Abstract
Many viruses infecting animals and plants share common cores of homologous genes involved in the key processes of viral replication. In contrast, genes that mediate virus – host interactions including in many cases capsid protein genes are markedly different. There are three distinct scenarios for the origin of related viruses of plants and animals: i) evolution from a common ancestral virus predating the divergence of plants and animals; ii) horizontal transfer of viruses, for example, through insect vectors; iii) parallel origin from related genetic elements. We present evidence that each of these scenarios contributed, to a varying extent, to the evolution of different groups of viruses.
Introduction
From a parochial perspective, animals and plants are the most conspicuous life forms due to their sheer size and central position in food chains. However, from a scientific standpoint, these complex, multicellular eukaryotes have lost their exclusive status thanks to the realization of the paramount roles in biodiversity, global ecology, and geochemical cycles played by microbes including bacteria, archaea, and unicellular eukaryotes. Furthermore, phylogenetic studies have yielded a consensus evolutionary tree of eukaryotes in which plants and animals comprise only two branches within two of the five supergroups[1, 2]. The remaining three supergroups as well as many relatives of plants and animals are diverse unicellular eukaryotes.
Another striking recent revelation brought about by environmental metagenomics is that the biosphere is literally awash in viruses [3, 4]. Numerically, viruses are the dominant life forms with a conservative estimate of about 10 virus particles per living cell. In terms of genetic diversity, the viral pangenome is probably more complex than the pangenome of cellular life forms [5].
Beyond the ubiquity and high abundance of viruses, the viromes of multicellular eukaryotes show dramatic differences from those of bacteria and archaea. In contrast to prokaryotes whose viromes are heavily dominated by viruses with double-stranded (ds) DNA genomes [6, 7], plants and animals harbor enormously diverse viromes enriched for RNA viruses and reverse-transcribing (retroid) viruses which are rare or not found in prokaryotes. The study of the viromes of unicellular eukaryotes is still in its infancy but recent data point to a considerable diversity of RNA viruses [8, 9]. The implication of these findings is that the shift from DNA- to RNA-dominated viromes occurred at the earliest stages of eukaryote evolution, perhaps concomitantly with eukaryogenesis, and might be related to the emergence of the cytosol, an ‘RNA compartment’ well suited for propagation of RNA viruses [10, 11].
Even with a correction for the bias toward sampling humans and ‘subservient’ agriculture-related organisms, the non-trivial global ecology of the major classes of viruses demands an explanation rooted in virus-host co-evolution or lack thereof. Here we approach this fundamental problem by surveying the results of viral phylogenomics with an emphasis on evolutionary connections between animal and plant viruses, and their relationships with the host organisms, as well as other viruses of eukaryotes and prokaryotes.
A global view of virus diversity
All cellular life forms share ~100 (nearly) universal genes that enable phylogeny-based classification[12]. Although the ultimate merit of a “universal tree of life” spanning the entire hierarchy of cellular organisms remains a matter of intense debate[13, 14], the common ancestry of all cells is broadly accepted. In contrast, all viruses and virus-like genetic elements collectively denoted the ‘virus world’ or the ‘virosphere’ are unlikely to have shared a single ancestor and accordingly are not conducive to a comprehensive phylogenetic classification. Indeed, not a single gene is universally conserved in all viruses although many virus groups are linked in a complex network of evolutionary relationships that become apparent through the conservation of ‘virus hallmark genes’ encoding key proteins required for viral replication and virion formation [10]. In sharp contrast to the uniformity of cellular dsDNA genomes, viral genomes are either DNA or RNA, and either double-stranded or single-stranded. Inevitably, a meaningful picture of virus evolution and classification should be based on multiple criteria, such as the chemical nature of the genome, genome replication-expression cycle, and conservation of genes and gene arrays. An attempt to apply this approach for viruses of eukaryotes with the emphasis on viruses infecting animals and plants is illustrated in Fig. 1.
Figure 1.
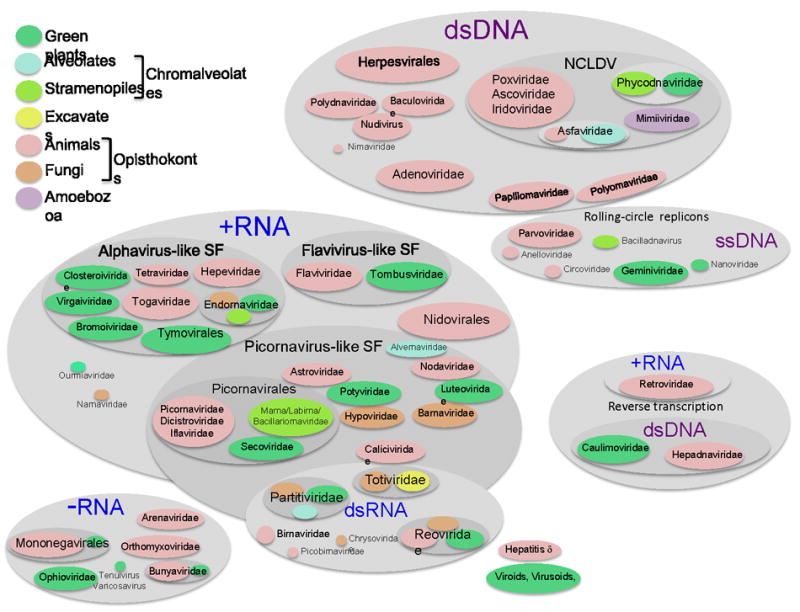
Global ecology of viruses.
