Background: Non-viral generation of induced pluripotent stem cells (iPSCs) in vitro is generally of low efficiency.
Results: In vivo expression of non-integrated transgenes Oct4, Sox2, and Klf4 efficiently reprograms muscle cells.
Conclusion: Reprogrammed, undifferentiated cells can be reliably and rapidly produced using naked DNA, exploiting synergy between muscle repair and reprogramming.
Significance: In vivo approach throws light on the molecular networks underlying reprogramming, suggesting alternate iPSC generation strategies.
Keywords: Cell Differentiation, Cell Proliferation, Induced Pluripotent Stem Cells, Reprogramming, Xenopus, Gene Delivery Systems in Vivo, Muscle Repair, Non-viral Somatic Gene Transfer, Pluripotency Transcription Factors
Abstract
Adult mammalian cells can be reprogrammed into induced pluripotent stem cells (iPSCs) by a limited combination of transcription factors. To date, most current iPSC generation protocols rely on viral vector usage in vitro, using cells removed from their physiological context. Such protocols are hindered by low derivation efficiency and risks associated with genome modifications of reprogrammed cells. Here, we reprogrammed cells in an in vivo context using non-viral somatic transgenesis in Xenopus tadpole tail muscle, a setting that provides long term expression of non-integrated transgenes in vivo. Expression of mouse mOct4, mSox2, and mKlf4 (OSK) led rapidly and reliably to formation of proliferating cell clusters. These clusters displayed the principal hallmarks of pluripotency: alkaline phosphatase activity, up-regulation of key epigenetic and chromatin remodeling markers, and reexpression of endogenous pluripotent markers. Furthermore, these clusters were capable of differentiating into derivatives of the three germ layers in vitro and into neurons and muscle fibers in vivo. As in situ reprogramming occurs along with muscle tissue repair, the data provide a link between these two processes and suggest that they act synergistically. Notably, every OSK injection resulted in cluster formation. We conclude that reprogramming is achievable in an anamniote model and propose that in vivo approaches could provide rapid and efficient alternative for non-viral iPSC production. The work opens new perspectives in basic stem cell research and in the longer term prospect of regenerative medicine protocols development.
Introduction
The ability to reprogram differentiated cells into induced pluripotent stem cells (iPSCs)6 via ectopic expression of the four transcription factors Oct4, Sox2, c-Myc, and Klf4 has revolutionized the concepts of pluripotency (1). Following this groundbreaking discovery, iPSCs have been generated in a number of mammalian species, from a wide range of different cell types with efficiencies depending on the differentiation status of the somatic cell source used (2, 3). To date, reprogramming to iPSC mainly relies on the use of retroviral delivery methods and is a gradual process occurring over 1–2 weeks in vitro (1, 4–6). However, the use of integrative viral vectors and c-Myc as a reprogramming factor is frequently associated with tumor formation in iPSC-derived chimeric mice (7). Attempts to overcome this problem, by elimination of c-Myc (8, 9) or by replacement of retroviruses with non-integrative vectors, including plasmids (2, 10–12), led to lower reprogramming efficiencies (12, 13).
Reversing terminally differentiated cells to pluripotency through reprogramming is not a new notion. It was first introduced in amphibians half a century ago when Sir J. Gurdon and his colleagues successfully cloned tadpoles from differentiated Xenopus cell nuclei transplanted into the cytoplasm of unfertilized eggs (14). Following this pioneering demonstration, nuclear reprogramming by somatic nuclear transfer has been achieved in many mammalian species (12, 15). More recently, reprogramming of mammalian nuclei to a pluripotent-like status by Xenopus oocyte cytoplasm demonstrated that the Xenopus oocyte can override the stability of mammalian cell differentiation (16). Nevertheless, whereas all vertebrates share pluripotency, most data on reprogramming comes from mammalian systems, mainly human and mouse. Moreover, the in vitro protocols used for iPSC generation do not take into account contexts that might impact on the reprogramming process and its efficiency at higher order levels (e.g. tissue, organ, system). Therefore, reprogramming approaches to generate iPSCs in vivo, which remain poorly investigated, would allow to explore the influence of the native environment of the cells to be reprogrammed.
We exploited the experimental advantages of the Xenopus model to explore the ability to reprogram Xenopus differentiated cells in vivo. We used in vivo non-viral somatic transgenesis that allows long lasting gene expression in live tadpoles (17, 18). We showed that combined transfection of mouse mOct4, mSox2, and mKlf4 (OSK) into Xenopus tadpole tail muscle led to proliferative cell clusters formation. Cells in these clusters expressed typical hallmarks of pluripotency, such as reactivation of endogenous pluripotent markers, and showed the capacity to differentiate into derivatives of all three germ layers. In vivo reprogramming occurred in every tadpole transfected, probably being facilitated by simultaneous muscle repair. We conclude that reprogramming can be efficiently obtained by non-viral methods in vivo and that Xenopus reprogrammed cells share properties with mammalian iPSCs.
EXPERIMENTAL PROCEDURES
Animals
Xenopus laevis tadpoles were raised as described (18) and staged according to Nieuwkoop and Faber (19). Sacrifices and animal studies were conducted according to the principles and procedures described in Guidelines for Care and Use of Experimental Animals.
Plasmid Injections
Somatic gene transfer was carried out as described previously using perchlorated tadpoles at stage NF55 (18). In brief, 1 μl of different plasmid mixes was injected intramuscularly at the concentrations indicated in the text. DNA constructs used were: peGFP-C1 (CMV-GFP) and pDsRed2-N1 (CMV-RFP) (Clontech); pGL3 (CMV-LUC) (Invitrogen); CMV-mOct4 and CMV-mSox2; SV40-LUC. Mouse Klf4 cDNA was PCR-amplified and cloned in the pCMV-3×FLAG plasmid (Sigma), giving CMV-mKlf4. Plasmids were purified using the QiaFilter kit (Qiagen). pCMV-3×FLAG was used as an empty vector to equalize the DNA amount for each injection.
