Background: p63DBD assembles into a tetramer upon binding to response elements.
Results: We determined crystal structures of p63DBD complexes with various DNAs.
Conclusion: Increased number of C/G base pairs abrogates the DNA superhelix. p63DBD tetramer binds to 1-bp overlapping response element.
Significance: p63DBD binding to superhelical DNAs suggests ability to bind within nucleosomes. Binding to overlapping response elements provides a mechanism for promoter specificity.
Keywords: Crystal Structure, p63, Protein-DNA Interaction, Surface Plasmon Resonance (SPR), X-ray Crystallography
Abstract
We show that changes in the nucleotide sequence alter the DNA conformation in the crystal structures of p63 DNA-binding domain (p63DBD) bound to its response element. The conformation of a 22-bp canonical response element containing an AT spacer between the two half-sites is unaltered compared with that containing a TA spacer, exhibiting superhelical trajectory. In contrast, a GC spacers abolishes the DNA superhelical trajectory and exhibits less bent DNA, suggesting that increased GC content accompanies increased double helix rigidity. A 19-bp DNA, representing an AT-rich response element with overlapping half-sites, maintains superhelical trajectory and reveals two interacting p63DBD dimers crossing one another at 120°. p63DBD binding assays to response elements of increasing length complement the structural studies. We propose that DNA deformation may affect promoter activity, that the ability of p63DBD to bind to superhelical DNA suggests that it is capable of binding to nucleosomes, and that overlapping response elements may provide a mechanism to distinguish between p63 and p53 promoters.
Introduction
Transcription factor p63 is a homologue of the p53 master tumor suppressor (1), yet these two transcription factors have distinct physiological roles. p53 is responsible for maintaining the genomic stability in somatic cells. Almost all cancers are related to point mutations in p53, most of which are located in the DNA-binding domain (DBD)2 (2). In contrast, p63 regulates development of epithelial tissues (3, 4), and point mutations in p63 are linked to genetic facial and limb syndromes (5). p63 is also involved in turmorigenesis of certain cancers, especially head and neck cancer, by overexpression of isoforms lacking the N-terminal transcription activation domain (6, 7).
p53 and p63 proteins have similar modular organization. Both contain an N-terminal transactivation domain followed by a DNA-binding domain and a tetramerization domain. p63 but not p53 contains an additional sterile α motif and a transcription inhibitory domain at the C terminus. The domains are linked by unstructured regions (8, 9). Point mutations, mostly in the p63DBD, are associated with ectrodactyly ectodermal dysplasia cleft syndrome (5). Mapping of the point mutations on the p63DBD-DNA crystal structure suggests that most point mutations destabilize the protein or impair protein-DNA interactions (10).
Numerous structures of p53DBD in complex with DNA response elements have been reported (11–16), and recently we have determined the structure of p63DBD in complexes with DNA (10). A canonical full binding site contains two tandem repeats of a 10-bp half-site with the sequence RRRCWWGYYY (where R = A or G; Y = T or C; and W = A or T), either continuously or separated by a spacer of up to 21 base pairs (17). The reported p53DBD-DNA structures contain CG-rich DNA sequences (11–15), whereas two p63DBD-DNA complexes determined in our laboratory contained AT-rich canonical DNA sequences, one with a 10-bp half-site and the second with a 22-bp (denoted 22-bp TA), including a 2-bp TA spacer (see Fig. 1A) (10). We selected these sequences based on the previous reports that p63 binds preferentially to AT-rich response elements (18, 19). p53DBD and p6DBD dimers bind to half-site response elements in the same manner, and key protein-DNA interactions are conserved. Moreover, the B-form DNA molecules assemble end-to-end along the crystal lattices, simulating continuous double strands. However, the DNA trajectories are different; in the p53DBD-DNA structures, the DNA adopts a straight trajectory (11, 13), whereas in both p63DBD-DNA complexes, the DNA mimics a superhelix conformation (10).
FIGURE 1.
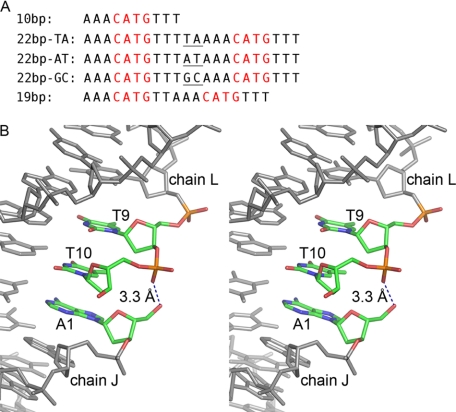
Response elements used in previous and current p63DBD-DNA crystallographic studies. A, from the top down: self-complimentary canonical 10-bp half-site (10), 22-bp canonical full response element with a 2-bp TA spacer (10), 22-bp canonical full response element with a 2-bp AT spacer, 22-bp canonical full response element with a 2-bp GC spacer, and 19-bp overlapping full response element. The CATG core motifs are colored in red, and the 2-bp spacers between the half-sites are marked with underlining. B, stereoscopic representation of the 10-bp DNA at the type III dimer-dimer junction (chains L and J of in the Protein Data Bank entry 3QYM). There is no room to insert a connecting phosphate between T10 of chain L and A1 of chain J, and the distance between T10 phosphate and A1 5′ hydroxyl group is 3.3 Å, suggesting that 19-bp overlapping response element can accommodate the type III p63DBD dimer-dimer interface.
DNA sequences and base pair steps affect DNA structures and the tendency of DNA molecules to deviate from ideal B-DNA conformation (20). Therefore, we hypothesized that the conformation of AT-rich DNA is more flexible, promoting the formation of superhelix (10). This ability of the DBD to bind deformed double-stranded DNA is relevant to all p53 family members because data have been accumulating showing that p53DBD binds to sites within the nucleosome, where the DNA adopts a superhelical conformation (21, 22).
The current study investigates further how modifying the sequences of response elements changes the DNA conformation in p63DBD-DNA structures. First, we examine changes in the 2-bp spacer sequence from TA to AT and GC (see Fig. 1A). Second, analysis of the p63DBD-10-bp structure (10) (Protein Data Bank code 3QYM) shows that the DNA superhelix mimicry may be attributed primarily to a pseudo unwinding of the DNA at the junction between the response elements half-sites, such that the first base of one response element half-site nearly stacks above the last base of the adjacent response element half-site. A unique p63DBD dimer-dimer interaction, referred to as the Type III tetramer (10), is associated with this arrangement where the corresponding p63DBD dimers orient at an angle of 120° to one another with only one dimer subunit interacting with its counterpart. This raised the question of whether crystal packing constraints produced this type of tetramer and the accompanied superhelical trajectory. Fortunately, we can address this issue because the same p63DBD-DNA arrangement can be designed by eliminating one of the stacked bases and using a continuous 19-bp full response element (see Fig. 1B), representing two half-sites that overlap by one base pair. The DNA sequence used in this complex eliminated the last T-A base pair of the first half-site (see Fig. 1A). Overlapping response elements half-sites have not been identified yet for p53; nevertheless they occur in whole genome sequences and have been described previously for the transcriptional regulator Cyp1p binding to its target site UAS1-B/CYC1 (23). Interestingly, the potential for an overlapping p63 response element was reported previously but has not been fully confirmed (24).