The shapes denoting virus taxa are color-coded in accordance with their host range as indicated in the figure. NCLDV, Nucleo-cytoplasmic large DNA viruses; SF, superfamily.
Based on the nature of the genome that correlates with fundamental features of replication and expression, three principal domains of the virus world are apparent: RNA viruses, DNA viruses, and retro-transcribing (retroid) viruses. The entire replication-expression cycle of the RNA viruses is based on RNA, with no DNA stage. The viruses in the second domain have DNA genomes whose expression occurs via mRNA transcription similar to cellular organisms. The replication cycle of retroid viruses includes both RNA and DNA phases. All these virus classes are represented in animals and plants but the relative abundances are dramatically different.
RNA viruses
Within the RNA domain, there are three classes that include viruses with positive-strand and negative-strand RNA genomes, as well as dsRNA viruses (Fig. 1). All these viruses encode RNA-dependent RNA polymerases (RdRp) that are responsible for genome replication and, in some cases, transcription, yielding subgenomic mRNAs. The RNA viruses evolve fast so that divergence often obscures evolutionary relationships. Nevertheless, amino acid sequences and 3D structures of RdRps are conserved not only among positive-strand RNA viruses but also in dsRNA viruses suggesting common roots of these virus classes [15, 16]. The evolutionary provenance of the RdRps of negative-strand RNA viruses is less clear in the absence of a solved 3D structure.
Positive-strand RNA viruses comprise the largest and most diverse class of RNA viruses. Phylogenetic analysis of the RdRps delineated three superfamilies of positive-strand RNA viruses: Picornavirus-like, Alphavirus-like, and Flavivirus-like [15]. Remarkably, each of these superfamilies includes multiple taxa of animal and plant RNA viruses (Fig. 1). In addition to the RdRp phylogeny, the monophyly of the picornavirus-like and alphavirus-like superfamilies is supported by the (partial) conservation of distinct gene arrays encoding the key proteins of virus reproduction [17-19]. These signature gene arrays consist of the superfamily 3 helicase (S3H), chymotrypsin-like protease, and RdRp in the picornavirus-like superfamily; and capping enzyme, superfamily 1 helicase (S1H), and RdRp and in the alphavirus-like superfamily (Fig. 2A). The Flavivirus-like superfamily remains somewhat tentative because it includes plant and animal viruses with dissimilar genome organizations that share only related RdRps.
Figure 2.
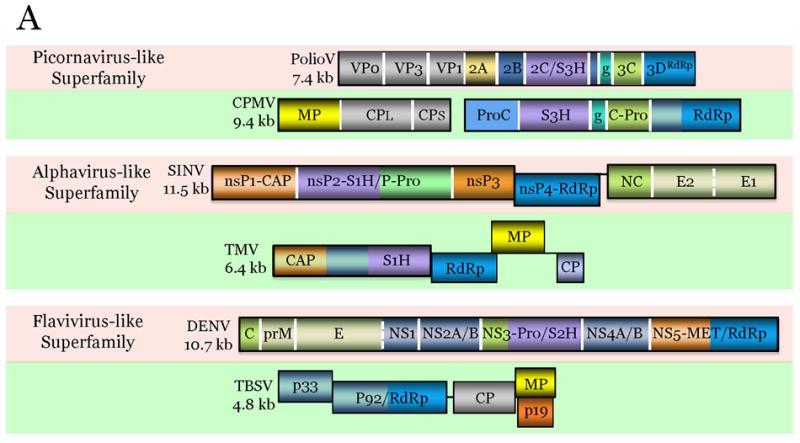
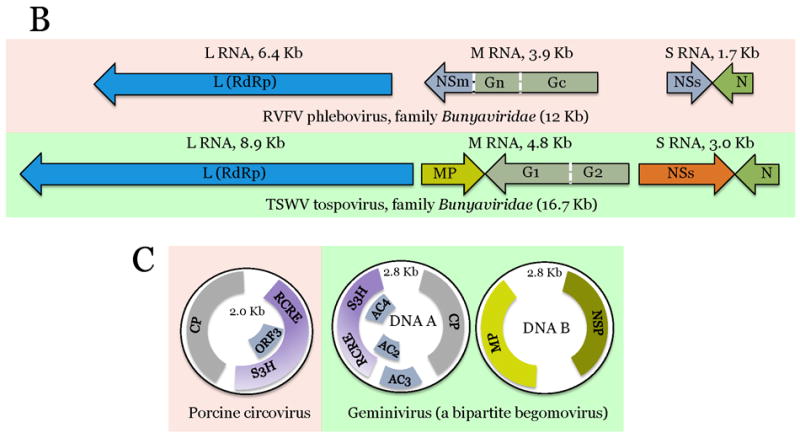
Comparison of genome architectures of related plant and animal viruses: The housekeeping and interactive gene modules.