Immunohistochemistry
In toto GFP reporter expression was monitored on living tadpoles before further analyses. Before being processed for immunohistochemical analyses, cell cultures were PFA-fixed (4% in PBS for 10 min at 4 °C), and injected tail muscles were dissected, PFA-fixed (4% in PBS for 3 h at 4 °C), and sectioned using a cryostat (14 μm). Immunodetection was carried out as described previously (20) on sections or fixed cell cultures, using the following primary antibodies: rabbit anti-phosphohistone H3 (1:300; Upstate Biotechnology), rabbit anti-active caspase 3 (1:250; BD Biosciences Pharmigen), rabbit anti-β-tubulin III (1:300; Sigma), mouse anti-MZ15 (1:500; DSHB), mouse anti-NCAM (1:300; DSHB), rabbit anti-mKlf4 (1:150; Santa Cruz Biotechnology), rabbit anti-mOct4 (1:400; Abcam), mouse anti-HA (1:100; Sigma), mouse anti-Pax7 (1:300; DSHB), rabbit anti-GFP (1:300; Invitrogen), and the appropriate secondary fluorescent antibodies (1:1500). 5-Bromo-2′-deoxyuridine (BrdU) labeling was carried out using BrdU Labeling and Detection kit I (Roche Applied Science) following intraperitoneal injection of 2 μl of BrdU (1 mg/ml) in NF55 perchlorated tadpoles every 2 days after transfection. Alkaline phosphatase activity was revealed using the AM0100–1KT kit (Sigma). All sections were DAPI (4′,6-diamidino-2-phenylindole)-counterstained and mounted in Moviol (Calbiochem) before observation under light and fluorescent microscopy.
Cell Culture
Cell cultures were derived from tadpole muscles at 7 or 12 days after transfection according to the previously reported protocol (21). Cells were cultured at 20 °C in 70% Leibovitz L-15 medium (Invitrogen) with 5× serum replacement (Sigma) supplemented with antibiotic (Invitrogen). Treatments (3 or 5 days) were carried out with retinoic acid (1 μm), recombinant mouse FGF-8b (50 ng/ml), recombinant mouse Noggin/Fc chimera (50 ng/ml), recombinant human/mouse/rat activin A (5, 50, or 100 ng/ml). All molecules were from R&D Systems, except retinoic acid (Sigma). Cultured cells were fixed with 4% PFA in PBS for 30 min at 4 °C, rinsed in PBS, and immunostained with corresponding antibodies, as described above.
Real-time Quantitative PCR Analyses
For each point monitored, RNA extractions were performed on three pooled muscle samples or trypsinized cell cultures. The protocol, primers, graphical representations, and statistical analyses are detailed in supplemental Experimental Procedures and supplemental Table S1.
Beads Transplanted in Muscle
Heparin-acrylic beads (Sigma) were incubated for 2.5 h at room temperature with FGF-8 (10 μg/ml), Noggin (10 μg/ml), or BSA 0.1% (Sigma), then washed in PBS. For each factor, one bead was implanted subcutaneously at 12 days post-injection (dpi) in each transfected myomere (see Fig. 1). After 5 days, muscles were fixed with 4% PFA in PBS for 3h at 4 °C, rinsed in PBS, then sectioned and immunostained with anti-β-tubulin III (1:300).
FIGURE 1.
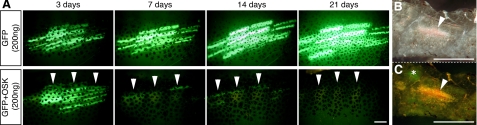
Expression of mouse OSK factors in tadpole muscle leads to GFP loss and cell mass appearance. A, somatic transgenesis in tadpole tail muscle of a GFP-reporter construct ± mouse OSK (200 ng/μl per plasmid). GFP expression was monitored at 3, 7, 14, and 21 dpi, increasing in controls (upper panels) but decreasing at 7 dpi in OSK-injected muscles (lower panels; arrowheads indicate the same myomeric units). Experiments were performed ≥5 times (n >10 for each time point), providing similar results. B, 200-μm-thick slice from OSK muscle at 11 dpi revealing a large, dense cell mass. C, residual GFP-positive fiber (*) near the mass (arrowhead). Scale bars, 500 μm.
Xenopus Transgenic Lines
Transgenic animals were obtained according to the modified Kroll and Amaya protocol (22). In the pNanog-GFP transgenic line, GFP reporter expression is placed under the control of the promoter sequence [−332/+50] of the mouse pluripotency gene mNanog (23). In the pCar-GFP transgenic line, GFP reporter expression is placed under the control of the actin cardiac promoter, leading to a specific GFP expression in cardiac and skeletal muscle (24).
Graft Transplant
OSK injection was performed in perchlorated NF55-staged pCar-GFP transgenic tadpoles. At 10 dpi, injected muscles were dissected and placed in cold filtered PBS (pH 7.5). The muscle region containing clusters were dissected into 0.4 mm2-faced cubes and placed in cold L-15 medium, with 1× serum replacement supplemented with antibiotic. Cubes were transplanted intramuscularly into perchlorated WT tadpoles at the same developmental stage (NF55). Two months later, tail muscles were fixed with 4% PFA in PBS for 3 h at 4 °C, rinsed in PBS, then cryosectioned (120 μm), and floating sections were immunostained with rabbit anti-GFP (1:300) and mounted in Moviol before observation under fluorescent microscopy.
RESULTS
Clusters of Small Cells Appear in Tadpole Tail Muscle following Somatic Injection of Mouse OSK Factors
Non-integrative, non-viral, and non-replicative expression vectors encoding mouse OSK and/or GFP, all under the transcriptional control of the CMV promoter, were injected in Xenopus pre-metamorphic tadpole tail muscles at various doses (see “Experimental Procedures,” Fig. 1A, and supplemental Fig. S1 for optimization experiments). This “naked” DNA-based gene delivery method is known to specifically transfect muscle fibers without integration (25) and, in that way, is used for human vaccination (26). The conditions used in somatic injection experiments yielded an average of 25 transfected muscle fibers per injected tadpole as estimated by GFP expression (data not shown). Three dpi, GFP was detectable only in differentiated muscle fibers of both OSK-injected and control animals. However, the GFP-expressing fibers were rapidly lost in OSK-transfected muscles concomitantly with the appearance of large, dense cell clusters (Fig. 1, B and C).
