Background: FBXL5 is required for proper regulation of cellular iron homeostasis.
Results: FBXL5 contains a structurally characterized hemerythrin-like domain.
Conclusion: The atypical features of the hemerythrin-like domain facilitate its role as a metabolic sensor.
Significance: FBXL5 is the only identified mammalian protein containing a hemerythrin-like domain. Resolution of the structure of the domain provides mechanistic insights into its iron and oxygen responsiveness.
Keywords: Crystal Structure, E3 Ubiquitin Ligase, Iron Metabolism, Metalloproteins, Oxygen Binding
Abstract
Mammalian cells maintain iron homeostasis by sensing changes in bioavailable iron levels and promoting adaptive responses. FBXL5 is a subunit of an E3 ubiquitin ligase complex that mediates the stability of iron regulatory protein 2, an important posttranscriptional regulator of several genes involved in iron metabolism. The stability of FBXL5 is regulated in an iron- and oxygen-responsive manner, contingent upon the presence of its N-terminal domain. Here we present the atomic structure of the FBXL5 N terminus, a hemerythrin-like α-helical bundle fold not previously observed in mammalian proteins. The core of this domain employs an unusual assortment of amino acids necessary for the assembly and sensing properties of its diiron center. These regulatory features govern the accessibility of a mapped sequence required for proteasomal degradation of FBXL5. Detailed molecular and structural characterization of the ligand-responsive hemerythrin domain provides insights into the mechanisms by which FBXL5 serves as a unique mammalian metabolic sensor.
Introduction
Because both iron deficiency and iron overload can be deleterious, it is imperative that cells maintain proper iron homeostasis (1). Cellular iron homeostasis is regulated in part by iron regulatory protein 2 (IRP2),5 which posttranscriptionally regulates a cohort of iron metabolism genes (2). IRP2 controls either the translation or the stability of its target mRNAs by binding to RNA hairpin structures, known as iron-responsive elements (3, 4). IRP2 accumulates under iron- or oxygen-deficient conditions but is polyubiquitinated and degraded by the proteasome when iron and oxygen are plentiful (5–7). The physiological importance of IRP2 is reflected in the observed phenotypes arising from targeted gene deletion, including anemia and mild neurodegeneration (8–11). Proper IRP2 regulation necessitates that cells sense both iron and oxygen availability and a means by which such information affects an appropriate change in IRP2 stability. However, the mechanisms by which IRP2 is regulated in an iron- and oxygen-dependent manner are only beginning to be delineated.
Recently, we and others reported that an E3 ubiquitin ligase complex containing F-box and leucine-rich repeat protein 5 (FBXL5) regulates IRP2 stability (12, 13). A reduction in FBXL5 expression leads to inappropriate accumulation of IRP2 and subsequent misregulation of IRP2 target genes in cultured cells, indicating that FBXL5 contributes to the maintenance of iron homeostasis. Notably, FBXL5 is regulated in an inverse fashion to IRP2 as it is stabilized under iron-replete conditions and preferentially degraded when iron or oxygen becomes limiting. Domain mapping studies revealed that the N terminus of FBXL5 is required for its iron-dependent regulation. Bioinformatic analysis predicted that the N-terminal region of FBXL5 encodes a hemerythrin-like domain (12, 13).
Hemerythrin (Hr) domains are characterized by an α-helical bundle fold that commonly contains a diiron center (14), although other metal centers are possible (Protein Data Bank (PDB) ID 2P0N) (16–18). These iron atoms are redox-active and can switch between the fully oxidized diferric (met) state, a partially reduced semi-met state, and a fully reduced diferrous (deoxy) state. In canonical Hr domains, one iron atom (Fe1) is coordinated by the imidazole side chains from three histidines, whereas the other iron (Fe2) is coordinated by two histidines. Typically, a glutamate and an aspartate provide carboxylate groups that bridge Fe1 and Fe2 (14). In the deoxy state, the iron atoms are also bridged by a μ-hydroxo species, ostensibly derived from the solvent. These hemerythrins often reversibly bind dioxygen to Fe2 with electron delocalization from both irons to the oxygen to form the oxy-state. A proton is also transferred from the bridging hydroxo to the resulting peroxo ligand. The hydroperoxo adduct is stabilized by a hydrogen bond to the μ-oxo bridge (19, 20). Although not previously identified in any vertebrate proteins, the putative ligand binding properties of the Hr domain make it an attractive candidate for a central iron- and oxygen-sensing module.
Here we report the crystal structure of the isolated FBXL5 Hr domain and characterization of key residues within this domain. These data reveal atypical features that allow FBXL5 to function via its N-terminal Hr domain as a ligand-dependent regulatory switch in the maintenance of iron homeostasis.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The N terminus of Homo sapiens FBXL5 (residues 1–161) was cloned into the pGST-parallel1 vector and expressed in Escherichia coli BL21 (DE3) cells grown in medium supplemented with 100 μm ferric ammonium citrate (FAC). Soluble FBXL5 Hr protein was purified using glutathione agarose (GE Healthcare). The GST fusion tag was cleaved upon incubation with tobacco etch virus protease for 6 h at 24 °C, and the liberated Hr domains were purified by anion exchange chromatography (HiTrap Q, GE Healthcare). Iron-loaded Hr was separated from apoHr by an additional anion exchange (Mono Q, GE Healthcare) purification step. Purified FBXL5 Hr used to generate Native form 1 crystals was exchanged into buffer composed of 24 mm Tris-HCl pH 8.0, 50 mm NaCl, 5 mm β-mercaptoethanol, and 0.5 mm tris(2-carboxyethyl) phosphine using a Centricon (Millipore 5 kDa molecular weight cut off) and passed through a 0.22-μm filter.
Crystallization and X-ray Diffraction Data Collection
Crystals of FBXL5 Hr (Native 1) were grown using the hanging drop vapor diffusion method from 1:1 (v/v) mixtures of protein (5 mg/ml) and reservoir solution (10% (w/v) PEG 6000, 0.1 m HEPES pH 6.5). Plate-like crystals appeared after 1 day at 20 °C and grew to their maximal extent (0.2 × 0.5 × 0.1 mm) by 2–3 days. Cryoprotection was performed by stepwise transfer of the crystals to a final solution of 25% (v/v) ethylene glycol, 10% PEG 6000, and 0.1 m HEPES, pH 6.5, followed by flash-cooling in liquid nitrogen.