EXPERIMENTAL PROCEDURES
Protein Production, Crystallization, and Structure Determination
Recombinant p63DBD was produced in Escherichia coli and purified to homogeneity as described previously (10). The protein was concentrated to 30 mg/ml in 10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm DTT, and 10 μm zinc acetate and stored in −80 °C. Lyophilized powders of synthetic DNA molecules were purchased from Invitrogen, and DNA double strands were prepared by annealing. p63DBD was mixed with double-stranded DNA at 1:0.5 molar ratio with the final protein concentration at 0.25 mm. The crystals were obtained by the vapor diffusion method in hanging drops at room temperature with the reservoir solution containing 0.2 m ammonium acetate, 0.1 m Bis-Tris, pH 6.8, and 14% polyethylene glycol 3350 for the p63DBD-22-bp AT DNA sample; 0.2 m ammonium phosphate monobasic, 0.1 m Bis-Tris, pH 6.8, and 14% polyethylene glycol 3350 for the p63DBD-22-bp GC DNA sample; and 0.2 m ammonium formate, 0.1 m Bis-Tris, pH 6.8, and 12% polyethylene glycol 3350 for the p63DBD-19-bp DNA sample.
The crystals were transferred into mother liquor cryoprotected with 5% polyethylene glycol 400 and immersed in liquid nitrogen. X-ray diffraction data were collected at the synchrotron Beamline 23-ID, General Medical Sciences and National Cancer Institute Collaboration Access Team, the Advanced Photon Source, Argonne National Laboratory. The beamline was equipped with a MARmosaic MX-300 detector (Marresearch). The p63DBD crystals in complexes with 22-bp AT, 22-bp GC, and 19-bp DNA molecules diffracted x-rays to 2.5, 2.8, and 4.2 Å resolutions, respectively. The data were processed with the computer program XDS (25). Structure factors were calculated using TRUNCATE (26) as implemented in CCP4 (27). 5% randomly selected reflections were set aside for calculation of the Rfree value (28) for the structures with 22-bp AT and 19-bp DNA and 9% for the 22-bp GC DNA. All of the structures were determined by Molecular Replacement with the program PHASER (29). The search model for the two structures with 22-bp DNA was a protein dimer of the p63DBD-22-bp TA complex including the associated 10-bp DNA (10) (Protein Data Bank accession code 3QYN). The search yielded two solutions for the p63DBD-22-bp AT structure and one solution for the p63DBD-22-bp GC structure, consistent with an asymmetric unit that contains a tetramer for p63DBD-22-bp AT structure and a dimer for p63DBD-22-bp GC structure, respectively. Manual model rebuilding was performed with the interactive computer graphics program XTALVIEW (30). Structures were refined with CNS (31) and PHENIX (32).
The p63DBD-19-bp diffraction data were initially processed at a 4.0 Å resolution limit and space group C222. A p63DBD dimer in complex with 8-bp DNA derived from half-site of the p63DBD-22-bp GC structure by removing one base pair from each end served as the search model for molecular replacement. The search yielded two solutions with log likelihood gains of 188 and 397 and translation Z scores of 10.7 and 16.0, consistent with the asymmetric unit containing a p63DBD tetramer. Next, a p63DBD-10-bp model was aligned on the two molecular replacement solutions, revealing a 1-bp overlap. The overlapping base pair was removed, and the geometry of a continuous DNA was regularized. However, attempts to refine this structure failed because both Rfree and Rwork remained greater than 0.38. Because the unit cell dimensions were similar to those of the p63DBD-10-bp C2 crystal, the data were reprocessed using the lower symmetry C2 space group. Molecular replacement with the p63DBD-19-bp complex as the search model yielded two solutions, related by a 2-fold rotation about the c unit cell axis and a slight translation of ∼0.5 Å along the b unit cell axis. The refinement included pseudo-merohedral twining as implemented in PHENIX, for which the twin law is a 2-fold rotation about the c axis. The twinning fraction was calculated to be 0.5. Rigid body refinement of tetramers was carried out first to preserve protein/DNA interface. Temperature factors refinement grouped each p63DBD and DNA double strand. Because of the low resolution (4.2 Å), positional refinement was carried out with tight restraints to the original model. Finally, TLS refinement was carried out with each tetramer as a TLS group, yielding final values of Rwork = 0.326 and Rfree = 0.334 at 4.2 Å. It is worth noting that the sequences of the two DNA strands in the 19-bp DNA differ. The 10th base is an adenine in one strand and a thymine in the second strand. Because of the low resolution, the strand sequences were assigned arbitrarily.
Structure quality was assessed with the program PROCHECK (33). Superposition of structures was carried out with PYMOL (34) or LSQKAB (35) as implemented in the CCP4 suite. PYMOL was also used to generate figures. The DNA global axis and parameters was calculated using the program CURVES+ (36). Surface areas were calculated by AREAIMOL as implemented in CCP4 (27).
Surface Plasmon Resonance
SPR binding assays were performed as described previously (10), using a Biacore T100 optical biosensor (GE Health Sciences) with buffer modification to include Mg2+, which improved specific DNA binding affinities. The buffer comprised 25 mm Hepes, pH 7.6, 100 mm potassium acetate, 10 mm magnesium acetate, and 0.005% Surfactant P20 at 25 °C. Biotinylated double-stranded DNA molecules containing p63 binding sites or nonspecific reference DNA were immobilized on streptavidin-coated sensor chips as described (10).
Kinetic data were analyzed using the BIAevaluation software, version 3.1 (BIAcore). All of the binding curves were corrected for background and bulk refractive index contribution using the curves from nonspecific DNA flow cells. Because of fast dissociation phase rates (koff), the Kd values were determined using a steady state model.
RESULTS
The Overall Structures
Co-crystals of the p63DBD and double-stranded DNA molecules were obtained with two self-complementary 22-bp DNA molecules with the sequences shown in Fig. 1A and with a 19-bp in which the two half-sites overlap by including 5 bp instead of 6 bp between the two CATG cores (Fig. 1A). For brevity, the structures corresponding to these three complexes will be termed p63DBD-22-bp AT, p63DBD-22-bp GC, and p63DBD-19-bp in the following discussion. The p63DBD-22-bp AT and p63DBD-22-bp GC crystals were grown in different conditions because attempts to grow crystals under the same conditions failed, suggesting that from the outset, the DNA structures in the two complexes had different structural properties. The space groups of these two crystals are also different; the crystal of the p63DBD-22-bp AT complex belongs to space group P63 and is isomorphous with that of the previously reported p63DBD-22-bp TA crystal (10). The space group of the p63DBD-22-bp GC crystal is C2.
