Background: Ppz1 orthologs, novel family of phosphatases, have a unique N-terminal non-catalytic domain.
Results: Ppz1 ortholog from halotolerant yeast Debaryomyces hansenii plays important role in salt tolerance, cell wall integrity, and cell growth through distinct mechanism.
Conclusion: Short serine arginine-rich motif in non-catalytic domain is essential for its role in salt tolerance.
Significance: This motif is conserved among orthologs and functionally important.
Keywords: Fungi, Molecular Biology, Mutagenesis Site-specific, Phosphatase, Yeast Physiology, Debaryomyces hansenii, Ppz1, Salt Tolerance, Ser/Arg-rich Motif
Abstract
PPZ1 orthologs, novel members of a phosphoprotein phosphatase family of phosphatases, are found only in fungi. They regulate diverse physiological processes in fungi e.g. ion homeostasis, cell size, cell integrity, etc. Although they are an important determinant of salt tolerance in fungi, their physiological role remained unexplored in any halotolerant species. In this context we report here molecular and functional characterization of DhPPZ1 from Debaryomyces hansenii, which is one of the most halotolerant and osmotolerant species of yeast. Our results showed that DhPPZ1 knock-out strain displayed higher tolerance to toxic cations, and unlike in Saccharomyces cerevisiae, Na+/H+ antiporter appeared to have an important role in this process. Besides salt tolerance, DhPPZ1 also had role in cell wall integrity and growth in D. hansenii. We have also identified a short, serine-arginine-rich sequence motif in DhPpz1p that is essential for its role in salt tolerance but not in other physiological processes. Taken together, these results underscore a distinct role of DhPpz1p in D. hansenii and illustrate an example of how organisms utilize the same molecular tool box differently to garner adaptive fitness for their respective ecological niches.
Introduction
Phosphoprotein phosphatases (PPP)4 constitute an important family of phosphatases that regulate a plethora of cellular processes in eukaryotes (1–4). Based on their primary sequences, they are further classified into a few distinct groups such as protein phosphatase 1 (PP1), PP2A, PP2B (commonly known as calcineurin), PP4, PP5, PP6, and PP7. Within the conserved catalytic domain (∼30 kDa), members of the PPP family contain three characteristics sequence motifs (GDXHG, GDXVDRG, and GNHE) (2, 4). Besides these, some groups also exhibit conserved sequence motifs outside the catalytic domain, e.g. Ca2+-calmodulin binding motif in calcineurin, TPR motif in PP5, etc., that are very important for the functionality of the respective members (2, 4, 5). Compared with the number of serine/threonine kinases, eukaryotic genomes contain fewer phosphatases that control specific dephosphorylation of thousands of phospho-protein substrates. Based on the evidences gathered from few well studied members, it appeared that the combinatorial associations with multiple regulatory subunits and other interacting proteins conferred potentially distinct substrate specificity to the catalytic subunits (4, 6, 7). Thus, the details of these interactions are of paramount importance in deciphering the biological roles of these phosphatases. PPP enzymes are among the ancient phosphatases, with at least one PPP-like member in all prokaryotes and eukaryotes. Fungi, in general, contain several PPP family of phosphatase. Although the corresponding orthologs for most of them could be found in higher eukaryotes, a few of them are quite unique and restricted to this kingdom of life only (8).
Ppz1p and Ppz2p are such type of novel members of PPP family of phosphatase, originally identified in Saccharomyces cerevisiae (9–11). Within the catalytic domain, Ppz1p shared ∼60% sequence identity to PP1 and ∼40% identity to PP2A, making them distinct from those of PP1. PPZ1 orthologs appeared to regulate diverse physiological processes in fungi, e.g. ion homeostasis, cell growth, cell size, and integrity (12). In S. cerevisiae, deletion of PPZ1 leads to hypertolerance phenotype toward sodium or lithium cations, which is more intensified by additional deletion of PPZ2. Deletion of PPZ2 alone has no effect on salt tolerance (12). Recent evidences indicated that Ppz1p affects cellular ion homeostasis primarily through the negative regulation of Trk1p, the major K+ transport system, and this has substantial effect on salt tolerance and intracellular pH (13, 14). Ppz1p had also been found to regulate the expression of Na+ efflux pump ENA1 (15). Functional overlap of Ppz1p with other members of PPP family has also been documented in S. cerevisiae. Sit4p, a PP2A phosphatase, and PPz1p play opposite roles in regulating G1/S transition (16). Complex genetic interaction of Ppz1p with Glc7p (an essential PP1 phosphatase) as well as with the calcineurin/Crz1 pathway have also been observed (17–19). Hal3p is one of the most important regulators of Ppz1p. Overexpression of Hal3p increases salt tolerance by suppressing the activity of Ppz1p (20). Hal3p has been shown to act as a negative regulator by virtue of it binding to the catalytic domain of Ppz1p. Interactions between Hal3p and Ppz1p are destabilized upon an increase in the intracellular pH. It has been suggested that the Hal3p-Ppz1p complex acts as a pH sensor within the cell (14). Besides Hal3p, physical interaction of Ppz1p with quite a few other proteins has also been demonstrated in S. cerevisiae (14, 21, 22). Some of them e.g. Trk1p, appeared to be its substrate, although others, e.g. Ypi1p, regulate phosphatase activity. However, the details of the interactions of Ppz1p with these proteins are not known. Ppz1p contains a large N-terminal non-catalytic extension that is a characteristic feature of this group of phosphatases. This region is rich in few amino acids e.g. serine and arginine residues, and has been shown to be functionally important (9, 12, 23).
PPZ1 is an important determinant of salt tolerance in yeast. Most of our understanding about this molecule is based on studying orthologs from very few species (24–26). Compared with these species, the yeast D. hansenii displays extreme halotolerance. In the recent past D. hansenii has been considered as a model organism to understand halotolerance in yeast (27–29). In this study we have identified and cloned a PPZ1 ortholog (DhPPZ1) from D. hansenii. Functional characterization revealed that DhPPZ1 played an important role in salt tolerance and cell wall integrity as well as in growth of D. hansenii through distinct mechanism. Most importantly, we have identified for the first time a 10-amino acid-long serine/arginine-rich motif in the N-terminal non-catalytic domain of DhPpz1p that was essential for its role in salt tolerance but not in other physiological processes. This motif was not only conserved among orthologs but also functionally important as revealed from mutational analysis of S. cerevisiae Ppz1p.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
Yeast strains and plasmids used in this study are listed in Table 1. D. hansenii strains were grown in YPD or SD minimal media at 28 °C. For dilution spotting, freshly grown cultures (12 h at 28 °C) were normalized to A600 1.0. Serial dilutions were made in sterile water, and 5 μl of each of the 10−1–10−4 dilutions were spotted onto YPD or SD plates with required supplements.
TABLE 1.