Crystals of the second form of FBXL5 Hr (Native 2) were obtained using an identical purification scheme as Native 1, except that recombinant protein was exchanged into buffer lacking reducing agents and allowed to incubate at 4 °C for 24 h. Native 2 crystals were grown using the hanging drop vapor diffusion method from 1:1 (v/v) mixtures of protein (5 mg/ml) and reservoir solution (0.1 m HEPES, pH 7.4, 25% (w/v) PEG 3350). Plate-like crystals appeared after 1 day at 20 °C and grew to their maximal extent by 2–3 days. Cryoprotection was performed by stepwise transfer of the crystals to a final solution of 0.1 m HEPES, pH 7.4, 25% PEG 3350, and 20% ethylene glycol prior to flash-cooling using liquid nitrogen. Native 2 crystals exhibited the symmetry of space group P21 with unit cell parameters of a = 76.3 Å, b = 54.4 Å, c = 78.2 Å, and β = 90.02° and contained four molecules of FBXL5 Hr per asymmetric unit. Native 2 crystals displayed moderate anisotropy and P222 pseudosymmetry, and diffracted to a dmin of 2.10 Å when exposed to synchrotron radiation.
Crystals of the third form of FBXL5 Hr (Native 3) were obtained using an identical purification scheme as Native 1 except that recombinant protein was exchanged into buffer containing 2 m equivalent Na2S2O4. Crystals were generated in an anaerobic chamber (Coy Laboratory Products Inc.) using the hanging drop vapor diffusion method from 1:1 (v/v) mixtures of protein (5 mg/ml) and reservoir solution (12% (w/v) PEG 6000, 0.1 m HEPES, pH 6.5). Prior to usage, all reagents were purged with nitrogen gas for 30 min followed by an overnight incubation in an anaerobic chamber. Plate-like crystals appeared after 1 day at room temperature and grew to their maximal extent by 2–3 days. Cryoprotection was performed by stepwise anaerobic transfer of the crystals to a final solution of 30% (v/v) ethylene glycol, 12% PEG 6000, and 0.1 m HEPES, pH 6.5, followed by flash-cooling in liquid nitrogen. Native 3 crystals exhibited the symmetry of space group C2 with unit cell parameters of a = 60.4 Å, b = 76.0 Å, c = 77.4 Å, and β = 112.1° and contained two molecules of FBXL5 Hr per asymmetric unit. Native 3 crystals diffracted isotropically to a dmin of 2.20 Å when exposed to synchrotron radiation. Data were indexed, integrated, and scaled using the HKL-3000 program package (21). Data collection statistics are provided in supplemental Table S1.
Structure Determination and Refinement
Phases for Native 1 crystals of FBXL5 Hr were obtained from a two-wavelength anomalous dispersion experiment using data to a resolution of 2.5 Å, collected near the iron absorption edge. Four iron and two sulfur sites were located using the program SHELXD (22); this represented two single-occupancy iron sites and one methionine sulfur site per FBXL5 Hr monomer. Phases were refined with the program MLPHARE (23), resulting in an overall figure of merit of 0.59 for data between 44.7 and 2.5 Å. Phases were further improved by density modification and 2-fold noncrystallographic averaging with the program DM (24), resulting in a figure of merit of 0.85. An initial model containing about 86% of all residues was automatically generated by alternating cycles of the programs ARP/warp (25). Additional residues were manually modeled in the program O (26). Refinement was performed with native data to a resolution of 1.85 Å using the program PHENIX (27), with a random 5.1% of all data set aside for Rfree calculations. All atoms in the protein were restrained in a similar manner, using restraints provided by the PHENIX refinement program. No explicit metal ligand restraints were added.
Phases for the Native 2 and Native 3 crystals of FBXL5 Hr were obtained via molecular replacement in the program PHASER (28) using the coordinates of Native 1 FBXL5 Hr as a search model. Model building and refinement were performed as described above. The current model for Native 2 crystals contains four FBXL5 Hr monomers; included are residues 5–80 and 83–159 in molecule A, residues 5–73, 77–80, and 83–159 in molecules B and C, residues 5–73, 77–80, and 84–160 in molecule D, and 202 waters. The Rwork is 0.197, and the Rfree is 0.270. A Ramachandran plot generated with Molprobity indicated that 98.6% of all protein residues are in the most favored regions, with the remaining 1.4% in additionally allowed regions. The current model for Native 3 crystals contains two FBXL5 Hr monomers; included are residues 4–80 and 83–159 in molecule A, residues 4–80 and 83–160 in molecule B, and 46 waters. The Rwork is 0.177, and the Rfree is 0.223. A Ramachandran plot generated with Molprobity indicated that 98.7% of all protein residues are in the most favored regions, with the remaining 1.3% in additionally allowed regions. Phasing and model refinement statistics are provided in supplemental Table S1.
EPR Spectroscopy
X-band EPR spectra were recorded using a Bruker Elexsys E-500 spectrometer equipped with an Oxford Instruments ESR-10 liquid helium cryostat, under the following conditions unless otherwise indicated: temperature 10 K, modulation amplitude 1 millitesla, and 5-milliwatt microwave power, with 200–500 μm FBXL5 Hr as estimated by a calculated molar extinction coefficient of 19.3 mm−1 cm−1 at 280 nm. Reduction of the FBXL5 hemerythrin domain was achieved through the anaerobic addition of 3 molar equivalents of Na2S2O4 in the presence of 20 μm methyl viologen in a Coy Laboratory Products anaerobic chamber. The samples were transferred into sealed quartz EPR tubes and frozen in liquid N2. EPR quantification was performed through comparison with a 1 mm copper perchlorate spin standard under nonsaturating conditions. Determinations of the exchange coupling constants of mixed valent FBXL5 Hr were conducted as described previously (29).
Stopped-flow Kinetics
Experiments were performed using an Applied Photophysics Ltd. SX.18.MV stopped-flow spectrophotometer. Following reduction, excess Na2S2O4 and methyl viologen were removed using a PD-10 desalting column (Amersham Biosciences) before loading into an anaerobic stopped-flow syringe. Reduced FBXL5 Hr (200 μm) was rapidly mixed with O2-saturated buffer at 4 °C (∼1.8 mm). The rate of FBXL5 oxidation at 340 nm under pseudo first order conditions was fit to a summed exponential expression using Pro-K software (Applied Photophysics).