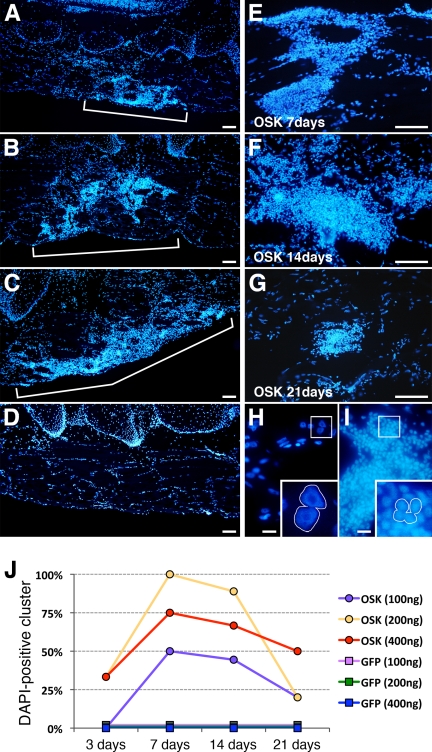
Clusters occurred at all tested plasmid concentrations (100, 200, and 400 ng/μl; Fig. 2, A–C and E–G), many filling the space previously occupied by a lost GFP-expressing fiber (supplemental Fig. S2, A–C). OSK-injected tadpoles frequently showed several clusters in the transfected area (Fig. 2, A–C and clearly seen in supplemental Fig. S2A). Furthermore, nuclei in the area surrounding the site of OSK injection were disorganized, contrasting with the regular nuclear distribution of GFP-injected control muscles (compare Fig. 2D and supplemental Fig. S2D). This disorganized nuclear distribution also clearly differed from that of apoptotic muscle fibers (supplemental Fig. S2, E and F), frequently seen following somatic transfection in muscle (18). Nuclei morphology also differed, with large nuclei present in intact fibers (Fig. 2H) and 2–3 times smaller nuclei in clusters (Fig. 2I). Using 200 ng/μl plasmid-injected concentration, the clusters were observed in 30% of OSK-transfected tadpoles at 3 dpi and in 100% at 7 dpi (Fig. 2J); at 21 dpi, smaller clusters were still observable in 20% of injected animals. Clusters never formed in control GFP-injected tadpoles (Fig. 2, D and J). Notably, cell clusters were also observed in muscles injected with individual transcription factors (O, S, or K alone); however, clusters were fewer than with OSK injection (supplemental Fig. S3). As 200 ng/μl OSK injection gave optimal cluster formation (Fig. 2J), this plasmid concentration was used in all following experiments.
FIGURE 2.
Cell cluster formation is dose- and time-dependent. A–D, nuclei, observed by DAPI-staining, are grouped in OSK clusters at 7 dpi (A, 100 ng/μl; B, 200 ng/μl; C, 400 ng/μl for each plasmid), whereas muscle fibers show normal peripheral nuclear organization in GFP-controls (D). E–G, representative clusters surround neighboring muscle fibers at 7, 14, and 21 dpi for the 200 ng/μl condition. H and I, magnifications show large nuclei in control fibers (H), whereas nuclei in OSK clusters are smaller (I). J, kinetics of cluster occurrence for GFP-controls and OSK-injected muscles shown at the 3 concentrations. For each point, independent 4 ≤ n ≤ 9 samples were pooled and the percentage of cluster-containing muscles calculated. Scale bars, 100 μm (A–G), 20 μm (H–I).
Expression of the Three Mouse Factors OSK Decreases Over Time
Given that mouse transcription factors were used, their expression can be followed unambiguously, unhampered by putative endogenous expression of the Xenopus homologues. Expression of exogenous reprogramming factors in OSK-transfected tadpoles was high at 3 and 7 dpi, then decreased strongly between 7 and 14 dpi, and was virtually undetectable by 21 dpi (supplemental Fig. S4A). Immunodetection revealed that, in OSK-transfected tadpoles, mouse proteins were detected in nuclei of GFP-positive fibers at 7 dpi, but not in cluster cells nuclei (supplemental Fig. S4, B–F). Furthermore, all mOct4-positive nuclei were Pax7-negative, implying that satellite cells are not transfected following OSK injection (supplemental Fig. S4G).
Cell Clusters Are Proliferative and Reexpress Pluripotent Marker Alkaline Phosphatase
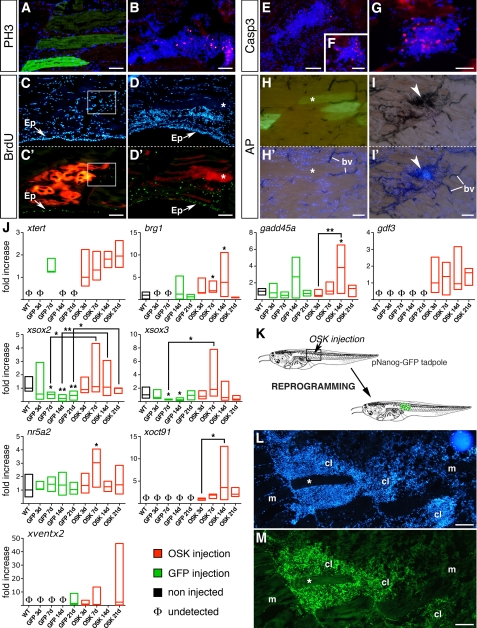
The proliferative status of clusters was analyzed using the mitotic marker PH3 and BrdU labeling. At 14 dpi, clusters contained numerous PH3-positive cells (Fig. 3B) whereas no PH3 signal was seen in GFP-controls (Fig. 3A) or in non-injected tadpole muscle (data not shown). BrdU labeling showed high numbers of labeled cells in clusters at 14 dpi (Fig. 3, D and D′), confirming their proliferative nature. In GFP-controls, only rare BrdU-positive cells were detected in the injected area (Fig. 3, C and C′). As expected, epidermal BrdU incorporation was similar in OSK and GFP-control tadpoles (Fig. 3, C′ and D′). Clusters proliferated up to 14 dpi. At 21 dpi, when cluster size decreased, PH3 labeling was lost concomitantly with an increase of caspase 3-positive cells, suggesting that apoptosis might be involved in cluster reduction (Fig. 3, E–G).
FIGURE 3.
