Background: The link between obesity and diabetes is poorly understood. Wnt signaling is implicated in adipogenesis and glucose metabolism.
Results: LRP6+/− mice are protected against diet-induced obesity and insulin resistance by regulation of genes involved in adipogenesis, metabolism, and insulin signaling.
Conclusion: This study identifies novel pathways that regulate metabolism.
Significance: LRP6 is a potential target for novel therapeutics for diabetes and obesity.
Keywords: Adipogenesis, Diabetes, Gluconeogenesis, Glucose Metabolism, Insulin Resistance, Metabolic Regulation, Metabolic Syndrome
Abstract
Body fat, insulin resistance, and type 2 diabetes are often linked together, but the molecular mechanisms that unify their association are poorly understood. Wnt signaling regulates adipogenesis, and its altered activity has been implicated in the pathogenesis of type 2 diabetes and metabolic syndrome. LRP6+/− mice on a high fat diet were protected against diet-induced obesity and hepatic and adipose tissue insulin resistance compared with their wild-type (WT) littermates. Brown adipose tissue insulin sensitivity and reduced adiposity of LRP6+/− mice were accounted for by diminished Wnt-dependent mTORC1 activity and enhanced expression of brown adipose tissue PGC1-α and UCP1. LRP6+/− mice also exhibited reduced endogenous hepatic glucose output, which was due to diminished FoxO1-dependent expression of the key gluconeogenic enzyme glucose-6-phosphatase (G6pase). In addition, in vivo and in vitro studies showed that loss of LRP6 allele is associated with increased leptin receptor expression, which is a likely cause of hepatic insulin sensitivity in LRP6+/− mice. Our study identifies LRP6 as a nutrient-sensitive regulator of body weight and glucose metabolism and as a potential target for pharmacological interventions in obesity and diabetes.
Introduction
Impaired insulin sensitivity plays a key role in the pathogenesis of metabolic syndrome and its sequelae by mechanisms that are largely unknown. Alterations in Wnt signaling have been associated with metabolic syndrome and type 2 diabetes both in human and animal models (1–5). A growing body of evidence has been emerging on the role of Wnt signaling in insulin secretion and beta cell function (6, 7). However, very little attention has been given to the effect of Wnt signaling on peripheral tissue insulin sensitivity, which is considered to be the basis for metabolic syndrome.
The canonical Wnt signaling pathway consists of cascades of events that follow the binding of Wnt proteins to their receptor frizzled and coreceptor LRP5/6,2 causing activation of the dishevelled family of proteins and inhibition of a protein complex that includes axin, GSK-3β, and APC (8–11). Furthermore, phosphorylated LRP6 directly inhibits β-catenin phosphorylation by GSK-3β independently of axin function (12, 13). These events stabilize the cytoplasmic pool of β-catenin and enhance its translocation to the nucleus to interact with other transcription regulators, including T-cell factor (TCF)/lymphoid enhancer factor (LEF) and FoxO1, triggering the expression of a variety of target genes that regulate gluconeogenesis, insulin secretion, and signaling (1–5, 7). The effects of Wnt signaling on insulin sensitivity and glucose homeostasis in peripheral tissues are emerging through recent studies in rodents. For instance, targeted knock-out of β-catenin in the liver reduces hepatic glucose production and promotes hepatic insulin sensitivity (14). In humans, variations of TCF7L2 have been linked to impaired peripheral insulin sensitivity (3, 6, 15). Most recently, a rare mutation in the Wnt coreceptor LRP6 was found to be associated with metabolic syndrome and diabetes (1). Finally, several genes involved in mitochondrial energy expenditure and glucose homeostasis have been identified as targets of LRP6/Wnt signaling (16, 17). Energy expenditure and fuel oxidation within the mitochondria of the brown adipose tissue (BAT) is primarily regulated by PGC1-α, which is inhibited by Wnt-10b (18–21). Wnt activates the key nutrient sensing pathway mTORC1 by inhibiting GSK-3β (22), an effect that leads to diminished IRS1 expression and function and perturbation of insulin signaling. Many of these findings are results of in vitro studies, and their in vivo correlates remain to be determined. In this study, we explored the role of Wnt coreceptor LRP6 in glucose metabolism and insulin sensitivity of LRP6+/− mice by physiological and molecular studies.
EXPERIMENTAL PROCEDURES
Generation of LRP6 Knock-out Mice
LRP6 knock-out mouse were generated from LST067 embryonic stem (ES) cells obtained from Bay Genomics. These ES cells are derived from the 129/Ola mouse strain electroporated with the secretory trap vector pGT2TMpfs harboring an insertion mutation in exon 2 of LRP6 gene. For confirmation, the complementary DNA was sequenced by rapid amplification of cDNA ends. ES cell clones were injected into C57BL/6 (albino) blastocysts. The resulting chimeras were backcrossed with C57BL/6 (albino) females for six generations. Animals were housed under controlled temperature (23 °C) and lighting (12 h of light, 6 a.m. to 6 p.m.; 12 h of dark, 6 p.m. to 6 a.m.) with free access to water and either standard mouse chow or Western diet according to the protocol. High fat diet (hfd) contained 55% fat (28% saturated, 30% trans, 28% monounsaturated, 14% polyunsaturated), 21% protein, and 24% carbohydrates (Harlan Teklad, Madison, WI).
Intraperitoneal Glucose Tolerance Test (IPGTT)
Study protocols were approved by the Institutional Animal Care and Use Committee at Yale. Eight male heterozygote LRP6 knock-out (F6) and 8 WT (age, 8 weeks) mice were placed on hfd for 6–8 weeks. Mice were transferred to clean cages without food at the end of the dark cycle (7 p.m.) at the Yale Metabolic Phenotyping Center. The next day, glucose and insulin concentrations were determined after 14 h of fasting from tail veins prior to an intraperitoneal injection of glucose (1 mg of glucose/g of body weight), and subsequent readings were taken at 15, 30, 45, 60, 90, and 120 min. After completion of the experiment, animals were anesthetized with ketamine/xylazine intraperitoneally (80–100 mg/kg and 10 mg/kg of body weight, respectively), and interscapular BAT, gastrocnemius muscle, liver, and abdominal white adipose tissues were freeze clamped, dissected, and stored at −80 °C until further analysis. This experiment was duplicated to test reproducibility.
Hyperinsulinemic-Euglycemic Clamp Experiments
Eight-week-old male LRP6+/− mice (F6) and age-, sex-, and weight-matched WT mice on hfd for 3 weeks underwent hyperinsulinemic-euglycemic clamp experiments. One week before the clamp experiment, permanent catheters were inserted into the left jugular vein under general anesthesia. All procedures were approved by the Yale University Animal Care and Use Committee and were carried out by the Yale Metabolic Phenotyping Center.