Cell Culture and Reagents
All cell lines were grown in Dulbecco's modified high glucose Eagle's medium (HyClone) supplemented with 10% fetal bovine serum (Atlanta Biologicals). Plasmid DNA and siRNA transient transfections were performed as described previously (12). Low O2 experiments were performed in a hypoxic incubator (Coy Laboratory Products) containing 1% O2 and 5% CO2 with the balance being N2. HEK 293 cells stably expressing the 3×FLAG-Hr-HA construct were generated over two rounds of clonal selection in the presence of 400 μg/ml G418 (Research Products Inc.). Ts-20 BALB/c 3T3 cells were kindly provided by H. Ozer.
Plasmids
Wild-type, H57A, and E61A human FBXL5 were cloned into the pCI FLAG vector (kindly provided by X. Wang) as described previously (12). The FBXL5 H15A construct was generated using a triple ligation approach. A 5′ DNA fragment containing the H15A mutation was purchased from IDT Technologies. This fragment was amplified using the oligonucleotide 5′-GGAGAGATCTCAGACCATTTATAATGTACATTCTGACAATAAACTC and a previously described WT FBXL5 forward primer (12). The remaining 3′ fragment was amplified from FBXL5 plasmid DNA using the oligonucleotide 5′-GGAGAGATCTCAGACCATTGCAGCTGCACATGCTGACAATAAACTCTCCGAGATGCTT and a previously described WT FBXL5 reverse primer (12). The resulting fragments were ligated together via an artificially inserted BglII site and inserted into the pCI FLAG vector at the BamHI and SalI sites. The oligonucleotide 5′-GAGGGTCGACTCAAGCGTAATCTGGAACATCGTATGGGTACTGAGAGCAGTGTTGTGC was used to incorporate a C-terminal HA tag into the FBXL5 1–161 pCI FLAG plasmid. The remaining FBXL5 mutant and deletion constructs were generated either by standard methods or by the PCR overlap extension method using the oligonucleotides as listed in supplemental Table S4. For constructs containing a V5 epitope tag, the pcDNA 3.1 vector (Invitrogen) was used, whereas constructs containing a FLAG epitope were generated in the pCI vector.
Immunoblot Analysis
Samples were resuspended in SDS sample buffer, and proteins were resolved by SDS-PAGE. Mouse monoclonal antibodies were obtained as follows: α-FLAG (Sigma, catalog number F3165), α-V5 (Invitrogen, catalog number R960-25), α-tubulin (Sigma, catalog number T6199), and α-ubiquitin (Santa Cruz Biotechnology, catalog number sc-8017). Quantitation of immunoblots for Figs. 3 and 4, E and F, and supplemental Fig. S4, B and C was performed using an Amersham Biosciences ImageScanner (Powerlook UDS 1120) and analyzed using the GE Healthcare ImageQuant software (version 5.2). Quantitation of immunoblots for Fig. 4, C and D, was performed using a Kodak Image Station (4000R Pro) and analyzed using Carestream molecular imaging software (version 5.0.2.30). Quantitative data of immunoblots are shown in supplemental Table S5.
FIGURE 3.
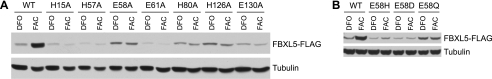
Regulation of FBXL5 is dependent on assembly of diiron center within hemerythrin domain. Eight hours after transfection, HEK 293T cells were treated with either 100 μm DFO or 100 μm FAC for 16 h, and FBXL5 accumulation was assessed by immunoblot analysis. A, analysis of mutations to residues comprising the primary iron coordination shell. B, analysis of FBXL5 Glu-58 mutations.
FIGURE 4.
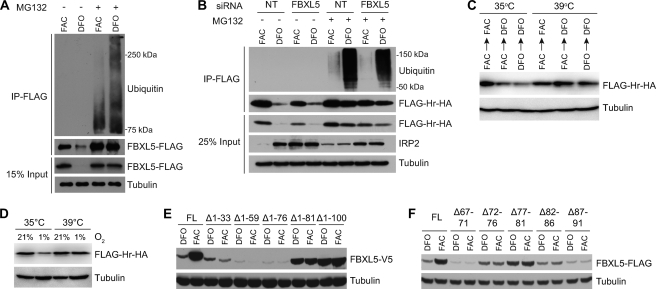
FBXL5 degradation under iron-deplete conditions requires functional ubiquitin-proteasome system and regulatory sequence. A, immunoblot analysis of immunoprecipitated (IP) FLAG-FBXL5 from stably transfected HEK 293T cells treated with FAC or DFO in the absence or presence of the proteasome inhibitor MG132. B, ubiquitination of FBXL5 Hr is not dependent on FBXL5. HEK 293 cells stably expressing the FBXL5 Hr domain were transfected with a negative control siRNA (NT) or siRNA targeting endogenous FBXL5 prior to treatment with FAC or DFO in the absence or presence of MG132. C, immunoblot analysis of FLAG-Hr-HA accumulation in ts-20 BALB/c 3T3 cells incubated with FAC or DFO for 6 h followed by a switch to either E1-permissive (35 °C) or E1-restrictive (39 °C) temperatures for 24 h in medium containing FAC or DFO as indicated. D, immunoblot analysis of FLAG-Hr-HA accumulation in ts-20 BALB/c 3T3 cells. Cells were incubated with FAC for 6 h and maintained in 1 or 21% O2 at 35 °C or 39 °C for 24 h. E and F, immunoblot analysis of FBXL5 accumulation from expression constructs having increasingly longer N-terminal deletions (E) or short internal deletions (F). FL, full length.
Ubiquitination Assays
HEK 293T cells stably expressing an N-terminal 3×FLAG-tagged FBXL5 construct (12) were treated with or without 30 μm MG132 (Boston Biochem) for 1 h followed by the addition of 50 μm FAC (Sigma) or 50 μm deferoxamine mesylate (DFO; Sigma). Cells were then incubated for 6 h. Cell extracts were prepared by adding lysis buffer containing 50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Triton X-100, 250 μm phenylmethylsulfonyl fluoride (Sigma), 1× protease inhibitor mixture (Sigma), and 10 mm N-ethylmaleimide. Lysates were incubated 20 min at 4 °C, and cysteine (0.1% final concentration) was added to neutralize the N-ethylmaleimide. Lysates were clarified by centrifugation at 17,000 × g for 20 min, and protein concentration was determined by the Bradford assay (Bio-Rad). Lysates (∼1 mg) were incubated with 12 μl of FLAG M2 resin (Sigma) overnight to immunoprecipitate FBXL5. Resin was washed three times with lysis buffer, and immunoprecipitated protein was eluted using FLAG peptide. Proteins were resolved by SDS-PAGE and analyzed by immunoblotting. The ubiquitination assay of the FBXL5 hemerythrin domain was carried out as described using HEK 293 cells stably expressing the N-terminal 3×FLAG- and C-terminal HA-tagged FBXL5 hemerythrin domain construct (HEK-Hr). To suppress endogenous FBXL5 expression, HEK-Hr cells were transfected with a FBXL5 siRNA (Dharmacon, catalog number D-012424-04) and a non-targeting siRNA (Dharmacon, catalog number D-001210-01) for 48 h.