The two p63DBD-22-bp structures were refined to the resolution limits of 2.5 and 2.8 Å, respectively (Table 1). Both 22-bp DNA molecules assume B-form conformation and pack end-to-end throughout the crystal lattices despite the different crystal space groups. An asymmetric unit of the p63DBD-22-bp AT structure contains four p63DBD molecules and one double-stranded DNA corresponding to an entire biological unit, whereas that of the p63DBD-22-bp GC structure contains two p63DBD molecules and a single-stranded DNA, with the complete biological unit comprising the double-stranded DNA and two dimers of p63DBD generated by the crystallographic 2-fold symmetry operator (supplemental Fig. S1A). The DNA electron density of the p63DBD-22-bp GC structure is depicted in supplemental Fig. S1B, showing the integrity of the double helix.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for highest resolution shell.
| Crystals | p63DBD/22AT | p63DBD/22GC | P63DBD/19bp |
|---|---|---|---|
| DNA sequence | 5′AAACATGTTTATAAACATGTTT3′ | 5′AAACATGTTTGCAAACATGTTT3′ | 5′AAACATGTTAAACATGTTT3′ |
| Data collection | |||
| Space group | P63 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 141.8, 141.8, 119.6 | 85.0, 101.2, 71.5 | 119.7, 180.5, 98.2 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 122.8, 90.0 | 90.0, 90.05, 90.0 |
| Resolution (Å) | 59.8-2.5 (2.56-2.5) | 19.6-2.8 (2.87-2.8) | 20.3-4.2 (4.3-4.2) |
| Rsym | 0.082 (0.451) | 0.149 (0.443) | 0.112 (0.453) |
| I/σI | 19.9 (3.5) | 8.5 (1.8) | 7.8 (1.6) |
| Completeness (%) | 99.9 (100) | 97.5 (95.3) | 96.8 (94.6) |
| Redundancy | 12.8 (9.6) | 3.1 (2.8) | 2.8 (2.3) |
| Refinement | |||
| Resolution (Å) | 59.8-2.5 (2.55-2.5) | 19.6-2.8 (2.93-2.8) | 60.0-4.2 |
| No. reflections | 47224 (2745) | 12335 (1494) | 14781 |
| No. of p63DBD/dsDNA | 4/1 | 2/0.5 | 8/2 |
| Rwork | 0.206 (0.385) | 0.201 (0.345) | 0.326 |
| Rfree | 0.239 (0.417) | 0.234 (0.345) | 0.334 |
| No. atoms | |||
| Protein | 5990 | 2914 | 11656 |
| DNA | 896 | 448 | 1546 |
| Zn2+ | 4 | 2 | 8 |
| Water | 394 | 174 | 0 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.011 | 0.013 | 0.018 |
| Bond angles (°) | 1.6 | 1.7 | 1.7 |
Interdimer contacts between two biological units in both structures are generated because of crystal packing, and they differ because of different DNA trajectories. For the p63DBD-22-bp AT crystal packing, two adjacent dimers orient parallel to generate the type II dimer-dimer interaction described for both p53DBD and p63DBD (10–13). For the p63DBD-22-bp GC, two dimers across biological units intersect one another at an angle of 120°, analogous to the type III assembly observed in p63DBD-10-bp DNA structure described previously (10).
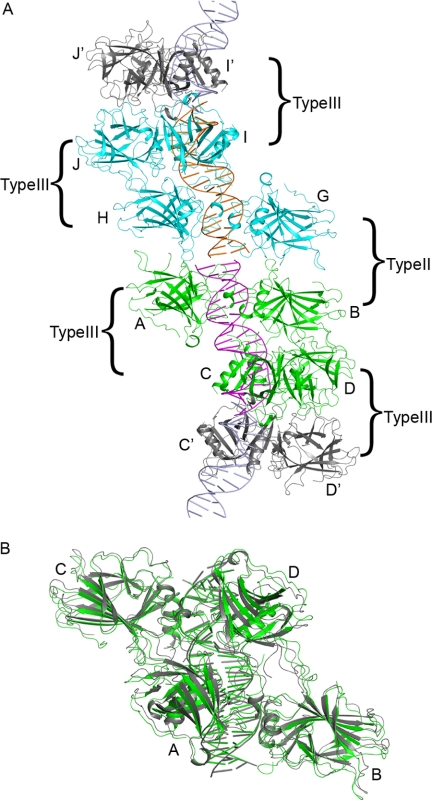
The p63DBD-19-bp crystal diffracted to a low resolution of 4.2 Å, prohibiting detailed structure analysis. Thus, only the gross protein/DNA mode of association and the packing of the molecules in the crystal unit cell can be reliably ascertained. The p63DBD-19-bp and p63DBD-10-bp crystals are similar (space group C2) but not isomorphous, and the former crystals grow with pseudo-merohedral twinning resembling the higher symmetry C222 space group. The asymmetric unit of the p63DBD-19-bp crystal contains two identical biological units, each of which comprises one 19-bp double-stranded DNA molecule and two p63DBD dimers. Because the sequences of the two DNA strands were not self-complementary (the 10th base is an adenine in one strand and a thymine in the other), two difference Fourier electron density maps accounting for the two possible orientations of the DNA double strand within the crystal were calculated, showing no discrimination between the two. Therefore, the two DNA strands were assigned arbitrarily. The two p63DBD dimers in the biological unit form a type III interface analogous to that observed between two p63DBD-10-bp complexes. Crystal packing generates a type II dimer-dimer interface and a type III dimer-dimer interface across the biological units (Fig. 2A). The two Type III tetramers of the p63DBD-19-bp and p63DBD-10-bp structures superpose well (Fig. 2B), and both DNAs exhibit continuous trajectory along the crystal lattice with intertwined superhelices (10). Therefore, the structure establishes the feasibility of formation of a type III dimer-dimer interface in the context of a full-length response element superhelix and shows that this arrangement of p63DBD dimers does not require a break between two response element half-sites.
FIGURE 2.