Conscious mice were placed in restraining tubes. During the first 120-min basal period, a primed-continuous [3-3H]glucose infusion (10-μCi bolus, 0.1 μCi/min) was used for the estimation of postabsorptive basal versus insulin-stimulated glucose turnover. Hyperinsulinemic-euglycemic clamps were carried out for 120 min with a 4-min primed (29 milliunits/kg) followed by a continuous (3 milliunits/kg-min) insulin infusion (Novolin, Novo Nordisk). Euglycemia was maintained by a variable glucose (D-20) infusion. A single 2-deoxy-d-[1-14C]glucose injection was administered at 75 min. To determine serum [3-3H]glucose, 3H2O, and 2-deoxy-d-[1-14C]glucose concentrations, blood samples were collected at 80, 85, 90, 100, 110, and 120 min of the clamp, and for measurement of basal [3-3H]glucose concentrations, blood samples were collected in the final 10 min of the basal period. A serum sample for determination of basal insulin levels was obtained during the final 10 min of the basal period, and for steady-state insulin levels, the sample was obtained at 120 min of the clamp. Infusions were performed using microdialysis pumps (CMA/Microdialysis, North Chelmsford, MA). Radioisotopes were purchased from PerkinElmer Life Sciences and American Radiolabeled Chemicals (St. Louis, MO).
In Vivo and in Vitro Signaling Studies
For Western blot analysis, samples were homogenized in radioimmune precipitation assay buffer containing protease inhibitor (Roche Applied Science) and Halt phosphatase inhibitor mixtures (Thermo Scientific). A total of 50 μg of protein was electrophoresed on 4–15% precast SDS-polyacrylamide gels and transferred onto a nitrocellulose membrane (Bio-Rad D102491). Blots were probed with antibodies for p-IRS1Tyr-1222, IRS1, p-AktSer-473, Akt, GSK-3βSer-9, GSK-3β, p-stat3Tyr-705, stat3, FoxO1, p-FoxO1Thr-24, LRP6, phosphoenolpyruvate carboxykinase, S6, p-S6Ser-235/236, p-S6KThr-389 (all purchased from Cell Signaling Technology), PGC1-α, UCP1 (Novus Biologicals), and leptin receptor (lepr) (Thermo Scientific), β-catenin, and glucose-6-phosphatase (Santa Cruz Biotechnology, Inc.). Results were detected by chemiluminescence (ECL, Amersham Biosciences), visualized on a Molecular Imager® ChemiDocTM XRS System, and analyzed by Bio-Rad Quantity One Analysis Software. An internal standard (wild-type mouse homogenate) was used to normalize results and to control for blot-to-blot variation.
To examine LRP6 regulation of mTOR signaling in vitro, serum-starved 3T3L1 cells with or without LRP6 knockdown were treated with 20 mm LiCl for 6 h or 150 ng/ml Wnt3a for 1 h. pS6 activation was examined by Western blot analysis. To assess whether increased expression levels of PGC1-α are caused by LRP6 deficiency, we prepared adenovirus expressing wild-type LRP6 from the CMV promoter and transiently expressed it in 3T3L1 cells. PGC1-α protein levels were determined by Western blotting. In separate experiments, native LRP6 was knocked down by RNA interference. Lentivirus containing LRP6-specific shRNA was purchased from Santa Cruz Biotechnology, Inc. 3T3L1 cells were infected by this vector, and expression levels of PGC1-α were determined by Western blotting.
Wnt signaling blocks brown adipogenesis in the early stages of differentiation and stimulates their conversion to white adipocytes via UCP1 inhibition (17). This could explain the development of the phenotypes that we will describe in the current study, particularly lean body weight and adaptive thermogenesis. Therefore, we studied the role of LRP6 in adipocyte differentiation and UCP1 expression during the process of adipocyte differentiation in 3T3L1 cells. 3T3L1 preadipocytes were differentiated to adipocytes by 3-isobutyl-1-methylxanthine in the presence or absence of the GSK-3β inhibitor LiCl according to standard protocol (23, 24). Expression levels of UCP1 and LRP6 were assessed by Western blotting at day 8 of adipocyte differentiation. Hepatic triglyceride contents and serum triglyceride, LDL, and HDL were analyzed at the Mouse Metabolic Phenotyping Center at Yale by COBAS Mira Plus (Roche Applied Science).
Preparation of Nuclear Fraction
The hepatic nuclear fractions were prepared with the Nuclear Extraction kit (Active Motif). Briefly, ∼100 mg of liver tissue was homogenized and lysed in the cytoplasmic extraction reagent for 15 min on ice. The extract was subsequently centrifuged to separate the nuclear pellet from the cytoplasmic supernatant. The nuclear pellet was resuspended in the Nuclear Extraction reagent, lysed for 30 min at 4 °C on a shaker at 150 rpm, and centrifuged to obtain the nuclear extract. 50 μg of nuclear extract was used for SDS-PAGE and subsequent Western blotting.
Quantitative Real Time PCR
Total RNA was extracted from homogenates using TRIzol reagent (Invitrogen) and an RNeasy Mini kit (Qiagen). First strand cDNA synthesis was performed using Omniscript reverse transcriptase (Qiagen). Quantitative PCR was carried out using iQTM SYBR® Green Supermix (Bio-Rad). Reactions were run in triplicate on an Eppendorf realplex, and the relative quantification of gene expression was performed using the comparative CΤ (ΔΔCΤ) method. mRNA expression levels of PGC1-α, UCP1, glucose-6-phosphatase, phosphoenolpyruvate carboxykinase, lepr, and LRP6 were assessed using specific primers.
ATP Content Measurement
ATP content (nmol/mg of protein) in the extracts of freeze clamped BAT was measured using HPLC analysis as described by Zhang et al. (25).
Affymetrix Exon Array Studies
RNA was extracted from the livers of four LRP6+/− mice and an equal number of WT mice using TRIzol (Invitrogen), and cDNA was produced using SuperScript III reverse transcription (Invitrogen) kits. Six to 8 μg of total RNA were processed, labeled, and hybridized to Affymetrix Human Exon ST 1.0 arrays by the Keck core facility at Yale. Exon-level expression values were derived from the CEL file probe-level hybridization intensities using the model-based RMA algorithm as implemented in the Affymetrix Expression ConsoleTM. The significance analysis of microarrays software (SAM) was used to determine potential signature genes. The MetaCoreTM biological network analysis platform was used to detect pathways that were differentially expressed between LRP6+/− mice and an equal number of WT mice.
Cold Exposure Protocol
LRP6+/− mice (n = 12) and their wild-type littermates (n = 12) were housed in individual cages with free access to standard diet and water on a 12:12-h artificial light-dark cycle. Animals were exposed to 4 ± 1 °C cold temperature without prior acclimation. Mice were placed within restrainers (transparent plastic tubes) to avoid extraneous activities. Rectal temperatures were measured every 10 min during cold exposure. Animals were immediately removed from the cold if there were significant signs of distress present.
Histological Studies
Mouse livers and interscapular tissues were fixed in 4% paraformaldehyde followed by tissue processing, cryoembedding, sectioning, and H&E and red oil O staining (Mass Histology Service, Worchester, MA). For imaging, a Nikon Eclipse E800 microscope (Nikon Instruments Inc., Melville, NY) fitted with a SpotTM digital camera (Diagnostic Instruments Inc., Sterling Heights, MI) was used.
Statistical Analysis
Statistical analysis was carried out with two-factor analysis of variance. p < 0.05 was considered statistically significant for all experiments. For all statistics, we used GraphPad Prism version 4.03 for Windows (GraphPad Software, San Diego, CA).