RESULTS
N Terminus of FBXL5 Adopts Hemerythrin Fold
Recombinant FBXL5 Hr was purified under ambient oxygen conditions (supplemental Fig. S1, A and B). The EPR spectrum of the as-isolated Hr (Fig. 1, spectrum 1) shows an S = 1/2 signal with g-values of 1.95, 1.80, and 1.67 (Fig. 1, inset), typical of diiron enzymes in the mixed valent state in which antiferromagnetic coupling of high spin FeIII (S = 5/2) and FeII (S = 2) can be observed (29–31). Reduction with excess sodium dithionite results in the disappearance of this signal, and no additional EPR signal can be observed in either perpendicular (Fig. 1, spectrum 2) or parallel modes (data not shown), consistent with formation of the diferrous state. Spin quantification shows that the EPR-active mixed redox state represents only a minor (5–10%) fraction of the total protein. The majority of the as-isolated FBXL5 Hr likely resides in the EPR-silent (S = 0) diferric resting state based on the intense optical feature at 340 nm (supplemental Fig. S1C) that originates from an oxo to ferric ion charge-transfer transition.
FIGURE 1.
EPR spectra of FBXL5 Hr domain. Spectra 1 and 2, the 20 K EPR spectrum of the Hr domain following purification under ambient oxygen conditions (spectrum 1) and upon the addition of excess reducing agent (spectrum 2). Comparison with a canonical Hr from P. gouldii is shown in the inset.
Crystals of the FBXL5 Hr were generated under ambient oxygen conditions. An x-ray fluorescence scan of these crystals displayed a maximum at an incident energy of 7.12 keV. This energy closely corresponds to the absorption edge of iron (32), implying that this metal is bound to the protein. These Native 1 crystals exhibited the symmetry of space group C2 with unit cell parameters of a = 60.1 Å, b = 75.8 Å, c = 78.7 Å, and β = 111.8° and contained two molecules of FBXL5 Hr per asymmetric unit. Native 1 crystals diffracted isotropically to a dmin of 1.85 Å when exposed to synchrotron radiation (Native form 1, supplemental Table S1). The current model for the Native 1 crystals contains two FBXL5 Hr monomers and includes residues 4–74, 77–80, and 84–159 from molecule A and residues 4–74, 77–80, and 84–160 from molecule B along with 168 waters. The Rwork is 0.170, and the Rfree is 0.211. A Ramachandran plot generated with Molprobity (33) indicated that 98.3% of all protein residues are in the most favored regions with the remaining 1.7% in additionally allowed regions.
Although the FBXL5 Hr up-down α-helical bundle featuring a diiron center is consistent with the tertiary structure of previously characterized hemerythrins (supplemental Fig. S2A), there are several unusual features present in this Hr, including a fifth helix packed against the apex of the canonical bundle (Fig. 2A). Overall, the FBXL5 Hr has a similar diameter (∼20 Å) but greater length (58 Å) relative to other hemerythrin structures (∼40–50 Å) (34, 35) and is most structurally homologous to the oxygen-responsive Hr from the bacterial DcrH protein (supplemental Fig. S3). In canonical Hr structures, each of the iron-ligating residues is positioned within an α-helix (14). However, in the FBXL5 Hr, helix α3 is relatively short and does not extend through His-80, one of the seven residues comprising the primary iron coordination shell. Instead, this residue exists in a small ordered region that is in a partially disordered loop preceding helix α3 (Fig. 2 and supplemental Fig. S3A). The coordination geometry of the FBXL5 Hr diiron site also differs from those of canonical hemerythrins. As noted in supplemental Table S2, the Fe-Fe distance is shorter in the FBXL5 Hr (3.12 Å versus ∼3.34–3.41 Å) than other hemerythrins (14, 35), and the FBXL5 Hr Fe-Oμ-Fe angle is more acute (98.1°) than those previously observed in invertebrate (116.7°) and bacterial (132.4°) hemerythrins (14, 35). Moreover, the core iron atoms are positioned closer to the apex of the bundle in the FBXL5 Hr (supplemental Fig. S3B).
FIGURE 2.
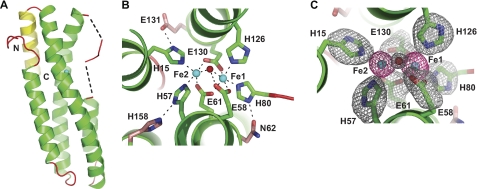
Crystal structure of FBXL5 hemerythrin domain. A, ribbon representation of the hemerythrin domain from H. sapiens FBXL5 (residues 5–159; PDB code 3V5X). Helices are shown in green, and loops are in shown in red. The additional fifth C-terminal helix found is shown in yellow. The dotted lines represent the disordered residues 75–76 and 81–83. B, the first coordination sphere iron ligands of the FBXL5 Hr diiron center are shown as green sticks, and the conserved second coordination sphere iron ligands are shown as pink sticks. Iron ligands and hydrogen bonds are shown as dotted black lines. C, diiron center of FBXL5 Hr Native 1, monomer A with superimposed Fo − Fc simulated annealing omit electron density map (gray, contoured at 3.0 σ) calculated for the bridging oxygen (red) and the side chains (green) of the first coordination sphere (atoms beyond the C-β for the amino acids were omitted). An anomalous difference Fourier electron density map (red, contoured at 15.0 σ) is shown for the iron atoms (cyan).
Iron Binding by Hemerythrin Domain Is Required for Stability of FBXL5
Examination of the amino acid side chains responsible for coordinating iron (Fig. 2, B and C) reveals differences between the FBXL5 diiron center and the consensus observed in the majority of hemerythrins. In place of an aspartate and glutamate to bridge the diiron center, the FBXL5 Hr employs two glutamates (Glu-61 and Glu-130). As most Hr domains studied to date employ five coordinating histidines, the most significant variation in the FBXL5 Hr diiron center is the presence of an additional glutamate (Glu-58) in lieu of a third histidine binding Fe1. Interestingly, the distance between Oϵ2 of Glu-58 and the bridging oxygen is ∼2.8 Å (supplemental Table S2) with an Oμ-Oϵ2-Cγ angle of ∼106°. This geometry is consistent with Glu-58 forming a hydrogen bond with a μ-oxygen species (supplemental Fig. S2B).