The virus genes are drawn as boxes approximately to scale. For each pair of animal (pink background) and plant (green background) viruses, the homologous genes are shown in the same color. Gene designations: VP, virus protein; S3H, superfamily 3 helicase; g, virus protein, genome-linked; RdRp, Rna-dependent RNA polymerase; MP, movement protein; CP, capsid protein; ProC, protease cofactor; C-Pro, cysteine protease; nsP, non-structural protein; CAP, capping enzyme; S1H, superfamily 1 helicase; P-Pro, papain-like protease; NC, nucleocapsid protein; E, envelope protein; C, nucleocapsid protein; prM, pre-membrane protein; NS, non-structural protein; Pro, protease; S2H, superfamily 2 helicase; MET; methyltransferase; G, glycoprotein; N, nucleocapsid protein; RCRE, rolling-circle replication endonuclease; NSP, nuclear shuttle protein. Virus abbreviations: PolioV, Poliovirus; CPMV, Cowpea mosaic virus; SINV, Sindbis virus; TMV, Tobacco mosaic virus; DENV Dengue fever virus; TBSV, Tomato bushy stunt virus; RVFV, Rift valley fever virus; TSWV, Tomato spotted wilt virus.
The dsRNA virus subdomain combines viruses with distinct evolutionary trajectories [20]. The families Hypoviridae, Totiviridae and Partitiviridae appear to have evolved from distinct branches of the Picornavirus-like superfamily (Fig. 1) [11]. The Birnaviridae family shares an unusual permuted RdRp, a genome-linked protein, and a distinct variant of the jelly-roll capsid protein with a subset of positive-strand RNA viruses in the Tetraviridae family, suggestive of common origin [21]. The viruses of the Reoviridae family which infect animals, fungi and plants possess unusual capsids formed by two concentric icosahedra similar to the capsids of the single known family of prokaryotic dsRNA viruses, Cystoviridae [22]. Thus, the dsRNA subdomain appears to be polyphyletic and combines viruses with roots in distinct lineages of eukaryotic positive-strand RNA viruses and dsRNA bacteriophages.
Negative-strand RNA viruses transcribe and replicate their genomes using an RdRp that is either unrelated to or is an extremely divergent derivative of RdRps of positive-strand RNA viruses [23]. A vast majority of known negative-strand RNA viruses infect either vertebrates or plants. For instance, families Rhabdoviridae (order Mononegavirales) and Bunyaviridae each include highly similar viruses infecting either vertebrate or plant hosts.
Reverse-transcribing (retroid) viruses
This domain of the virus world relies on RNA-dependent DNA polymerase, or reverse transcriptase (RT), to go through a replication cycle that involves both RNA and DNA. Bona fide viruses are in the minority in the reverse-transcribing domain: the majority belongs to diverse parasitic retroelements and retrotrasposons that are abundant in eukaryotes and many bacteria (e.g., mammalian LINE elements or bacterial Group II introns) [24, 25]. The true retroid viruses come in two flavors: i) animal Retroviridae with positive-strand RNA genomes that are copied into dsDNA by RT, integrated into host chromosomes, and then transcribed by host DNA-dependent RNA polymerase (DdRp) to produce viral mRNAs and genomes; and ii) pararetroviruses with dsDNA genomes that are transcribed into RNA and then reverse transcribed during replication in the host cells. No retroviruses proper are known in plants although plant genomes are packed with various retroelements[26]. Pararetroviruses are known in plants (Caulimoviridae, Badnaviridae) and animals (Hepadnaviridae) (Fig. 1). Notably, retroid viruses have not been detected in any hosts other than plants and animals. Moreover, the RT is the only shared gene between retroviruses and pararetroviruses.
DNA viruses
The third and largest domain of the virus world includes viruses with dsDNA and ssDNA genomes. All DNA viruses share the basic genome replication-expression cycle with cellular life forms. The required DNA-dependent DNA polymerases (DdDp) and DdRps are either virus-encoded (in dsDNA viruses with large genomes) or are borrowed from the host by dsDNA and ssDNA viruses with small genomes.
Most of the ssDNA viruses have circular genomes and a peculiar Rolling Circle Replication (RCR) cycle that involves nicking one of the strands in a dsDNA intermediate by a virus-encoded endonuclease. This key enzyme, the RCR endonuclease (RCRE), is conserved not only in eukaryotic ssDNA viruses, including the plant families Geminiviridae and Nanoviridae, and the animal families Parvoviridae and Circoviridae, but also in ssDNA bacteriophages and a variety of capsid-less RCR replicons including bacterial and archaeal plasmids, and animal polinton/helitron transposons [27, 28]. Small animal dsDNA viruses of the Polyomaviridae and Papillomaviridae families encode an inactivated homolog of the RCRE which is involved in RNA-primed genome replication [29]. This link along with the conservation of a Superfamily 3 helicase reveals direct evolutionary connections between ssDNA and dsDNA viruses with small genomes.
The largest monophyletic group of eukaryotic dsDNA viruses, the Nucleocytoplasmic Large DNA Viruses (NCDLV), includes the giant viruses of the Mimiviridae family, with genomes over 1 Mb that exceed in size many bacterial genomes, along with several families of smaller viruses, in particular the Poxviridae [30, 31]. The NCLDV and other groups of large dsDNA viruses, such as Herpesvirales, Baculoviridae and Adenoviridae, show evolutionary connections with different groups of bacterial and archaeal viruses through several shared hallmark genes[31-33]. The core gene sets of these large viruses are further complemented with genes acquired from bacteria and from their eukaryotic hosts.
