Background: The channelrhodopsins are widely used for optogenetic application, whereas a structural model was not available before now.
Results: Our modeled structure identifies remarkable structural motifs and elucidates important electrophysiological properties of Channelrhodopsin-2.
Conclusion: Channel function relies on the unusual properties of helices 1 and 2.
Significance: The atom level structure promotes further understanding of function and may guide the engineering of channelrhodopsins for novel optogenetic applications.
Keywords: Chlamydomonas, Electrophysiology, Homology Modeling, Optogenetics, Photoreceptors, Potassium Channels, Quantum Chemistry, Rhodopsin, Sodium Channels, Molecular Dynamics Simulation
Abstract
Channelrhodopsins (ChRs) are light-gated cation channels that mediate ion transport across membranes in microalgae (vectorial catalysis). ChRs are now widely used for the analysis of neural networks in tissues and living animals with light (optogenetics). For elucidation of functional mechanisms at the atomic level, as well as for further engineering and application, a detailed structure is urgently needed. In the absence of an experimental structure, here we develop a structural ChR model based on several molecular computational approaches, capitalizing on characteristic patterns in amino acid sequences of ChR1, ChR2, Volvox ChRs, Mesostigma ChR, and the recently identified ChR of the halophilic alga Dunaliella salina. In the present model, we identify remarkable structural motifs that may explain fundamental electrophysiological properties of ChR2, ChR1, and their mutants, and in a crucial validation of the model, we successfully reproduce the excitation energy predicted by absorption spectra.
Introduction
The channelrhodopsins ChR13 and ChR2 were originally found in the green alga, Chlamydomonas reinhardtii, where they serve as sensory photoreceptors for phototaxis and phobic responses (1–4). The demonstration that channelrhodopsins act as light-gated ion channels was followed by major advances in neuroscience, where ChRs are used for neuronal depolarization and firing of action potentials simply via delivery of short light flashes (5–8). ChRs also suggest hope for certain clinical conditions. For example, ChRs might be employed clinically for recovery of photosensitivity in blind mice with degenerated photoreceptor cells, for deep brain stimulation to treat Parkinson disease, for peripheral nerve stimulation and treatment of heart pacing failure, and for modulating synaptic depression and associated fear (9). Interestingly, although most experiments have been conducted with ChR2 because of superior expression and 2-fold higher cation selectivity, ChR1 is in fact the dominant photoreceptor of the alga. It shows a strongly pH-dependent absorption, reduced inactivation, and a faster photocycle. Because of these specific differences, the analysis of both ChRs is of high interest.
ChRs are thought to belong to the protein family of microbial rhodopsins, which include rhodopsins of eubacteria, archaea, cyanobacteria, algae, and fungi. Although much knowledge has been gained by recent experimental studies, a deeper understanding of ChR function, including light gating, ion conduction, and color-tuning mechanisms, clearly requires the knowledge of the three-dimensional structure. The only experimental structure available so far has been resolved by electron crystallography at 6 Å resolution (10) and revealed that ChR2 consists of seven transmembrane helices similar to other microbial rhodopsins. However, because of the low resolution, precise understanding of the position and orientation of each residue is still not available. In fact, the amino acid residues constituting the rhodopsin helices C–G have been identified in the sequences of ChRs. However, the amino acid sequence that constitutes the first two helices has been much less tractable to structural prediction, because sequence alignment of ChRs with other microbial rhodopsins could not be determined uniquely (2–4).
The disagreement about the arrangement of helices A and B is not only caused by the lower homology with other microbial rhodopsins but also by the charged residues intruding into the transmembrane region. The sequence around the location of the first two helices contains a very high amount of charged amino acids, which are usually thought to be unlikely to be found in a transmembrane helix region. Although such charged residues seem to characterize the channel, they make it difficult to apply a generalized method for structure prediction. In fact, structure prediction programs as SOSUI (11), which is one of the most accurate algorithms for the secondary structure prediction of membrane proteins with accuracy of 80–90%, predict ChRs as proteins with only four or five transmembrane helices.
