.,31 through which mammalian cultured fibroblasts could be induced to synchronously express clock genes in a circadian fashion in response to a short treatment with high concentrations of serum (entrainment). In this study, we slightly modified this method to demonstrate, first, that the circadian oscillator of human mammary epithelial cells can be entrained in culture, and second, that the pattern of circadian mRNA accumulation of both clock genes and ERA is different between human mammary epithelial cells and breast cancer cells. Upon entrainment of the inner clock of the human mammary epithelial cell line HME1, key clock genes are transcribed in a circadian fashion. In contrast, entrainment of ERA-positive and ERA-negative breast cancer epithelial cells shows lack of circadian oscillation of key clock gene expression. Interestingly, we also show that circadian accumulation of ERA mRNA occurs in HME1 cells but not in ERA-positive breast cancer cells.
Results
ERA-positive human mammary epithelial HME1 cells are endowed with an inner functional clock.
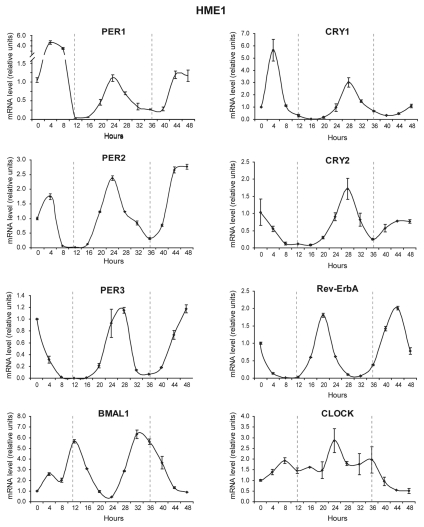
Circadian oscillation of clock genes was first demonstrated in cultured fibroblasts by using a serum shock method.31 To assess whether human mammary epithelial cells display circadian oscillation of clock genes, we used a slight modification of that method to entrain HME1, a human mammary cell line derived from a mammoplasty and immortalized with telomerase. HME1 cells have many features of normal mammary epithelial cells: they are ERA-positive, capable of acinar morphogenesis when grown in three-dimensional (3D) culture on reconstituted basement membrane and do not form tumors when xenografted in nude mice (Table 2). Upon HME1 entrainment, we first measured the mRNA accumulation of core circadian genes, PER1, PER2, PER3, CRY1, CRY2, Rev-ErbA, BMAL1 and CLOCK, over a 48 h period and plotted this as a function of time. PER1, PER2 and PER3 transcription showed oscillations that peaked at 24 and 48 h. The mRNA levels of Rev-ErbA peaked at 20 and 44 h. CRY1 and CRY2 both had initial peaks at 28 h and began increasing again at approximately 48 h. In contrast, BMAL1 mRNA oscillated approximately antiphase to the Period genes with peaks at approximately 12 and 36 h. CLOCK mRNA did not show any circadian oscillation (Fig. 2). Based on these observations, apparently HME1 cells have conserved a functional inner clock that can be entrained in vitro.
Table 2.
Molecular and biological features of the cell lines used in this study
| HME1 | T47D | MCF7 | MCF10A | HS578T | MB-MDA-231 | |
| ERA-positive | Yes | Yes | Yes | No | No | No |
| Immortal | Yes | Yes | Yes | Yes | Yes | Yes |
| Capable of proper acinar morphogenesis in 3D culture on reconstituted basement membrane | Yes | No | No | Yes | No | No |
| Capable of anchorage-independent growth in semisolid medium | No | Yes | Yes | No | Yes | Yes |
| Tumorigenic | No | Yes | Yes* | No | No | Yes |
| Invasive | No | No | Yes* | No | No | Yes |
In the presence of estrogen.
Figure 2.
ERA-positive human mammary epithelial HME1 cells are endowed with an inner functional clock. Quantitative RT-PCR analysis on cultured HME1 cells at 4-h intervals after entrainment via serum shock shows that the mRNA of the clock genes PER1, PER2, PER3, CRY1, CRY2, RevErbA and BMAL1 accumulates in a circadian fashion, while the mRNA of CLOCK does not show any circadian oscillation.
Entrainment of ERA-negative human mammary epithelial MCF10A cells detects a defective clock.