The dsDNA part of the virus world reveals an obvious bias toward animal and unicellular eukaryotic hosts at the expense of plants (Fig. 1). Actually, there are no known dsDNA viruses infecting multicellular (vascular) plants. Given that large dsDNA viruses of the NCLDV class (Phycodnaviridae) infect unicellular green algae [34], the ancestors of the vascular plants, the basic green plant cell biology is compatible with dsDNA virus reproduction. The problem seems to be the symplastic organization of vascular plants. All plant cells are encased by thick cell walls that are impenetrable for viruses. Viruses can colonize plants only by moving through the plasmodesmata, thin cytosolic sleeves used for intercellular communications [35]. Because the known plant DNA viruses move their genomes between cells in the form of ssDNA (Geminiviridae) or sneak dsDNA between cells within small virions (Caulimoviridae), it seems that unencapsidated dsDNA cannot pass through plasmodesmata [35, 36]. Due to the small size of plasmodesmata, large virions of adenoviruses let alone herpesviruses or NCDLV simply could not pass providing a mechanistic cause behind the absence of large DNA viruses in plants. It is unclear why small dsDNA viruses, e.g. similar to polyomaviruses, are not found in plants. The possibilities are that such viruses will yet be discovered or that their absence in plants might be a “frozen accident” given the low diversity of small dsDNA viruses in animals.
Comparative viromics of plants and animals
Summarizing the data on the compositions of the plant and animal viromes, plants are not known to support reproduction of dsDNA viruses and retroviruses. All other major classes of viruses are represented by multiple evolutionarily related families in both kingdoms of multicellular eukaryotes. However, the relative contributions of different classes of viruses to the viromes substantially differ (Fig. 3). The plant virome is heavily dominated by positive-strand RNA viruses, especially those of the alphavirus-like superfamily. The animal virome shows a greater overall diversity and a more uniform distribution of viral groups; in contrast to plants, the picornavirus-like superfamily is the dominant group of RNA viruses (Fig. 3). As discussed above, the absence of large dsDNA viruses in plants finds a plausible explanation in the inability of these viruses to pass through plasmodesmata. The differences in the abundance of other virus classes also might have to do with idiosyncrasies of plant and animal biology but identification of the relevant differences remains a challenge for future research.
Figure 3.
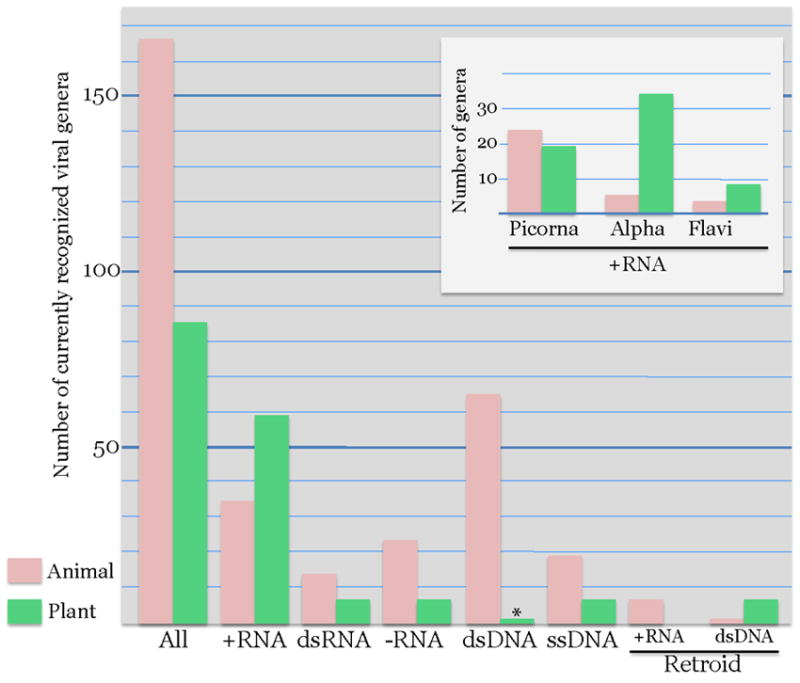
Prevalence of major groups of viruses in plant and animal viromes.
The bar graphs show the number of virus genera infecting plants and animals for each major group of viruses. The asterisks indicates that the only known group of typical dsDNA viruses in the plant kingdom, the Phycodnaviridae, infects green algae rather than land plants. Virus taxonomy was from the NCBI Taxonomy Browser: http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&name=Viruses&lvl=3&srchmode=1&keep=1&unlock
Plant and animal viruses: common cores of replication genes and distinct gene suits for virus-host interaction
Comparison of the genome organizations of related plant and animal viruses reveals a recurrent theme with an apparent simple “evolutionary logic”. Virus genes can be roughly partitioned into the “housekeeping (replication) module” that includes genes essential for virus genome replication and expression, and the “interactive module” comprised of genes involved in virus-host interactions. The replication modules are conserved between viruses of plants and animals whereas the interactive modules are host-specific (Fig. 2).