Strains and plasmids used in this study
| Strain/Plasmid | Descriptions | Source/Reference |
|---|---|---|
| Strain | ||
| D. hansenii | ||
| DBH 9 | dhhis4 | Ref. 30 |
| DBH93 | dhhis4 dharg1 | This study |
| DBH91 | dhhis4 dhppz1::DhHIS4 | This study |
| DBH936 | dhhis4 dharg1 dhppz1::DhHIS4 | This study |
| DBH932 | dhhis4 dharg1 dhmpk1::DhHIS4 | This study |
| S. cerevisiae | ||
| BY4742 | MAT α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 | Euroscarf |
| Y10557 | MAT α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; ppz1::kanMX4 | Euroscarf |
| EGY 48 | MATα; trp1; his3; ura3; 6ops-LEU2 | Ref. 33 |
| Plasmids | ||
| pDH4 | DhHIS4 CfARS16 | Ref. 30 |
| pDH11 | DhHIS4 CfARS16 RFP (under DhTEF promoter) | Ref. 30 |
| pAN4 | DhHIS4 CfARS16 DhPPZ1-RFP | This study |
| pAN5 | DhARG1 CfARS16 DhPPZ1 | This study |
| pAN6 | DhARG1 CfARS16 DhPPZ1 (under DhTEF promoter) | This study |
| pAN51 | DhARG1 CfARS16 DhPPZ1 (Δ27–36) | This study |
| pAN52 | DhARG1 CfARS16 DhPPZ1 (Δ59–75) | This study |
| pAN53 | DhARG1 CfARS16 DhPPZ1 (Δ81–104) | This study |
| pAN54 | DhARG1 CfARS16 DhPPZ1 (Δ106–120) | This study |
| pAN55 | DhARG1 CfARS16 DhPPZ1 (Δ162–183) | This study |
| pAN56 | DhARG1 CfARS16 DhPPZ1 (Δ241–254) | This study |
| pAN9 | DhARG1 CfARS16 DhPPZ1 (Δ279–572) | This study |
| pAN10 | DhARG1 CfARS16 DhPPZ1 (Δ1–278) | This study |
| pDA1 | DhARG1 CfARS16 | This study |
| pRS-PPZ1 | HIS3 2μ PPZ1 | This study |
| PPZ1 Δ43–52 | HIS3 2μ PPZ1(Δ43–52) | This study |
| pEG202- DhPPZ1 | HIS3 2μ DhPPZ1 | This study |
| pEG202- DhPPZ1Δ27–36 | HIS3 2μ DhPPZ1(Δ27–36) | This study |
| pJG4-5-DhHAL3 | TRP1 2μ DhHAL3 | This study |
| pEG202 (bait vector) | HIS3 2μ | Ref. 33 |
| pJG4-5 (prey vector) | TRP1 2μ | Ref. 33 |
Construction of Recombinant Plasmids Carrying DhPPZ1 and Its Mutants
Plasmids pAN5 was constructed to express DhPpz1p from its native promoter in D. hansenii. For this, fragments of sizes ∼0.8 kb (corresponding to the upstream of the ATG codon) and 1.7 kb (corresponding to DhPPZ1 ORF) were amplified from genomic DNA of CBS767 using primer pairs DhPPZ1_F/DhPPZ1_int R1 and DhPPZ1_int F1/DhPPZ1_R. The amplified fragments were digested with BamH1/Nco1 and Nco1/Sac1, respectively, and cloned at the BamH1-Sac1 site of plasmid pAG-CfARS to obtain pPA1. Subsequently, a ∼1.9-kb fragment corresponding to DhARG1 gene was cloned at Cla1, digested, and Klenow-treated pPA1 to obtain pAN5. To construct pAN6, a 610-bp fragment corresponding to DhTEF promoter was PCR-amplified from genomic DNA using the forward DhTEF_F and DhTEF_R primers. This fragment was digested with BamH1 and Nco1 and cloned at BamH1-Nco1 site in plasmid pAN5 replacing DhPPZ1 promoter.
pAN4 plasmids was constructed to express DhPpz1p-RFP fusion protein in D. hansenii. A 2.5-kb fragment (corresponding to DhPPZ1 ORF along with its native promoter) was amplified using the forward DhPPZ1_Full F and reverse DhPPZ1_ORF RFPR primers and cloned at Sal1/Xho1 sites in plasmid pDH11 (30) to obtain pAN4.
For constructing different N-terminal mutants (pAN51, pAN52, pAN53, pAN54, pAN55, and pAN56) the PCR-based overlap extension method was followed (31). Two separate PCR reactions were performed by utilizing the primer pairs, forward DhPPZ1_int F1 with corresponding reverse mutagenic primer and the corresponding forward mutagenic primer with reverse DhPPZ1_int R2 primers to obtain overlapping mutated products. These products were combined in the third PCR reaction using primers DhPPZ1_int F1 and DhPPZ1_int R2. The amplified fragment (∼950 bp) was digested with Nco1 and BsrG1 and cloned into Nco1- and BsrG1-digested pAN5. Similarly pAN52, pAN53, pAN54, pAN55, and pAN56 were made by using the primers listed in supplemental Table 1.
The constructs 4SA, 3RA, 3RE, SRAA, SRAE, S27A, S30A, S33A, S36A, R29A, R32A, and R34A were made by introducing point mutations at different serine and arginine residues in the conserved Ser/Arg rich motif. For this, ∼305-bp fragments carrying different point mutations were made by PCR-based overlap extension method and used to replace corresponding NcoI-HindIII fragment in wild type DhPPZ1. Plasmid pRS-PPZ1 was constructed by cloning a 2.7-kb PCR-amplified fragment consisting of the ORF of PPZ1 gene of S. cerevisiae along with upstream promoter region (620 bp) into vector pRS423 at Sph1-Kpn1 site. Subsequently, PPZ1 Δ43–52 was constructed by overlapping extension PCR. All the mutations were subsequently confirmed by DNA sequencing.
Construction of DhPPZ1 and DhMPK1 Knock-out Strains
For constructing dhppz1 mutant strains, a 2.8-kb fragment harboring DhPPZ1 was amplified by PCR and cloned into plasmid pGEM7Z (Promega) at BamH1-Xho1 site to obtain pAN2. The disruption plasmid pDhPPZ1-HIS4 was constructed by inserting a 3.2-kb DhHIS4 gene at the EcoRV site in pAN2. Subsequently a ∼6.0-kb BamHI/XhoI fragment from pDhPPZ1-HIS4 containing the DhHIS4 gene flanked by 1946 and 923 bp of the DhPPZ1 gene sequence on either side was transformed into DBH9 and DBH93 to obtain DBH91 and DBH936. The disruption of DhPPZ1 locus in DBH91 and DBH936 was confirmed by PCR.
To make deletion in DhMPK1, a 2.2-kb DNA fragment-harboring DhMPK1 gene was PCR-amplified using the forward primer DhMPK1_F and reverse primer DhMPK1_R using genomic DNA from D. hansenii strain CBS767 as template. The PCR fragment was cloned at BamHI/XhoI sites in pGEM7Z vector to obtain plasmid pAN7. Subsequently, the disruption cassette was constructed by replacing a 351-bp EcoR1/EcoRV fragment of DhMPK1 ORF with 3.2-kb DhHIS4 gene. The resultant plasmid (pAN7-HIS4) was then digested with BamHI/XhoI to isolate a ∼5.2-kb disruption cassette. This cassette was transformed into DBH93, and the transformants obtained on SD plates supplemented with arginine were analyzed by PCR to confirm deletion in DhMPK1. One of the transformants DBH932 was used subsequently.
Determination of Intracellular Li+ and K+
To determine the intracellular concentration of Li+ and K+ in D. hansenii, the method described by Posas et al. (12) was essentially followed. Briefly, the cells from 100-ml culture at early logarithmic phase (A600 ∼ 1.0) were harvested by centrifugation, quickly washed twice with chilled MilliQ water, and finally resuspended in 25 ml of incubation buffer (10 mm Tris-HCl, 50 mm MgCl2) supplemented with 200 mm LiCl salt. Cells (4 ml) were collected at different time intervals (0, 60, 90, and 120 min) on 0.45-μm membrane filters. The filters were washed twice with chilled buffer (20 mm MgCl2, 1 m sorbitol). Cation was extracted from the filters by incubating them in 10 ml of extraction buffer (10 mm MgCl2, 0.2 m HCl) at room temperature for 20 h. Concentration of cation in the filtered extracts was determined by atomic emission spectrophotometer AA-6800 (Shimadzu). Steady state concentration of cation (Li+int and K+int) was expressed as ppm/A600 of cells.
Quantitative Real-time PCR (qPCR) Assay
The RT-qPCR was performed with the SYBR® Green one-step RT-qPCR kit (Invitrogen). Each reaction was performed with 10 μl of 2× SYBR® Green reaction mix, 200 nm gene-specific forward primer, 400 nm gene-specific reverse primer (supplemental Table 1), 100 nm fluorescein (reference dye), 0.8 units of SuperScriptTM III RT/Platinum® Taq mix, and 500–1000 ng of RNA template in a 20-μl total reaction volume. The PCR reaction was carried out in i-cycler iQTM (Bio-Rad). Reaction conditions for one-step RT-PCR include initial cDNA synthesis at 55 °C for 45 min 15 s followed by initial denaturation at 95 °C for 2 min and 40 cycles at 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 20 s. Amplification specificity was assessed using melting curve analysis. Relative expression levels were normalized using DhGPDH as an internal control. The average relative expression levels for each gene in wild type and dhppz1 mutant was calculated using the ΔΔCt method and expressed as fold difference. The RNA was made free of DNA contamination by in-column DNase I (Invitrogen) digestion. The absence of the DNA contamination in the RNA samples was confirmed by a reaction with platinum Taq DNA polymerase without reverse transcriptase.