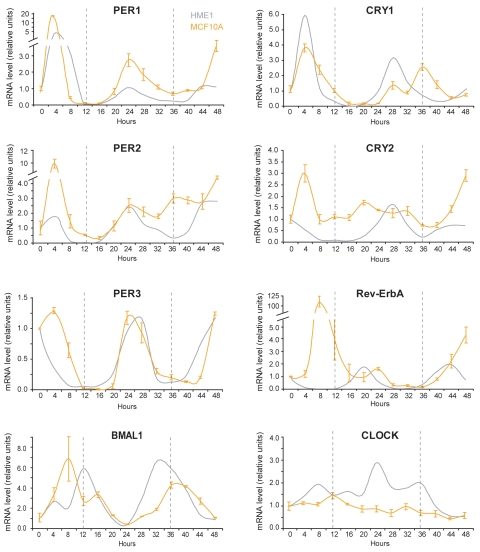
Next, we entrained an immortal, non-tumorigenic human mammary epithelial cell line, MCF10A, which was derived from a fibrocystic breast tissue, and is ERA-negative. MCF10A cells are capable of acinar morphogenesis in 3D culture on reconstituted basement membrane and do not form tumors when xenografted in nude mice (Table 2). We found that MF10A cells show rhythmic oscillations of clock gene expression, albeit more irregular ones than the ones detected in HME1 cells. Specifically, while the oscillations of PER1 and PER3 mRNA were circadian and showed peaks at 24 and 48 h, the oscillation of PER2 showed a less clear circadian pattern, with an increase in mRNA between the 24 and 48 h peaks. A circadian oscillation was detectable also in Rev-ErbA mRNA, with peaks (24 h and 48 h) delayed approximately 4 h relative to HME1 and operating anti-phase to BMAL1. CRY1 and CRY2 mRNAs oscillate irregularly and do not show a circadian pattern. CLOCK mRNA did not oscillate (Fig. 3). There is no way for us to conclude whether the MCF10A circadian oscillation pattern is a reflection of loss of ERA expression. However, we cannot discount that the defective clock may be consequent to lack of transactivation of PER2, a downstream ERA target gene,22 due to lack of estrogen-ERA signaling in MCF10A cells.
Figure 3.
Entrainment of ERA-negative human mammary epithelial MCF10A cells detects a defective clock. Quantitative RT-PCR at 4 h time points after entrainment shows that in cultured MCF10A cells, only PER1 and PER3 mRNAs have a circadian oscillation comparable with HME1 cells, while the other clock genes display either an imperfect circadian oscillation (CRY1, PER2, RevErbA, BMAL1) or no circadian oscillation (CRY2, CLOCK).
ERA-negative human breast cancer cells show a disrupted inner clock.
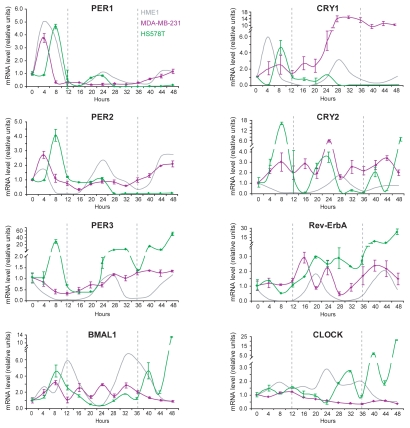
To gather additional insights as to whether ERA-negative breast cancer cell lines always display a defective clock, we entrained two ERA-negative breast cancer cell lines, HS578T and MDA-MB-231. Analysis of the mRNA oscillation of HS578T clock genes showed that PER1 and PER2 mRNAs, after an initial peak following the serum shock, slightly increased at 24 h but sharply declined and remained constant thereafter. PER3 mRNA showed the lowest levels at approximately 16 h and 36 h, while it accumulated mostly at 28–32 h and at 48 h. CRY1 mRNA underwent an oscillation similar to PER1, with a weak peak at 24 h then declining and remaining constant thereafter. CRY2 showed non-circadian oscillations with multiple peaks (8 h, 24 h, 40 h, 48 h) within the 48 h. Rev-ErbA mRNA increased over time and peaked at 48 h. Similarly, BMAL1 mRNA did not accumulate in a circadian fashion and reached the highest level at 48 h. CLOCK mRNA did show some oscillations, although not circadian, with peaks at 32, 40 and 48 h (Fig. 4). In entrained MDA-MB-231 cells, PER1, PER2 and PER3 mRNA remained fairly stable and did not show any oscillation. CRY1 mRNA remained constant until 24 h then increased and reached a plateau between 28 h and 48 h. Interestingly, CRY2, Rev-ErbA and BMAL1 appear to have oscillations with a much greater frequency than in HME1 cells, although we could not recognize a defined pattern. CLOCK mRNA did not show any oscillation (Fig. 4).