The three-gene housekeeping modules of the picornavirus-like and alphavirus-like superfamilies of positive-strand RNA viruses are discussed above (Fig. 2A). The conservation of these modules, typically including the gene order, strengthens the case for the origin of the respective lineages of viruses from a single ancestral virus. In the case of ssDNA viruses, the conserved module encompasses the RCRE-S3H, two genes for interacting replication proteins that are either fused (Fig. 2C) or separate. In other cases, the housekeeping module is limited to a single gene such as the RdRp in the flavivirus-like superfamily (Fig. 2A), RdRp-containing L-protein in Bunyaviridae (Fig. 3B), and the RT in the retroid viruses.
The host-specific interactive modules include genes for movement proteins (MPs) of plant viruses that facilitate the virus passage through the plasmodesmata [35]. Strikingly, homologous MPs are shared by viruses from all three major viral domains, namely positive-strand and negative-strand RNA viruses, ssDNA viruses (Fig. 2), and pararetroviruses [37, 38]. By contrast, no counterpart to these MPs is detectable in animal viruses. This pattern suggests, first, that the MP genes have spread horizontally among diverse plant viruses, and second, that acquisition of a MP gene is a condition for the emergence of a new, evolutionarily competitive group of plant viruses. Due to a powerful selection to retain the MPs, viruses with otherwise different histories share this element of the interactive module. Other, more variable parts of the interactive modules consist of genes encoding proteins involved in counter-defense, such as diverse RNAi suppressors of plant viruses [39, 40]. The extreme structural and evolutionary variability of these proteins is not surprising given their involvement in virus-host arms race, a powerful accelerator of evolution [41]. The RNAi suppressors are less common in animal viruses [40], which encode instead a variety of “security proteins” that suppress animal-specific defense pathways of innate and adaptive immunity [42, 43].
The genes for capsid proteins (CPs) that form virions but also interact with host cells form an interface between the housekeeping and interactive modules. The CPs are conserved across a broad range of hosts in some groups of viruses but not in others [44]. For example, most viruses of the picornavirus-like superfamily share homologous jelly-roll CPs that form icosahedral capsids [11]. By contrast, the majority of plant viruses of the alphavirus-like superfamily encode CPs that form elongated (rod-like or filamentous) virions [45] and often aid MPs in facilitating virus passage through the plasmodesmata[46]. The spread of these plant virus-specific CPs could have contributed to the evolutionary success of the alphavirus-like superfamily in plants; the filamentous particle proteins even have invaded the picornavirus-like superfamily by replacing the icosahedral particle proteins in potyviruses, the largest plant virus family.
In large dsDNA viruses, that failed to make it to vascular plants, similar trends are observed on a different scale. In the NCLDV, housekeeping modules include up to 50 genes whereas interactive modules can include hundreds of genes [31]. Once again, viruses infecting vertebrates with their distinct adaptive immunity systems and viruses of unicellular eukaryotes have completely different interactive modules.
Three scenarios for the evolution of related viruses in plants and animals: common ancestry, virus transfer and convergence
The existence of numerous virus groups that include related viruses infecting animals and plants demands an evolutionary explanation. From general principles, there appear to be three major scenarios (Fig. 4):
Evolution of related plant and animal viruses from a common ancestral virus that predates the divergence of plants and animals;
Horizontal virus transfer (HVT) between plants and animals;
Parallel evolution of related viruses from related ancestral genetic elements.
Figure 4.
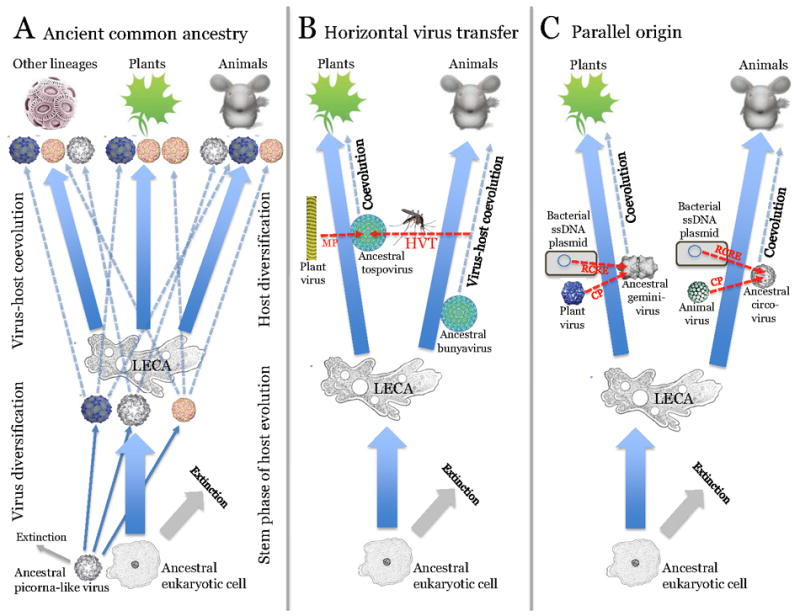
The three scenarios for the origin of related viruses in plants and animals: Common ancestry, horizontal virus transfer and parallel evolution.
For each scenario, examples of virus groups to which it most likely applies are given (see text). Abbreviations: LECA, Last eukaryotic common ancestor; MP, movement protein; HVT, horizontal virus transfer; RCRE, rolling-circle replication endonuclease; CP, capsid protein.