RESULTS
LRP6+/− Mice on High Fat Diet Exhibit Lower Base-line Serum Glucose and Enhanced Adipose Tissue and Hepatic Insulin Sensitivity
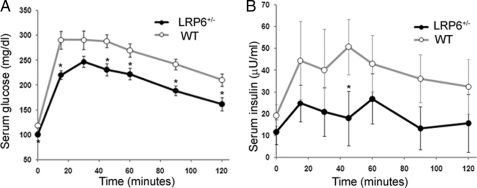
The homozygote LRP6 knock-out mice were either embryonically lethal or died shortly after birth as reported previously (26). Gross morphological abnormalities in these mice included midline defects and curved tails. The LRP6+/− mice had normal appearance and behavior. Expression levels of the LRP6 mRNA and protein in different tissues of LRP6+/− mice, including liver and BAT, were about 50% lower than in WT littermates (supplemental Fig. S1, A and B). Base-line serum glucose levels were lower in LRP6+/− mice on hfd (Fig. 1A) but not on chow diets (data not provided) compared with the WT littermates. This finding prompted us to carry out an IPGTT in male LRP6+/− mice 14 ± 2 weeks age on hfd and age- and gender-matched wild-type littermates. LRP6+/− mice exhibited enhanced glucose tolerance compared with their wild-type littermates during the IPGTT. After 15 min, serum glucose levels rose in WT mice by 60 ± 15 mg/dl higher than in LRP6+/− mice and remained higher for the entire duration of the test (Fig. 1A). LRP6+/− mice displayed reduced serum insulin responses despite lower serum glucose levels compared with WT mice (Fig. 1B). These findings indicated higher insulin sensitivity of LRP6+/− mice compared with the WT mice.
FIGURE 1.
IPGTT (A and B). Ten- to 12-week-old male LRP6+/− mice and age-matched WT littermates (n = 8 in each group) were fed hfd for 4 weeks and subsequently received a loading dose of glucose (1 mg/g of body weight), and the serum glucose levels were monitored at the indicated times. Intraperitoneally stimulated glucose levels (A) and glucose-induced insulin levels (B) in LRP6+/− mice and age-matched WT littermates are shown (data are mean ± S.E.; *, p < 0.05 by analysis of variance).
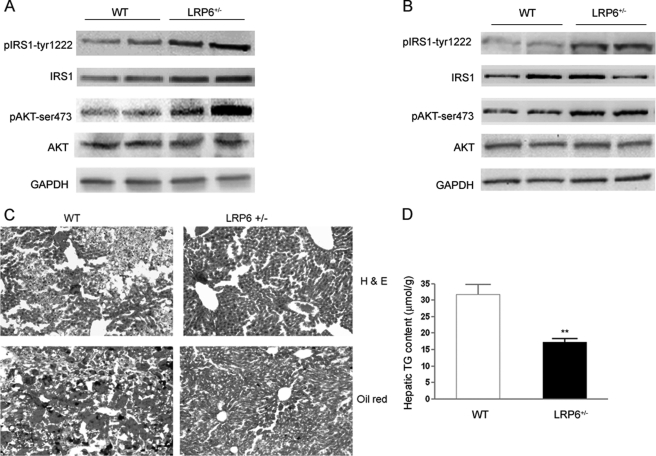
This finding prompted examination of the insulin signaling in insulin sensitive tissues of LRP6+/− mice and their wild-type littermates on hfd. Insulin signaling was enhanced in the BAT (Fig. 2A) and liver (Fig. 2B) but not in the skeletal muscles (supplemental Fig. S2A) of LRP6+/− mice compared with the wild-type littermates. Interestingly, LRP6+/− mice had increased IRS1 activity and expression of BAT. This particular finding prompted examination of the mTOR pathway for reasons that will be discussed. Consistent with increased hepatic insulin sensitivity, LRP6+/− mice on hfd exhibited considerably lower hepatic fat in histological examination with red oil and had lower hepatic triglyceride contents compared with WT mice (Fig. 2, C and D).
FIGURE 2.
Brown adipose tissue and hepatic insulin signaling of LRP6+/− and WT mice (A–D). Mice were sacrificed after IPGTT, and proteins were extracted from tissues and analyzed for the total and phosphorylated forms of each peptide using three independent measurements for each time point. Expression levels of insulin-stimulated p-IRS1Tyr-1222 and p-AktSer-473 were significantly higher in the BAT (A) and liver (B) of the LRP6+/− compared with WT mice (mean ± S.E. by densitometry; p < 0.05). Frozen sections of liver from LRP6+/− and WT mice were stained with oil red O. Hepatic steatosis was significantly less present in LRP6+/− compared with WT mice (C). Hepatic triglyceride (TG) content was significantly lower in LRP6+/− compared with WT mice (D) (mean ± S.E.; **, p < 0.01).
LRP6+/− Mice Are Protected against Diet-induced Obesity
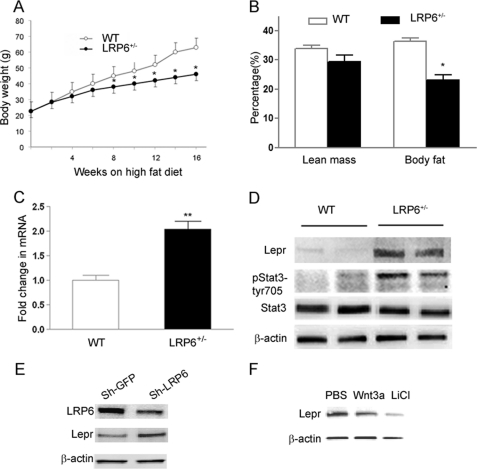
A noticeable phenotype of the LRP6+/− mice was their tendency toward lower body weights compared with their age- and gender-matched littermates. This prompted a systematic evaluation of body weight and total body fat. Six- to 8-week-old male LRP6+/− mice and age-matched male WT mice were placed on hfd, and their body weights and food consumption were measured over a period of 16 weeks. Reduced body weight in LRP6+/− mice was not due to decreased energy intake because LRP6+/− mice showed only a small but statistically not significant trend toward reduced food consumptions compared with WT littermates. Similarly, stool weights were not significantly different between the two groups during this period. Both mouse groups gained substantial body weight throughout the course of high fat feeding. However, LRP6+/− mice had significantly lower weights after the 8th week of hfd compared with WT mice (Fig. 3A). Analysis of body fat content using a dual energy x-ray absorptiometry scanner revealed that LRP6+/− mice had significantly lower body fat than WT littermates after 16 weeks of high fat feeding (Fig. 3B). LRP6+/− mice on chow diet had slightly lower total body weights but similar body fat content compared with their wild-type littermates (supplemental Fig. S3A). There were no major differences in lipid profiles between LRP6+/− mice and their littermates regardless of their diet (supplemental Table S1). Taken together, these findings suggested reduced adiposity of the LRP6+/− mice on hfd that may contribute to their enhanced insulin sensitivity compared with WT littermates.
FIGURE 3.