To account for the channel function, however, the situation may be quite different. It is interesting to note that there is a sequence pattern, a 7-residue periodicity, which allows the building of helices with the charged/polar side chains aligned along a quasi one-dimensional chain inside the protein, creating a hydrophilic environment that seems very likely to establish the channel. This sequence pattern could determine the structure of the first two helices, and further help for the alignment comes from the recently discovered sequence of a novel channelrhodopsin (DChR) of the halophilic alga Dunaliella salina, which constitutes an evolutionary link between bacterial rhodopsins as Anabaena sensory rhodopsin (ASR) and bacteriorhodopsin (BR), and the ChRs. With the help of these two findings, we are able to propose a three-dimensional structural model of ChR1 and ChR2 based on new experimental data and computational analysis.
In the first step, we build a homology model using the DChR sequence and the characteristic sequence pattern of helices A and B. Homology modeling has been successfully applied for structure prediction when the template sequence has high homology with that of the target protein, as recently shown for G protein-coupled receptors (12, 13), which have seven transmembrane helices similar to the microbial rhodopsins. Based on the ASR structural template, we construct a three-dimensional structural model, which is subjected to extensive molecular dynamics (MD) simulations to examine its stability and dynamical features. This model allows interpretation of recent experimental findings and has stimulated further site-directed mutational studies in our group, to examine the role of important amino acids in the active site in more detail. The characteristic three-dimensional structure of ChRs (especially in the region of helices A and B) also allows study of the distribution of internal water molecules, which traverse the ChR itself and may be responsible for channel function. Finally, we are able to localize ion binding sites and propose a model for the active site based on combined quantum mechanics/molecular mechanics (QM/MM) methods that allow computational reproduction of the excitation energy for ChR1 and ChR2 when applying high level post Hartree-Fock methods (multi-reference configuration interaction) for the QM region.
EXPERIMENTAL PROCEDURES
Generation of Three-dimensional Structural Model
In a first step, we constructed a homology model of ChR2 using the SWISS-MODEL web interface (14) based on the three-dimensional structural template of the ASR x-ray structure (Protein Data Bank code 1xio) (15). The residue correspondence between ChR2 and ASR was based on the sequence alignment shown in supplemental Fig. S1. Although the cRNA of ChR2 encodes in total 737 amino acids, we made use only of the amino acid residues 49–273 of ChR2, i.e. only those who have a counterpart in the ASR sequence. After the replacement of internal water molecules, the protein and water complex was embedded in 1-palmitoyl-2-oleoylphosphatidylcholine lipid bilayer. The modeling details are described in the supplemental Methods.
pKa Calculations
Because the pKa calculations do not necessarily lead to unambiguous results, we used two different models, the multi-conformer continuum electrostatics (16) model using two different values of the dielectric constant (ϵ = 4.0 and ϵ = 8.0) and the empirical predictions from the PROPKA server (17). The determination of the protonation state of the titratable residues is detailed in the supplemental Methods and supplemental Table S1.
Molecular Dynamics Simulations
The resulting structural model was subjected to extended MD simulations for structural sampling. To avoid an accidental sampling bias, we performed three 50-ns MD simulations with different initial velocities. As a result, we effectively obtained a 150-ns trajectory for the structural analysis and the subsequent QM/MM simulation. All of the MM calculations were performed using GROMACS 4.5 (18), which adopted CHARMM27 potential energy function (19). The protocol and the setup of the MD simulations are detailed in the supplemental Methods and supplemental Fig. S10.
QM/MM Simulation
In the QM/MM simulations, all lipid and bulk solvent molecules were removed, and harmonic constraints were imposed on the backbone Cα atoms to suppress global structural disorder.