The principal criteria for distinguishing between these routes of evolution include the host range of a virus group, in particular whether it contains viruses infecting unicellular eukaryotes, in addition to those infecting plants and animals; phylogenies of conserved genes – whether or not plant and animal viruses cluster together; the comparative diversity of the virus group in plants and animals; and the level of similarity between the sequences and genome organizations of members of the groups infecting plants and animals. Application of these criteria to the diverse groups of viruses suggests that all three routes have been important, their relative contributions varying between groups of viruses.
Common ancestry of plant and animal viruses is linked to the current view of the early stages in the evolution of eukaryotes. Although animals and plants are the most complex multicellular eukaryotes, there is no evidence that they are monophyletic. On the contrary, animals and plants form distinct branches within two of the five (or possibly six) supergroups of eukaryotes, and all recent attempts to root the eukaryote tree place the root between the supergroups that include, respectively, animals and plants. Thus, the common origin scenario would imply that the common ancestors of virus groups shared by animals and plants already existed at the stage of the Last Eukaryotic Common Ancestor (LECA), perhaps evolving concomitantly with eukaryogenesis.
The primary showcases for common ancestry come from the opposite poles of the virus world: some of the simplest viruses, the picornavirus-like superfamily of positive-strand RNA viruses (Fig. 4A) [11], and the most complex viruses, the NCLDV [31]. Indeed, among the viruses of diverse unicellular eukaryotes discovered by both traditional and metagenomic methods, the picornavirus-like viruses and the NCLDV are by far the most abundant groups among the RNA and DNA viruses, respectively [5, 9, 47]. There is a many to many mapping of the major branches of the respective viruses onto the supergroups of eukaryotes: viruses of the same branch infect hosts from different supergroups, and conversely, each supergroup hosts viruses from different branches. This pattern suggests early radiation of the major branches within the picornavirus-like superfamily and within the NCLDV, with subsequent assortment of viruses from pre-existing ancestral pools to the emerging supergroups of eukaryotes (Fig. 4A) [11, 31]. Thus, within the picornavirus-like superfamily, four of the six major branches include both plant and animal viruses, and viruses of unicellular eukaryotes, the implication being that ancestors of these groups of viruses have already diverged at the stage of LECA. Similar conclusions have been reached in the phylogenomic study of the NCLDV that probably were represented by several ancestral viruses at the stage of LECA. At the divergence of the supergroups, the animal lineage inherited the iridoviruses and the common ancestors of poxviruses and asfarviruses, whereas green algae were infected by phycodnaviruses, later excluded from the land plant lineage.
The case for a likely transfer of viruses between plants and animals is presented by the negative strand RNA viruses such as rhabdoviruses and bunyaviruses (Fig. 4B). There is no evidence that any negative strand RNA viruses infect unicellular eukaryotes, so origin of related plant and animal viruses from a common ancestor antedating LECA is unlikely. Evolution via HVT is compatible with the high similarity between the protein sequences and genome architectures of plant and animal viruses in these families (Fig. 2B). Furthermore, vehicles for transfer are available: invertebrate parasites of animals and plants. Strikingly, viruses of the order Mononegavirales and the family Bunyaviridae that infect plants and vertebrates can also reproduce in their arthropod vectors [48, 49]. The discovery of negative strand RNA viruses, some of which are related to animal and others to plant viruses, in plant parasitic nematodes, the most abundant animals on earth, suggests that HVT could be even more opportune than currently appreciated [50]. Furthermore, the direction of HVT can be inferred with considerable confidence: from animals to plants, given the much greater diversity of negative strand RNA viruses in animals and the fact that all suspected vectors are animals.
The ssDNA viruses of plants and animals present a story of apparent parallel evolution from related prokaryotic genetic elements that replicate via the RCR mechanism (Fig. 4c) [27]. Although animal circoviruses and plant geminiviruses show similar organizations of the replication and CP modules (Fig. 2C), sequence similarity between the respective proteins is low, and virion architectures are different, a single and a ‘siamese twins’ icosahedra, respectively [51, 52]. A handful of known ssDNA viruses from unicellular eukaryotes, diatoms, have distinct genome organization and share little sequence similarity with either circoviruses or geminiviruses [53]. In contrast, a close evolutionary relationship appears to exist between replication proteins of geminiviruses and ssDNA plasmids from phythopathogenic bacteria both of which reproduce within plant phloem cells [54]. Given that at least some geminiviruses can replicate in bacteria [55], the origin of this plant virus family could involve horizontal transfer of the replicase gene module from a bacterial plasmid with concomitant acquisition of the CP and MP genes from pre-existing plant virus(es) [54, 56]. Although with less certainty, a potential circovirus ancestor was also proposed to be a bacterial ssDNA plasmid [54]. Thus, it appears likely that geminiviruses and circoviruses evolved in parallel and via similar scenarios, namely recombination between a plasmid from a bacterial parasite and a plant or animal virus, respectively. Certainly, conclusions on the routes of virus evolution have to be taken cautiously because new discoveries of ecological genomics have the potential to change the scenarios. A case in point is the single-cell genomic study of picobiliphytes, a group of marine picoeukaryotes distantly related to green algae and plants, in which a putative virus related to plant nanoviruses has been discovered [57]. Numerous homologs of this novel virus are detectable in marine metagenomic samples, suggesting the existence of abundant nanovirus-like agents in unicellular eukaryotes and implying an ancient origin of this family of ssDNA viruses. Similar discoveries on other virus groups by no means can be ruled out.