Figure 4.
ERA-negative human breast cancer cells show a disrupted inner clock. Quantitative RT-PCR shows that entrainment of the ERA-negative breast cancer cell lines HS578T and MDA-MB-231 cannot induce clock gene mRNA circadian oscillation, as it does in HME1 cells. See the Results section for a more detailed description of the single gene oscillations in each cell line.
From this qualitative analysis, it is apparent that both ERA-negative breast cancer cell lines have a disrupted inner clock. Again, we cannot conclude whether this disruption is a consequence of loss of ERA expression.
ERA-positive human breast cancer cells also display a disrupted inner clock.
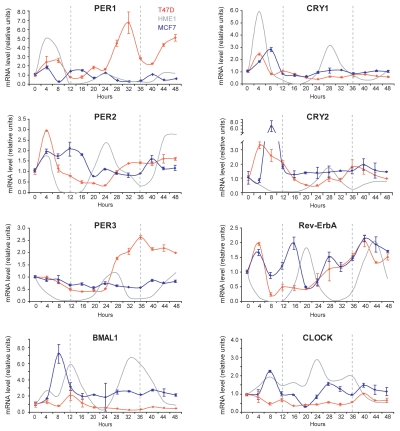
Next, we set out to test whether the circadian oscillation of clock gene expression is present in two ERA-positive breast cancer cell lines, T47D and MCF7, as it is in ERA-positive HME1 cells. As shown in Figure 5, both cell lines do not display circadian oscillations in any of the clock genes analyzed. Specifically, in T47D cells, PER1 mRNA levels peaked at 32 h and 48 h but showed no circadian rhythm. PER2 and PER3 mRNAs show an increase between 24 h and 48 h but do not oscillate. CRY1 and CRY2 mRNAs, after an initial peak following the serum shock, remained fairly constant throughout the 48 h. Rev-ErbA mRNA showed a steady increase from 8 h to 40 h, and did not show any circadian rhythm. BMAL1 mRNA, after a faint peak at 12 h, decreased and remained constant thereafter. CLOCK showed no oscillation (Fig. 5). In MCF7 cells, PER1 and PER2 mRNAs underwent irregular oscillations with an increased frequency relative to HME1, while PER3 mRNA did not show any oscillation. CRY1 and CRY2 mRNAs, after an initial increase at 8 h after the serum shock, decreased and remained constant. Rev-ErbA mRNA showed oscillations with increased frequency relative to HME1 (period of 12 h rather than 24 h). BMAL1 and CLOCK mRNAs did not oscillate (Fig. 5).
Figure 5.
ERA-positive human breast cancer cells also display a disrupted inner clock. Quantitative RT-PCR at different time points after entrainment by serum shock shows that the circadian oscillation of clock genes mRNA is lost in ERA-positive T47D and MCF7 breast cancer cells. See the Results section for a more detailed description of the single gene oscillations in each cell line.
From this overall analysis, we found of interest that, in contrast to what observed in ERA-positive HME1 cells, PER2 mRNA (like the mRNAs of the other clock genes) does not show a circadian oscillation in the ERA-positive breast cancer cell lines T47D and MCF7. Since PER2 transcription is regulated, at least in part, by ERA, we set out to test whether the difference in PER2 mRNA oscillation between these cell lines was correlated with a difference in ERA mRNA oscillation.
ERA mRNA oscillates in a circadian fashion in ERA-positive HME1 cells but not in ERA-positive breast cancer cells.