To confirm the critical roles of the observed iron-ligating residues to iron-dependent regulation of FBXL5, each residue was individually mutated to an alanine. The resulting expression constructs were transfected into HEK 293T cells prior to treatment with the iron chelator DFO or the iron species FAC. In contrast to wild-type (WT) FBXL5, which preferentially accumulates under iron-replete conditions, all seven mutant proteins displayed constitutively low accumulation levels (Fig. 3A) that could be rescued by the addition of the proteasome inhibitor MG132 (supplemental Fig. S4A). In addition to these direct iron-binding ligands, the FBXL5 Hr structure indicated that residues Asn-62, Glu-131, and His-158 form hydrogen bonds with the primary coordination shell residues His-80, His-15, and His-57, respectively (Fig. 2B). We postulated that these secondary shell interactions might be crucial for the stable formation of the diiron center by bringing the imidazole moieties of residues His-80, His-15, and His-57 into their optimal conformations to coordinate iron. Constructs containing mutations to secondary shell residues (N62A, E131A, and H158A) were transiently transfected into HEK 293T cells, and they, too, displayed lowered protein levels under both iron-replete and iron-deficient conditions (supplemental Fig. S4B). Loss of the additional fifth helix (Δ143–161), found in few other hemerythrin-like structures (PDB ID 2P0N), also led to constitutive destabilization of FBXL5 (supplemental Fig. S4C). This result was expected, given the importance of the secondary coordination shell residue His-158 within helix α5.
In the FBXL5 Hr domain, Fe1 is ligated by two histidines (His-126 and His-80) and a glutamate (Glu-58) that appears to make a hydrogen bond with the bridging, solvent-derived oxygen atom. To directly interrogate whether this unique ligand environment alters the electronics of the diiron site, we determined the exchange coupling constant J of the mixed valent FBXL5 Hr to be 120 cm−1 (supplemental Fig. S5). This large value is suggestive of an oxo bridge, rather than a hydroxo bridge (J < 20 cm−1), as observed for mixed valent forms of the canonical Hr (30, 31) (supplemental Table S3). Hence, unlike Hr and other diiron enzymes, in which reduction from the diferric to the mixed valent state is accompanied by protonation of the single atom bridge, the bridge of FBXL5 is likely retained as an oxo ligand.
To determine whether this unusual arrangement was required for Hr function, FBXL5 Glu-58 was mutated to a histidine (E58H). As observed for the E58A variant, E58H FBXL5 was constitutively unstable following both FAC and DFO treatment (Fig. 3B). Likewise, when residue Glu-58 was mutated to aspartate (E58D) or glutamine (E58Q), FBXL5 was again unstable and unresponsive to iron (Fig. 2C). The failure of histidine or aspartate to functionally substitute for Glu-58 could be due to steric constraints. Modeling indicates that there is an uncommon, high energy His rotamer that could place the Nδ1 atom within 2.8 Å of the iron. However, a more common, low energy rotamer position for His-58 would place the Nϵ2 atom 3.7 Å from Fe1, too far away to function as an iron ligand. Similarly, aspartate at position 58 would place a carboxylate oxygen ∼2.6 Å from Fe1, which is still rather far for an iron ligand, whereas the second carboxylate oxygen is also too far from the μ-hydroxo atom to function as a hydrogen bond acceptor. Moreover, a glutamine at position 58 also fails to bind iron (data not shown) and restore proper iron responsiveness (Fig. 3B), although its carboxamide oxygen can on rare occasions ligate iron. Together, these data suggest that FBXL5 induction in response to increased iron bioavailability requires an intact Hr domain capable of assembling a diiron center with unusual ligands.
FBXL5 Is Polyubiquitinated and Degraded by Proteasome under Iron-deplete Conditions
Destabilization of FBXL5 upon iron depletion is dependent on the proteasome (12). To determine whether the proteasome sensitivity of FBXL5 is accompanied by polyubiquitination in an iron-dependent fashion, HEK 293T cells stably expressing FBXL5-FLAG were treated with either FAC or DFO in the absence or presence of MG132, and FBXL5-FLAG was immunoprecipitated. As shown in Fig. 4A, FBXL5 is polyubiquitinated under iron-deficient conditions. However, when iron is plentiful, the extent of the laddering characteristic of heterogeneous polyubiquitination is diminished (Fig. 4A). These data are consistent with bioavailable iron levels regulating FBXL5 via its polyubiquitination status.
Similar to full-length FBXL5, the isolated FBXL5 Hr domain is preferentially polyubiquitinated under iron-deficient conditions (Fig. 4B). To further confirm that ubiquitination is required for FBXL5 Hr degradation, a murine ts-20 BALB/c 3T3 fibroblast cell line featuring a temperature-sensitive E1 variant (36, 37) was transfected with a FLAG-Hr-HA construct. At the E1-permissive temperature (35 °C), FLAG-Hr-HA levels diminish upon a shift from iron-replete conditions to iron-deplete conditions (Fig. 4C). At the restrictive temperature (39 °C), loss of E1 activity globally compromises polyubiquitination and degradation of substrates by the ubiquitin-proteasome system (37, 38). Under these conditions, FLAG-Hr-HA protein accumulation levels remain high, even upon the addition of DFO (Fig. 4C). Likewise, a functional ubiquitin-proteasome system is required for FBXL5 Hr degradation under low O2 conditions (Fig. 4D).
As the F-box subunits of Skp1/Cul1/F-box (SCF) E3 ubiquitin ligase complexes can be subjected to autoubiquitination (39), we suspected that SCFFBXL5 might itself be the responsible Hr E3 ligase. However, the isolated Hr lacks a F-box domain and thus cannot assemble into an SCF E3 ligase complex, arguing against an autoubiquitination model. To exclude the possibility that SCFFBXL5 could ubiquitinate the FBXL5 Hr domain in trans, endogenous FBXL5 was depleted from HEK-Hr cells using an siRNA selectively targeting the full-length FBXL5 mRNA. Although FBXL5 knockdown was sufficient to stabilize IRP2, no effect on accumulation or ubiquitination status of the isolated Hr domain was observed (Fig. 4B). These data suggest that another, hitherto unidentified E3 ubiquitin ligase activity is required for FBXL5 regulation conferred through its Hr domain.