We employ the CHARMM27 force field as implemented in the CHARMM 36a2 (20) program for the MM part and apply the approximate density functional theory method SCC-DFTB (21) for the QM region. Self-consistent-charge density-functional tight-binding (SCC-DFTB) has been carefully benchmarked and widely applied in our previous studies to retinal proteins (see the supplemental Methods). To mimic the screening effect of bulk solvent and membrane, the Poisson-Boltzmann charge scaling scheme proposed by Dinner et al. (22) was adopted, where the partial charges of the atoms exposed to bulk phase were scaled. The QM region consists of the retinal, the side chains of Lys-257, Glu-123, Asp-253, and Lys-103 and the closest three water molecules to the Schiff base. As a starting point for the QM/MM geometry optimization and MD simulations, we chose the average structure from the classical MD simulations (see the supplemental Methods).
Excitation Energy Calculation
For the vertical excitation energy calculations, we applied multi-reference configuration interaction method called spectroscopy-oriented configuration interaction (SORCI) method (23) implemented in ORCA 2.8 program. In the calculation, the first order interaction space is divided automatically into weakly and strongly interaction spaces. The former is subjected to a variational treatment, whereas the latter is taken into account using perturbation theory. In the SORCI calculations, Ahlrich's SV(P) basis set was employed, and the threshold parameters Tpre = 10−3, Tsel = 10−6, and Tnat = 10−6 were used.
Electrophysiology
DNA mutations were generated by site-directed mutagenesis (QuikChange II; Agilent Technologies) on an hChR2 (amino acids 1–309)-T159C-mCherry vector and confirmed by sequencing. HEK293 cell culturing and whole cell patch clamp measurements were done as earlier described by Yizhar et al. (24). Transient transfection was performed with FuGENE HD (Roche Applied Science) 20–28 h prior to the electrical recordings. The external measurement solution contained 140 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, 2 mm KCl, 10 mm HEPES (pH 7.2). The internal solution was composed of 110 mm NaCl, 10 mm EGTA, 2 mm MgCl2, 1 mm CaCl2, 5 mm KCl, 10 mm HEPES (pH was adjusted to 7.2 either using CsOH or HCl). HEK cell whole cell patch clamping was performed with an EPC 7 (HEKA, Elektronik GmbH, Lambrecht, Germany) amplifier, whereas a Polychrome V unit (TILL Photonics, Planegg, Germany) was used as activation light source, resulting in a final photon density of ∼1 × 1022 photons m−2 s−1 at 460 nm on the coverslip. For recording the action spectra, only 50% of the light intensity was used. Excitation wavelength for amplitude determination was 460 nm for all constructs.
For experiments in Xenopus laevis oocytes, we used the plasmid DNA of pGEM-ChR11–346 (25). cRNA of ChR11–346 (amino acids 1–346) and mutants were synthesized in vitro from NheI-linearized pGEM-ChR1 plasmid by using T7 RNA polymerase (mMessage mMachine; Ambion) (1). Oocytes from female X. laevis were injected with 50 nl of cRNA (0.5–1 μg/μl) and incubated in the dark at 18 °C in ORI solution (96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm MOPS-NaOH, pH 7.5) in the presence of 5 mm sodium pyruvate, 50 μg/ml gentamicin, and 1 μm all-trans-retinal (Sigma).
Two-electrode voltage clamp measurements on X. laevis oocytes were performed under conditions described before (26) by using a Turbo Tec-03X (NPI Electronic, Tamm, Germany). Data acquisition and light triggering were controlled with pCLAMP software via DigiData 1322A interfaces (Molecular Devices, Sunnyvale, CA). The microelectrodes were fabricated by pulling borosilicate glass capillaries (1.5-mm outer diameter and 1.17-mm inner diameter) using a micropipette puller (model P-97; Sutter Instruments, Novato, CA) and filled with 3 m KCl. The resistances of microelectrodes were 0.5–1.5 MΩ. A polychrome2 (TILL Photonics, Gräfelfing, Germany) was used as tunable light source. The light was applied to the oocytes by using a light guide (diameter of 2 mm). The light intensity was 1.5 × 1022 photons s−1 m−2 at the surface of the oocyte. The data obtained were averages of more than three experiments. The solution contained 100 mm NaCl, 1 mm MgCl2, 0.1 mm CaCl2, and 5 mm citrate (pH 5.0).