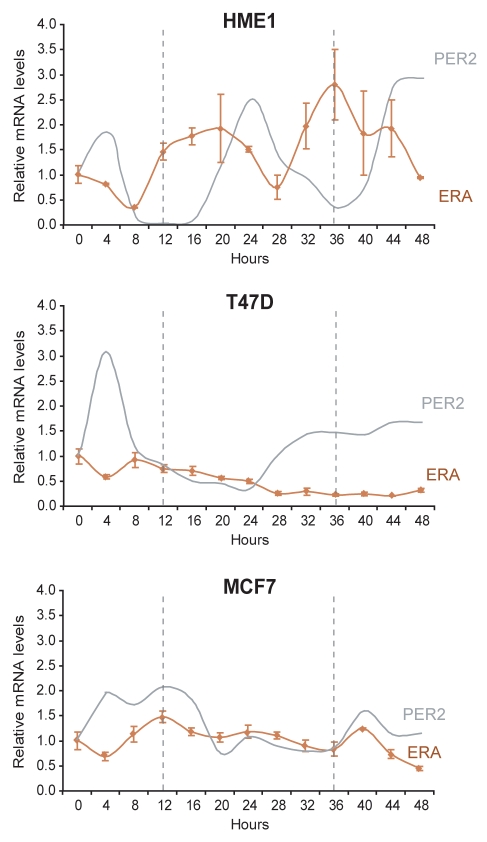
As shown in Figure 6, we found that in HME1 cells ERA mRNA oscillates with a period of 20–24 h, with broad peaks starting at 12 h and 32–36 h and declining at 24–28 h and 44–48 h. Interestingly, ERA mRNA accumulation precedes, of approximately 8 h, the PER2 mRNA accumulation, suggesting that ERA and PER2 circadian oscillations are coordinated (Fig. 6, top). In contrast, T47D and MCF7 breast cancer cells, which did not show PER2 mRNA circadian oscillation, also did not display any ERA mRNA circadian rhythmicity (Fig. 6). In T47D, ERA mRNA levels steadily decreased after a faint peak at 8 h (Fig. 6, middle), while in MCF7, ERA mRNA, after an initial peak at 12 h, did not significantly oscillate until declining at 48 h.
Figure 6.
ERA mRNA oscillates in a circadian fashion in ERA-positive HME1 cells but not in ERA-positive breast cancer cells. Quantitative RT-PCR at 4 h time points after entrainment by serum shock shows that ERA mRNA oscillates in a circadian fashion, and its accumulation precedes the circadian accumulation of PER2 mRNA in ERA-positive, mammary epithelial HME1 cells. In contrast, ERA-positive T47D and MCF7 breast cancer cells, which do not show PER2 circadian expression, also do not show any ERA mRNA circadian rhythmicity.
From these findings, we conclude that ERA mRNA displays a circadian expression in ERA-positive mammary epithelial cells (HME1) but not in ERA-positive breast cancer cells (T47D and MCF7) and correlates with PER2 mRNA circadian oscillation.
Discussion
The possible link between disruption of circadian rhythms and breast cancer was hypothesized far before the discovery of the molecular clock.32 Among all environmental cues, light is the most powerful circadian synchronizer.5 Indeed, epidemiological studies suggested that women experiencing disruption of the circadian clock, such as night-shift workers constantly exposed to artificial light or subjects with sleep deprivation, have an increased risk of developing breast cancer.25,27,33–35 Disruption of circadian rhythms has been found associated with breast tumor progression as well. Indeed, patients with metastatic breast cancer, who had impaired circadian rhythms, had earlier mortality than patients with an intact rhythm.36 This observation is further supported by animal studies showing that mammary tumors grow more rapidly, and overall survival decreases, in rodents subjected either to constant light or constant darkness.37,38
In this report, we discuss for the first time that entrainment of human mammary epithelial cells in culture represents an additional strategy that takes into account the biological time component to advance breast cancer research. Based on the observation that rhythmic expression of core circadian genes is observed in most peripheral tissues39 and can be induced in cultured fibroblasts by serum shock,31 we set out to test whether serum-shocked cultured human mammary epithelial cells display an autonomous functional inner clock. We found that HME1 cells, which are immortal, non-tumorigenic and capable, like normal mammary epithelial cells, of proper 3D acinar morphogenesis, display circadian gene expression of key clock genes upon entrainment by serum shock. HME1 cells are ERA-positive and, upon estrogen treatment, express canonical downstream ERA targets, including progesterone receptor (our unpublished observations). Apparently, ERA mRNA oscillates in a circadian fashion in HME1 cells entrained in culture. This observation is consistent with previous in vivo studies showing that ERA is a nuclear receptor under circadian control.20 ERA is a transcriptional regulator of the PER2 gene,22 and estradiol induces PER2 mRNA in HME1 cells (our unpublished observation). The accumulation of ERA mRNA in entrained HME1 cells precedes the peaks of PER2 mRNA accumulation by approximately 8 h, suggesting that ERA and PER2 circadian oscillations are coordinated. In contrast, in the human mammary epithelial cell line MCF10A, which is ERA-negative, we found that the circadian oscillation of many clock genes, including PER2, is partially or completely lost. We do not know yet whether lack of ERA expression, by abrogating ERA-mediated estrogen transactivation of PER2, had contributed to the development of a defective clock in MCF10A cells. Regardless, neither the lack of ERA nor the defective clock seem to have had an effect on the process of acinar morphogenesis of MCF10A cells, which maintain the ability to form normal, hollow acini lined with polarized cells when grown in 3D culture on basement membrane.40,41 Apparently, the microenvironment-dependent morphological architecture is “dominant” over loss of ERA and the defective clock in MCF10A.42