Structure of p63DBD-19-bp. A, an asymmetric unit of the C2 space group crystal includes two biological units, each consisting of 19-bp DNA (colored magenta and orange) and a type III p63DBD tetramer (10), labeled A–D (colored green) and G–J (colored cyan). Among the two tetramers, chain B and chain H interact with chain C and I, respectively. The dimers A/B and G/H form a type II interface between the two tetramers. Crystal packing generates type III tetramers between dimers C/D or I/J with their counterparts in the neighboring asymmetric unit, C′/D′ and I′/J′, respectively. The protomers D and J interact with their counterparts D′ and J′, respectively, whereas protomers C and I do not interact with their counterparts. B, superposition of two type III p63DBD tetramers: one in complex with the 19-bp DNA (green) and the other in complex with two 10-bp DNA half-sites reported previously (gray) (10). The structures are aligned based on the protein tetramer backbone, yielding a root mean square deviation value of 1.7 Å.
p63DBD-22-bp GC and p63DBD-22-bp AT Structures Exhibit Different Interdimer Orientations
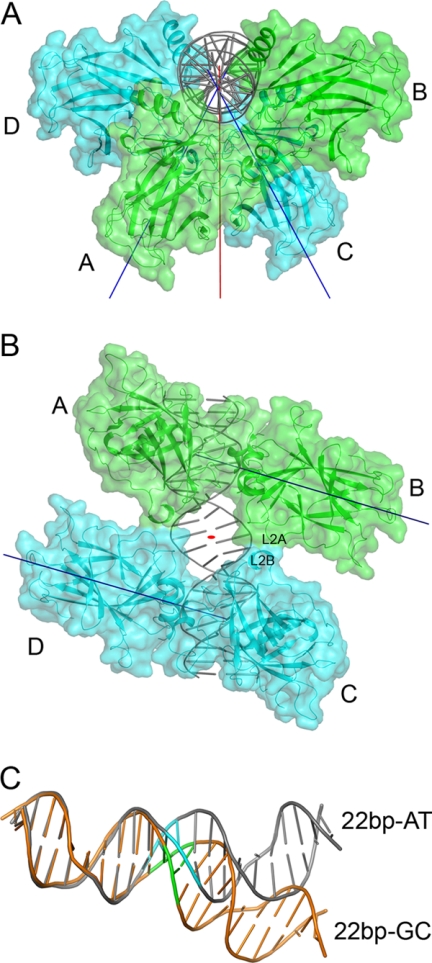
The structure of p63DBD-22-bp AT (supplemental Fig. S2A) is isomorphous with that of the reported p63DBD-22-bp TA (10). In both structures, the two dimers do not contact one another. There are only minor structural differences caused by the different 2-bp spacer sequences (supplemental Fig. S2B). In contrast, the p63DBD-22-bp GC exhibits architecture markedly different from that of p63DBD-22-bp AT and p63DBD-22-bp TA complexes as a consequence of its different DNA conformation, which is described later. The two p63DBD dimers orient at 56° to one another (Fig. 3A) rather than at 48° as seen in the complexes with response elements containing AT and TA spacers. Both angles deviate substantially from the 72° expected for a twist angle of a 2-bp spacer in an ideal DNA double helix because of bending of the 22-bp GC DNA (see below). As with the p63DBD-22-bp TA complex (10), there is no contact between the two protein dimers in the p63DBD-22-bp TA complex, whereas a minor interdimer interface of 130 Å2 embedded surface area is formed in the p63DBD-22-bp GC complex because of the DNA bending, consisting of two identical patches (Fig. 3B). Each patch involves the L2A loop (residues 193–204) of one molecule and the L2B loop (residues 211–225) of another as indicated in Fig. 3B. SPR experiments using 20-bp and spacer-containing 22-bp response elements showed that p63DBD binding to the response element in solution is stronger in the presence of interdimer interaction (Ref. 10 and this work). As discuss below, the SPR experiments revealed no significant improvement of specific binding to the 22-bp GC sequence over nonspecific binding, indicating that the contribution of 130 Å2 embedded surface area to binding affinity is small.
FIGURE 3.
Structure of p63DBD-22-bp GC. A, overall structure of the biological unit viewed down the DNA axis, showing the two dimers that are oriented at 56°S to one another. The protein molecules are shown in cartoon and surface models, and the DNA is shown in a cartoon model. The T-axis is colored in red, and the D-axis is in blue (10). B, viewed down the T-axis. The small interdimer interface revealed by surface representation consists of two identical patches related by a crystallographic 2-fold symmetry: one between chain A and chain D and another between chain B and chain C. C, superposition of 10-bp half-sites of 22-bp AT and 22-bp GC depicts the deviation in the trajectories of the two DNA molecules. The superposed half-sites have a root mean square deviation value of 0.7 Å. Superposition using two half-sites yields a root mean square deviation value of 3.4 Å. The 22-bp GC DNA is colored orange with the GC spacer in red, and the 22-bp AT DNA is colored gray with the AT spacer in cyan.
The 22-bp GC and 22-bp AT DNA Molecules Assume Different Conformations
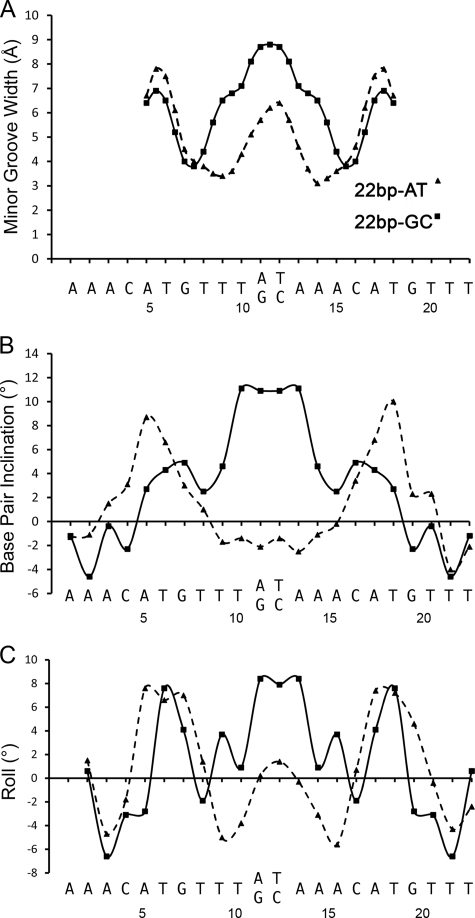
The DNA trajectories of the 22-bp AT (10) and 22-bp TA along the crystal lattices adopt superhelical conformation. With the GC 2-bp spacer, the crystal lattice also generates continuous DNA trajectory; however, it does not form a superhelix (supplemental Fig. S1A). The difference between the two DNA conformations is dramatic, as depicted in Fig. 3C. The superposition of the 22-bp AT and 22-bp GC half-sites shows that the deviation between the two DNAs begins close to the spacer region. Structure analyses using CURVES+ (36) shows that the 22-bp GC DNA axis bends by a total of 14.5°, whereas that of the 22-bp AT DNA bends by 22.2°. The inter-base pair parameters of the 22-bp GC and 22-bp AT DNA molecules are summarized in Table 2. The width of the minor groove, the inclination of base pairs, and the roll at the spacer regions of the two DNA structures exhibit the most prominent differences (Fig. 4).
TABLE 2.