Cell clusters are proliferative and express endogenous pluripotency markers. Using PH3-antibody (A and B) and BrdU labeling (C′ and D′), cell proliferation was compared at 14 dpi between GFP (A) or RFP (C and C′) controls and OSK-injected muscles (B, D, and D′). PH3-positive cells (B) and strong BrdU labeling (D′) occurred in OSK clusters, but not in controls (A and C′). Note that BrdU labeling shows similar proliferative status of epidermis (Ep) in both conditions. Apoptosis was followed using active caspase 3 antibody in OSK-transfected muscles at 14 dpi (E and F) and 21 dpi (G), showing an increase of apoptotic cells in clusters. AP activity was strong in OSK-injected muscles at 14 dpi and co-localized with DAPI-positive clusters (I and I′, arrows), whereas GFP-controls (G and G′) showed no AP labeling except in blood vessels (bv). Each labeling was performed ≥3 times. Scale bars, 100 μm (A–E and H–I′); 50 μm (F and G). J, expression of xtert, brg1, and gadd45a (epigenetic and chromatin-remodeling markers) and nr5a2, xoct91, gdf3, xsox3, xsox2, and xventx2 (pluripotency factors) at 3, 7, 14, and 21 dpi in GFP-controls (green boxes) and OSK-transfected muscles (red boxes), using real-time quantitative PCR. For each gene, basal levels in non-injected muscles are indicated (WT, black boxes) and absence of transcript noted as Φ. Samples were from independent experiments, with three muscles pooled per sample (4 ≤ n ≤ 8 samples per group). mRNA levels were expressed as relative to WT except when expression was not detected, then mRNA levels were expressed as relative to the first value observed for OSK. Boxes represent minimum and maximum values around the median. Non-parametric Mann-Whitney U test was used to assess statistical differences versus WT, except when indicated by black bars: **, p < 0.01; *, p < 0.05. K, schematic representation of the protocol used in reprogramming experiment performed with the pNanog-GFP transgenic tadpoles. L and M, GFP immunolabeling of a pNanog-GFP transgenic Xenopus tadpole injected with OSK shows a strong and specific GFP expression in clusters (cl) at 7 dpi, whereas muscle fibers (m and *) are GFP-negative. Scale bars, 100 μm (K–L).
Alkaline phosphatase (AP) activity, routinely used as a pluripotency marker for embryonic stem cells (ESCs) and iPSCs (27), was strongly detected in OSK clusters (Fig. 3, I and I′). No AP activity was observed in GFP-controls (Fig. 3, H and H′), except in blood vessels, which are known to be AP-positive (28). AP expression suggests a return to an undifferentiated state. To assess further this possibility, reactivation of a pluripotency program in OSK-injected muscles was evaluated using different markers.
Xenopus Pluripotency Genes Are Reactivated in OSK-injected Tadpoles
Following injection, myogenic markers were similarly reactivated in GFP-controls and OSK-injected muscles (supplemental Fig. S5); however, significant differences were observed for endogenous homologues of mammalian markers of pluripotency (Fig. 3J). Notably, OSK-injected muscle showed specific activation of xoct91, the functional homologue of mammalian Oct4 (29), of nr5a2, a regulator of Oct4 expression in pluripotency (30), and of gdf3, a TGF-β family member involved in regulation of the pluripotent state (31) and in iPSCs reprogramming (32, 33). As for xoct91, gdf3 expression was specific to OSK-injected muscles and maintained for 3 weeks. Expression of xsox2 and xsox3, two genes related to mammalian Sox2 (34), was significantly down-regulated in GFP-controls and up-regulated in OSK-injected tadpoles as the reprogramming process progressed. We also examined reactivation of xventx2, previously described as expressed in multipotent stem cells in Xenopus (35) and proposed to share a functional relationship with Nanog (36).7 As for xoct91, xventx2 was precociously activated in OSK samples. xventx2 expression was totally down-regulated at 14 dpi, suggesting that reprogrammed cells at this time might be responsive to differentiation cues. Intriguingly, we observed reexpression of xventx2 in both OSK and GFP-control muscles at 21 dpi, suggesting an unknown role for this factor during late muscle repair.
To validate further the establishment of a transcriptional state similar to that of mammalian pluripotent cells in these clusters, a transgenic Xenopus line expressing GFP under the promoter of the mouse pluripotency factor mNanog (pNanog-GFP) was used. Similar readouts, using transgenic cell lines expressing GFP under the control of either the Oct4 or the Nanog promoters (7, 11, 37–39), are commonly used in mammalian systems to define whether cultured somatic cells transfected with reprogramming factors have reached full iPSC (i.e. pluripotent) status. Thus, using pNanog-GFP transgenic tadpoles, the OSK plasmid mix was injected into the muscle without the CMV-GFP construct. In this case, GFP expression therefore reflected activation of the mouse Nanog promoter integrated in the genome. As shown in Fig. 3L, OSK injection resulted in GFP expression specifically in clusters, but not in non-transfected muscle cells.
Epigenetic Factors Are Up-regulated in OSK-reprogrammed Cells
Reprogramming is facilitated if epigenetic switches and chromatin remodeling are activated (40, 41). We thus monitored expression of brg1 and gadd45a, two factors essential for nuclear reprogramming in Xenopus (42, 43), and that show, respectively, a key role in chromatin remodeling and in active DNA demethylation. Both factors were detectable in uninjected (WT) muscle but were specifically up-regulated in OSK-injected muscles (Fig. 3J). Reactivation of brg1 expression was observed from 14 dpi in GFP-controls and might be involved in MyoD-mediated differentiation during muscle repair (44). Telomerase activity induction is another important feature of iPSCs (45). Expression of the telomerase reverse transcriptase gene (xtert) was markedly increased in OSK samples (Fig. 3J). Altogether, activation of brg1, gadd45a, and xtert suggests that epigenetic changes occurred in OSK-injected muscles, favoring reprogramming.
OSK-reprogrammed Cells Differentiate in Vitro into Derivatives of Three Germ Layers
GFP-control or OSK-injected muscles were cultured to test their differentiative potential (supplemental Fig. S6A). After 24 h, only in OSK-derived cultures we observed the presence of AP-positive clones with characteristic ESC-like morphology (supplemental Fig. S6, B–H). When cultured for 5 days in normal medium these clones progressively acquired various differentiated morphologies (supplemental Fig. S7), suggesting spontaneous differentiation.