Conclusions
The existence of multiple related groups of viruses in plants and animals was a startling discovery at the dawn of viral genomics. These findings withstood the test of subsequent genome sequencing of hundreds of new plant and animal viruses. It became clear that all major divisions of the virus world include viruses infecting both plants and animals, with the single exception of dsDNA viruses that are missing in plants, likely due to their inability to move between plant cells. The genome architectures of all related viruses of plants and animals follow the same simple principle: a conserved housekeeping (replication) module is combined with a host-specific interaction module. Clearly, the ancestors of each group of plant and animal viruses evolved via recombination between a pre-existing selfish element (virus or plasmid) that provided the replication module with another virus already adapted to the respective host or with host genes from which the interaction module is derived. These formative stages of virus evolution do not fit into a virus-host co-evolution pattern, whereas the following diversification of novel virus classes occurs in accord with this paradigm.
Traditional virology focused on viruses that infect a few model animals, plants and bacteria. A recent breakthrough was the discovery and exploration of numerous viruses infecting diverse hosts, particularly unicellular eukaryotes, by means of direct virus isolation and metagenomics. Ecological genomics revealed unexpected aspects of virus distribution among hosts such as the apparent heavy dominance of picornavirus-like viruses in unicellular eukaryotes. These findings allow one to distinguish, even if tentatively, between the three logically possible evolutionary scenarios for related plant and animal viruses: origin from a common viral ancestor antedating the divergence of the hosts, horizontal virus transfer and parallel evolution. Nevertheless, the current sampling of viruses from diverse hosts and environments remains sparse. Further, extensive studies on global ecology of viruses should allow a comprehensive reconstruction of evolutionary relationships and could overturn some of the current views.
Highlights.
>In this article we present an overview of the evolutionary relationship between plant and animal viruses.
>In many virus families, related plant and animal viruses share suits of homologous genes involved in genome replication but possess distinct virus-host interaction modules
>Three major processes appear to have contributed to the evolution of related viruses in plants and animals: i) evolution from a common ancestral virus predating the divergence of plants and animals; ii) horizontal transfer of viruses, for example, through insect vectors; iii) parallel origin from related genetic elements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keeling PJ. Genomics. Deep questions in the tree of life. Science. 2007;317:1875–6. doi: 10.1126/science.1149593. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses - major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- ••4.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. This overview of the results of marine virology characterizes viruses as a major biogeochemical factor that affects the environment on the one hand by killing cells and releasing their content, and on the other hand, by enabling cells to occupy new ecological niches using genes transferred by viruses. [DOI] [PubMed] [Google Scholar]
- ••5.Kristensen DM, Mushegian A, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. An analytic review of virus metagenomics showing how metagenomics changes the prevailing ideas on the structure of the virus world. Statistical analysis of viral metagenomic sequences suggests that the marine DNA virome could be dominated by virus-like particles encapsidating host DNA, known as Gene Transfer Agents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatful GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011 doi: 10.1111/j.1574-6976.2011.00280.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–8. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- •9.Lang AS, Rise ML, Culley AI, Steward GF. RNA viruses in the sea. FEMS Microbiol Rev. 2009;33:295–323. doi: 10.1111/j.1574-6976.2008.00132.x. An overview of RNA viruses isolated from marine organisms, primarily unicellular eukaryotes, with an emphasis on the dominance of picorna-like viruses. [DOI] [PubMed] [Google Scholar]
- 10.Koonin EV, Senkevich TG, Dolja VV. The ancient virus world and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. The big bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 12.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–36. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci U S A. 2007;104:2043–9. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Malley MA, Koonin EV. How stands the Tree of Life a century and a half after The Origin? Biol Direct. 2011;6:32. doi: 10.1186/1745-6150-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72(Pt 9):2197–206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 16.Ng KKS, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haseloff J, Goelet P, Zimmern D, et al. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci USA. 1984;81:4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franssen H, Leunissen J, Goldbach R, et al. Homologous sequences in nonstructural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984;3:855–861. doi: 10.1002/j.1460-2075.1984.tb01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 20.Koonin EV. Evolution of double-stranded RNA viruses: a case for polyphyletic origin from different groups of positive-stranded RNA viruses. Seminars in Virology. 1992;3:327–339. [Google Scholar]
- 21.Gorbalenya AE, Pringle FM, Zeddam JL, et al. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–63. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. Embo J. 1989;8:3867–74. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 25.Goodier JL, Kazazian HH. Retrotransposons revisited:the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 26.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–34. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–85. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novick RP. Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem Sci. 1998;23:434–8. doi: 10.1016/s0968-0004(98)01302-4. [DOI] [PubMed] [Google Scholar]