To map an element within the FBXL5 Hr domain that is required for targeting the protein for degradation, a series of progressively longer FBXL5 N-terminal truncation constructs was generated. Because such large truncations were expected to compromise assembly of the diiron center, and consequently Hr integrity, it was postulated that all deletions would produce iron-insensitive and constitutively unstable products. Although the Δ1–33, Δ1–59, and Δ1–76 proteins were predictably unstable, the Δ1–81 and Δ1–100 proteins accumulated to high levels following both FAC and DFO treatment (Fig. 3C), suggesting that there is a regulatory sequence required for FBXL5 degradation located C-terminal to residue Ile-76. To better delineate this sequence, we generated a series of FBXL5 constructs containing adjacent 5-amino acid deletions in the region spanning residues 67–91. Interestingly, deletion of residues 77–81 was sufficient to confer a substantial constitutive increase in FBXL5 accumulation (Fig. 4E) despite the fact that loss of His-80 should preclude any iron binding. We propose that these residues are a required part of a regulatory sequence mediating FBXL5 proteasomal degradation. Notably, this sequence resides in the unusual extended loop joining helices α2 and α3 (Fig. 2A and supplemental Fig. S3A).
Oxygen-dependent Regulation of FBXL5
IRP2 degradation is induced by both high iron and high oxygen availability (5, 6) and is proposed to be largely conferred through the effects of iron and oxygen on FBXL5 (40). Similar to ectopically expressed FBXL5 (12), endogenous FBXL5 accumulation in the presence of excess iron is attenuated in a proteasome-dependent manner when cells are incubated under low O2 conditions.6 The isolated FBXL5 Hr domain also directly responds to O2 (Fig. 4D), suggesting that oxygen availability affects the stability of FBXL5 via polyubiquitination and proteasomal degradation mediated by its Hr domain.
The majority of previously characterized hemerythrins have been shown to reversibly bind oxygen at the diiron center (34, 41, 42). Although FBXL5 and its Hr domain are clearly regulated in an oxygen-dependent manner, no ordered electron density corresponding to O2 was observed at the diiron center of the FBXL5 Hr crystal structure (Fig. 2B). Interestingly, neither FBXL5 Hr structures derived from crystals grown in the absence of reducing agent (Native form 2, supplemental Table S1) nor structures derived under anoxic conditions in the presence of excess dithionite reductant (Native form 3, supplemental Table S1) were appreciably different (supplemental Fig. S6A).
Closer examination of the FBXL5 Hr diiron center reveals that molecular oxygen would not be able to bind to the diiron center in these structures. When comparing FBXL5 Hr and the O2-binding DcrH Hr, the spatial conservation of the iron-ligating residues with respect to the diiron center is remarkably similar, although there are significant spatial variations in the hydrophobic amino acids lining the putative O2-binding pockets (Fig. 5A). In the FBXL5 Hr, Phe-123 is positioned toward the center of the helical bundle and in close proximity (∼4 Å) to the bridging μ-oxygen atom. Met-127 is also positioned closely (∼4 Å) to both the bridging oxygen atom and Fe2. In the DcrH Hr crystal structure (35), Trp-114, the corresponding hydrophobic residue to Phe-123, is near the periphery of the hydrophobic core adjacent to Fe1. The analogous residue to Met-127 in the DcrH Hr, Leu-115, is more distal (∼6 Å) to both Fe2 and the bridging oxygen. Consequently, the binding pocket in the DcrH Hr has much larger dimensions than the FBXL5 Hr site and can accommodate an O2 molecule. In fact, there is not even sufficient room to model a molecule of water solvent in the Fe2 coordination site. Thus, substantial rearrangements to the FBXL5 Hr core would be required for ligand binding.
FIGURE 5.
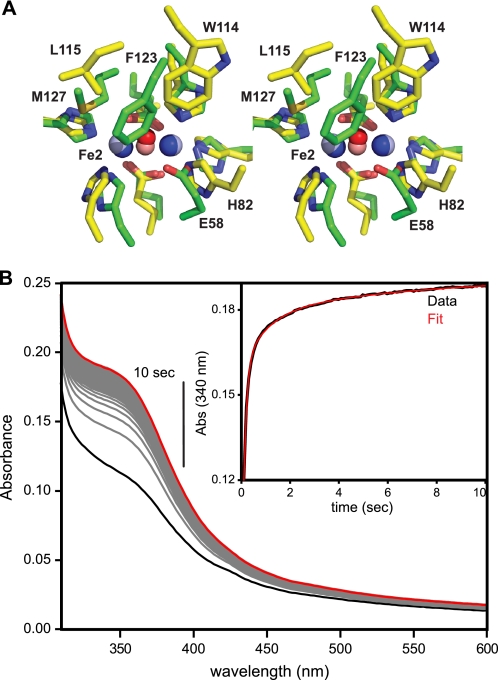
FBXL5 Hr domain does not appear to bind oxygen ligand. A, comparison of the FBXL5 and DcrH diiron centers. Shown in stereo is a superposition of the bound iron atoms (dark blue balls), bridging oxygen (dark red ball), and first coordination sphere iron ligands (green) of FBXL5 Hr (PDB code 3V5Y) with corresponding atoms (pale balls) and ligands (yellow) of deoxy-DcrH Hr (PDB code 2AWC). B, stopped-flow kinetics of reduced anaerobic FBXL5 Hr rapidly mixed with O2 saturated buffer. Fitting of the reoxidation of the Hr domain at 340 nm required three summed exponentials to adequately account for the observed data (inset), indicating a complex oxidation process. Abs, absorbance.
Rapid mixing of diferrous FBXL5 with saturating dioxygen results in the rapid reappearance (t½∼250 ms) of the Fe-oxo charge transfer band at 340 nm with no other transient intermediate accumulating prior to reoxidation to the diferric resting state (Fig. 5B). In particular, no oxygenated form of the domain, such as an end-on hydroperoxo moiety observed in Phascolopsis gouldii Hr (ϵ ∼2000 m−1cm−1 at 500 nm) or a μ-1,2-peroxo adduct (typically observable at ∼700 nm), can be observed following dioxygen addition. Thus, although the oxidation state of the FBXL5 Hr diiron center responds to O2 availability, its degron accessibility is not likely to be regulated by conformational changes stemming from reversible binding of O2.