Body weight and fat mass and hepatic leptin signaling of LRP6+/− mice (A–F). Growth curves of 8-week-old male LRP6+/− (filled circles) and WT mice (unfilled circles) on hfd are shown. Mice were weighed once every 2 weeks at the same time during the day for 16 weeks (data are mean ± S.E.; *, p < 0.05) (A). A comparison of the percentage of total body fat in LRP6+/− and WT mice on hfd for 8 weeks by dual energy x-ray absorptiometry scanning (n = 12 mice per group; data are mean ± S.E.; *, p < 0.05) is shown (B). Hepatic leptin receptor mRNA (n = 8; mean ± S.E.; **, p < 0.001) (C) and protein levels (D) in LRP6+/− mice compared with their wild-type littermates are shown. Activation (phosphorylation) of the hepatic leptin receptor downstream protein stat3 in LRP6+/− versus WT mice (n = 8; mean ± S.E. by densitometry; p < 0.001) is shown. Leptin receptor protein expression in HepG2 after knockdown of LRP6 by RNA interference compared with scrambled shRNA (Sh-LRP6) (n = 4; mean ± S.E. by densitometry; p < 0.001) (E) and after treatment of the cells with Wnt and GSK-3β inhibitor LiCl (F) is shown.
Enhanced Expression of Hepatic Leptin Receptor in LRP6+/− Mice
The question of whether the enhanced hepatic insulin sensitivity is caused by cross-talk between the adipose tissue and liver led to measurement of circulating adipokines. Serum adiponectin levels were only modestly (∼1.2-fold) higher in LRP6+/− mice compared with WT littermates (supplemental Fig. S4A). Serum leptin levels were not significantly different (supplemental Fig. S4B).
We then examined the global hepatic gene expression profiling of LRP6+/− mice and their WT littermates using Affymetrix expression arrays. Strikingly, the hepatic lepr in LRP6+/− mice was among the 15 highest up-regulated genes compared with their wild-type littermates (supplemental Table S2); the finding was confirmed by RT-PCR (Fig. 3C). At the protein level, the difference was more significant (5-fold increase in LRP6+/− compared with WT mice; p ≤ 0.005; Fig. 3D). We then examined whether the changes of the hepatic lepr levels in LRP6+/− mice alters the activity of its downstream targets (27–29). Phosphorylation of the hepatic stat3, a downstream target of leptin, was significantly higher in LRP6+/− versus WT mice (Fig. 3D). lepr expression is inversely related to body adiposity in both rodents and human. However, the mechanisms underlying the link between adipose tissue and hepatic lepr expression and insulin sensitivity are not well understood. Our finding raised the question of whether Wnt/LRP6 negatively regulates lepr expression and functions as a link between body fat and hepatic insulin sensitivity. To address this question, LRP6 was knocked down in HepG2 cells by RNA interference. Knockdown of LRP6 by an LRP6-specific shRNA in these cells decreased LRP6 expression by 50% and increased lepr protein levels by 2-fold compared with GFP shRNA (Fig. 3E). Wnt stimulation of HepG2 cells resulted in reduced expression of lepr (Fig. 3E), indicating that LRP6 mediates the effect of Wnt on lepr expression. Lithium, a GSK-3β inhibitor, dramatically reduced lepr protein expression (Fig. 3F), indicating that lepr is regulated by the canonical Wnt pathway. Taken together, these findings suggest that the haploinsufficiency of LRP6 in the LRP6+/− mouse diminishes the inhibitory effect of Wnt on lepr transcription, an effect that likely contributes to increased hepatic insulin sensitivity of LRP6+/− mice.
Enhanced Brown Adipose Tissue Insulin Sensitivity Is Associated with Impaired Activity of mTORC1 Pathway
The differential response of LRP6+/− mice to diet and their increased adipose tissue expression of IRS1 compared with their littermates suggested the involvement of the mTORC1 pathway. mTOR is a key nutrient-sensing kinase that regulates adipogenesis, body weight, and insulin response to caloric intake. mTOR activation deranges cellular metabolism, triggers degradation of IRS, and leads to impairment of insulin sensitivity (30). At least three transgenic mouse models with disruption of mTORC1 activity have displayed phenotypes that precisely match those in LRP6+/− mice (22, 31, 32).
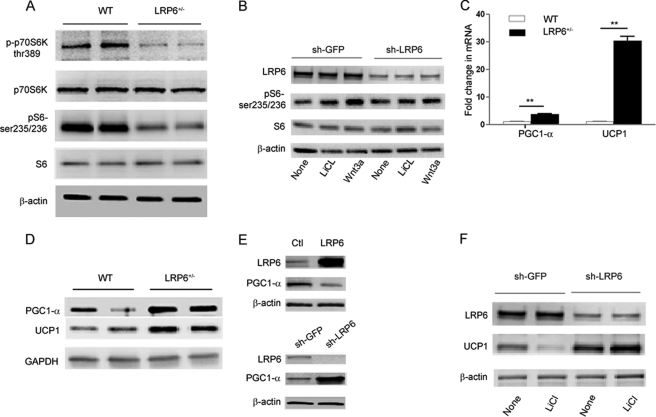
Examination of insulin-sensitive tissues showed that mTORC1 activity in BAT but not in the skeletal muscle or liver of LRP6+/− mice is significantly reduced compared with their wild-type littermates. Both BAT S6K and S6 phosphorylations were lower in LRP6+/− mice compared with control littermates (Fig. 4A, data only shown for BAT). This finding strongly correlates with the higher IRS1 expression and activity and subsequent insulin sensitivity of the BAT in LRP6+/− mice (Fig. 3A).
FIGURE 4.
Brown adipose tissue mTOR activity and expression levels of energy metabolism genes (A–F). A, mTOR activity of the BAT in LRP6+/− compared with WT mice. B, mTOR activation in response to GSK-3β inhibition by LiCl and Wnt3a in 3T3L1 cells before and after LRP6 knockdown. PGC1-α and UCP1 mRNA (C) and protein (D) expression in BAT of LRP6+/− compared with WT mice is shown. **, p < 0.01. E, PGC1-α expression in 3T3L1 preadipocytes overexpressing LRP6 compared with control adenovirus (Ctl) (top) and in 3T3L1 preadipocytes after LRP6 knockdown by LRP6-specific shRNA (sh-LRP6) compared with the scrambled shRNA (sh-GFP) (bottom; data are mean ± S.E. by densitometry; p < 0.05). F, UCP1 expressions in 3T3L1 preadipocytes with and without LRP6 knockdown undergoing adipogenic transformation in response to Wnt agonist LiCl.
We then examined the role of LRP6 in Wnt activation of mTOR in 3T3L1 cells. Earlier studies have shown that canonical Wnt activates mTOR by inhibiting GSK-3β. LiCl is an inhibitor of GSK-3β. We treated 3T3L1 cells with lithium and Wnt3a with and without LRP6 knockdown. Although LiCl and Wnt3a increased S6 phosphorylation in wild-type cells, S6 phosphorylation was marginally increased in LRP6 knockdown cells (Fig. 4B). Taken together, these findings strongly suggest that Wnt activation of mTOR is LRP6-dependent. Diminished mTOR activity due to loss of LRP6 allele is likely a major cause of enhanced brown adipose tissue insulin sensitivity.