RESULTS
Alignment
Throughout this study the residue numbering of ChRs follows that of ChR2. Correspondences among ChRs based on homology and alignment are shown in supplemental Fig. S1.
In the last years, several proposals for sequence alignments of ChRs with BR have been put forward (1, 3, 4, 27). Although the position of the helices C–G can be determined quite well because of reasonable sequence homology, the position of the first two helices has remained unresolved because the alignment is highly underdetermined when based on the BR sequence. To include more information for a reasonable modeling, we included the sequences of sensory rhodopsin of the cyanobacterium Anabaena PCC 7120 (ASR) and the only recently identified channelrhodopsin sequences of Mesostigma (28) and the halophilic alga D. salina.4 Cyanobacteria are considered evolutionary precursors of green algae and as an evolutionary link between eubacteria and eukaryota (29). Moreover, the intracellular part of ASR is more hydrophilic and contains more water than other microbial rhodopsins (15), which might serve as a structural link between light-driven pumps and light-gated channels. DChR in turn might serve as an evolutionary link between algae and halophilic archaea, which lives in symbiosis with Dunaliella supplemental Table S4. Finally, Mesostigma ChR is of interest because Mesostigma is evolutionary distant from Chlamydomonas, Volvox, and Dunaliella and therefore could highlight residues that are essential for channel function.
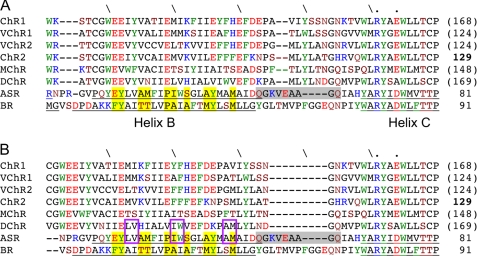
The most interesting feature of the ChR sequences is the large number of charged residues in the potential region of the first two helices, which are Glu-82, Glu-83, Glu-90, Lys-93, Glu-97, His-100, Glu-101, Asp-103, and Glu-104 in ChR1 and ChR2. Glu-82, Glu-90, and Glu-101 are completely conserved in all six known ChRs (Fig. 1). These residues posed a large problem for many of the earlier alignments, because they were either placed in a loop or inside the helices, being partially oriented toward the membrane. However, their spacing shows a very interesting periodicity: charged residues appear at every seventh amino acid site in the series Glu-83, Glu-90, Glu-97, and Glu-104 (and for Lys-93 and His-100 as well).
FIGURE 1.
Two alignment candidates for helix B. Helical regions in ASR and BR are underlined. Positively and negatively charged residues are colored blue and red, respectively. Aromatic residues are colored green. Yellow highlighting means the amino acid site where the side chains are oriented inside protein. Homologies between DChR and ASR in helix C are highlighted with violet squares.
In an α-helix this periodicity locates the residues on top of each other, forming a chain of charged residues; because the α-helix contains 3.6 residues per turn, each amino acid residue rotates 100° in the helix, and the side chains of every seventh residue in an α-helix roughly point in the same direction. This remarkable feature strongly suggests a functional significance. First, it allows all charged side chains to have an inward orientation that is clearly favored from an energetic perspective. Charges pointing toward the hydrophobic lipid environment can be assumed to be highly unfavorable. This is highlighted in supplemental Fig. S2, where a helical wheel representation is given. Second, the inward oriented chain of charged groups creates a hydrophilic environment, which is likely to be solvated with water molecules. In fact, as will be shown below, such a structure is readily flooded by waters in a MD simulation, forming a water channel that connects the extracellular with the intracellular bulk phase and is only interrupted in the dark state by the retinal moiety. Note, that such a periodicity can also be observed for the aromatic residues, Tyr-43, Tyr-57, Tyr-64, and Phe-71 in BR, and these first three residues belong to helix B. These residues in BR can then be thought of preventing such a water file in BR, which can be seen as a major difference between the pump BR and the channels ChR1 and ChR2.