DISCUSSION
Here we employed structural and molecular approaches to characterize the Hr domain that mediates regulation of the E3 ligase subunit, FBLX5. X-ray crystallography demonstrated that the isolated FBXL5 N terminus adopts a hemerythrin-like α-helical bundle fold stabilized by a diiron center. Nevertheless, this mammalian Hr contains several unique attributes that could contribute to its switch-like properties including 1) a required fifth helix contributing a residue in the secondary iron coordination shell, 2) a noncanonical primary iron ligand that also makes an apparent hydrogen bond contact with the bridging solvent-derived oxygen atom, 3) crowded side-chain packing within the presumptive O2-binding site, and 4) a truncated helix α3 preceded by several disordered amino acids and an extended loop that contains an important regulatory element.
Under iron- and oxygen-replete conditions, both FBXL5 and its isolated Hr are resistant to polyubiquitination and proteasomal degradation. Compromising formation of the diiron center, either through chelation of bioavailable iron or by mutation of any residue within the primary or secondary iron coordination spheres, leads to increased degradation of FBXL5. The requirement for E1 activity suggests that ubiquitination is required for FBXL5 degradation. Although the responsible E3 ligase has not yet been identified, residues 77–81 appear to act as part of a functional degron as deletion of this region stabilizes the protein, despite the inability of the Hr domain to bind iron upon removal of His-80. We propose that in the absence of iron binding, this regulatory sequence becomes fully accessible, promoting a large induction in the extent of polyubiquitination and proteasomal degradation.
Despite our efforts to crystallize both oxidized and fully reduced forms of FBXL5 Hr, the resulting structures do not display statistically significant differences in bond distances or angles of the diiron centers. Given the observed Fe1-Fe2 distances (supplemental Table S2) and preparation conditions, it is most likely that these structures all represent a mixed valent or diferric state. However, the observation of a putative hydrogen bond between Glu-58 and the bridging oxygen atom is difficult to reconcile if the FBXL5 is present in the met or semi-met state in the crystal. Under the working buffer conditions (pH 6.5 and 7.4), the probability that Glu-58 is protonated and serving as a hydrogen bond donor to a μ-oxo bridge is small, particularly as it is serving as an iron ligand. Only the reduced diferrous Hr could provide a requisite H-bond donating μ-hydroxyl group as both the diferric and the mixed valent semi-met states feature a μ-oxo bridge. As crystallization could trap a minor reduced species, or the oxidation state of the Hr could change during crystal manipulations (15), we cannot unambiguously assign the oxidation states of our crystals.
Among the Hr domain structures solved to date, the FBXL5 Hr is most structurally homologous to the bacterial DcrH Hr, an O2 sensor within a bacterial chemotaxis protein (35, 42). The DcrH Hr contains a putative substrate channel that is thought to facilitate diffusion of O2 to the diiron center. Oxygen binding to the DcrH Hr causes a subtle conformational change in an N-terminal loop, which may drive further conformational changes within the full-length DcrH protein to govern its aerotactic signaling function (35).
Instead of a channel, there is a large solvent-accessible surface near Fe1 of the FBXL5 Hr diiron center facilitating oxygen exposure (supplemental Fig. S7). However, FBXL5 does not have an analogous N-terminal loop and, based on the structures presented here, it would have to undergo a substantial conformational change displacing residues Phe-123 and Met-127 to accommodate an O2 ligand at the Fe2 site. Another potential hindrance to O2 binding the diiron center is residue Glu-58, which appears to engage the bridging oxygen in a hydrogen bond interaction. Moreover, no evidence for transient O2 binding was observed during reoxidation experiments or in the structures themselves, although oxygen was present throughout two of the three protein and crystal preparations. Alternatively, the O2 responsiveness observed for the isolated Hr domain in vitro and in cell culture studies could originate from electron transfer through outer sphere amino acid ligands.
Overall, the data presented here are consistent with a model in which the FBXL5 N-terminal Hr domain senses both iron and oxygen to regulate FBXL5 stability. When intracellular iron levels are abundant, the Hr domain binds iron and adopts a conformation in which a degron containing residues 77–81 is sequestered. Notably, in these crystal structures, the loop from residues 74–83 is not well ordered as evidenced by the high atomic displacement parameters for this region (supplemental Fig. S6B). This feature is unique to the FBXL5 Hr domain and overlaps a key regulatory element required for degradation. It is tempting to hypothesize that low oxygen conditions facilitate a physiologically relevant increase in accessibility of helix α3, predisposing the FBXL5 Hr domain to enhanced degradation.
Supplementary Material
Acknowledgments
The structure shown in this report is derived from work performed on beamlines 19-BM and 19-ID at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. UChicago Argonne, LLC, operates Argonne for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program (Grant C06 RR 15437-01) from the National Center for Research Resources, National Institutes of Health.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1–S7 and Tables S1–S5.
The atomic coordinates and structure factors (codes 3V5X, 3V5Y, and 3V5Z) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
S. Chollangi, J. W. Thompson, J. C. Ruiz, and R. K. Bruick, manuscript in preparation.
- IRP2
- iron regulatory protein 2
- FBXL5
- F-box and leucine-rich repeat protein 5
- Hr
- hemerythrin
- FAC
- ferric ammonium citrate
- DFO
- deferoxamine mesylate
- SCF
- Skp1/Cul1/F-box.