Enhanced Expression of Genes Involved in Mitochondrial Biogenesis, Energy Dissipation, and Adoptive Thermogenesis in LRP6+/− Mice
Insulin sensitivity and reduced body fat/body weight in mice deficient for genes in the mTOR pathway have been linked to increased PGC1-α/UCP1 activity (22, 31, 32). These findings were greatly relevant because Wnt activates mTORC1 (22) and similarly inhibits PGC1-α and UCP1 (17). Thus, reduced adiposity in LRP6+/− mice could be a consequence of reduced Wnt-dependent activation of mTOR, up-regulation of PGC1-α and UCP1, and increased mitochondrial respiration. Consequently, we examined PGC1-α/UCP1 expression in the BAT of LRP6+/− mice and their wild-type littermates.
The mRNA and protein expression levels of PGC1-α and its downstream gene UCP1 were considerably higher in the BATs of LRP6+/− mice compared with their weight-, age-, and gender-matched wild-type littermates (Fig. 4, C and D). Strikingly, the UCP1 content of the abdominal white adipose tissue, which under normal conditions is undetectable, was significantly increased in the LRP6+/− mice (supplemental Fig. S5, A and B).
The negative regulation of PGC1-α by LRP6 was further confirmed by overexpression of wild-type LRP6 in 3T3L1 preadipocytes. In these cells, PGC1-α was expressed at significantly lower levels compared with cells transfected with the vehicle alone (Fig. 4E). Accordingly, LRP6 knockdown in these cells led to higher PGC1-α expression (Fig. 4E). UCP1 was not expressed in wild-type or LRP6 knockdown preadipocytes.
It is well established that UCP1 is expressed when preadipocytes are differentiated to mature adipocytes. We examined the effect of LRP6 on UCP1 expression during differentiation in wild-type and LRP6 knockdown 3T3L1 cells. 3T3L1 cells were either treated with PBS or LiCl. Lithium inhibits brown adipocyte differentiation (23). LRP6 knockdown resulted in significant up-regulation of UCP1 (Fig. 4F) compared with cells infected with a GFP shRNA. LiCl treatment of 3T3L1 inhibited UCP1 expression (Fig. 4F). However, cells with LRP6 knockdown continued with transformation despite treatment with LiCl and expressed UCP1 at levels that were similar to those in untreated cells. This finding underscores the critical role of LRP6 in regulation of adipogenesis and expression of UCP1.
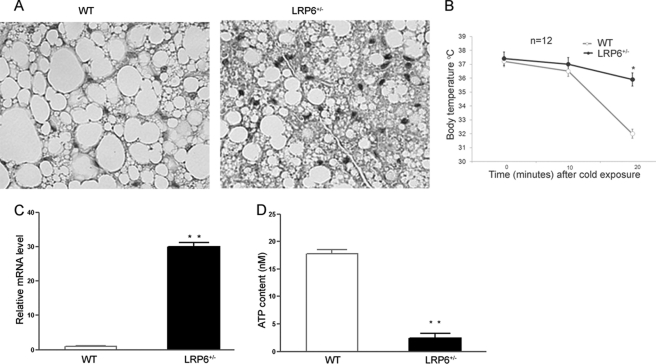
Increased Mitochondrial Biogenesis and Augmented Energy Expenditure in LRP6+/− Mice
We examined the functional consequences of increased PGC1-α/UCP1 expressions in LRP6+/− mice. Although the crude masses of the BAT between the two mouse groups were not significantly different, brown adipocytes were more abundant in the interscapular BAT of LRP6+/− mice versus their wild-type littermates (Fig. 5A). A key component of energy expenditure is adaptive thermogenesis. We examined whether the up-regulation of the BAT UCP1 in LRP6+/− mice enhances their thermogenic response compared with their littermates. We exposed male LRP6+/− and weight- and gender-matched WT mice to standardized cold challenges. Resting body temperatures were similar between LRP6+/− and the WT mice (Fig. 5B). Eight-week-old mice on hfd were immobilized and exposed to 4 °C, and their rectal temperatures were monitored. For the purpose of standardization, mice were kept immobilized in transparent plastic tubes. Cold exposures within 25 min resulted in significant distress in WT mice, prompting termination and limitation of this experiment to 20 min according to the approved protocol by the Institutional Animal Care and Use Committee. LRP6+/− mice displayed enhanced adaptation to the cold challenges and a lesser decrease in body temperatures compared with WT mice.
FIGURE 5.
Comparison of brown adipocyte activity between LRP6+/− mice and wild-type littermates (A–D). A, H&E-stained sections of interscapular BAT from LRP6+/− and WT mice on hfd. B, enhanced adaptive thermogenesis in LRP6+/− (n = 12) compared with WT (n = 12) mice. Mice were exposed to 4 °C for 20 min while immobilized in restrainers (data are mean ± S.E.; *, p < 0.05 by analysis of variance. BAT mitochondrial transcriptional factor A (TFA) (C) and ATP contents (D) are shown. (Data are mean ± S.E. **, p < 0.01).
Increased expression of PGC1-α is associated with enhanced mitochondrial biogenesis and metabolism. Accordingly, there was increased BAT mitochondrial transcriptional factor A indicative of enhanced mitochondrial biogenesis in LRP6+/− mice compared with WT littermate (Fig. 5C). In contrast, the ATP content in BAT of LRP6+/− mice compared with WT littermates was significantly lower (Fig. 5D). This finding was suggestive of increased energy dissipation in the mutant mice due to excessive expression of UCP1. Taken together, these findings indicated augmented functional activity of the BAT in LRP6+/− mice and its contribution to enhanced energy expenditure and diminished body adiposity compared with WT mice.
Reduced Endogenous Hepatic Glucose Output in LRP6+/− Mice
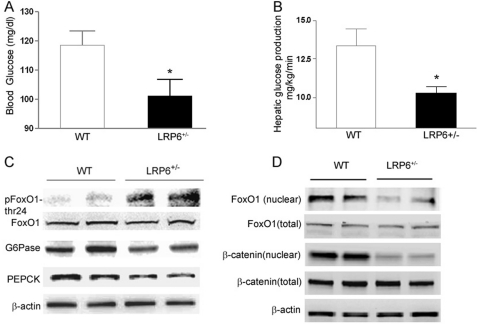
The question of whether enhanced insulin sensitivity in LRP6+/− mice on hfd is only accounted for by their lower body adiposity compared with WT mice prompted further investigations in the mice prior to their differentiated weight gain. Eight- to 9-week-old male LRP6+/− mice and weight- and age-matched male wild-type littermates were placed on hfd for 3 weeks. The basal glucose levels were lower in LRP6+/− compared with WT mice despite their equal weights (Fig. 6A). All mice were then subjected to hyperinsulinemic-euglycemic clamp studies. Insulin infusion during the hyperinsulinemic-euglycemic clamp similarly raised serum insulin levels in both LRP6+/− and WT mice. Glucose disposal rates were not significantly different between the two groups. However, the endogenous hepatic glucose output was significantly lower in LRP6+/− versus WT mice (Fig. 6B). This finding suggested that the enhanced insulin sensitivity of the LRP6+/− mice is linked to their lower body weight. In contrast, lower endogenous glucose output in LRP6+/− mice was independent of the body fat mass and the likely cause of lower base-line serum glucose compared with WT littermates.
FIGURE 6.