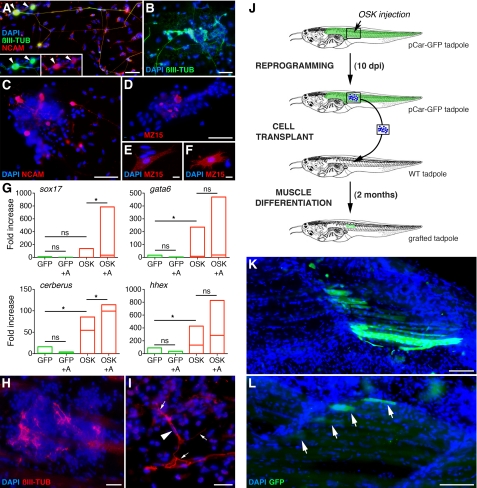
OSK-derived cultures were then treated with different factors to induce differentiation. FGF-8 + Noggin treatments (46) resulted in differentiation toward a neurectodermal fate (Fig. 4, A–C). Retinoic acid with FGF-8 + Noggin induced differentiated neurons (Fig. 4A). No neuronal differentiation was seen in similarly treated GFP-control cultures. To differentiate OSK-derived cells toward a mesodermal fate, low activin concentrations were used (47). Colonies and individual cells labeled with the MZ15 antibody that recognizes mesodermal-derived notochord cells were obtained (Fig. 4, D--F), indicating that OSK-derived cells can give rise to cells with a different mesodermal precursor origin from that of muscle. Finally, we used Noggin and high doses of activin, known to promote endodermal differentiation of Xenopus animal caps (47). Real-time quantitative PCR was used to monitor the expression of endodermal specification markers sox17, gata6, cerberus, and hhex (Fig. 4G). These markers were weakly expressed or absent from controls. In contrast, gata6, cerberus, and hhex mRNAs increased significantly in non-treated OSK-derived cultures, suggesting that spontaneous differentiation to endoderm occurred. In treated cells, sox17 and cerberus increased significantly, demonstrating enhanced endodermal differentiation. The mesodermal marker myf5 was down-regulated by high dose activin, favoring an endodermal fate (supplemental Fig. S8). Furthermore, spontaneous mesodermal and neurectodermal differentiation was revealed using the xnot and xsox2 markers, respectively (supplemental Fig. S8). Thus, OSK cultures can differentiate spontaneously into the three germ layers, and activin can enhance endoderm differentiation.
FIGURE 4.
OSK-reprogrammed cells can differentiate into derivates of the three embryonic lineages in vitro and into neurons and muscles in vivo. A–C, neuroectodermal differentiation with FGF-8 + Noggin + retinoic acid (A) or FGF-8 + Noggin (B and C) treatment of 12 dpi OSK-derived cultures, shows tubulin III labeled neurons after 5 days (A and B), and NCAM-positive cells at 3 days (C). D–F, mesodermal differentiation was obtained with low activin treatment of 12 dpi OSK-derived cultures and revealed with a notochord (MZ15) antibody, showing MZ15-positive cells after 3 days (D, activin 5 ng/ml), or individual labeled cells at 3 days (F, activin 50 ng/ml) or 5 days (E, activin 5 ng/ml). G, endodermal differentiation was obtained with high activin (100 ng/ml) + Noggin treatments of 12 dpi OSK-derived cultures and revealed by real-time quantitative PCR using endodermal markers (sox17, gata6, cerberus, and hhex). After 3 days, spontaneous as well as induced endoderm induction was observed in non-treated (OSK) and treated (OSK+A) cultures, but not in 12 dpi GFP-control cultures (GFP and GFP+A). Treatments were performed >6 times. Quantitative PCR data are represented as described in Fig. 3 and mRNA levels expressed as relative to control cultures (GFP): *, p < 0.05; not significant (ns) p > 0.1. H and I, OSK-injected muscles labeled with anti-tubulin III antibody at 14 dpi showed spontaneous neuronal differentiation in cell clusters (H). I, differentiated neuron, with axonal network (arrows) and its cell body (arrowhead indicates nucleus), was obtained following 5 days treatment of 12 dpi OSK-injected tadpoles with FGF-8 + Noggin. J, schematic represents transplant protocol used to follow reprogrammed cell fate reversion in muscle tissue. OSK was injected in a pCar-GFP transgenic tadpole, at 10 dpi the reprogrammed cells were transplanted in a WT tadpole, and the muscle of grafted tadpoles was observed 2 months later. K and L, two examples of grafted tadpoles show the presence of numerous (K) and at a lesser extent (L, arrows) GFP-positive muscle fibers, indicating the reversion of pCar-GFP reprogrammed cells toward a muscle fiber phenotype after transplantation. Scale bars, 50 μm (A–D), 10 μm (E and F), 25 μm (H and I), 100 μm (K and L).
Xenopus-iPS Cells Differentiate into Neuronal and Muscle Phenotypes in Vivo
Teratoma formation is another stringent method for assessing iPSC pluripotency (27). We explored the possibility that Xenopus-reprogrammed cells in clusters share the ability to differentiate toward non-mesodermal derivatives directly in vivo. We observed neurons within OSK-induced clusters (Fig. 4H), showing that spontaneous neurectodermal differentiation occurs in vivo. Furthermore, when neurectodermal differentiation in vivo was forced by FGF-8 + Noggin treatment, large and fully differentiated neurons were observed within muscle (Fig. 4I). No neurons were induced when treating GFP-control muscle.
Using a transplant protocol, we also demonstrated that OSK-reprogrammed cells retain the ability to redifferentiate into muscle fibers. Indeed, pCar-GFP transgenic tadpoles were injected with OSK and at 10 dpi, dissected pieces of tissue containing the reprogrammed cells were transplanted into WT non-transgenic tadpoles. Two months later, we observed the presence of GFP-positive fibers in tails of transplanted tadpoles, their origin necessarily arising from reprogrammed fibers of pCar-GFP animals (Fig. 4, J–L). Conversely, as a control, when the graft originated from pCar-GFP tadpoles injected with the pCMV-3×FLAG empty vector instead of the reprogramming factors, no GFP-expressing fibers were observed in transplanted WT tadpoles.
DISCUSSION
A fundamental goal in stem cell biology is to understand better the regulation of the transcription factor networks that control pluripotent cell identity and fate decisions in vivo. This goal is related to the need to understand the capacity of a cell to reprogram in tissue- and stage-specific contexts. However, the mechanisms that favor or limit the reprogramming process are not well understood. For instance, most studies on iPSC reprogramming are performed in vitro and show that reprogramming efficiency depends mainly on the differentiation status of the cell source used (12, 48, 49), the cellular context greatly influencing factors required to generate iPSCs (50). In addition, one of the main aims of iPSC studies is to facilitate regenerative medicine, hence the importance of placing iPSC generation in a physiological perspective. Our hypothesis is that working in an in vivo context allows the physiological setting and higher order regulations to be taken into account. In our experiments we used the living Xenopus tadpole to explore the transcription factor-mediated reprogramming ability of differentiated cells in vivo.