Based on the differences of the circadian oscillator between HME1 and MCF10A cells, we further tested whether there is an association between loss of ERA expression and defective circadian oscillation of clock genes in breast cancer cells as well. Entrainment of two ERA-negative breast cancer cell lines, HS578T and MDA-MB-231, and two ERA-positive breast cancer cell lines, T47D and MCF7, showed an erratic mRNA oscillation of core clock genes in both ERA-negative and ERA-positive cells. In ERA-positive cancer cell lines, with a disrupted clock, we found that ERA mRNA does not undergo circadian oscillation, as it does in HME1 cells with a functional clock. Based on the ERA expression level alone, we always assumed that ERA-mediated estrogen signaling is not impaired in ERA-positive breast cancer cells. However, it is possible that, when ERA mRNA does not accumulate in a circadian fashion, the estrogen signaling is somewhat impaired. This observation might be relevant for hormone therapy of ERA-positive cancer. Current hormone therapies targeting ERA-positive breast cancer have been based on the assumption that ERA is constantly expressed. Our data suggest that drugs targeting ERA may be less efficient in breast cancer cells that are ERA-positive yet incapable of accumulating ERA in a circadian fashion.
The circadian clock integrity is deemed to be important for tumor suppression in vivo.5 However, the intricacy of multiple regulatory mechanisms makes it cumbersome to use in vivo models to discriminate whether breast cancer initiation is triggered, either by genetic/epigenetic mutations/variants of clock components, which are capable of influencing ERA oscillation, or by genetic/epigenetic loss of ERA, which regulates inner clock components. To answer these questions requires testing numerous parameters in a mechanistic and circadian fashion. Entrainment of human breast (cancer) epithelial cells provides a convenient tool to perform this type of studies. One of our ongoing studies, using entrained HME1 cells, stems both from published data, showing that women with variants in the CLOCK gene have a higher risk of breast cancer and that CLOCK expression is higher in ERA-negative tumors vs. ERA-positive tumors,43 and from our observation that ERA contains in the first intron an E-box. Thus, ERA could be potentially affected by a dysfunctional CLOCK/BMAL1 heterodimer. Another ongoing study aims at deciphering, mechanistically, the influence of an impaired PER2-ERA “regulome” on breast cancer initiation. The rationale is provided by a wealth of observations, including loss of PER2 expression in breast cancer,44,45 evidence of ERA-PER2 mutual regulation22 and PER2's ability to coordinate circadian outputs via protein-protein interaction with ERA22 and other nuclear receptors46 (Fig. 7). The expected outcome of these investigations is to understand whether loss of ERA expression can lead to a disrupted clock due to lack of proper estrogen-mediated PER2 transactivation, or whether the development of a disrupted clock is a consequence of an impaired estrogen-ERA signaling. Unraveling these mechanisms might improve preventative and therapeutic strategies for breast cancer.
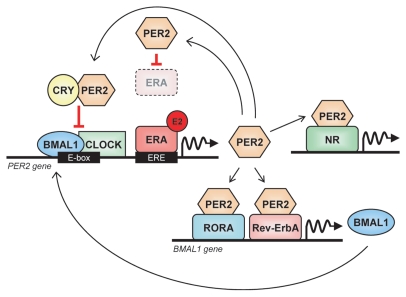
Figure 7.
The core clock gene PER2 links the circadian oscillator with ERA signaling. PER2 transcription can be induced by estrogen (E2)-ERA signaling; second, PER2 physically interacts with ERA and leads to its degradation; finally, PER2 interacts with RevErbA, RORA and other nuclear receptors (NR) to modulate the transcription of NR-target genes, including BMAL1. Based on references 22 and 46.
Future studies using entrained normal and breast cancer cells will enable us to focus on understanding whether components of the circadian oscillators play a role in breast cancer by influencing hormone regulation or the other way around. It will be possible to learn from these studies whether breast cancer patients receiving hormone treatment would benefit the most by receiving treatment when ERA expression is least abundant in normal cells and most abundant in cancer cells. Ultimately, we expect that model cell systems that facilitate circadian analysis will accelerate the progress of breast cancer “chronotherapy.” Chronotherapy is a strategy that takes advantage of the asynchronies between normal and malignant tissues by administering therapy at a specific time of the day.