REFERENCES
- 1. Andrews N. C., Schmidt P. J. (2007) Iron homeostasis. Annu. Rev. Physiol. 69, 69–85 [DOI] [PubMed] [Google Scholar]
- 2. Hentze M. W., Muckenthaler M. U., Galy B., Camaschella C. (2010) Two to tango: regulation of mammalian iron metabolism. Cell 142, 24–38 [DOI] [PubMed] [Google Scholar]
- 3. Muckenthaler M. U., Galy B., Hentze M. W. (2008) Systemic iron homeostasis and the iron-responsive element/iron regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 28, 197–213 [DOI] [PubMed] [Google Scholar]
- 4. Rouault T. A. (2006) The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2, 406–414 [DOI] [PubMed] [Google Scholar]
- 5. Guo B., Phillips J. D., Yu Y., Leibold E. A. (1995) Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 270, 21645–21651 [DOI] [PubMed] [Google Scholar]
- 6. Hanson E. S., Rawlins M. L., Leibold E. A. (2003) Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 278, 40337–40342 [DOI] [PubMed] [Google Scholar]
- 7. Samaniego F., Chin J., Iwai K., Rouault T. A., Klausner R. D. (1994) Molecular characterization of a second iron-responsive element-binding protein, iron regulatory protein 2: Structure, function, and posttranslational regulation. J. Biol. Chem. 269, 30904–30910 [PubMed] [Google Scholar]
- 8. Galy B., Ferring D., Minana B., Bell O., Janser H. G., Muckenthaler M., Schümann K., Hentze M. W. (2005) Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood 106, 2580–2589 [DOI] [PubMed] [Google Scholar]
- 9. LaVaute T., Smith S., Cooperman S., Iwai K., Land W., Meyron-Holtz E., Drake S. K., Miller G., Abu-Asab M., Tsokos M., Switzer R., 3rd, Grinberg A., Love P., Tresser N., Rouault T. A. (2001) Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 27, 209–214 [DOI] [PubMed] [Google Scholar]
- 10. Meyron-Holtz E. G., Ghosh M. C., Iwai K., LaVaute T., Brazzolotto X., Berger U. V., Land W., Ollivierre-Wilson H., Grinberg A., Love P., Rouault T. A. (2004) Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 23, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cooperman S. S., Meyron-Holtz E. G., Olivierre-Wilson H., Ghosh M. C., McConnell J. P., Rouault T. A. (2005) Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron regulatory protein 2. Blood 106, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salahudeen A. A., Thompson J. W., Ruiz J. C., Ma H. W., Kinch L. N., Li Q., Grishin N. V., Bruick R. K. (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vashisht A. A., Zumbrennen K. B., Huang X., Powers D. N., Durazo A., Sun D., Bhaskaran N., Persson A., Uhlen M., Sangfelt O., Spruck C., Leibold E. A., Wohlschlegel J. A. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stenkamp R. E. (1994) Dioxygen and hemerythrin. Chem. Rev. 94, 715–726 [Google Scholar]
- 15. Carugo O., Djinoviç Carugo K. (2005) When X-rays modify the protein structure: radiation damage at work. Trends Biochem. Sci. 30, 213–219 [DOI] [PubMed] [Google Scholar]
- 16. Traverso M. E., Subramanian P., Davydov R., Hoffman B. M., Stemmler T. L., Rosenzweig A. C. (2010) Identification of a hemerythrin-like domain in a P1B-type transport ATPase. Biochemistry 49, 7060–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J. H., Kurtz D. M., Jr. (1992) Metal substitutions at the diiron sites of hemerythrin and myohemerythrin: contributions of divalent metals to stability of a four-helix bundle protein. Proc. Natl. Acad. Sci. U.S.A. 89, 7065–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Demuynck S., Li K. W., Van der Schors R., Dhainaut-Courtois N. (1993) Amino acid sequence of the small cadmium-binding protein (MP II) from Nereis diversicolor (annelida, polychaeta): evidence for a myohemerythrin structure. Eur. J. Biochem. 217, 151–156 [DOI] [PubMed] [Google Scholar]
- 19. Kurtz D. M., Jr. (1999) Oxygen-carrying proteins: three solutions to a common problem. Essays Biochem. 34, 85–100 [DOI] [PubMed] [Google Scholar]
- 20. Wirstam M., Lippard S. J., Friesner R. A. (2003) Reversible dioxygen binding to hemerythrin. J. Am. Chem. Soc. 125, 3980–3987 [DOI] [PubMed] [Google Scholar]
- 21. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 22. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z. (1991) in Proceedings of the CCP4 Study Weekend (Wolf W., Evans P. R., Leslie A. G. W. eds) pp. 80–86, Science and Engineering Research Council, Cambridge, UK [Google Scholar]
- 24. Cowtan K., Main P. (1998) Miscellaneous algorithms for density modification. Acta Crystallogr. D Biol. Crystallogr. 54, 487–493 [DOI] [PubMed] [Google Scholar]
- 25. Morris R. J., Zwart P. H., Cohen S., Fernandez F. J., Kakaris M., Kirillova O., Vonrhein C., Perrakis A., Lamzin V. S. (2004) Breaking good resolutions with ARP/wARP. J. Synchrotron Radiat. 11, 56–59 [DOI] [PubMed] [Google Scholar]
- 26. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 27. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 28. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Makris T. M., Chakrabarti M., Münck E., Lipscomb J. D. (2010) A family of diiron monooxygenases catalyzing amino acid β-hydroxylation in antibiotic biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 15391–15396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCormick J. M., Reem R. C., Solomon E. I. (1991) Chemical and spectroscopic studies of the mixed valent derivatives of the non-heme iron protein hemerythrin. J. Am. Chem. Soc. 113, 9066–9079 [Google Scholar]
- 31. Reem R. C., Solomon E. I. (1987) Spectroscopic studies of the binuclear ferrous active site of deoxyhemerythrin: coordination number and probable bridging ligands for the native and ligand-bound forms. J. Am. Chem. Soc. 109, 1216–1226 [Google Scholar]
- 32. Bearden J. A., Burr A. F. (1967) Reevaluation of x-ray atomic energy levels. Rev. Mod. Physics 39, 125–142 [Google Scholar]
- 33. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holmes M. A., Le Trong I., Turley S., Sieker L. C., Stenkamp R. E. (1991) Structures of deoxy and oxy hemerythrin at 2.0 Å resolution. J. Mol. Biol. 218, 583–593 [DOI] [PubMed] [Google Scholar]
- 35. Isaza C. E., Silaghi-Dumitrescu R., Iyer R. B., Kurtz D. M., Jr., Chan M. K. (2006) Structural basis for O2 sensing by the hemerythrin-like domain of a bacterial chemotaxis protein: substrate tunnel and fluxional N terminus. Biochemistry 45, 9023–9031 [DOI] [PubMed] [Google Scholar]
- 36. Jha K. K., Siniscalco M., Ozer H. L. (1980) Temperature-sensitive mutants of BALB/3T3 cells. III. Hybrids between ts2 and other mouse mutant cells affected in DNA synthesis and correction of ts2 defect by human X chromosome. Somatic Cell. Genet. 6, 603–614 [DOI] [PubMed] [Google Scholar]
- 37. Chowdary D. R., Dermody J. J., Jha K. K., Ozer H. L. (1994) Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14, 1997–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 39. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 40. Salahudeen A. A., Bruick R. K. (2009) Maintaining mammalian iron and oxygen homeostasis: sensors, regulation, and cross-talk. Ann. N.Y. Acad. Sci. 1177, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kao W. C., Wang V. C., Huang Y. C., Yu S. S., Chang T. C., Chan S. I. (2008) Isolation, purification, and characterization of hemerythrin from Methylococcus capsulatus (Bath). J. Inorg. Biochem. 102, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 42. Xiong J., Kurtz D. M., Jr., Ai J., Sanders-Loehr J. (2000) A hemerythrin-like domain in a bacterial chemotaxis protein. Biochemistry 39, 5117–5125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.