Taken together, our findings show that reprogrammed cells can be obtained in vivo by ectopic expression of mouse mOct4, mSox2, and mKlf4 in tadpole muscle (Fig. 5). Importantly, the gene delivery method used, naked DNA injection in muscle tissue, specifically transfects muscle fibers without integration of foreign DNA (25). Our laboratory has developed and used for decades such a technique in Xenopus (see for instance Refs. 17, 18), showing that transfection of muscular cell types other than fibers was not observed. Further, in the experiments described here, CMV-driven expression of GFP and of OSK was only detected in muscle fibers (Fig. 1 and supplemental Fig. S1 and Fig. S4) and was not detected in satellite cells (supplemental Fig. S4G). The transfected fibers remained stable in GFP-controls, whereas they gradually disappeared from OSK-injected muscles (Fig. 1 and supplemental Fig. S1) to be replaced by clusters of proliferative cells (Fig. 2 and Fig. 3, A–D, and supplemental Fig. S2). Remarkably, each OSK-transfected muscle showed the formation of at least one cluster at 7 dpi (Fig. 2 and supplemental Fig. S2), giving 100% efficiency in the generation of reprogrammed cells in vivo. We observed that cluster size increased up to 14 dpi and then decreased to be undetectable after 21 dpi (Fig. 2), at least in part involving apoptosis (Fig. 3, E--G). The transient occurrence of cluster in muscle can be explained by the fact that the non-replicative and non-integrative plasmid vectors will be lost by dilution during cell division. Thus, the absence of further source of reprogramming factors after 14 days (supplemental Fig. S4) will no longer support cluster formation in muscle. Indeed, the reprogrammed cells at this time will be responsive to differentiation cues, particularly those of muscle tissue, as shown by muscle redifferentiation of reprogrammed cells after transplant (Fig. 4, J–L).
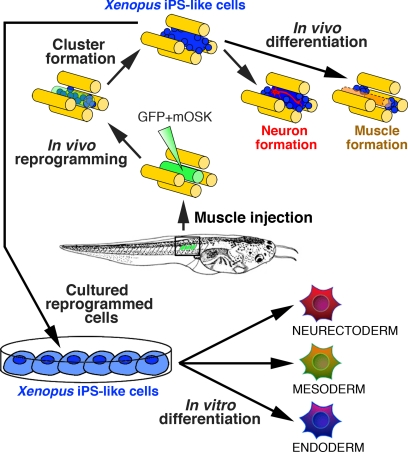
FIGURE 5.
Generation of Xenopus-iPS-like cells in vivo. Model for in vivo non-viral induction of Xenopus reprogrammed cells is shown. Following co-injection of mOSK with a GFP-reporter in tadpole tail (mOSK+GFP), GFP-transfected fibers (in green) dedifferentiate into proliferative undifferentiated Xenopus iPS-like cells (in blue). Reprogrammed cells of mesodermal origin possess an increased developmental potential, differentiating in vitro toward derivatives of the three embryonic lineages and into neurons and muscle fibers in vivo.
Four arguments demonstrate that OSK-injected muscle contain reprogrammed cells with properties reminiscent to that frequently reported for iPSC generated in mammals. First, clusters contain AP-positive cells (Fig. 3I), a pluripotent marker currently used to identify ESCs or iPSCs (27). Second, three bona fide markers of reprogramming are specifically expressed in OSK-transfected muscle: xoct91, xventx2, and gdf3 (Fig. 3J). In particular, xoct91 and xventx2 are, respectively, the functional homologues of Oct4 (29) and Nanog (35, 36),7 reactivation of which in iPSCs are common readouts (37–39) to identify mammalian reprogrammed cells, and considered by some as a “gold standard” (27). Indeed, xoct91 and xventx2 reexpression could distinguish pluripotent versus differentiated cells in Xenopus tadpoles and thus represent bona fide markers of reprogramming in this model. Third, activation of the epigenetic modifiers brg1, gadd45a, and xtert (Fig. 3J) could confer epigenetic plasticity to OSK-injected muscles, enhancing reprogramming, as seen by overexpression of Brg1 in mouse fibroblasts (41). Finally, the reprogrammed cells can differentiate into derivatives of the three germ layers in vitro (Fig. 4, A–G, and supplemental Fig. S7 and Fig. S8) and in vivo into muscle fibers (Fig. 4, J–L), as well as neurons, thus contributing to production of cell lineages other than the original mesodermal muscle (Fig. 4, H–I). In line with these observations, we show that the mouse Nanog promoter is activated in cell clusters (Fig. 3L), as previously observed in mammalian nuclei reprogrammed either by transcription factors or Xenopus oocyte extract (7, 11). Together, these findings strongly argue for a pluripotent status of the reprogrammed cells present in OSK-generated clusters.
In our experiments, myogenesis was reactivated in response to injection injury. A rapid down-regulation of myoD expression was observed (supplemental Fig. S5). Recent findings show that suppression of MyoD expression by Oct4 is required to initiate the iPSC reprogramming from mouse myoblasts in culture (51). Thus, in our experiments, decreased myoD could catalyze reprogramming in OSK-transfected muscle cells. Cell division can also accelerate iPSC formation (52). During amphibian tail regeneration, dedifferentiation of injured fibers has been reported to support the muscle repair process. Interestingly, muscle dedifferentiation leads to cell cycle reactivation, polynucleated fiber fragmentation into mononucleated cells, and proliferation (53). In our in vivo model, a similar process involving dedifferentiation should occur during muscle repair in response to injection injury, and cell proliferation could contribute to reprogramming efficiency. The data thus show that reprogramming factors might synergize with the repair process to generate reprogrammed cells in vivo.
Two further observations bolster the argument that in injected muscle the repair process enhances reprogramming. First, injection of each mouse reprogramming factor separately also induced cluster formation, but at a lower frequency (supplemental Fig. S3). Therefore, as for neural stem cells, which can be reprogrammed using Oct4 alone (54), we suggest that factors present in Xenopus tadpole muscle facilitate reprogramming. Second, pluripotent-related genes xsox2, xsox3, nr5a2 (Fig. 3J), or xoct25 and foxD3 (data not shown) are expressed in uninjected tadpole muscle, a feature arguing for a privileged ground state to reprogramming that could exist in muscle.