Interbase pair DNA parameters of 22-bp GC and 22-bp AT
The parameters were calculated with the computer program Curve. The parameters corresponding to the spacer base pairs are highlighted in bold.
| Number | Base | Shift |
Slide |
Rise |
Tilt |
Roll |
Twist |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GC | AT | GC | AT | GC | AT | GC | AT | GC | AT | GC | AT | ||
| 2 | A | −0.29 | 0.21 | 0.1 | 0.22 | 3.07 | 3.24 | −7.2 | −0.5 | 0.6 | 1.5 | 42.2 | 39.6 |
| 3 | A | 0.05 | 0.36 | −0.38 | −0.19 | 3.34 | 3.35 | 0 | 0.9 | −6.6 | −4.7 | 35.9 | 34.6 |
| 4 | C | 0.3 | 0.63 | −0.98 | −1.19 | 3.42 | 3.35 | −3.5 | 0.8 | −3.1 | −1.8 | 33.3 | 33.1 |
| 5 | A | 0.79 | 1.15 | 0.25 | 0.24 | 3.21 | 3.52 | 4.8 | 5.2 | −2.8 | 7.6 | 41.9 | 43 |
| 6 | T | 0.02 | −0.52 | −0.35 | 0.06 | 3.09 | 3.04 | 1 | −2.2 | 7.6 | 6.6 | 24.2 | 23 |
| 7 | G | −0.48 | −0.78 | 0.72 | 0.66 | 3.45 | 3.32 | −2.1 | −2.3 | 4.1 | 7 | 43.3 | 38.3 |
| 8 | T | −0.54 | −0.34 | −1.18 | −0.84 | 3.38 | 3.32 | −4.1 | −0.7 | −1.9 | 1.4 | 32.6 | 37.6 |
| 9 | T | −0.56 | −0.31 | 0.08 | −0.63 | 3.39 | 3.25 | −1.9 | −2 | 3.7 | −5 | 35.2 | 31.6 |
| 10 | T | 0.11 | −0.01 | 0.07 | −0.53 | 3.05 | 3.19 | 2.2 | 0.6 | 0.9 | −3.8 | 37.1 | 37.3 |
| 11 | G/A | 0.11 | 0.16 | 0.4 | −0.27 | 3.2 | 3.18 | −1.8 | −0.9 | 8.4 | 0.2 | 31.6 | 34.4 |
| 12 | C/T | 0 | −0.57 | −0.45 | −0.67 | 3.18 | 3.0 | 0 | 0.9 | 7.9 | 1.4 | 25.4 | 32.2 |
| 13 | A | −0.11 | 0.1 | 0.4 | −0.25 | 3.2 | 3.4 | 1.8 | −0.5 | 8.4 | −0.3 | 31.6 | 35.6 |
| 14 | A | −0.11 | −0.21 | 0.07 | −0.29 | 3.05 | 3.12 | −2.2 | 0.7 | 0.9 | −3.1 | 37.1 | 35 |
| 15 | A | 0.56 | 0.44 | 0.08 | −0.18 | 3.39 | 3.31 | 1.9 | −0.5 | 3.7 | −5.6 | 35.2 | 34.2 |
| 16 | C | 0.54 | 0.42 | −1.18 | −0.89 | 3.38 | 3.28 | 4.1 | 2.1 | −1.9 | 0.7 | 32.6 | 37.9 |
| 17 | A | 0.48 | 0.69 | 0.72 | 0.61 | 3.45 | 3.37 | 2.1 | 2.4 | 4.1 | 7.4 | 43.3 | 38.4 |
| 18 | T | −0.02 | 0.35 | −0.35 | −0.06 | 3.09 | 2.94 | −1 | 3.1 | 7.6 | 7.2 | 24.2 | 21.5 |
| 19 | G | −0.79 | −0.65 | 0.25 | 0.12 | 3.21 | 3.61 | −4.8 | −6.4 | −2.8 | 4.6 | 41.9 | 42.6 |
| 20 | T | −0.3 | −0.53 | −0.98 | −0.91 | 3.42 | 3.35 | 3.6 | 2 | −3.1 | −0.4 | 33.3 | 33.4 |
| 21 | T | −0.05 | −0.51 | −0.38 | −0.15 | 3.34 | 3.37 | 0 | −3.3 | −6.6 | −4.3 | 35.9 | 36.7 |
| 22 | T | 0.29 | −0.43 | 0.1 | 0.41 | 3.07 | 3.15 | 7.2 | 4.8 | 0.6 | −2.4 | 42.2 | 42.1 |
| Average | 0.00 | −0.02 | −0.14 | −0.23 | 3.26 | 3.27 | 0.00 | 0.2 | 1.4 | 0.7 | 35.2 | 35.3 | |
| Standard | 0.41 | 0.52 | 0.57 | 0.51 | 0.15 | 0.16 | 3.5 | 2.7 | 4.9 | 4.5 | 6.0 | 5.4 | |
FIGURE 4.
Comparison of the DNA parameters of the 22-bp AT and 22-bp GC DNA molecules. The DNA parameters are plotted against the DNA sequences. A, the width of the minor grooves. B, base pair inclinations. C, base pair rolls. These panels highlight the different shapes of the two 22-bp DNA molecules. The GC spacer exhibits wider minor groove and more inclination and roll compared with the AT spacer.
Crystal Packing Reveals Type II and Type III Interface Interactions
The p63DBD-22-bp GC crystal packing generates a type III interface across asymmetric units, and that of the p63DBD-22-bp AT and p63DBD-22-bp TA generates a type II interface. These structures were determined at higher resolution than those of the p63DBD-10-bp and p63DBD-19-bp complexes, and thus they allow a more detailed comparison of the types II and III interfaces even though these interfaces correspond to a dimer-dimer interaction across rather than within a biological unit. The type III interface involves one p63DBD molecule from each dimer and embeds 1370 Å2 surface area (supplemental Fig. S3). The assemblies of DBD tetramers are similar, with the two dimers oriented at 120° to one another. At the interface, direct protein/protein contacts include only van der Waals interactions and lack hydrogen bonds or salt bridges. There are also four internal water molecules that form a network of hydrogen bonds by interacting with the amino groups of the two symmetry-related Lys168.