Methods
Cells and cell culture.
The human, telomerase-immortalized breast epithelial cell strain h-TERT-HME1 (hereafter called HME1) (Clontech) was grown in mammary epithelial growth medium (MEGM) supplemented with bovine pituitary extract (Lonza). The human, non-tumorigenic epithelial cell line MCF-10A (ATCC, CRL-10317) derived from fibrocystic breast tissue was grown as described in reference 47, in DMEM/F12 supplemented with 5% horse serum, 20 ng/ml EGF, 0.5 µg/ml Hydrocortisone, 100 ng/ml cholera toxin and 10 µg/ml insulin and sterile filtered. The human breast cancer cell lines MDA-MB-231 (ATCC, HTB-26), T47D (ATCC, HTB-133), HS578T (ATCC, HTB-126) and MCF7 (ATCC, HTB-22) were grown in DMEM supplemented with 5% fetal bovine serum (FBS) (Invitrogen). All cell lines were grown in a humidified incubator at 37°C and 5% CO2.
Entrainment of cells by serum shock.
Cells were entrained by using a slightly modified serum shock procedure based on the method originally proposed by Balsalobre et al.31 Briefly, cells were seeded into 6-well plates at a density of 1.5 × 105 cells per well and allowed to grow to confluence. After being confluent for 2–3 d, cells were washed twice in PBS (Invitrogen) to remove residual growth factors and starved overnight. For HME1, the starvation medium was mammary epithelial basal medium (MEBM) (Lonza). For MCF10A, the starvation medium was DMEM/F12 (Invitrogen). All other cells were starved in DMEM without FBS. Thereafter, cells were serum shocked using their respective growth media supplemented with 50% horse serum for 2 h. Subsequently, the cells were washed in PBS and returned to starvation conditions. The first time point was taken prior to the serum shock (t = 0) and every 4 h thereafter until 48 h.
RNA extraction and cDNA synthesis.
Total RNA was extracted by using Trizol Reagent (Invitrogen) according to the manufacturer's instructions. Briefly, cells grown in 6-well plates were lysed with 1 ml Trizol/well and incubated for 5 min to allow for complete dissociation of the nucleoprotein complexes. After addition of 200 µl chloroform per 1 ml Trizol, samples were vortexed and centrifuged at 12,000x g for 15 min at 4°C to separate the upper aqueous phase, containing the RNA, from the lower phase. RNA was precipitated from the aqueous phase by addition of 500 µl of isopropyl alcohol and centrifugation at 12,000x g for 10 min. The RNA pellet was then washed with 1 ml 75% ethanol, briefly dried and resuspended in water. The RNA concentration was measured spectrophotometrically by using the Nanodrop (Thermo Scientific). One µg total RNA was retro-transcribed into cDNA by using the QuantiTect Reverse Transcription Kit (Qiagen), according to the manufacturer?fs instructions. Briefly, RNA was first incubated with a DNase-containing buffer (gDNA wipeout buffer, Qiagen) to remove possible genomic DNA contamination, then retrotranscribed with random primers in an iCycler thermal cycler (BioRad).
Real-time PCR.
Real-time PCR analyses were performed in 96-well plates. The reaction mix (20 µl/well) consisted of 1 µl cDNA (derived from 25 ng RNA), 10 µl iQ SYBR Green Supermix (BioRad), 0.2 µM of each forward and reverse primer specific for each gene of interest (see Table 1), and molecular grade water to reach a final volume of 20 µl. After brief centrifugation, the plates were placed in an iCycler thermal cycler equipped with a MyiQ Real-Time PCR detection system (Bio.Rad) programmed with an initial denaturation step at 95.0°C for 3 min, followed by 45 cycles of 95.0°C for 20 sec, 60.0°C for 30 sec and 72.0°C for 30 sec, with data collection during the extension step at 72.0°C. After the reaction, a melting curve analysis was performed to ascertain that a single-band PCR product was produced. The transcript levels of the genes of interest were quantified by the Delta delta Ct method, using the housekeeping gene GAPDH for normalization. The amplification efficiency, evaluated from the sample slopes, was similar for all the samples analyzed in the same experiment. The data, corresponding to the mean of at least three independent determinations ± standard deviation, were reported on the charts by setting the transcript level of the first time point (t = 0) as 1 for each gene and each cell line.
Table 1.