Nuclear transplantation of mammalian somatic nuclei into amphibian oocytes induces transcriptional reactivation of Oct4, Sox2, and Nanog, demonstrating that Xenopus oocyte determinants can trigger dedifferentiation of adult mouse nuclei (55). Reciprocally, here we demonstrate that mammalian factors can reprogram Xenopus somatic cells to generate undifferentiated Xenopus cells (Fig. 5), demonstrating that transcription factor-mediated reprogramming can be induced in groups as distant as mammals and amphibians. These data underline the usefulness of the Xenopus model for deciphering pluripotency and point to conserved fundamental principles for the regulatory gene networks known to control pluripotency in vertebrates. This notion is reinforced by the observation that the Xenopus Pou5f1 factors (xoct25, xoct91, and xoct60) can maintain pluripotency in mouse Oct4-deficient ESCs, and reciprocally mOct4 can rescue Xenopus Pou5f1s knocked-down embryos (29).
In conclusion, we show that Xenopus muscle is amenable to transcription factor-mediated reprogramming in vivo, yielding cells that seem comparable with Yamanaka's iPSCs (1). However, we cannot exclude that our reprogrammed cells have not reached the so-called “ground state pluripotency” because we did not verify germ line competence. One might wonder whether ground state pluripotency is achievable in Xenopus because germ line establishment relies on germ plasm in this organism (56). Nevertheless, our data unambiguously demonstrate that in vivo reprogrammed Xenopus cells have reached a “somatically primed” pluripotent state as recently defined by Johnson et al. (56). Therefore, we provide two entirely original demonstrations: first, that induced reprogramming can be achieved in an anamniote model and second, reprogrammed cells can be produced in an in vivo context without viral vectors. The fact that reprogramming can be induced and studied in a versatile developmental model could provide new leads for translational research. Furthermore, the finding that reprogramming is facilitated by repair could open new perspectives in basic stem cell research and regenerative medicine.
Supplementary Material
Acknowledgments
We thank G. Levi and L. Kodjabachian for constructive criticism of the manuscript, F. Ishikawa (Kyoto University, Japan) for providing the CMV-mOct4 and CMV-mSox2 plasmids, A. Nishikawa (Shimane University, Japan) for providing the cell culture protocol, L. Sachs for providing SV40-LUC, A. Mazabraud (CNRS UMR 8080, France) for providing the pCar-GFP transgenic line, G. Benisti and J.P. Chaumeil for Xenopus breeding and care.
This work was supported by European Union grant Crescendo and the French Programme National de Recherche sur les Perturbateurs Endocriniens program.

This article contains supplemental Figs. S1–S8, Table S1, supplemental Experimental Procedures, and additional references.
P. Scerbo, F. Girardot, C. Vivien, G. V. Markov, G. Luxardi, B. A. Demeneix, L. Kodjabachian, and L. Coen, unpublished data.
- iPSC
- induced pluripotent stem cell
- AP
- alkaline phosphatase
- dpi
- days post-injection
- OSK
- mOct4, mSox2, and mKlf4
- PFA
- paraformaldehyde.
REFERENCES
- 1. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 2. Ho R., Chronis C., Plath K. (2011) Mechanistic insights into reprogramming to induced pluripotency. J. Cell. Physiol. 226, 868–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tan K. Y., Eminli S., Hettmer S., Hochedlinger K., Wagers A. J. (2011) Efficient generation of iPS cells from skeletal muscle stem cells. PLoS One 6, e26406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaenisch R., Young R. (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stadtfeld M., Hochedlinger K. (2010) Induced pluripotency: history, mechanisms, and applications. Genes Dev. 24, 2239–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamanaka S., Blau H. M. (2010) Nuclear reprogramming to a pluripotent state by three approaches. Nature 465, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okita K., Ichisaka T., Yamanaka S. (2007) Generation of germ line-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 8. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 9. Wernig M., Meissner A., Cassady J. P., Jaenisch R. (2008) c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2, 10–12 [DOI] [PubMed] [Google Scholar]
- 10. Okita K., Hong H., Takahashi K., Yamanaka S. (2010) Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat. Protoc. 5, 418–428 [DOI] [PubMed] [Google Scholar]
- 11. Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. (2008) Generation of mouse induced pluripotent stem cells without viral vectors. Science 322, 949–953 [DOI] [PubMed] [Google Scholar]
- 12. Okita K., Yamanaka S. (2011) Induced pluripotent stem cells: opportunities and challenges. Philos. Trans. R Soc. Lond. B Biol. Sci. 366, 2198–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayden E. C. (2011) Stem cells: The growing pains of pluripotency. Nature 473, 272–274 [DOI] [PubMed] [Google Scholar]
- 14. Gurdon J. B. (2006) From nuclear transfer to nuclear reprogramming: the reversal of cell differentiation. Annu. Rev. Cell Dev. Biol. 22, 1–22 [DOI] [PubMed] [Google Scholar]
- 15. Hochedlinger K., Jaenisch R. (2006) Nuclear reprogramming and pluripotency. Nature 441, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 16. Gurdon J. B., Melton D. A. (2008) Nuclear reprogramming in cells. Science 322, 1811–1815 [DOI] [PubMed] [Google Scholar]
- 17. de Luze A., Sachs L., Demeneix B. (1993) Thyroid hormone-dependent transcriptional regulation of exogenous genes transferred into Xenopus tadpole muscle in vivo. Proc. Natl. Acad. Sci. U.S.A. 90, 7322–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rowe I., Le Blay K., Du Pasquier D., Palmier K., Levi G., Demeneix B., Coen L. (2005) Apoptosis of tail muscle during amphibian metamorphosis involves a caspase 9-dependent mechanism. Dev. Dyn. 233, 76–87 [DOI] [PubMed] [Google Scholar]
- 19. Nieuwkoop P. D., Faber J. (1994) Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis, Garland Publishing, New York [Google Scholar]
- 20. Coen L., Le Blay K., Rowe I., Demeneix B. A. (2007) Caspase-9 regulates apoptosis/proliferation balance during metamorphic brain remodeling in Xenopus. Proc. Natl. Acad. Sci. U.S.A. 104, 8502–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shibota Y., Kaneko Y., Kuroda M., Nishikawa A. (2000) Larval-to-adult conversion of a myogenic system in the frog, Xenopus laevis, by larval-type myoblast-specific control of cell division, cell differentiation, and programmed cell death by triiodo-l-thyronine. Differentiation 66, 227–238 [DOI] [PubMed] [Google Scholar]