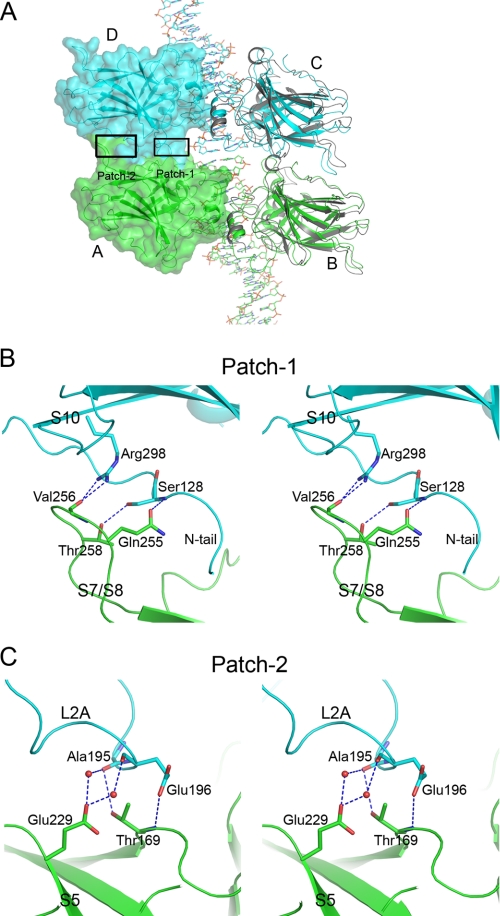
The p63DBD-22-bp AT crystal packing generates a type II interface that embeds 1770 Å2 surface area. The p63 type II interface involves loop regions analogous to those seen in p53DBD-DNA structures; however, the specific interactions differ as they do between different p53DBD-DNA complexes that exhibit type II tetramers (11, 12). The four p63DBD monomers are labeled A–D in Fig. 5A. The p63DBD type II dimer-dimer interface consists of two identical protein/protein contact regions, one involving molecules A and D and the other involving molecules B and C. Each contact contains two patches (labeled Patch-1 and -2 in Fig. 5A) involving networks of hydrogen bonds. Patch-1 is proximal to the DNA and involves loop S7/S8 (residues 251–260) of one molecule and the N terminus (residues 127–137) of the partner molecule (Fig. 5B). The main chain carbonyl group of Val256 forms bifurcated hydrogen bonds with the guanidinium group of Arg298. Side chain hydroxyl groups of Ser128 and Thr258 are hydrogen-bonded. Gln255 forms a hydrogen bond with the main chain amide group of Ser128. Patch-2 is distal to the DNA and involves loop L2A (residues 193–204) and loop S5/S6 (residues 230–234) (Fig. 5C). Glu229 side chain forms two hydrogen bonds with two water molecules, which in turn interact with the main chain carbonyl oxygen atoms of Ala195 and Glu196. The main chain carbonyl oxygen atom of Ala195 also forms a hydrogen bond with the hydroxyl group of Thr169. The carboxyl group of Glu196 forms hydrogen bond with the main chain amide group of Thr169. The comparison between the two modes of p63DBD dimer-dimer interactions suggests that the type II mode enhances tetramerization more than the type III does because the former involves larger surface areas and more specific interactions.
FIGURE 5.
The type II interface of p63DBD dimers across biological units in p63DBD-22-bp AT structure. A, the interface in the context of the overall structure. The A and B p63DBD molecules correspond to a dimer in one crystallographic asymmetric unit, and the C and D molecules correspond to a dimer in the adjacent asymmetric unit. The interface contains two identical protein/protein contacts, one between molecules A and D and the other between molecules B and C. Each of the interfaces consists of two patches, labeled Patch-1 and Patch-2. A surface representation of molecules A and D shows the intermolecular contact. A p53DBD type II monomer-monomer association (Protein Data Bank entry code 3KZ8) is shown (in gray) by superpositioning the molecules on the p63DBD molecules B and C, yielding a root mean square deviation value of 1.7 Å for backbone atoms. B and C, stereoscopic representation of the hydrogen bond networks of Patch-1 and Patch-2, respectively. Chain A is colored in green, and chain D is colored in cyan as in A.
Binding Affinities
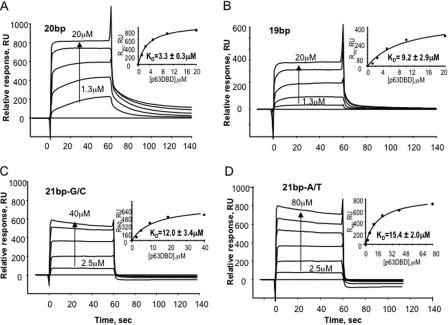
Previous p63DBD-DNA binding studies using SPR with immobilized biotinylated response element DNAs and protein analyte established that the binding is highly cooperative, with a Kd value of 11.7 μm for the 20-bp response element and a Kd value that was too weak to be measured for a 22-bp response element containing the TA spacer, i.e. within or above the 100 μm range (10). In the current study, we used the same approach, except that the buffer was modified to include Mg2+, which improved the p63DBD binding affinity to the 20-bp response element and decreased the Kd value from 11.7 to 3.3 ± 0.3 μm. The p63DBD binding affinities to both 22-bp response elements containing 2-bp spacers were still indistinguishable from that of a nonspecific 22-bp DNA. Using the same buffer, the Kd for the p63DBD-19-bp interaction, corresponding to formation of a type III dimer-dimer interface, was 9.2 ± 2.9 μm (Fig. 6). The lower p63DBD-19-bp binding affinity compared with the protein affinity toward a 20-bp response element is consistent with the 1370 Å2 embedded surface area of the type III interface, which is smaller than the 1770 Å2 embedded surface area of the type II interface.
FIGURE 6.
SPR assays of p63DBD binding to 20-bp, 19-bp, and 1-bp spacer containing 21bp response elements. The sensorgrams were corrected for background and bulk refractive index contribution using a nonspecific DNA as the reference. The Kd values were determined by using a steady state model, where the response signal at equilibrium was fitted as a function of protein concentration (shown in the insets). The low and high p63DBD concentrations are indicated on the sensorgrams. Intermediate concentrations correspond to 2-fold dilutions. A, 20-bp. B, 19-bp. C, G/C 1-bp spacer. D, A/T 1-bp spacer.
To further investigate the relationship of the interdimer interface and the length of the spacer, we tested two 21-bp DNA molecules: one with an A/T base pair spacer and the other with a G/C base pair spacer. The p63DBD-21-bp DNA binding affinities were weaker than that with 20-bp, exhibiting Kd = 15.3 ± 2.0 μm for the 21-bp A/T DNA and Kd = 12.0 ± 3.4 μm for the 21bp G/C DNA (Fig. 6). Taken together, the SPR experiments with 21-bp response elements suggest that a single base pair spacer allows formation of a substantial p63DBD dimer-dimer interface, presumably because of the bending capacity of the DNA. Note, however, that the presence of the p63 tetramerization domain enhances binding affinities (19), as discussed below.
DISCUSSION
Relationship between Conformation and Sequence of the 22-bp DNA Molecules
The impact of nucleotide sequence and the base pair steps on conformation of unbound DNA is well documented (37, 38). The structural differences between the two 22-bp DNA molecules are obviously due to the different spacers. However, it remains unclear whether the difference is induced by p63DBD binding because the structures of the unbound DNA molecules are not available. We previously proposed that the A/T-rich DNA is more elastic and tends to bend, based on the superhelical conformation of two p63DBD-DNA crystal structures (10). In the current study, we examined whether the change of spacer sequence alters the DNA conformation. Substitution of the TA by AT preserved the A/T content and accordingly did not change the DNA conformation or superhelical packing. In contrast, substitution by a GC spacer abolished the superhelical DNA trajectory and yielded DNA that packed in a straight pattern along the crystal lattice. Within the functional unit, the axis of the 22-bp GC DNA is bent, with the largest departures from ideal double helix conformation in the vicinity of the spacer region. Interestingly, deviations from ideal double helix are also present in the vicinity of the response element cores of all reported p53DBD and p63DBD complexes. In these cases, the contribution of the G/C geometrical preferences cannot be separated from the contribution of the protein-DNA interactions. The ability of p63DBD to bind to perturbed B-DNA, in particular to a superhelix, is functionally important because it implies that the protein may bind to DNA packed in nucleosomes, as reported recently for p53 (21).