Two-hybrid Assay
The two-hybrid assay was carried out as described earlier (32). Plasmids pEG202 and pJG4-5 were used for constructing the bait and prey, respectively (33). DhHAL3 ORF (1.77 kb) was PCR-amplified using primers DhHAL3 ORFf and DhHAL3 ORFr and cloned in pJG4-5 at EcoR1 (blunt-ended) and Xho1 site. DhPPZ1 and DhPPZ1Δ27–36 was cloned in bait vector pEG202 at BamH1-Xho1 site. S. cerevisiae strain EGY 48 was co-transformed with bait and prey constructs and selected on minimal media with 2% glucose and without tryptophan and histidine. Co-transformed cells were grown up to A600 ∼ 1.0, and 5 μl of the cultures (normalized to A600 1.0) were spotted onto minimal media with 2% galactose, 1% raffinose, and without tryptophan, histidine, and leucine. Plates were incubated for 4 days at 30 °C. For liquid growth assays, EGY48 carrying DhHAL3 as prey and DhPPZ1 or its mutant as bait were grown overnight in minimal media with 2% raffinose (without tryptophan and histidine). The cultures were re-inoculated in minimal media with 1% raffinose and 2% galactose (without tryptophan, histidine, and leucine) at an initial A600 of ∼0.10 and grown for 41 h. Experiments were carried out twice in duplicate with a pool of four independent transformants.
RESULTS
Identification and Cloning of PPZ1 Ortholog from D. hansenii
To identify the gene encoding PPZ1 ortholog in D. hansenii, a TBLASTN search (www.ncbi.nlm.nih.gov) was carried out using protein sequences of S. cerevisiae Ppz1p (692 amino acids) and Ppz2p (710 amino acids) as query against the available genome sequence data base of D. hansenii strain CBS767. A single 572-amino acid-long hypothetical protein was identified as putative PPZ1 phosphatase in D. hansenii. DhPPZ1 ORF sequence was found to be located at chromosome E, and its length was 1719 bp. Predicated amino acid sequence of DhPpz1p showed 54.8 and 54.2% identity with Ppz1p and Ppz2p, respectively. Furthermore, like a typical Ppz1p ortholog, DhPpz1p also contains a large, N-terminal non-catalytic region consisting of 254 amino acids and C-terminal 318 amino acids corresponding to the catalytic domain. C-terminal catalytic region of DhPpz1p showed 85.2 and 83.4% identity with that of Ppz1p and Ppz2p, respectively (supplemental Fig. S1). In the N-terminal half, DhPpz1p exhibited only 25.0 and 35.4% identity with that of Ppz1p and Ppz2p of S. cerevisiae, respectively. However, like Ppz1p and Ppz2p, the N-terminal non-catalytic region of DhPpz1 is also rich in serine (63 residues) and asparagine (45 residues).
Ppz1p is a membrane protein in S. cerevisiae and its membrane localization is due to the myristoylation at Gly-2 residue (14). This residue appears to be conserved in DhPpz1p. Therefore, intracellular localization of DhPpz1p was determined. For this purpose, the plasmid pAN4 was made to express DhPpz1p as the RFP fusion protein and transformed into D. hansenii strain DBH9. The transformants were grown to logarithmic phase and examined by fluorescence microscopy. In the cells expressing the DhPpz1p-RFP fusion protein, RFP fluorescence was visible throughout the cytoplasm, which indicated that unlike Ppz1p, DhPpz1p in D. hansenii was localized in the cytosol (supplemental Fig. S2).
DhPPZ1 Knock-out Strain Shows Increased Tolerance to Toxic Cations
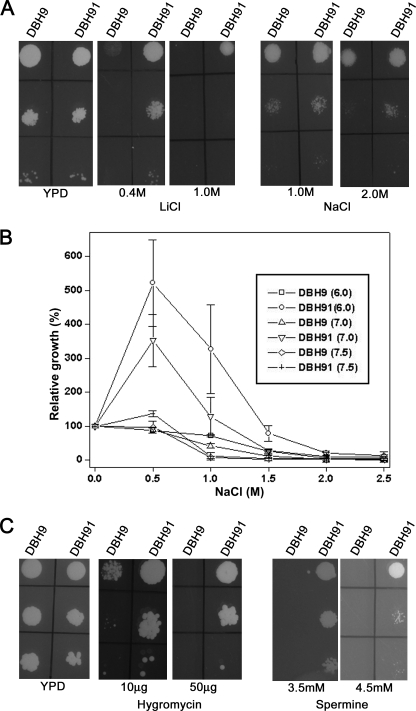
PPZ1 homolog has been shown to be an important determinant of salt tolerance in S. cerevisiae, and the deletion of this gene resulted in the increased tolerance to salt in this species (12). We have identified only one PPZ1 homolog in D. hansenii. To determine the role of this gene in salt tolerance, D. hansenii strain (DBH91)-carrying deletion in DhPPZ1 gene was created, and the growth pattern of this strain was checked on plates containing different concentrations of LiCl and NaCl. Compared with the control strain DBH9, which did not grow beyond 100 mm LiCl, DBH91 showed prominent growth even at 1 m LiCl. However, on plates containing 1 m or 2 m NaCl, no perceptible difference in growth was observed between these two strains (Fig. 1A). The effect of DhPPZ1 mutation on salt tolerance was also measured in liquid medium. For this, DBH9 and DBH91 strains were grown in YPD media or YPD media supplemented with different concentrations of salt for 24 h. Relative growth of the strain at each concentration of salt was expressed as the percentage of the growth (A600) observed in the absence of salt. It was apparent from the results that DBH91 grew better than DBH9 at all concentrations of LiCl tested. In the presence of NaCl, DBH91 exhibited 4–6 times more growth at 0.5 and 1.0 m NaCl, but at higher concentrations the difference was negligible (data not shown). Thus, the deletion of DhPPZ1 also conferred higher salt tolerance in D. hansenii. Next, we checked whether the pH of the growth medium had any effect on this increased resistance to salt stress. For this, DBH9 and DBH91 strains were re-inoculated in 50 ml of YPD medium containing the indicated concentrations of NaCl with the pH adjusted to 6.0, 7.0, and 7.5. After 24 h of incubation at 28 °C, A600 of these cultures was measured, and the data were expressed as relative growth. Growth (A600) in YPD medium without salt at the respective pH values was used as control (100%). It was observed that when pH of the medium was shifted to higher values from pH 6, the favorable effect of 0.5 m NaCl on the growth of the mutant strain started decreasing, and it almost diminished at pH 7.5 (Fig. 1B). A similar effect of pH on sodium tolerance was also observed at higher salt concentrations (Fig. 1B).
FIGURE 1.
Growth of dhppz1 mutant in the presence of toxic cations. A, 5 μl of 10-fold serial dilutions of logarithmic phase culture of D. hansenii strains DBH9 (wild type) and DBH91 (dhppz1) were spotted on YPD plates supplemented with different concentrations of LiCl and NaCl. Growth on plates was observed after 2 days of incubation at 28 °C. B, saturated cultures of DBH9 and DBH91 grown in YPD medium were re-inoculated (at initial A600 of 0.025) into 50 ml of YPD medium at different pH values containing different concentrations of NaCl. After 24 h of growth at 28 °C, the A600 of these cultures was measured. Relative growth of the culture at each concentration of salt was expressed as the percentage of the growth measured in the absence of added salt at same pH. Data presented are the results of three independent experiments (mean ± S.D.). C, shown is dilution spotting of DBH9 and DBH91 on YPD plates containing different amounts of hygromycin and spermine. Results are representative of three independent experiments.
Besides alkali metal cations, ppz1 mutant in S. cerevisiae strain can also tolerate high concentration of other toxic cations such as spermine, tetramethylammonium, and hygromycin B (13). To check whether the D. hansenii mutant also exhibits similar phenotype, growth of DBH91 and DBH9 strain on YPD plates supplemented with different concentrations of hygromycin and spermine were checked by dilution spotting. Our results showed that the DBH91 strain could grow in the presence of 50 μg of hygromycin or 4.5 mm spermine. In contrast, DBH9 strain failed to grow above 10 μg of hygromycin or 2.5 mm spermine-containing plates (Fig. 1C). The dhppz1 mutant DBH936 also showed a similar phenotype.