- 22. Coen L., du Pasquier D., Le Mevel S., Brown S., Tata J., Mazabraud A., Demeneix B. A. (2001) Xenopus Bcl-X(L) selectively protects Rohon-Beard neurons from metamorphic degeneration. Proc. Natl. Acad. Sci. U.S.A. 98, 7869–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005) Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Das B., Schreiber A. M., Huang H., Brown D. D. (2002) Multiple thyroid hormone-induced muscle growth and death programs during metamorphosis in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 99, 12230–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Braun S. (2008) Muscular gene transfer using nonviral vectors. Curr. Gene. Ther. 8, 391–405 [DOI] [PubMed] [Google Scholar]
- 26. Dueñas-Carrera S. (2004) DNA vaccination against hepatitis C. Curr. Opin. Mol. Ther. 6, 146–150 [PubMed] [Google Scholar]
- 27. Smith K. P., Luong M. X., Stein G. S. (2009) Pluripotency: toward a gold standard for human ES and iPS cells. J. Cell. Physiol. 220, 21–29 [DOI] [PubMed] [Google Scholar]
- 28. Chan J., Serluca F. C. (2004) Chemical approaches to angiogenesis. Methods Cell Biol. 76, 475–487 [PubMed] [Google Scholar]
- 29. Morrison G. M., Brickman J. M. (2006) Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development 133, 2011–2022 [DOI] [PubMed] [Google Scholar]
- 30. Heng J. C., Orlov Y. L., Ng H. H. (2010) Transcription factors for the modulation of pluripotency and reprogramming. Cold Spring Harbor Symp. Quant. Biol. 75, 237–244 [DOI] [PubMed] [Google Scholar]
- 31. Liu N., Lu M., Tian X., Han Z. (2007) Molecular mechanisms involved in self-renewal and pluripotency of embryonic stem cells. J. Cell. Physiol. 211, 279–286 [DOI] [PubMed] [Google Scholar]
- 32. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., Qin D., Feng S., Song H., Yang D., Zhang B., Zeng L., Lai L., Esteban M. A., Pei D. (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 33. Samavarchi-Tehrani P., Golipour A., David L., Sung H. K., Beyer T. A., Datti A., Woltjen K., Nagy A., Wrana J. L. (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 [DOI] [PubMed] [Google Scholar]
- 34. Chambers I., Tomlinson S. R. (2009) The transcriptional foundation of pluripotency. Development 136, 2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Papalopulu N., Kintner C. (1996) A Xenopus gene, Xbr-1, defines a novel class of homeobox genes and is expressed in the dorsal ciliary margin of the eye. Dev. Biol. 174, 104–114 [DOI] [PubMed] [Google Scholar]
- 36. Cao Y., Knöchel S., Donow C., Miethe J., Kaufmann E., Knöchel W. (2004) The POU factor Oct-25 regulates the Xvent-2B gene and counteracts terminal differentiation in Xenopus embryos. J. Biol. Chem. 279, 43735–43743 [DOI] [PubMed] [Google Scholar]
- 37. Silva J., Chambers I., Pollard S., Smith A. (2006) Nanog promotes transfer of pluripotency after cell fusion. Nature 441, 997–1001 [DOI] [PubMed] [Google Scholar]
- 38. Dixon J. E., Allegrucci C., Redwood C., Kump K., Bian Y., Chatfield J., Chen Y. H., Sottile V., Voss S. R., Alberio R., Johnson A. D. (2010) Axolotl Nanog activity in mouse embryonic stem cells demonstrates that ground state pluripotency is conserved from urodele amphibians to mammals. Development 137, 2973–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tat P. A., Sumer H., Jones K. L., Upton K., Verma P. J. (2010) The efficient generation of induced pluripotent stem (iPS) cells from adult mouse adipose tissue-derived and neural stem cells. Cell Transplant. 19, 525–536 [DOI] [PubMed] [Google Scholar]
- 40. De Carvalho D. D., You J. S., Jones P. A. (2010) DNA methylation and cellular reprogramming. Trends Cell Biol. 20, 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singhal N., Graumann J., Wu G., Araúzo-Bravo M. J., Han D. W., Greber B., Gentile L., Mann M., Schöler H. R. (2010) Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141, 943–955 [DOI] [PubMed] [Google Scholar]
- 42. Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S. K., Handa V., Döderlein G., Maltry N., Wu W., Lyko F., Niehrs C. (2007) Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445, 671–675 [DOI] [PubMed] [Google Scholar]
- 43. Hansis C., Barreto G., Maltry N., Niehrs C. (2004) Nuclear reprogramming of human somatic cells by xenopus egg extract requires BRG1. Curr. Biol. 14, 1475–1480 [DOI] [PubMed] [Google Scholar]
- 44. Albini S., Puri P. L. (2010) SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: it's time to exchange! Exp. Cell Res. 316, 3073–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang J., Wang F., Okuka M., Liu N., Ji G., Ye X., Zuo B., Li M., Liang P., Ge W. W., Tsibris J. C., Keefe D. L., Liu L. (2011) Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 21, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delaune E., Lemaire P., Kodjabachian L. (2005) Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development 132, 299–310 [DOI] [PubMed] [Google Scholar]
- 47. Okabayashi K., Asashima M. (2003) Tissue generation from amphibian animal caps. Curr. Opin. Genet. Dev. 13, 502–507 [DOI] [PubMed] [Google Scholar]
- 48. Blelloch R., Wang Z., Meissner A., Pollard S., Smith A., Jaenisch R. (2006) Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells 24, 2007–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hanna J., Markoulaki S., Schorderet P., Carey B. W., Beard C., Wernig M., Creyghton M. P., Steine E. J., Cassady J. P., Foreman R., Lengner C. J., Dausman J. A., Jaenisch R. (2008) Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133, 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. (2008) Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watanabe S., Hirai H., Asakura Y., Tastad C., Verma M., Keller C., Dutton J. R., Asakura A. (2011) MyoD gene suppression by Oct4 is required for reprogramming in myoblasts to produce induced pluripotent stem cells. Stem Cells 29, 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A., Jaenisch R. (2009) Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Straube W. L., Tanaka E. M. (2006) Reversibility of the differentiated state: regeneration in amphibians. Artif. Organs 30, 743–755 [DOI] [PubMed] [Google Scholar]
- 54. Kim J. B., Sebastiano V., Wu G., Araúzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B. K., Zaehres H., Schöler H. R. (2009) Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411–419 [DOI] [PubMed] [Google Scholar]
- 55. Jullien J., Astrand C., Halley-Stott R. P., Garrett N., Gurdon J. B. (2010) Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc. Natl. Acad. Sci. U.S.A. 107, 5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johnson A. D., Richardson E., Bachvarova R. F., Crother B. I. (2011) Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction 141, 291–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.