Overlapping Half Sites as a Determinant for p63 Specificity
The 19-bp p63 response element contains two half-sites with one overlapping base pair, such that the two CATG cores flank 5 bp instead of the 6 bp characteristic of two half-sites lined up continuously. Based on previous structural studies (10, 14, 15), the protein-DNA interactions of p53 family members are quite conserved. A characteristic recognition feature is a bidentate hydrogen bond between the guanidinium group of a conserved arginine (Arg311 in p63 and Arg280 in human p53) and the guanine base in the CATG core, an invariant guanine in the consensus motif of the response elements. The requirement to form simultaneously four such interactions in a full response element, together with the DBD molecular dimension, defines the minimal DNA length between the two response element cores. DNA bending and formation of the type III p63DBD dimer-dimer interface allow the a protein tetramer to fit on the 19-bp DNA, where the loops S7/S8 (residues 251–260), L2B (residues 211–225), and S5/S6 (residues 230–234) interdigitate to prevent steric collisions (10). The interactions within this interface of 1370 Å2 embedded surface area not as extensive as those formed by the type II interface, consistent with the SPR binding assays. Nevertheless, the full-length p63 contains the tetramerization domain, which mediates the oligomerization of the functional unit and enhances the cooperative binding affinity (19).
Modeling of p53DBD in the context of a type III dimer-dimer assembly showed abrogated interface because of the difference in the length and sequence of the p53DBD L2B loop, thus providing a structural basis for a p63 specificity determinant (10). Therefore, in the presence of the tetramerization domain, the p63DBD unique type III dimer-dimer assembly may provide a mechanism to discriminate between p53 and p63.
A number of p53 studies determined that the DBD binds to a consensus motif comprising two repeats of RRRCWWGYYY (R = A or G; Y = T or C; and W = A or T) separated by up to 21 base pairs and never reported less than six base pairs between the two CWWG cores (17, 39, 40). Additionally, this minimal number of base pairs is consistent with more than 100 confirmed p53 response elements. The consensus motif of p63 response elements is less well understood; nevertheless, a genomic study suggested a consensus motif similar to that of p53 (41). To our knowledge, only one p63 study implicates a response element with overlapping half-sites in the EVPL (Envoplakin) promoter (24). The EVPL protein is highly expressed in epithelial cells of skin and pharynx in p63+/+ mice, and its presence is undetectable in the tissues of p63−/− mice. The EVPL promoter contains two response elements for p63 and p53, RE1 and RE2, in which the RE1 is activated by both p53 and p63, whereas the RE2 is p63-specific. The RE2 contains three cores, of which the second core confers p63 specificity (24). There are five base pairs between the first and second cores, corresponding to overlapping half-sites, and six base pairs between the second and third cores, corresponding to two continuous half-sites. Because all three half-sites are consistent with the consensus motif of p53 and p63, the p63 specificity determinant is likely the two overlapping half-sites. Such 19-bp DNA allows accommodation of the type III dimer-dimer assembly and suggests a possible mechanism for p63 promoter specificity. This mechanism needs to be validated by mutating the third RE2 core to confirm that p63 can still recognize the response element.
Crystal Contact Effects and Biological Relevance
The DNA and protein concentrations used in the crystallization were over ∼20-fold higher than the Kd values, and in addition, the crystal packing forces may also enhance the p63DBD-DNA binding affinity. Nevertheless the p63-DNA interactions are definitely specific, and it is their robustness that facilitates crystal growth. We have now determined five p63DBD-DNA crystal structures using five different DNA sequences, which crystallized in three different crystal forms, yet the interactions of the p63DBD with the DNAs are always the same. First, the p63DBD interactions with the response element core (CATG) are conserved in all crystal structures, and these interactions are very similar to those of p53DBD with its response element. This would not be expected from a nonspecific interaction. Second, the p63DBD intradimer interactions are also the same and involve the same protein region as that mediating the p53DBD intradimer interactions. Third, only two p63DBD dimer-dimer modes of interaction have been observed: The type II mode seen also in several p53DBD-DNA complexes and the p63DBD unique type III. This is not surprising, because complementary protein surfaces that embed as extensive surface areas as observed in the types II and III dimer-dimer interfaces (>1700 and >1300 Å2, respectively) do not form accidentally. This is well documented. For example, a survey of 78 crystal structures showed that relative to physiological protein-protein interfaces, crystal contact surfaces were generally quite small; 45% had a surface less than 100 Å2, and only 8% were larger than 500 Å2 (42).
The continuous DNA trajectories seen in the response element complexes with both p63DBD and p53DBD are mediated by base pair stacking of adjacent DNA molecules, which is an excellent example of biologically important interactions that facilitate crystal growth. The collection of structures show clearly the sequence/conformation relationship of the response elements used to obtain the crystals of the p63DBD and p53DBD complexes. The cores of the response elements, CATG, are the same, but the remaining DNA nucleotides used to obtain the p53DBD crystals were rich in cytosines and guanines. In contrast, the corresponding response element sequences used to obtain the crystals of the p63DBD complexes were exclusively adenines and thymines, except for the sequence that included a GC spacer intended to test our hypothesis about DNA flexibility. Accordingly, p53-DNA complexes exhibit a straight DNA double helix trajectory, and the p63-DNA complexes exhibit a DNA superhelical trajectory, except for the sequence that contain a GC spacer, which resulted in abolishing the superhelix.
The DBD of p53 family members binds DNA, whereas the tetramerization domain does not (43). The tetramerization domain is linked to the DBDs by long unstructured peptides. Therefore, it is unlikely to affect the DNA conformation. Nevertheless, the tetramerization domain contributes to the DNA binding affinity, presumably by increasing the local concentration of the DBDs (avidity effect). For p53, it has been shown that mutations that impair oligomerization reduce p53-DNA affinity (44), whereas such studies have not been reported for p63. Klein et al. (45) showed that the binding affinity of p53DBD alone toward its response element is stronger than that of p63DBD. These investigators detected specific p53DBD-DNA binding using both gel shift assays and fluorescence correlation spectroscopy but did not detect p63DBD-DNA binding by these methods, suggesting that the binding affinity of p63DBD alone toward the response element is greater than ∼1 μm. Consistently, we did not detect p63DBD-DNA binding by using nitrocellulose filter DNA binding assay but could determine Kd values at the μm range by the more sensitive SPR method (Ref. 10 and this work). In contrast, Klein et al. showed that p63DBD fused to glutathione S-transferase-formed dimers in solution, which dramatically enhanced binding to response elements and yielded apparent dissociation constants comparable with those of p53DBD, i.e. in the nanomolar range.