DhPPZ1 Knock-out Strain Accumulates Lower Amount of Li+ Intracellularly
As mentioned above, dhppz1 mutant strain exhibited higher tolerance to toxic cation. This phenotype could arise if it accumulates less amount of toxic ion in comparison to parent strain. To examine this possibility, a steady state concentration of Li+ ions inside the cells (Li+int) was determined. dhppz1 mutant (DBH936) cells at early logarithmic phase was exposed to 200 mm LiCl at indicated pH. Samples were withdrawn at different time intervals, and the total intracellular Li+ ion content of the cells was measured by atomic absorption spectrophotometer as described under “Materials and Methods.” From our results it was apparent that dhppz1 mutant strain accumulated lower steady state concentration of lithium ion compared with the parent strain both at pH 6.0 and at pH 7.0 (Fig. 2A).
FIGURE 2.
Effect of DhPPZ1 knock out on cation ion transport in D. hansenii. A and B, shown is intracellular concentration of Li+ and K+ in wild type D. hansenii (DBH93) and dhppz1 mutant (DBH936) at different pH values. D. hansenii cells at early log phase were exposed to 200 mm LiCl in incubation buffer at different pH values. Cells were collected at different time points by filtration, and the total intracellular concentration (ppm per A600 of the cells) of Li+ and K+ ion in these cells were measured by atomic absorption spectrophotometer as described under “Materials and Methods.” Data (mean ± S.D.) from duplicate biological samples with three replicates each are shown. C–E, shown is a comparison of the levels of DhENA1, DhTRK1, and DhNHA1 transcript in wild type and dhppz1 strains as analyzed by RT-qPCR. Total RNA was isolated from logarithmic phase cultures of wild type (DBH9) and dhppz1 mutant (DBH91) strain grown in YPD medium or after exposing them to 200 mm LiCl for 20 min. The RT-qPCR was performed with one step SYBR-I Green reaction kit (Invitrogen) using gene specific primers. Relative expression levels were normalized using DhGPDH as an internal control. The average relative expression levels for each gene in wild type and dhppz1 mutant was calculated using the ΔΔCt method and expressed as -fold difference (mean ± S.D.).
In S. cerevisiae, Ppz1p modulates the activity of K+ transporter Trk1p. Increase in the activity of Trk1p in the absence of Ppz1p results in the accumulation of higher K+ ion concentration inside the cell, which indirectly affects the salt tolerance (13, 34). The increased resistance to LiCl exhibited by dhppz1 mutants could also be due to a higher accumulation of K+ intracellularly. To check this, the intracellular level of K+ ion was determined in dhppz1 mutant strain (described under “Materials and Methods”). DBH936 as well as parent DBH93 cells at early logarithmic phase were exposed to 200 mm LiCl at different pH. The cells were harvested by vacuum filtration at different time points, and the intracellular K+ concentration in these cells was measured by atomic absorption spectrophotometer. Results presented in Fig. 2B showed that dhppz1 strain accumulated higher K+ compared with the parent strain both at pH 6 and pH 7.
Activity of various transporters directly regulates the intracellular concentration of different cations. The above-mentioned difference in Li+ and K+ observed in dhppz1 mutant could be a result of differences in the level of cation transporters. It is to be noted that the loss of Ppz1p activity induces ENA1 transcription in S. cerevisiae (13). Therefore, the level of expression of major cation transporters DhENA1, DhTRK1, and DhNHA1 was compared in dhppz1 mutant and parent strain using real time PCR method. Under the salt stress, dhppz1 mutant showed 10-fold higher DhNHA1 expression compared with the parent strain used as wild type control (Fig. 2E). In comparison, little or no difference in the level of expression of DhENA1 and DhTRK1 could be observed between dhppz1 mutant and parent strain under these conditions (Fig. 2, C and D).
DhPPZ1 Is Crucial for Normal Cell Growth in D. hansenii
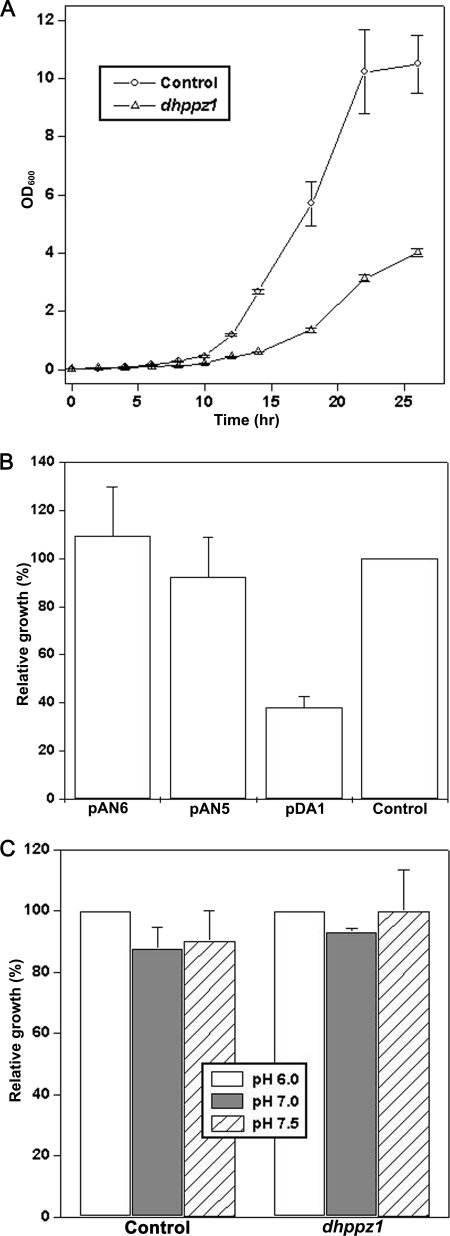
PPZ1 plays a regulatory role in the number of divergent pathways such as salt stress, cell wall remodeling, and cell cycle regulation in yeast. Disruption of DhPPZ1 showed that it was not an essential gene in D. hansenii; however, it did not rule out its vital role in normal cellular processes. To check this, growth pattern of the dhppz1 strain was compared with the parent strain. Saturated cultures of D. hansenii strains (DBH936 and DBH93) were re-inoculated into 100 ml of YPD medium at an initial A600 of 0.05 and incubated further at 28 °C with vigorous shaking (200 rpm). Growth of the cultures was monitored by measuring A600 at regular intervals. It was evident that dhppz1 mutant (DBH936) grew significantly slower than the parent strain DBH93 used as control (Fig. 3A). The growth defect exhibited by dhppz1 mutant of D. hansenii could be abrogated by expressing DhPPZ1 either under its own promoter (pAN5) or a stronger DhTEF promoter (Fig. 3B). Moreover, the growth of the strain expressing DhPPZ1 under DhTEF promoter was quite similar to DBH93, thereby clearly indicating that the overexpression of DhPPZ1 had a negligible effect on the growth of D. hansenii. Contrasting this, an excess of Ppz1p in S. cerevisiae is known to have an adverse effect on growth by affecting G1/S transition, bud emergence, and DNA synthesis (9, 11, 16). Although S. cerevisiae strains lacking both PPZ1/2 show normal growth in YPD medium, with the increase in the pH of the medium it exhibits a slow-growth phenotype (13). Therefore, the growth of the dhppz1 mutant strain was checked in YPD media at pH 6.0, 7.0, and 7.5. As shown in Fig. 3C, the growth pattern of dhppz1 mutant was quite similar at pH 6.0, 7.0, and 7.5, which indicated that the deletion of DhPPZ1 in D. hansenii had no significant effect on growth under alkaline conditions.
FIGURE 3.
DhPPZ1 knock-out affects growth of D. hansenii. A, growth curve of dhppz1 mutant (DBH936) and control parental (DBH93) D. hansenii strains are shown. Saturated cultures DBH936 and DBH93 were re-inoculated 100 ml of YPD medium at initial A600 of 0.05. Growth of these cultures was monitored by measuring A600 at regular intervals. B, shown is reversal of slow growth phenotype of dhppz1 mutant by expressing DhPPZ1 from plasmid. Saturated cultures of DBH936 strains transformed with indicated plasmids were re-inoculated into 25 ml of SD medium (at initial A600of 0.025). After 24 h of growth at 28 °C, A600 of these cultures was measured. DBH93 strain transformed with plasmids pDA1 and pDH4 was used as control. Data were expressed as their relative growth with respect to the control. C, relative growth of dhppz1 mutant at different pH values is shown. Saturated cultures of DBH936 and DBH93 (control) were re-inoculated (initial A600 0.025) into 50 ml of YPD medium buffered to different pH values. After 24 h of growth at 28 ºC, A600 of the culture was measured. The growth at pH 6.0 for the respective culture was taken 100%. Data presented are the results of three independent experiments (mean ± S.D.).