Hepatic glucose output in LRP6+/− mice compared with wild-type littermates (A–D). A, base-line serum glucose in LRP6+/− compared with WT mice (mean ± S. E.; *, p < 0.05). B, base-line hepatic glucose output during hyperinsulinemic-euglycemic clamp studies in LRP6+/− compared with WT mice (mean ± S.E.; *, p < 0.05). C, expression of the gluconeogenic enzymes glucose-6-phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK) and phosphorylation of FoxO1 in LRP6+/− mice compared with WT mice (n = 8; mean ± S.E. by densitometry; p < 0.05). D, hepatic nuclear localization of β-catenin and FoxO1 in LRP6+/− mice compared with the wild-type littermates (n = 4; data are mean ± S.E. by densitometry; p < 0.05).
Reduced endogenous hepatic glucose output in LRP6+/− mice correlated with reduced expression levels of the hepatic rate-limiting gluconeogenic enzymes glucose-6-phosphatase and phosphoenolpyruvate carboxykinase (Fig. 6C). Hepatic FoxO1, an upstream regulator of the gluconeogenic enzymes, was more phosphorylated and hence less active in LRP6+/− mice compared with the wild-type littermates (Fig. 6C). β-Catenin, a downstream transcription coactivator in the Wnt/LRP6 pathway, activates FoxO1 and promotes its nuclear translocation and transcriptional activity in vitro (14, 33). The nuclear localization of the hepatic β-catenin was significantly reduced in LRP6+/− versus WT mice, indicating impairment of the canonical Wnt signaling (Fig. 6D). This was associated with reduced activation and nuclear localization of FoxO1 in liver cells from LRP6+/− mice compared with their wild-type littermates. These findings suggested that the impaired LRP6-dependent β-catenin activity in the liver is accountable for decreased hepatic gluconeogenesis and lower base-line glucose levels in LRP6+/− mice.
DISCUSSION
Obesity is a major component of metabolic syndrome and an established risk factor for diabetes (34). The susceptibly to obesity is inherited; however, its underlying genetic causes have remained vastly unknown. Canonical Wnt signaling has been implicated in regulation of glucose homeostasis and adipogenesis (17). In this study, we demonstrate the crucial role of the Wnt coreceptor LRP6 in the regulation of body fat mass and glucose homeostasis in tight association with the mTOR nutrient sensing signaling network.
LRP6+/− mice on high fat diet are protected against diet-induced obesity and insulin resistance. These phenotypes are linked to inactivation of the BAT mTOR pathway and enhanced expression of UCP1 and PGC1-α. Mice deficient for the mTORC1 component raptor are resistant to diet-induced obesity and have enhanced glucose tolerance. Adipose tissue in raptor-deficient mice displays enhanced expression of UCP1 and PGC1-α (31). Mice deficient for mTOR substrate S6K1 are similarly protected against obesity, which is also associated with increased expression of mitochondrial genes UCP1 and PGC1-α. S6K−/− mice are sensitive to insulin due to increased IRS1 expression and activity (32). 4ebp1−/− mice manifest markedly smaller white adipose tissue, higher energy expenditure, and increased expression of white adipose tissue PGC1-α and UCP1 (35). These in vivo models suggest that the mTORC1 signaling pathway negatively regulates PGC1-α and UCP1.
UCP1 is a mitochondrial carrier that causes dissipation of the mitochondrial gradient and thus uncouples oxidative phosphorylation from ATP synthesis, leading to increased thermogenesis (21). As a consequence, ATP synthesis requires increased metabolism of fuel substrates. Mice overexpressing UCP1 in the adipose tissue (aP2-Ucp1) are protected against diet-induced obesity (36). Conversely, Ucp1−/− mice exhibit increased sensitivity to cold, diet-induced obesity, fatty liver, and impaired insulin sensitivity with aging (37). PGC1-α is a major regulator of brown adipogenesis, mitochondrial biogenesis, and metabolism. Several studies have indicated the relevance of PGC1-α and UCP1 to human diseases. Both PGC1-α and UCP1 have been linked to basal metabolic rate and heat dissipation in humans (20). A common loss of function mutation of the PGC1-α gene has been associated with an increased risk of type 2 diabetes (38). Lower expression levels of PGC1-α in the skeletal muscle are associated with an increased risk for diabetes in the general population by mechanisms that are not understood (39).
Brown adipogenesis and its key proteins PGC1-α and UCP1 are also negatively regulated by Wnt (40–42). UCP1-Wnt10b transgenic mice, which are deficient for PGC1-α and UCP1 in the interscapular tissue, convert brown to white adipocytes (17). Strikingly, pharmacological inhibition of Wnt signaling with harmine delays diabetes in ob/ob mice by enhancing mitochondrial biogenesis, augmenting white adipose tissue PGC1 and UCP1 expression and enhancing energy expenditure (43). Interestingly, Wnt activates mTORC1 signaling by inhibiting GSK-3β (30). Association among Wnt/LRP6, mTOR activity, and UCP1 and PGC1-α expression is best demonstrated in LRP6+/− mice in which loss of LRP6 allele results in reduced mTOR activity, excessive expression of UCP1 and PGC1-α, reduced adiposity, and insulin sensitivity. Our in vitro experiments using Wnt3a and LiCl suggest that Wnt activation of mTOR, inhibition of adipogenic differentiation, and negative regulation of UCP1/PGC1-α are all entirely LRP6-dependent. The LRP6+/− mouse model provides a cascade of molecular interactions that links a signal (Wnt) to its receptor (LRP6), the pathway with which it interacts (mTOR), and their final targets (UCP1/PGC1). These molecular interactions provide an intriguing explanation for a lean body and insulin sensitivity in the LRP6+/− mouse and mice with diverse defects in the mTOR pathway. Furthermore, reduced mTOR activity of the BAT in the LRP6+/− mouse explains its insulin sensitivity compared with wild-type littermates.
Enhanced hepatic insulin action in LRP6+/− mice was also associated with increased transcription of the hepatic leptin receptor. Our studies in HepG2 cells suggested that LRP6 mediates Wnt regulation of the hepatic leptin receptor via the canonical pathway. Wnt inhibition of the hepatic lepr may occur by its methylation or more likely at the transcriptional level through microRNAs or transcription suppressors. A detailed understanding of the molecular mechanism of this action is quite important and should be the focus of a future investigation.
Leptin regulates hepatic triglyceride metabolism (44); diminishes the synthesis of the key enzyme of gluconeogenesis, phosphoenolpyruvate carboxykinase; and reduces hepatic glucose production (45). Overexpression of the hepatic lepr in rodents results in enhanced insulin sensitivity (29, 45). In diet-induced obese rats, hepatic lepr expression is significantly reduced compared with wild-type littermates. Impaired lepr-dependent intracellular signaling in these rats is associated with increased levels of the gluconeogenic enzymes (46–49). Hepatic lepr expression also inversely correlates with obesity in humans (46–49). Furthermore, lepr expression levels are lower in liver and skeletal muscles of patients with type 2 diabetes compared with the general population (47). Taken together, our findings in the LRP6+/− mouse identify LRP6 as a molecular link between body fat and hepatic insulin sensitivity.