The ability of both p63DBD and p53DBD to interact with a superhelical DNA is biologically important in light of the recent publications by Prives's group (21) and Zhurkin's group (22) reporting that p53 binds to intact nucleosomes. Clearly, the cellular DNA superhelical conformation is determined by the DNA interactions with the histones, whereas it is the A/T-enriched composition of the response element that apparently confers the tendency to form the DNA superhelical trajectory in the crystals. Nevertheless, our studies suggest that there is no intrinsic requirement for the DNA to relax into a straight conformation prior to either p63 or p53 transcription factors binding.
Supplementary Material
Acknowledgments
We are grateful to the staff at Advanced Photon Source of the General Medical Sciences and National Cancer Institute Collaboration Access Team for help with data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM087922. This work was also supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38.

This article contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3US0, 3US1, and 3US2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- DBD
- DNA-binding domain
- SPR
- surface plasmon resonance
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxy- methyl)propane-1,3-diol.
REFERENCES
- 1. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 2. Vousden K. H., Lane D. P. (2007) p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8, 275–283 [DOI] [PubMed] [Google Scholar]
- 3. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 [DOI] [PubMed] [Google Scholar]
- 4. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 [DOI] [PubMed] [Google Scholar]
- 5. Rinne T., Brunner H. G., van Bokhoven H. (2007) p63-associated disorders. Cell Cycle 6, 262–268 [DOI] [PubMed] [Google Scholar]
- 6. Candi E., Dinsdale D., Rufini A., Salomoni P., Knight R. A., Mueller M., Krammer P. H., Melino G. (2007) TAp63 and ΔNp63 in cancer and epidermal development. Cell Cycle 6, 274–285 [DOI] [PubMed] [Google Scholar]
- 7. Flores E. R. (2007) The roles of p63 in cancer. Cell Cycle 6, 300–304 [DOI] [PubMed] [Google Scholar]
- 8. Joerger A. C., Fersht A. R. (2008) Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 [DOI] [PubMed] [Google Scholar]
- 9. Wells M., Tidow H., Rutherford T. J., Markwick P., Jensen M. R., Mylonas E., Svergun D. I., Blackledge M., Fersht A. R. (2008) Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 105, 5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen C., Gorlatova N., Kelman Z., Herzberg O. (2011) Structures of p63 DNA binding domain in complexes with half-site and with spacer-containing full response elements. Proc. Natl. Acad. Sci. U.S.A. 108, 6456–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kitayner M., Rozenberg H., Rohs R., Suad O., Rabinovich D., Honig B., Shakked Z. (2010) Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol. 17, 423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y., Dey R., Chen L. (2010) Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure 18, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malecka K. A., Ho W. C., Marmorstein R. (2009) Crystal structure of a p53 core tetramer bound to DNA. Oncogene 28, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kitayner M., Rozenberg H., Kessler N., Rabinovich D., Shaulov L., Haran T. E., Shakked Z. (2006) Structural basis of DNA recognition by p53 tetramers. Mol. Cell 22, 741–753 [DOI] [PubMed] [Google Scholar]
- 15. Ho W. C., Fitzgerald M. X., Marmorstein R. (2006) Structure of the p53 core domain dimer bound to DNA. J. Biol. Chem. 281, 20494–20502 [DOI] [PubMed] [Google Scholar]
- 16. Cho Y., Gorina S., Jeffrey P. D., Pavletich N. P. (1994) Crystal structure of a p53 tumor suppressor-DNA complex. Understanding tumorigenic mutations. Science 265, 346–355 [DOI] [PubMed] [Google Scholar]
- 17. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 18. Perez C. A., Pietenpol J. A. (2007) Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle 6, 246–254 [DOI] [PubMed] [Google Scholar]
- 19. Ortt K., Sinha S. (2006) Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Lett. 580, 4544–4550 [DOI] [PubMed] [Google Scholar]
- 20. Olson W. K., Gorin A. A., Lu X. J., Hock L. M., Zhurkin V. B. (1998) DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U.S.A. 95, 11163–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laptenko O., Beckerman R., Freulich E., Prives C. (2011) p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 108, 10385–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cui F., Sirotin M. V., Zhurkin V. B. (2011) Impact of Alu repeats on the evolution of human p53 binding sites. Biol. Direct 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naït-Kaoudjt R., Guiard B., Gervais M. (1998) Evidence of an overlap between the two half-sites of UAS1-B/CYC1. A new model for Cyp1p (Hap1p) DNA binding. Eur. J. Biochem. 254, 111–116 [DOI] [PubMed] [Google Scholar]
- 24. Osada M., Park H. L., Nagakawa Y., Yamashita K., Fomenkov A., Kim M. S., Wu G., Nomoto S., Trink B., Sidransky D. (2005) Differential recognition of response elements determines target gene specificity for p53 and p63. Mol. Cell. Biol. 25, 6077–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kabsch W. (2010) XDS. Acta Crystallogr. D 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. French S., Wilson K. (1978) On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 [Google Scholar]
- 27. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 28. Brünger A. T. (1992) Free R value. A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McRee D. E. (1999) XtalView/Xfit-A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 31. Brünger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX. Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 33. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 34. DeLano W. L. (2002) PyMOL, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 35. Kabsch W. (1976) A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 36. Lavery R., Moakher M., Maddocks J. H., Petkeviciute D., Zakrzewska K. (2009) Conformational analysis of nucleic acids revisited. Curves+. Nucleic Acids Res. 37, 5917–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Svozil D., Kalina J., Omelka M., Schneider B. (2008) DNA conformations and their sequence preferences. Nucleic Acids Res. 36, 3690–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. el Hassan M. A., Calladine C. R. (1996) Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J. Mol. Biol. 259, 95–103 [DOI] [PubMed] [Google Scholar]
- 39. Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., Ruan Y. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 40. Wang B., Xiao Z., Ren E. C. (2009) Redefining the p53 response element. Proc. Natl. Acad. Sci. U.S.A. 106, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang A., Zhu Z., Kapranov P., McKeon F., Church G. M., Gingeras T. R., Struhl K. (2006) Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell 24, 593–602 [DOI] [PubMed] [Google Scholar]
- 42. Carugo O., Argos P. (1997) Protein-protein crystal-packing contacts. Protein Sci. 6, 2261–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pavletich N. P., Chambers K. A., Pabo C. O. (1993) The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 7, 2556–2564 [DOI] [PubMed] [Google Scholar]
- 44. Weinberg R. L., Veprintsev D. B., Fersht A. R. (2004) Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 341, 1145–1159 [DOI] [PubMed] [Google Scholar]
- 45. Klein C., Georges G., Künkele K. P., Huber R., Engh R. A., Hansen S. (2001) High thermostability and lack of cooperative DNA binding distinguish the p63 core domain from the homologous tumor suppressor p53. J. Biol. Chem. 276, 37390–37401 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.