Effect of DhPPZ1 Mutation on Cell Wall Remodeling in D. hansenii
To check the involvement of DhPPZ1 in cell wall remodeling in D. hansenii, the sensitivity of dhppz1 mutant toward cell wall-destabilizing agents, such as caffeine, was determined by dilution spotting. The strain DBH93, which was used as the control, grew very well on plates containing 5 mm caffeine. In contrast, DBH936 did not grow at all on this plate. However, with the addition of 1 m sorbitol as osmotic stabilizer, the growth of DBH936 in the presence of 5 mm caffeine could be seen (supplemental Fig. S3A). Mutation in dhppz1 gene also conferred temperature-sensitive phenotype in D. hansenii, as DBH936 failed to grow on YPD plates incubated at a temperature of 32 °C and above. This defect could also be alleviated by the addition of 1 m sorbitol as the osmotic stabilizer (supplemental Fig. S3B). These results clearly indicated an important role of DhPPZ1 in cell wall integrity in D. hansenii as well.
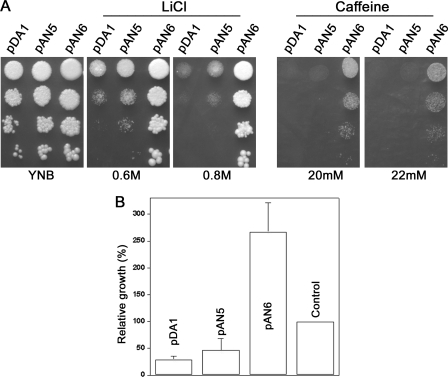
In yeast, a signal transduction pathway comprising mitogen-activated protein kinase MPK1 plays pivotal role in mediating cell wall remodeling under different physiological conditions including stress (35). Genetic interaction of this pathway with PPZ1 has been reported in S. cerevisiae earlier (11, 34). Through BLAST analysis of the genome sequence data base, we identified a putative MPK1 ortholog (DhMPK1) in D. hansenii. To determine the genetic interaction between this pathway and DhPPZ1, we constructed a DBH932 strain-carrying deletion in DhMPK1 gene. DBH932 displayed a temperature-dependent cell lysis defect that could be abrogated by the addition of 1 m sorbitol (supplemental Fig. S4). These phenotypes suggested that DhMPK1 was a typical member of the cell wall integrity MAPK pathway in D. hansenii. We next transformed DBH932 strain with pAN5 (DhPPZ1 gene under its native promoter) and pAN6 (DhPPZ1 gene under strong DhTEF promoter), and transformants were selected on SD plates. Phenotype analysis of the transformants showed that only the expression of DhPpz1p from a strong promoter but not from its own promoter could suppress caffeine and salt sensitivity of DBH932 (Fig. 4A). Similarly, the growth defect of dhmpk1 deletion could also be abrogated only by overexpressing DhPpz1p (Fig. 4B). Thus, like that in S. cerevisiae, DhPPZ1 behaved like a multicopy suppressor of the growth defect associated with the deletion of the cell wall integrity pathway component (11).
FIGURE 4.
Overexpression of DhPpz1p suppresses phenotypic defects of dhmpk1 deletion in D. hansenii. A, suppression of the toxic cation and caffeine sensitivity of dhmpk1 mutant is shown. DBH932 harboring plasmids pAN5, pAN6, or pDA1 (vector control) were grown in SD medium to logarithmic phase (A600 ∼ 1.0). 10-Fold serial dilutions of these cultures were on SD plates containing LiCl or caffeine as indicated. B, shown is relative growth of dhmpk1 mutant harboring different plasmids as indicated. A600 after 24 h of growth for control (DBH93 harboring pDA1 and pDH4) was taken 100%. Data presented are results of three independent experiments (mean ± S.D.).
N-terminal Non-catalytic Region Is Essential for Functionality of DhPpz1p
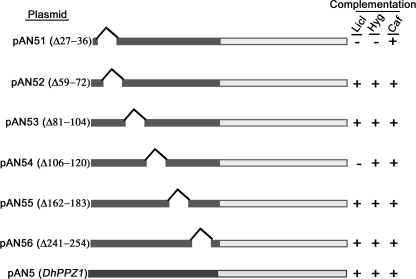
Ppz1p represents a novel phosphatase family with a conserved, catalytic C-terminal region and a large non-catalytic N-terminal region rich in few amino acids e.g. serine, asparagine, arginine residues (8, 23). The C-terminal catalytic domain shares substantial sequence homology among Ppz1p orthologs as well with other PPP members. In comparison, the N-terminal regions are quite diverged in their primary sequences. The N-terminal region is a unique feature of Ppz1p orthologs and appeared to have a regulatory role in the functionality of this phosphatase. Surprisingly, not much information is available about this region. Analysis of DhPpz1p sequence showed that the N-terminal region comprised of amino acid residues 1–278 and the catalytic domain was from amino acid residues 279–572. Phenotypic analysis showed that either the N-terminal (pAN9) or C-terminal catalytic (pAN10) region was non-functional as they failed to suppress LiCl and hygromycin tolerance or caffeine-sensitive phenotype of dhppz1 mutant (data not shown). Besides these, none of these constructs could rescue the slow growth phenotype exhibited by dhppz1 mutant (data not shown). Together these results validated the importance of both halves for the functionality of DhPpz1p. Primary sequence clearly indicated that N terminus of DhPpz1p was also rich in serine and asparagine residues, as it contained 63 serine (22.58%) and 45 asparagine (16.13%) residues. Analysis of the primary sequence of the N-terminal region (1–278 amino acids) of DhPpz1p with the help of bioinformatics tools such as SMART (Simple Modular Architecture Research Tool at EMBL) and PROSITE (data base of protein families and domains) did not indicate the presence of any known domain or motif in this region. However, it showed a six-sequence motif of low complexity that was rich in serine, asparagine, or arginine residues (Fig. 5). To delineate the role of these regions in the functionality of DhPpz1p, six mutant constructs were generated by a PCR-based method by deleting the amino acid residues corresponding to 27–36 (pAN51), 59–75 (pAN52), 81–104 (pAN53), 106–120 (pAN54), 162–183 (pAN55), and 241–254 (pAN56). All these constructs were transformed in D. hansenii strain DBH936. Transformants were selected on SD plates and assessed for their ability to complement phenotype associated with dhppz1 mutation. For this, serial dilutions of log phase cultures of DBH936 cells harboring pAN51, pAN52, pAN53, pAN54, pAN55, and pAN56 plasmid were spotted on SD plates supplemented with different concentrations of LiCl, hygromycin, or caffeine. Growth on plates was observed after 3–4 days of incubation at 28 °C (Fig. 5). DBH936 strains transformed with plasmids pAN51 and pAN54 remained LiCl-resistant similar to DBH936 (dhppz1) strain, although all other mutant constructs fully reverted to lithium-sensitive phenotypes similar to DBH936-expressing wild type DhPpz1p. On hygromycin plates, DBH936 transformed with only pAN51 showed a hygromycin-resistant phenotype similar to that of dhppz1 mutant, whereas with all other constructs, hygromycin-sensitive phenotype was observed. These results thus indicated that pAN51 encoded a non-functional allele of DhPpz1p with respect to this phenotype. D. hansenii strain DBH936 was highly sensitive to caffeine, and therefore, it did not grow on plates containing caffeine. In contrast, DBH936 strain transformed with all the mutant constructs were able to grow on these plates, which clearly indicated that all the mutant constructs behaved like wild type DhPpz1p in this regard (Fig. 5).
FIGURE 5.
Structure-function analysis of N-terminal domain of DhPpz1p. Schematics depicting wild type and mutant DhPpz1p are shown. C-terminal catalytic domain (light box), N-terminal non-catalytic domain (dark box), and the position of different deletions are shown. The right panel shows phenotypic complementation of dhppz1 mutation in DBH936 by different plasmid constructs as determined by dilution spotting on SD plates containing LiCl, hygromycin (Hyg), and caffeine (Caf). + indicates complementation, and − indicates no complementation. Experiments were repeated three times with similar results.