List of primers used for real time PCR
| Primer name | Orientation | Sequence |
| BMAL1 | sense | 5′-AGG ATG GCT GTT CAG CAC ATG A-3′ |
| antisense | 5′-CAA AAA TCC ATC TGC TGC CCT G-3′ | |
| CLOCK | sense | 5′-AAG TTA GGG CTG AAA GAC GAC GA-3′ |
| antisense | 5′-GAA CTC CGA GAA GAG GCA GAA G-3′ | |
| CRY1 | sense | 5′-GAC AAG ATC ATA GAA CTC AAT GGT-3′ |
| antisense | 5′-AAG CAG AGA ATT CGC ATT CAT TCG-3′ | |
| CRY2 | sense | 5′-TCC CTA GCA TGT CAG CCC GTT-3′ |
| antisense | 5′-AGG ATT TGA GGC ACT GTT CCG A-3′ | |
| ERA | sense | 5′-AAG AGC TGC CAG GCC TGC C-3′ |
| antisense | 5′-TTG GCA GCT CTC ATG TCT CC-3′ | |
| GAPDH | sense | 5′-GAA GGT GAA GGT CGG AGT C-3′ |
| antisense | 5′-GAA GAT GGT GAT GGG ATT TC-3′ | |
| PER1 | sense | 5′-GCT GAT TGC AGA GCG CAT CCA-3′ |
| antisense | 5′-GAT GCA GGA ACA GGA GCA CTG-3′ | |
| PER2 | sense | 5′-AAG CAG GTG AAA GCC AAT GAA GA-3′ |
| antisense | 5′-CCA CCG CAA ACA TAT CGG CAT T-3′ | |
| PER3 | sense | 5′-TGC AGG GCA TCC TCC CTT TGA-3′ |
| antisense | 5′-TCC GGC TCC AGG GAT TCA CAA-3′ | |
| Rev-ErbA | sense | 5′-GAG CAC CAG CAA CAT CAC CAA G-3′ |
| antisense | 5′-TCT TGA AGC GAC ATT GCT GGC A-3′ |
Acknowledgements
This work is part of the Master of Science thesis submitted by JE to the Faculty of the Graduate School of the University at Buffalo. We are grateful to Dr. Marina Antoch, Roswell Park Cancer Institute, for critical comments. This work was supported by the National Cancer Institute (NCI) grant P30 CA016056 and by the Department of Defense (DOD) Concept Award W81XWH0610657 to N.S.
Abbreviations
- 3D culture
three dimensional culture
- CCG(s)
clock controlled gene(s)
- CKIE
casein kinase Iε
- CRY1
cryptochrome 1
- CRY2
cryptochrome 2
- ERA
estrogen receptor alpha
- ERE
estrogen response element
- PER1
period 1
- PER2
period 2
- PER3
period 3
- qRT-PCR
quantitative RT-PCR
- Rev-ErbA
Rev-Erbalpha
- RORA
RAR-related orphan receptor alpha
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Pando MP, Sassone-Corsi P. Signaling to the mammalian circadian clocks: in pursuit of the primary mammalian circadian photoreceptor. Sci STKE. 2001;2001:16. doi: 10.1126/stke.2001.107.re16. [DOI] [PubMed] [Google Scholar]
- 3.Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Moser M, Schaumberger K, Schernhammer E, Stevens RG. Cancer and rhythm. Cancer Causes Control. 2006;17:483–487. doi: 10.1007/s10552-006-0012-z. [DOI] [PubMed] [Google Scholar]
- 5.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 6.Canaple L, Kakizawa T, Laudet V. The days and nights of cancer cells. Cancer Res. 2003;63:7545–7552. [PubMed] [Google Scholar]
- 7.Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- 8.Sahar S, Sassone-Corsi P. Circadian clock and breast cancer: a molecular link. Cell Cycle. 2007;6:1329–1331. doi: 10.4161/cc.6.11.4295. [DOI] [PubMed] [Google Scholar]
- 9.Brown SA, Schmitt K, Eckert A. Aging and circadian disruption: causes and effects. Aging (Albany NY) 2011;3:813–817. doi: 10.18632/aging.100366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 11.Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- 12.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/S0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi JS. Finding new clock components: past and future. J Biol Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson CH. Circadian clocks and cell division: what's the pacemaker? Cell Cycle. 2010;9:3864–3873. doi: 10.4161/cc.9.19.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 17.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/S0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 18.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/S0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 19.Ripperger JA, Schmutz I, Albrecht U. PERsuading nuclear receptors to dance the circadian rhythm. Cell Cycle. 2010;9:2515–2521. doi: 10.4161/cc.9.13.12075. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 21.Gery S, Koeffler HP. Circadian rhythms and cancer. Cell Cycle. 2010;9:1097–1103. doi: 10.4161/cc.9.6.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 23.Bocchinfuso WP, Korach KS. Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol Neoplasia. 1997;2:323–334. doi: 10.1023/A:1026339111278. [DOI] [PubMed] [Google Scholar]