Another critical finding of our study is the role of LRP6 in regulating glucose homeostasis of the liver. LRP6+/− mice had significantly lower levels of the hepatic rate-limiting gluconeogenic enzymes phosphoenolpyruvate carboxykinase and glucose-6-phosphatase and had lesser base-line glucose output compared with WT mice prior to differential weight changes. This was due to reduced nuclear localization of β-catenin and greater inactivation of FoxO1. These results confirm in vitro findings that β-catenin interacts with FoxO1 and enhances its nuclear localization and transcriptional activity (14, 33). Our findings also expand upon the results of a recent study in which targeted hepatic knock-out of β-catenin reduced FoxO1 nuclear localization and glucose-6-phosphatase activity (14, 33). Thus, the Wnt/LRP6 signaling pathway has an important emerging role in the regulation of hepatic gluconeogenesis.
Taken together, our findings suggest that the impairment of Wnt signaling in LRP6+/− mice results in reduced body fat mass, diminished hepatic gluconeogenesis, and enhanced BAT and hepatic insulin sensitivity. These developments culminate in enhanced total body insulin sensitivity. Results of our study raise the broader question of whether inherited or acquired gain of function in the Wnt signaling pathway is a feature that links obesity and type 2 diabetes in the general population. In summary, our study provides insight into pathways that underlie the association of body weight and hepatic insulin response and identifies LRP6 as its molecular link and a potential target for novel pharmacotherapy of diabetes and obesity.
Supplementary Material
Acknowledgments
We thank the Yale Liver Center for help in histological preparation and microscopy of mice liver. We thank Dr. Lawrence L. Young and Michael Simons for valuable input and thorough review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL094784-01 and R01HL094574-03 (to A. M.).

This article contains supplemental Figs. S1–S5 and Tables S1 and S2.
- LRP
- LDL receptor-related protein
- BAT
- brown adipose tissue
- hfd
- high fat diet
- IPGTT
- intraperitoneal glucose tolerance test
- lepr
- leptin receptor
- mTOR
- mammalian target of rapamycin
- IRS
- insulin receptor substrate
- S6K
- S6 kinase.
REFERENCES
- 1. Mani A., Radhakrishnan J., Wang H., Mani A., Mani M. A., Nelson-Williams C., Carew K. S., Mane S., Najmabadi H., Wu D., Lifton R. P. (2007) LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science 315, 1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fujino T., Asaba H., Kang M. J., Ikeda Y., Sone H., Takada S., Kim D. H., Ioka R. X., Ono M., Tomoyori H., Okubo M., Murase T., Kamataki A., Yamamoto J., Magoori K., Takahashi S., Miyamoto Y., Oishi H., Nose M., Okazaki M., Usui S., Imaizumi K., Yanagisawa M., Sakai J., Yamamoto T. T. (2003) Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc. Natl. Acad. Sci. U.S.A. 100, 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang J., Kuusisto J., Vänttinen M., Kuulasmaa T., Lindström J., Tuomilehto J., Uusitupa M., Laakso M. (2007) Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia 50, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 4. Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Balkau B., Heude B., Charpentier G., Hudson T. J., Montpetit A., Pshezhetsky A. V., Prentki M., Posner B. I., Balding D. J., Meyre D., Polychronakos C., Froguel P. (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885 [DOI] [PubMed] [Google Scholar]
- 5. Scott L. J., Mohlke K. L., Bonnycastle L. L., Willer C. J., Li Y., Duren W. L., Erdos M. R., Stringham H. M., Chines P. S., Jackson A. U., Prokunina-Olsson L., Ding C. J., Swift A. J., Narisu N., Hu T., Pruim R., Xiao R., Li X. Y., Conneely K. N., Riebow N. L., Sprau A. G., Tong M., White P. P., Hetrick K. N., Barnhart M. W., Bark C. W., Goldstein J. L., Watkins L., Xiang F., Saramies J., Buchanan T. A., Watanabe R. M., Valle T. T., Kinnunen L., Abecasis G. R., Pugh E. W., Doheny K. F., Bergman R. N., Tuomilehto J., Collins F. S., Boehnke M. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lyssenko V., Lupi R., Marchetti P., Del Guerra S., Orho-Melander M., Almgren P., Sjögren M., Ling C., Eriksson K. F., Lethagen A. L., Mancarella R., Berglund G., Tuomi T., Nilsson P., Del Prato S., Groop L. (2007) Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Investig. 117, 2155–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Z., Habener J. (2010) Wnt signaling in pancreatic islets. Adv. Exp. Med. Biol. 654, 391–419 [DOI] [PubMed] [Google Scholar]
- 8. Bilic J., Huang Y. L., Davidson G., Zimmermann T., Cruciat C. M., Bienz M., Niehrs C. (2007) Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316, 1619–1622 [DOI] [PubMed] [Google Scholar]
- 9. Schweizer L., Varmus H. (2003) Wnt/Wingless signaling through β-catenin requires the function of both LRP/Arrow and frizzled classes of receptors. BMC Cell Biol. 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tamai K., Semenov M., Kato Y., Spokony R., Liu C., Katsuyama Y., Hess F., Saint-Jeannet J. P., He X. (2000) LDL-receptor-related proteins in Wnt signal transduction. Nature 407, 530–535 [DOI] [PubMed] [Google Scholar]
- 11. Zeng X., Huang H., Tamai K., Zhang X., Harada Y., Yokota C., Almeida K., Wang J., Doble B., Woodgett J., Wynshaw-Boris A., Hsieh J. C., He X. (2008) Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development 135, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu G., Huang H., Garcia Abreu J., He X. (2009) Inhibition of GSK3 phosphorylation of β-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One 4, e4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cselenyi C. S., Jernigan K. K., Tahinci E., Thorne C. A., Lee L. A., Lee E. (2008) LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3's phosphorylation of β-catenin. Proc. Natl. Acad. Sci. U.S.A. 105, 8032–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H., Fergusson M. M., Wu J. J., Rovira I. I., Liu J., Gavrilova O., Lu T., Bao J., Han D., Sack M. N., Finkel T. (2011) Wnt signaling regulates hepatic metabolism. Sci. Signal. 4, ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saxena R., Gianniny L., Burtt N. P., Lyssenko V., Giuducci C., Sjögren M., Florez J. C., Almgren P., Isomaa B., Orho-Melander M., Lindblad U., Daly M. J., Tuomi T., Hirschhorn J. N., Ardlie K. G., Groop L. C., Altshuler D. (2006) Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes 55, 2890–2895 [DOI] [PubMed] [Google Scholar]
- 16. Yoon J. C., Ng A., Kim B. H., Bianco A., Xavier R. J., Elledge S. J. (2010) Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 24, 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kang S., Bajnok L., Longo K. A., Petersen R. K., Hansen J. B., Kristiansen K., MacDougald O. A. (2005) Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1α. Mol. Cell. Biol. 25, 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 19. Jitrapakdee S., Wutthisathapornchai A., Wallace J. C., MacDonald M. J. (2010) Regulation of insulin secretion: role of mitochondrial signalling. Diabetologia 53, 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crowley V., Vidal-Puig A. (2001) Mitochondrial uncoupling proteins (UCPs) and obesity. Nutr. Metab. Cardiovasc. Dis. 11, 70–75 [PubMed] [Google Scholar]