Next we determined whether these mutant constructs could rescue the slow growth phenotype of dhppz1 mutant observed earlier. For this, saturated cultures of DBH936 strain transformed with all these constructs were re-inoculated into 25 ml of SD medium at initial A600 of 0.025. After 24 h of growth at 28 °C, A600 of each culture was measured, and the data were expressed as their relative growth compared with the parent DBH93 strain used as control. Our results revealed that all mutant constructs could suppress the growth defect of dhppz1 strain, indicating their functional activity in vivo. Interestingly, DBH936 harboring pAN51 exhibited 2-fold more growth than the parent strain DBH93 used as controls in this experiment (supplemental Fig. S5).
Conserved Serine/Arginine-rich Motif in N-terminal Non-catalytic Region Is Important for Cation Tolerance
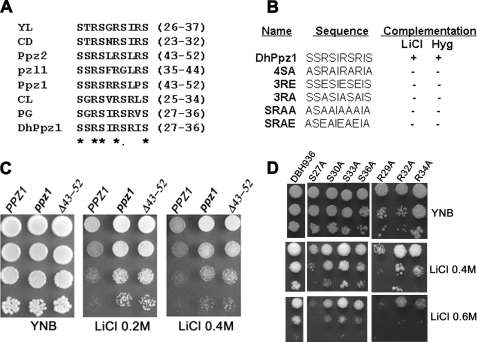
From the above analysis, it thus appeared that none of the motifs was important for DhPpz1p toward its role in cell wall remodeling or cell growth. However, the short sequence motif corresponding to 27–36 amino acid residues was very important for both lithium and hygromycin tolerance phenotype and appeared to be indispensable for DhPpz1p toward its role in ion homeostasis. This motif was rich in serine and arginine residues, and it contained five serine and three arginine residues. Analysis of the sequence of few other orthologs from different yeast and fungal species clearly showed the presence of this motif, and thus it appeared to be evolutionary conserved among the members of this group (Fig. 6A). The sequence alignment of this motif showed that the residues Ser-27, Ser-30, Ser-36, Arg-29, and Arg-32 were present in all. However, Ser-33 appeared to be conserved only in yeast and not in filamentous fungi. Interestingly, the eighth and ninth positions were always occupied by an arginine along with a bulky hydrophobic residue. To confirm the functional importance of the conserved residues, we have created few mutant constructs by site-directed mutagenesis. In these mutants serine residues were changed to alanine where as the arginine residues were changed to either alanine or glutamic acid. The plasmids carrying these mutants were transformed in D. hansenii strain DBH936. The growth patterns of the transformants were assessed by dilution spotting on SD plates containing different concentrations of LiCl or hygromycin. The result of this experiment was summarized in Fig. 6B. Mutation of conserved serine and arginine residues together (SRAA or SRAE) could not suppress the lithium- or hygromycin-resistant phenotype of the dhppz1 mutant. The mutation of only serine or arginine residues alone (4SA, 3RE, or 3RA) also could not suppress this phenotype. Replacement of arginine residues with either alanine or glutamic acid (3RA or 3RE) had similar effects. These results clearly indicated that both the serine and arginine residues have crucial roles in the functionality of DhPpz1p. This motif is a common feature among Ppz1p orthologs (Fig. 6A). To check the functional importance of this motif in other orthologs, we constructed a mutant of S. cerevisiae PPZ1 (PPZ1Δ43–52) that will express a Ppz1p allele devoid of this motif. Transformation of this construct into ppz1 mutant of S. cerevisiae failed to suppress the salt hypertolerant phenotype of this strain (Fig. 6C). This result clearly indicated the functional importance of this motif in other orthologs also.
FIGURE 6.
Mutational analysis serine/arginine-rich motif in DhPpz1p and Ppz1p. A, sequence alignment of the serine/arginine-rich motif present in different Ppz1p orthologs from different species is shown: YL, Yarrowia lipolytica XP_504610.2; CD, Candida dubliniensis XP_002422157.1); Pzl1, Neurospora crassa AF071752; CL, Clavispora lusitaniae XP_002619471; PG, Pichia guilliermondii EDK41476.2; Ppz1, S. cerevisiae NP_013696.1; Ppz2, S. cerevisiae NP_010724.1; DhPpz1, D. hansenii XP_459586.2. Conserved residues are shown by asterisks. B, phenotypic complementation of wild type DhPpz1p and mutants carrying point mutations in serine/arginine-rich motif along with the sequence of respective mutated region are shown. + indicates complementation, and − indicates not complementation. C, dilution spotting of S. cerevisiae strains expressing wild type Ppz1p or its mutants on SD plate containing LiCl is shown. PPZ1 (parental strain BY4742/pRS423), ppz1 mutant (Y10557/pRS423), and Δ43–52 (Y10557/PPZ1-Δ43–52) were grown overnight on SD without histidine before serial dilution. Representative data of three independent experiments are shown. D, effect of mutation in individual serine and arginine residues of Ser/Arg motif on salt tolerance exhibited by dhppz1 mutant is shown. 10-Fold serial dilution of the DBH936 strain expressing different point mutations was spotted on SD plates containing 0.4 and 0.6 m LiCl. Plates were incubated at 28 ºC for 3–4 days before being photographed. A culture of DBH936 harboring empty vector pDA1 was used as the control. Representative data of two independent experiments are shown.
To assess the contribution of each serine and arginine residue in this motif, individual residues were mutated to alanine separately. The growth pattern of DBH936 carrying these mutants (S27A, S30A, S33A, S36A, R29A, R32A, and R34A) was checked on lithium chloride-containing plates. Among the serine residues, Ser-30, -33, and -36 were important, whereas Ser-27 was not essential as the later mutation completely suppressed the salt hypertolerant phenotype of DBH936 (Fig. 6D). Similarly, Arg-29 also appeared to be dispensable, as this mutant failed to grow on LiCl plates. Surprisingly, mutants R32A and R34A showed partial complementation on LiCl plates. It is thus possible that Arg residues create a positive charge environment in the vicinity that is important. In such a scenario, individual residues could be dispensable as their absence could be compensated by others in the vicinity.
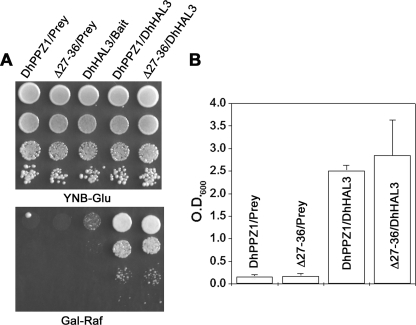
In S. cerevisiae, Hal3p is an important negative regulator of Ppz1p. Hal3p has been shown to interact physically with the catalytic moiety of Ppz1p and inhibit its activity (18, 20). Similar to the deletion of PPZ1, overexpression of Hal3p also increases salt tolerance through the inhibition of Ppz1p. An ortholog of Hal3p (DhHal3p, GenBankTM accession number XP_459184) is also present in D. hansenii. It is possible that the Ser/Arg-rich motif could play a regulatory role, and thus the absence of this motif could increase the interaction of DhPpz1p with DhHal3p, resulting in the higher salt resistance phenotype. To check this possibility, we performed a yeast two-hybrid interaction. As shown in Fig. 7A, DhHal3p strongly interacted with both DhPpz1p and Ser/Arg motif mutants. Comparison of these two-hybrid interactions by liquid growth assay showed that there were no significant differences between DhPpz1p and its mutant (Fig. 7B). Thus, the Ser/Arg-rich motif does not modulate Hal3p binding.
FIGURE 7.
Role of Ser/Arg-rich motif in the interaction of DhPpz1p with DhHal3p. A, two-hybrid assay for interactions between DhHal3p and DhPpz1p. Wild type DhPPZ1 or its mutant having a deletions in the Ser/Arg motif (Δ27–36) cloned in pEG202 vector was used as bait, whereas DhHAL3 cloned in plasmid pJG4-5 was used as prey. Bait and prey constructs (in pairs as indicated) or in combination with empty prey or bait (as control) were transformed into S. cerevisiae strain EGY48. Growth of the transformants on Gal-Raf minimal medium is shown after dilution spotting. Experiments were repeated twice with pool of four independent transformants. Representative data are shown. B, growth of EGY48 harboring DhPPZ1 and Δ27–36 bait along with empty prey or DhHal3 in prey vector were grown overnight in minimal media with 2% raffinose (without tryptophan and histidine). The cultures were re-inoculated in minimal media with 1% raffinose and 2% galactose (without tryptophan, histidine, and leucine) at A600 ∼0.10 and grown further for 41 h. Data (mean ± S.D.) of two independent experiments, each performed in duplicate with a pool of four different transformants, is shown here.