- 24.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 25.Hansen J. Risk of breast cancer after night- and shift work: current evidence and ongoing studies in Denmark. Cancer Causes Control. 2006;17:531–537. doi: 10.1007/s10552005-9006-5. [DOI] [PubMed] [Google Scholar]
- 26.Haus E, Smolensky M. Biological clocks and shift work: circadian dysregulation and potential long-term effects. Cancer Causes Control. 2006;17:489–500. doi: 10.1007/s10552-005-9015-4. [DOI] [PubMed] [Google Scholar]
- 27.Davis S, Mirick DK. Circadian disruption, shift work and the risk of cancer: a summary of the evidence and studies in Seattle. Cancer Causes Control. 2006;17:539–545. doi: 10.1007/s10552-005-9010-9. [DOI] [PubMed] [Google Scholar]
- 28.Bullough JD, Rea MS, Figueiro MG. Of mice and women: light as a circadian stimulus in breast cancer research. Cancer Causes Control. 2006;17:375–383. doi: 10.1007/s10552-005-0574-1. [DOI] [PubMed] [Google Scholar]
- 29.Stevens RG. Artificial lighting in the industrialized world: circadian disruption and breast cancer. Cancer Causes Control. 2006;17:501–507. doi: 10.1007/s10552-005-9001-x. [DOI] [PubMed] [Google Scholar]
- 30.Stevens RG. Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology. 2005;16:254–258. doi: 10.1097/01.ede.0000152525.21924.54. [DOI] [PubMed] [Google Scholar]
- 31.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/S0092-8674(00)81199-X. [DOI] [PubMed] [Google Scholar]
- 32.Stevens RG. Electric power use and breast cancer: a hypothesis. Am J Epidemiol. 1987;125:556–561. doi: 10.1093/oxfordjournals.aje.a114569. [DOI] [PubMed] [Google Scholar]
- 33.Hansen J. Increased breast cancer risk among women who work predominantly at night. Epidemiology. 2001;12:74–77. doi: 10.1097/00001648-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 34.Davis S, Mirick DK, Stevens RG. Night shift work, light at night and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–1562. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 35.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses' health study. J Natl Cancer Inst. 2001;93:1563–1568. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 36.Sephton SE, Sapolsky RM, Kraemer HC, Spiegel D. Diurnal cortisol rhythm as a predictor of breast cancer survival. J Natl Cancer Inst. 2000;92:994–1000. doi: 10.1093/jnci/92.12.994. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Dauchy RT, Tirrell PC, Wu SS, Lynch DT, Jitawatanarat P, et al. Light at night activates IGF-1R/PDK1 signaling and accelerates tumor growth in human breast cancer xenografts. Cancer Res. 2011;71:2622–2631. doi: 10.1158/0008-5472.CAN-10-3837. [DOI] [PubMed] [Google Scholar]
- 38.Mormont MC, Levi F. Circadian-system alterations during cancer processes: a review. Int J Cancer. 1997;70:241–247. doi: 10.1002/(SICI)1097-0215(19970117)70:2<241::AID-IJC16>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 39.Fukuhara C, Tosini G. Peripheral circadian oscillators and their rhythmic regulation. Front Biosci. 2003;8:642–651. doi: 10.2741/1042. [DOI] [PubMed] [Google Scholar]
- 40.Rossetti S, Hoogeveen AT, Esposito J, Sacchi N. Loss of MTG16a (CBFA2T3), a novel rDNA repressor, leads to increased ribogenesis and disruption of breast acinar morphogenesis. J Cell Mol Med. 2010;14:1358–1370. doi: 10.1111/j.1582-4934.2009.00982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/S0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 42.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, et al. CLOCK in breast tumorigenesis: genetic, epigenetic and transcriptional profiling analyses. Cancer Res. 2010;70:1459–1468. doi: 10.1158/0008-5472.CAN-09-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM. Period-2: a tumor suppressor gene in breast cancer. J Circadian Rhythms. 2008;6:4. doi: 10.1186/1740-3391-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 46.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]