To build a structural model for the ChRs based on this finding, we identified several amino acids in helix B of the BR and ASR template structures that point toward the protein interior and are separated by six residues. These are Glu-36, Tyr-37, Ala-40, Ile-43, Pro-44, Ser-47, Ala-50, Tyr-51, and Met-54 in the ASR structure and Phe-42, Tyr-43, Thr-46, Val-49, Pro-50, Ala-53, Met-56, and Tyr-57 in the BR structure. Interestingly, only two alignments satisfy the condition that the charged sites in ChRs coincide with the inside-orientated sites in ASR (Fig. 1, models A and B). However, it is difficult to decide which of the two alignments, which differ by a sequence shift of seven residues, is a better candidate for helix B. Therefore, we have constructed two model structures using both alignments for further investigation.
In a second step, we considered a model for helix A that shows a reduced homology between DChR and ASR compared with helix B. Although it contains no prominent charged residues, one still can identify a seven-residue periodicity for polar amino acid residues between residues 52 and 73 of ChR2, which we propose to constitute helix A. There are two groups exhibiting such a periodicity: the first one consisting of Ser-52, Ala-59 (threonine in ChR1), Leu-66 (glutamine in DChR2), and Gln-73 and the second one consisting of Gln-56, Ser-63, and Tyr-70 (see supplemental Fig. S1). The residues should prefer an inward orientation similar to the charged ones in helix B. The side chains of helix A in BR with inward orientation are numbered as: 9, 13, 16, 20, 23, 24, 27, and 30 (4, 8, 11, 15, 18, 19, 22, and 25 in ASR). With this information, the alignment of helix A can be determined uniquely because the two groups of residues in ChRs can be nicely matched with the corresponding ones from BR and ASR.
Three-dimensional Structure of the First Two Helices
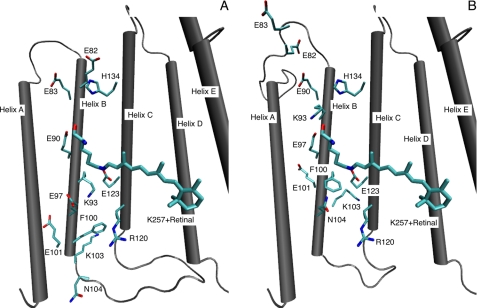
Based on the ASR crystal structure as template, we have built two three-dimensional structural models of ChR2 based on the two different alignments for helix B (Fig. 2 and supplemental Fig. S3).
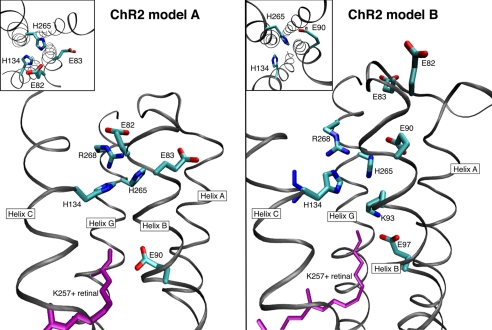
FIGURE 2.
Three-dimensional structure of the ChR2 homology models A and B with focus on the active site region.
Model A at first sight seems favorable compared to model B because the gap region between helix B and helix C is smaller, which is generally preferred by conventional alignment techniques and all glutamic acid residues from Glu-82 to Glu-104 reside in the helical region, which is not the case for alignment B (Fig. 2 and supplemental Fig. S3). Further, Glu-82, Ile-89, and Asp-103 would be conserved for all ChRs (except VChR2 with Val-89) and ASR when using model A.