DISCUSSION
PPZ1 orthologs constitute unique members of PPP family of serine threonine phosphatases that are present only in the fungal world. They are involved in a variety of cellular processes that includes salt tolerance, ion homeostasis, maintenance of cell integrity, and cell cycle regulation (13, 14). Although they are an important regulator of salt tolerance, their role in the physiology of any halotolerant organism remained unexplored. To this effect, we undertook molecular characterization of PPZ1 ortholog from D. hansenii, which is one of the most halotolerant species of yeast and is considered as a model organism to understand halotolerance in yeast (27–29). Interestingly, this report is also the first documentation of functional characterization of any D. hansenii gene in its native host.
Analysis of sequence data base identified only one PPZ1 ortholog among the eight members of PPP family that are present in D. hansenii genome. This organism harbors fewer PPP phosphatases than S. cerevisiae, which contains 12 such proteins. Except for the PPH3 ortholog, D. hansenii contains at least one ortholog of each group that is present in S. cerevisiae, and redundant or partially redundant isoforms are notably absent in this species.5 From the primary sequence, DhPpz1p appeared to have all the features typical to this type of proteins. To determine its role in D. hansenii, phenotypic analysis of DhPPZ1 knock-out strains (DBH91 and DBH936) was carried out. dhppz1 mutant exhibited a high level of resistance toward different toxic cations such as Li+, hygromycin, and spermine (Fig. 1). For NaCl, better growth of the dhppz1 mutant was apparent only at lower concentrations, and no significant difference in growth was observed at 2.0 m or higher concentration of NaCl (Fig. 1B). dhppz1 accumulated a less amount of Li+ compared with the parent strain both at pH 6.0 and 7.0. A lower steady state concentration of toxic cation in the cell was expected to lead to higher resistance toward these ions. Besides this, the intracellular K+ concentration appeared to be elevated in the mutant (Fig. 2). Higher K+ in the cell could also affect the accumulation of toxic cation. In the case of S. cerevisiae ppz1 mutant, a lower intracellular Li+ and increased level of intracellular K+ was also observed. This was achieved through two distinct mechanisms. One is through the transcriptional up-regulation of ENA1 encoding Na-H+ ATPase, the major Na+ efflux pump in S. cerevisiae. As a substrate, the major K+ transporter Trk1p is also regulated by Ppz1p. In the absence of Ppz1p, the increase in the activity of Trk1p resulted in the accumulation of K+ and thereby affected the membrane potential and accumulation of toxic cation in the cell. In S. cerevisiae, interaction of Ppz1p with Trk1p has been demonstrated through immunoprecipitation (14). Both Ppz1p and Trk1p have been shown to be associated with the membrane as observed by GFP fusion as well as other methods, and their physical proximity in the cell has been suggested to be crucial for Ppz1p to utilize Trk1p as substrate. Our results showed that DhPpz1p-RFP fusion protein was present in the cytosol, and therefore, the scenario was quite different in D. hansenii. Whether DhPpz1p could still modulate the activity of K+ transporter DhTrk1p in D. hansenii thus remains an open question.
We had observed that the mutation in DhPPZ1 did not significantly affect the level of DhENA1 transcript in D. hansenii which indicated the role of Ena1 independent mechanism in the salt tolerance of D. hansenii dhppz1 mutant. Earlier studies showed that the extent of salt tolerance displayed by S. cerevisiae ppz1 mutant remained unaffected by raising the pH of the medium to 8.5 (12). Contrasting this, the salt tolerance of D. hansenii mutant diminished at higher pH (Fig. 1B). This observation indicated a possible role of Na+/H+ antiporter DhNha1p. Our observation that under salt stress DhNHA1 expression is strongly up-regulated in dhppz1 mutant further supports this view (Fig. 2E). In yeast, the role of Nha1p orthologs in the regulation of cell cycle, cell volume, membrane potential, and ion homeostasis has been demonstrated. Although both S. cerevisiae and D. hansenii accumulate less toxic cation in the absence of PPZ1, this could, however, be achieved through distinct mechanisms.
DhPPZ1 is not an essential gene in D. hansenii. However, its deletion caused a dramatic decrease in the growth of the mutant, indicating its vital role in the cellular physiology in this species. Earlier studies showed that an excess of Ppz1p confers slow growth phenotype in S. cerevisiae by affecting G1/S transition and bud emergence (9, 11, 16). In contrast, the overexpression of DhPPZ1 had a negligible effect on the growth of D. hansenii. In S. cerevisiae, strains lacking PPZ1/2 showed pH-dependent growth; a relatively slow-growth phenotype was observed on medium with increasing pH (13). Our result showed no significant changes in the growth of dhppz1 mutant with the increase in the pH of the medium (Fig. 3C). DhPpz1p appeared to have a functional link with the cell wall integrity pathway in D. hansenii. Like a mutant having a defect in the cell wall integrity pathway, dhppz1 strains also showed a cell lysis defect at higher temperature and sensitivity toward cell wall destabilizing agents, such as caffeine. Both these defects could be suppressed by the addition of 1 m sorbitol to the medium (supplemental Fig. S3). In S. cerevisiae, PPZ1 acts as a multicopy suppressor of the defect associated with the mutation in MPK1 (11). A similar phenotype was also observed with DhPPZ1 in D. hansenii. Taken together, these results underscore a distinct role of DhPpz1p in D. hansenii despite having some functional overlaps with its ortholog in S. cerevisiae. Both of these species live in a different ecological niche. Thus it is not strange that they have evolved with different survival strategies using the same molecular elements but in distinct ways.
Like a typical member of its group, DhPpz1p possesses a large N-terminal non-catalytic region that is rich in a few amino acid residues such as serine. Earlier studies with Ppz1p showed that it is essential for the functionality. Deletion of this region in DhPpz1p also rendered the molecule non-functional. To have structure-function insight about this region, we undertook bioinformatics analysis, which did not show the presence of any known domain or motif but identified six regions of short sequence motif with low complexity. To check the role of these motifs, deletion constructs (pAN51, pAN52, pAN53, pAN54, pAN55, and pAN56) were created and subjected to phenotype analysis (Fig. 5 and supplemental Fig. S5). All the mutants could complement the caffeine sensitivity and growth defect of dhppz1 mutation similar to that of wild type DhPpz1p. Compared with other constructs, copious growth was observed with pAN51. Therefore, all these motifs were dispensable for the role of DhPpz1p in cell integrity and cell growth. Surprisingly, pAN51 failed to suppress the lithium and hygromycin resistance, indicating motif 27–36 could be indispensable for salt tolerance. The growth of pAN54 on LiCl but not on a hygromycin-containing plate indicated a subtle, yet unexplained, difference in these two as toxic cations. Nevertheless, analysis of these constructs clearly demonstrated that the participation of DhPpz1p in salt tolerance, cell integrity pathway, or cell growth occurred through distinct molecular events. The sequence motif 27–36 was rich in serine and arginine residues that are indispensable for its function in salt tolerance. The discovery that this motif is highly conserved among the Ppz1 orthologs also suggests its importance. Mutational analysis of S. cerevisiae Ppz1p further supports this view (Fig. 6). Previous studies suggested that Hal3p binds to the catalytic moiety of Ppz1p in S. cerevisiae and the N-terminal non-catalytic half of Ppz1p might play a protective function against the inhibition by Hal3p (18, 20). The Ser/Arg-rich motif identified in this study is present in the N-terminal region of DhPpz1p, which is far outside of the catalytic domain and, therefore, likely to have, if any, a built-in switch-like function in preventing the interaction with DhHal3p. However, the yeast two-hybrid assay clearly showed that this motif had no role in binding DhHal3p (Fig. 7). Recently, serine/arginine-rich motifs have been shown to be involved in protein-protein interactions (36, 37). It is unclear at present how this motif could function. As a member of PPP family of phosphatases, the spectrum of activities exhibited by DhPpz1p is regulated by its interactions with different cellular proteins. It is plausible that some of these interactions are mediated by the serine-arginine-rich motif.
Supplementary Material
Acknowledgments
We are thankful to R. Sharma and D. Bhatt for excellent technical assistance and A. Sukla for atomic absorption spectroscopy.