- 29.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–96. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–34. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–292. doi: 10.1159/000312913. The latest description and phylogenomic analysis of the NCLDV presenting argument that the divergence of the major groups of the NCLDV antedates the radiation of the eukaryote supergroups. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–85. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Khayat R, Tang L, Larson ET, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson WH, Van Etten JL, Allen MJ. The phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol. 2009;328:1–42. doi: 10.1007/978-3-540-68618-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benitez-Alfonso Y, Faulkner C, Ritzenthaler C, Maule AJ. Plasmodesmata: gateways to local and systemic virus infection. Mol Plant Microbe Interact. 2010;23:1403–1412. doi: 10.1094/MPMI-05-10-0116. [DOI] [PubMed] [Google Scholar]
- 36.Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. Exploiting clinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol. 2005;43:361–394. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- 37.Koonin EV, Mushegian AR, Ryabov EV, Dolja VV. Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J Gen Virol. 1991;72:2895–2903. doi: 10.1099/0022-1317-72-12-2895. [DOI] [PubMed] [Google Scholar]
- 38.Melcher U. The ‘30K’ superfamily of viral movement proteins. J Gen Virol. 2000;81:257–66. doi: 10.1099/0022-1317-81-1-257. [DOI] [PubMed] [Google Scholar]
- 39.Ding SW, Vionnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •40.Wu Q, Wang X, Ding SW. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe. 2010;8:12–15. doi: 10.1016/j.chom.2010.06.009. An up to date survey of proteins suppressors of RNA interference that are encoded by plant and less commonly animal viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••41.Forterre P, Prangishvili D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann N Y Acad Sci. 2009;1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x. A conceptually important article that presents arguments for the arms race between viruses and cellular hosts being one of the principal driving forces of all evolution. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi O, Akira S. Innate immunity to viral infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••43.Agol VI, Gmyl AP. Viral security proteins: counteracting host defences. Nat Rev Microbiol. 2010;8:867–78. doi: 10.1038/nrmicro2452. This review article develops the concept of “security proteins”, dedicated virus-encoded counter-defense devices. Using primarily picornavirus security proteins as examples, the article shows how otherwise related viruses employ completely different counter-defense strategies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krupovic M, Bamford DH. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 45.Dolja VV, Boyko VP, Agranovsky AA, Koonin EV. Phylogeny of capsid proteins of rod-shaped and filamentous plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology. 1991;184:79–86. doi: 10.1016/0042-6822(91)90823-t. [DOI] [PubMed] [Google Scholar]
- •46.Niehl A, Heinlein M. Cellular pathways for viral transport through plasmodesmata. Protoplasma. 2011;248:75–99. doi: 10.1007/s00709-010-0246-1. An up to date review describing how the interaction between viral proteins, including dedicated movement proteins and capsid proteins, with plant cell components provides for virus passage through plasmodesmata. [DOI] [PubMed] [Google Scholar]
- 47.Monier A, Larsen JB, Sandaa RA, et al. Marine mimivirus relatives are probably large algal viruses. Virol J. 2008;5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogenhout SA, Ammar ED, Whitefield AE, Redibaugh MG. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol. 2008;46:327–359. doi: 10.1146/annurev.phyto.022508.092135. [DOI] [PubMed] [Google Scholar]
- 49.Weaver SC. Evolutionary influences in arboviral disease. Curr Top Microbiol Immunol. 2006;299:285–314. doi: 10.1007/3-540-26397-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••50.Bekal S, Domier LL, Niblack TL, Lambert KN. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J Gen Virol. 2011;92:1870–9. doi: 10.1099/vir.0.030585-0. This article describes negative-strand RNA viruses in nematodes, the most abundant group of animals on earth in which typical viruses have not been previously discovered. This discovery reveals a plausible path of virus transfer between animals and plants. [DOI] [PubMed] [Google Scholar]
- 51.Finsterbusch T, Mankertz A. Porcine circoviruses--small but powerful. Virus Res. 2009;143:177–83. doi: 10.1016/j.virusres.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 52.Jeske H. Geminiviruses. Curr Top Microbiol Immunol. 2009;331:185–226. doi: 10.1007/978-3-540-70972-5_11. [DOI] [PubMed] [Google Scholar]
- 53.Tomaru Y, Takao Y, Suzuki H, et al. Isolation and Characterization of a Single-Stranded DNA Virus Infecting Chaetoceros lorenzianus Grunow. Appl Environ Microbiol. 2011 doi: 10.1128/AEM.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••54.Krupovic M, Ravantti JJ, Bamford DH. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol Biol. 2009;9:112. doi: 10.1186/1471-2148-9-112. This article demonstrates a specific evolutionary relationship between the replication proteins of geminiviruses and plasmids of phytopathogenic bacteria, as well as between capsid proteins of geminiviruses and plant RNA viruses. The results suggest an origin of geminiviruses from plasmid rolling circle replicons through acquisition of the capsid protein gene from a virus, in agreement with the hypothesis proposed in Ref. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selth LA, Randles JW, Rezaian MA. Agrobacterium tumefaciens supports DNA replication of diverse geminivirus types. FEBS Lett. 2002;516:179–82. doi: 10.1016/s0014-5793(02)02539-5. [DOI] [PubMed] [Google Scholar]
- 56.Koonin EV, Ilyina TV. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J Gen Virol. 1992;73(Pt 10):2763–6. doi: 10.1099/0022-1317-73-10-2763. [DOI] [PubMed] [Google Scholar]
- ••57.Yoon HS, Price DC, Stepanauskas R, et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–7. doi: 10.1126/science.1203163. This article employs single-cell genomics to describe an abundant ssDNA virus of picobiliphytes, a group of uncultivated marine picoeukaryotes. The results imply an ancient origin of nanoviruses and more generally show the possibility of existence of unsuspected groups of viruses, particularly in uncultivated organisms. [DOI] [PubMed] [Google Scholar]