- 21. Barbera M. J., Schluter A., Pedraza N., Iglesias R., Villarroya F., Giralt M. (2001) Peroxisome proliferator-activated receptor α activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 276, 1486–1493 [DOI] [PubMed] [Google Scholar]
- 22. Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., Guan K. L. (2006) TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968 [DOI] [PubMed] [Google Scholar]
- 23. Rodríguez de la Concepción M. L., Yubero P., Iglesias R., Giralt M., Villarroya F. (2005) Lithium inhibits brown adipocyte differentiation. FEBS Lett. 579, 1670–1674 [DOI] [PubMed] [Google Scholar]
- 24. Student A. K., Hsu R. Y., Lane M. D. (1980) Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J. Biol. Chem. 255, 4745–4750 [PubMed] [Google Scholar]
- 25. Zhang L., He H., Balschi J. A. (2007) Metformin and phenformin activate AMP-activated protein kinase in the heart by increasing cytosolic AMP concentration. Am. J. Physiol. Heart Circ. Physiol. 293, H457–H466 [DOI] [PubMed] [Google Scholar]
- 26. Pinson K. I., Brennan J., Monkley S., Avery B. J., Skarnes W. C. (2000) An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407, 535–538 [DOI] [PubMed] [Google Scholar]
- 27. Mancuso P., Peters-Golden M., Goel D., Goldberg J., Brock T. G., Greenwald-Yarnell M., Myers M. G. (2011) Disruption of leptin receptor-STAT3 signaling enhances leukotriene production and pulmonary host defense against pneumococcal pneumonia. J. Immunol. 186, 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uchiyama T., Takahashi H., Sugiyama M., Sakai E., Endo H., Hosono K., Yoneda K., Yoneda M., Inamori M., Nagashima Y., Inayama Y., Wada K., Nakajima A. (2011) Leptin receptor is involved in STAT3 activation in human colorectal adenoma. Cancer Sci. 102, 367–372 [DOI] [PubMed] [Google Scholar]
- 29. Uotani S., Abe T., Yamaguchi Y. (2006) Leptin activates AMP-activated protein kinase in hepatic cells via a JAK2-dependent pathway. Biochem. Biophys. Res. Commun. 351, 171–175 [DOI] [PubMed] [Google Scholar]
- 30. Tremblay F., Marette A. (2001) Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J. Biol. Chem. 276, 38052–38060 [DOI] [PubMed] [Google Scholar]
- 31. Polak P., Cybulski N., Feige J. N., Auwerx J., Rüegg M. A., Hall M. N. (2008) Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 8, 399–410 [DOI] [PubMed] [Google Scholar]
- 32. Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 33. Essers M. A., de Vries-Smits L. M., Barker N., Polderman P. E., Burgering B. M., Korswagen H. C. (2005) Functional interaction between β-catenin and FOXO in oxidative stress signaling. Science 308, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 34. Vaidya D., Mathias R. A., Kral B. G., Yanek L. R., Becker L. C., Becker D. M. (2010) Independent metabolic syndrome variants predict new-onset coronary artery disease. Diabetes Care 33, 1376–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsukiyama-Kohara K., Poulin F., Kohara M., DeMaria C. T., Cheng A., Wu Z., Gingras A. C., Katsume A., Elchebly M., Spiegelman B. M., Harper M. E., Tremblay M. L., Sonenberg N. (2001) Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7, 1128–1132 [DOI] [PubMed] [Google Scholar]
- 36. Rossmeisl M., Flachs P., Brauner P., Sponarova J., Matejkova O., Prazak T., Ruzickova J., Bardova K., Kuda O., Kopecky J. (2004) Role of energy charge and AMP-activated protein kinase in adipocytes in the control of body fat stores. Int. J. Obes. Relat. Metab. Disord. 28, Suppl. 4, S38–S44 [DOI] [PubMed] [Google Scholar]
- 37. Kontani Y., Wang Y., Kimura K., Inokuma K. I., Saito M., Suzuki-Miura T., Wang Z., Sato Y., Mori N., Yamashita H. (2005) UCP1 deficiency increases susceptibility to diet-induced obesity with age. Aging Cell 4, 147–155 [DOI] [PubMed] [Google Scholar]
- 38. Hara K., Tobe K., Okada T., Kadowaki H., Akanuma Y., Ito C., Kimura S., Kadowaki T. (2002) A genetic variation in the PGC-1 gene could confer insulin resistance and susceptibility to Type II diabetes. Diabetologia 45, 740–743 [DOI] [PubMed] [Google Scholar]
- 39. Patti M. E., Butte A. J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., Landaker E. J., Goldfine A. B., Mun E., DeFronzo R., Finlayson J., Kahn C. R., Mandarino L. J. (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U.S.A. 100, 8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bowers R. R., Lane M. D. (2008) Wnt signaling and adipocyte lineage commitment. Cell Cycle 7, 1191–1196 [DOI] [PubMed] [Google Scholar]
- 41. Takada I., Kouzmenko A. P., Kato S. (2009) Wnt and PPARγ signaling in osteoblastogenesis and adipogenesis. Nat. Rev. Rheumatol. 5, 442–447 [DOI] [PubMed] [Google Scholar]
- 42. Takada I., Kato S. (2008) Molecular mechanism of switching adipocyte/osteoblast differentiation through regulation of PPAR-γ function. Clin. Calcium 18, 656–661 [PubMed] [Google Scholar]
- 43. Waki H., Park K. W., Mitro N., Pei L., Damoiseaux R., Wilpitz D. C., Reue K., Saez E., Tontonoz P. (2007) The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARγ expression. Cell Metab. 5, 357–370 [DOI] [PubMed] [Google Scholar]
- 44. Huang W., Dedousis N., Bandi A., Lopaschuk G. D., O'Doherty R. M. (2006) Liver triglyceride secretion and lipid oxidative metabolism are rapidly altered by leptin in vivo. Endocrinology 147, 1480–1487 [DOI] [PubMed] [Google Scholar]
- 45. Anderwald C., Müller G., Koca G., Fürnsinn C., Waldhäusl W., Roden M. (2002) Short-term leptin-dependent inhibition of hepatic gluconeogenesis is mediated by insulin receptor substrate-2. Mol. Endocrinol. 16, 1612–1628 [DOI] [PubMed] [Google Scholar]
- 46. Brabant G., Müller G., Horn R., Anderwald C., Roden M., Nave H. (2005) Hepatic leptin signaling in obesity. FASEB J. 19, 1048–1050 [DOI] [PubMed] [Google Scholar]
- 47. Koch A., Weiskirchen R., Zimmermann H. W., Sanson E., Trautwein C., Tacke F. (2010) Relevance of serum leptin and leptin-receptor concentrations in critically ill patients. Mediators Inflamm. 2010, 473540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Le D., Marks D., Lyle E., Corless C. L., Diggs B. S., Jobe B. A., Kay T., Deveney C. W., Wolfe B. M., Roberts C. T., Jr., O'Rourke R. W. (2007) Serum leptin levels, hepatic leptin receptor transcription, and clinical predictors of non-alcoholic steatohepatitis in obese bariatric surgery patients. Surg. Endosc. 21, 1593–1599 [DOI] [PubMed] [Google Scholar]
- 49. Liu Z. J., Bian J., Liu J., Endoh A. (2007) Obesity reduced the gene expressions of leptin receptors in hypothalamus and liver. Horm. Metab. Res. 39, 489–494 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.