This article contains supplemental Table 1 and Figs. 1–5.
A. Minhas, A. Sharma, H. Kaur, Y. Rawal, K. Ganesan, and A. K. Mondal, unpublished observation.
- PPP
- phosphoprotein phosphatase
- PP
- protein phosphatase
- qPCR
- quantitative real-time PCR
- RFP
- red fluorescent protein
- SD
- synthetic dextrose
- YPD
- yeast extract/peptone/dextrose.
REFERENCES
- 1. Cohen P. T. (1997) Novel protein serine/threonine phosphatases. Variety is the spice of life. Trends Biochem. Sci. 22, 245–251 [DOI] [PubMed] [Google Scholar]
- 2. Cohen P. T. (2004) Protein Phosphatases (Arino J., ed.) pp. 1–20, Springer, Berlin [Google Scholar]
- 3. Moorhead G. B., De Wever V., Templeton G., Kerk D. (2009) Evolution of protein phosphatases in plants and animals. Biochem. J. 417, 401–409 [DOI] [PubMed] [Google Scholar]
- 4. Shi Y. (2009) Serine/threonine phosphatases. Mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 5. Blatch G. L., Lässle M. (1999) The tetratricopeptide repeat. A structural motif mediating protein-protein interactions. Bioessays 21, 932–939 [DOI] [PubMed] [Google Scholar]
- 6. Barford D., Das A. K., Egloff M. P. (1998) The structure and mechanism of protein phosphatases. Insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 [DOI] [PubMed] [Google Scholar]
- 7. Ceulemans H., Bollen M. (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 8. Ariño J. (2002) Novel protein phosphatases in yeast. Eur. J. Biochem. 269, 1072–1077 [DOI] [PubMed] [Google Scholar]
- 9. Posas F., Casamayor A., Morral N., Ariño J. (1992) Molecular cloning and analysis of a yeast protein phosphatase with an unusual amino-terminal region. J. Biol. Chem. 267, 11734–11740 [PubMed] [Google Scholar]
- 10. Hughes V., Müller A., Stark M. J, Cohen P. T. (1993) Both isoforms of protein phosphatase Z are essential for the maintenance of cell size and integrity in Saccharomyces cerevisiae in response to osmotic stress. Eur. J. Biochem. 216, 269–279 [DOI] [PubMed] [Google Scholar]
- 11. Lee K. S., Hines L. K., Levin D. E. (1993) A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol. Cell. Biol. 13, 5843–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Posas F., Camps M., Ariño J. (1995) The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J. Biol. Chem. 270, 13036–13041 [DOI] [PubMed] [Google Scholar]
- 13. Yenush L., Mulet J. M., Ariño J., Serrano R. (2002) The Ppz protein phosphatases are key regulators of K+ and pH homeostasis. Implications for salt tolerance, cell wall integrity, and cell cycle progression. EMBO J. 21, 920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yenush L., Merchan S., Holmes J., Serrano R. (2005) pH-responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell. Biol. 25, 8683–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruiz A., Yenush L., Ariño J. (2003) Regulation of ENA1 Na+-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot. Cell 2, 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clotet J., Garí E., Aldea M., Ariño J. (1999) The yeast Ser/Thr phosphatases sit4 and ppz1 play opposite roles in regulation of the cell cycle. Mol. Cell. Biol. 19, 2408–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferrando A., Kron S. J., Rios G., Fink G. R., Serrano R. (1995) Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol. Cell. Biol. 15, 5470–5481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Nadal E., Clotet J., Posas F., Serrano R., Gomez N., Ariño J. (1998) The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc. Natl. Acad. Sci. U.S.A. 95, 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Venturi G. M., Bloecher A., Williams-Hart T., Tatchell K. (2000) Genetic interactions between GLC7, PPZ1, and PPZ2 in Saccharomyces cerevisiae. Genetics 155, 69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muñoz I., Ruiz A., Marquina M., Barceló A., Albert A., Ariño J. (2004) Functional characterization of the yeast Ppz1 phosphatase inhibitory subunit Hal3. A mutagenesis study. J. Biol. Chem. 279, 42619–42627 [DOI] [PubMed] [Google Scholar]
- 21. García-Gimeno M. A., Muñoz I., Ariño J., Sanz P. (2003) Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J. Biol. Chem. 278, 47744–47752 [DOI] [PubMed] [Google Scholar]
- 22. Ruiz A., Muñoz I., Serrano R., González A., Simón E., Ariño J. (2004) Functional characterization of the Saccharomyces cerevisiae VHS3 gene. A regulatory subunit of the Ppz1 protein phosphatase with novel, phosphatase-unrelated functions. J. Biol. Chem. 279, 34421–34430 [DOI] [PubMed] [Google Scholar]
- 23. Clotet J., Posas F., de Nadal E., Ariño J. (1996) The NH2-terminal extension of protein phosphatase PPZ1 has an essential functional role. J. Biol. Chem. 271, 26349–26355 [DOI] [PubMed] [Google Scholar]
- 24. Balcells L., Gómez N., Casamayor A., Clotet J., Ariño J. (1997) Regulation of salt tolerance in fission yeast by a protein phosphatase-Z-like Ser/Thr protein phosphatase. Eur. J. Biochem. 250, 476–483 [DOI] [PubMed] [Google Scholar]
- 25. Szöor B., Fehér Z., Zeke T., Gergely P., Yatzkan E., Yarden O., Dombrádi V. (1998) pzl-1 encodes a novel protein phosphatase-Z-like Ser/Thr protein phosphatase in Neurospora crassa. Biochim. Biophys. Acta 1388, 260–266 [DOI] [PubMed] [Google Scholar]
- 26. Vissi E., Clotet J., de Nadal E., Barceló A., Bakó E., Gergely P., Dombrádi V., Ariño J. (2001) Functional analysis of the Neurospora crassa PZL-1 protein phosphatase by expression in budding and fission yeast. Yeast. 18, 115–124 [DOI] [PubMed] [Google Scholar]
- 27. Prista C., Loureiro-Dias M. C., Montiel V., García R., Ramos J. (2005) Mechanisms underlying the halotolerant way of Debaryomyces hansenii. FEMS Yeast Res. 5, 693–701 [DOI] [PubMed] [Google Scholar]
- 28. Aggarwal M., Mondal A. K. (2006) Role of N-terminal hydrophobic region in modulating the subcellular localization and enzyme activity of the bisphosphate nucleotidase from Debaryomyces hansenii. Eukaryot. Cell 5, 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aggarwal M., Mondal A. K. (2009) Biotechnology: Diversity and Applications (Kunze G., Satyanarayana T., ed) pp. 65–84, Springer Science+Business Media B.V [Google Scholar]
- 30. Minhas A., Biswas D., Mondal A. K. (2009) Development of host and vector for high efficiency transformation and gene disruption in Debaryomyces hansenii. FEMS Yeast Res. 9, 95–102 [DOI] [PubMed] [Google Scholar]
- 31. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 32. Meena N., Kaur H., Mondal A. K. (2010) Interactions among HAMP domain repeats act as an osmosensing molecular switch in group III hybrid histidine kinases from fungi. J. Biol. Chem. 285, 12121–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Golemis E. A., Brent R. (1997) The Yeast Two-hybrid System (Bartel P. L., Fields S., ed.) pp. 43–72 Oxford University Press, Oxford, UK [Google Scholar]
- 34. Merchan S., Bernal D., Serrano R., Yenush L. (2004) Response of the Saccharomyces cerevisiae Mpk1 mitogen-activated protein kinase pathway to increases in internal turgor pressure caused by loss of Ppz protein phosphatases. Eukaryot. Cell 3, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levin D. E. (2005) Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Briknarová K., Nasertorabi F., Havert M. L., Eggleston E., Hoyt D. W., Li C., Olson A. J., Vuori K., Ely K. R. (2005) The serine-rich domain from Crk-associated substrate (p130cas) is a four-helix bundle. J. Biol. Chem. 280, 21908–21914 [DOI] [PubMed] [Google Scholar]
- 37. Jalota A., Singh K., Pavithra L., Kaul-Ghanekar R., Jameel S., Chattopadhyay S. (2005) Tumor suppressor SMAR1 activates and stabilizes p53 through its arginine-serine-rich motif. J. Biol. Chem. 280, 16019–16029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.