Considering an evolutionary relationship between ASR and DChR, the latter may be considered as a link between the ChRs and ASR. This relation would support model B: DChR then would have a high homology with the ChRs but also substantial homology with ASR identified in three doublets Leu-91/Val-92, Ile-98/Trp-99, and Ala-106/Met-107 in DChR and Leu-38/Val-39, Ile-45/Trp-46, and Ala-53/Met-54 in ASR (Fig. 1).
Global Structure and Active Site
From the homology model, we generated a three-dimensional structural model embedded into a model membrane surrounded by bulk water, and MD simulations have been performed to probe the dynamics and stability of the model structures (for details see the supplemental Methods). The most obvious residue to study is certainly Glu-123, which is close to the Schiff base in both alignments. Glu-123 is the substitute for D85 in BR, which is the proton acceptor during deprotonation of the Schiff base. Glu-123 has been replaced in earlier studies by the protonated glutamine (E123Q), by the hydrophobic alanine (E123A), or by the threonine (E123T). These mutations accelerate the photocycle and shift the action spectrum to longer wavelengths by 20 nm for Thr and Gln and 9 nm for Ala (30, 31), which is in line with both arrangements of Fig. 2. Next to Glu-123, a positively charged residue is located close to the Schiff base but different at different places in both models. It is Lys-93 in the model A and His-100/Lys-103 (in ChR1/ChR2) for model B.
In model A (Fig. 2), the active sites are equal in ChR1 and ChR2, consisting of Glu-90, Glu-123, Lys-93, and, a little further apart, Glu-97, whereas they are very different according to model B (for ChR1, see supplemental Fig. S3). Here, for ChR1 the protonated His-100 in the center is surrounded by the amino acids Glu-97, Glu-101, Lys-103, and Glu-104 on helix B and Glu-123 on helix C, as shown in supplemental Fig. S3. This structure can be seen in the three other channelrhodopsins ChR2, VChR1, and VChR2 as well, although Glu-104 is mutated into serine in VChR1, and the positively charged site shifts from His-100 to Lys-103 in ChR2. This highly polar structural motif would be critical for channel gating and ion conductance but may also contribute to the blue shifted absorption of both ChR1 and ChR2 when compared with BR. The latter is investigated in more detail below.
To understand this residue network in more detail, we mutated several amino acid residues of ChR2 and tested the photocurrents in HEK cells. Mutations were made on the ChR2-T159C background because the currents were very large (31), and 159 is far from the active site (discussed later in Fig. 5).
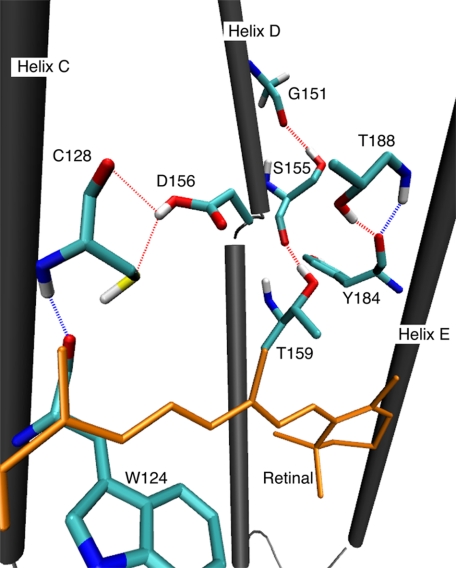
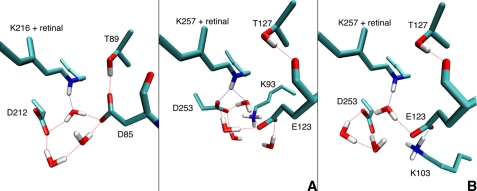
FIGURE 5.
Threonines on the helix C, D, and E with hydrogen bonds. The structure was optimized by QM/MM extracted from the trajectory of QM/MM simulation.
First, we considered three mutations, namely F100H, K103D, and N104E. According to model B, this would convert the ChR2 active site into the ChR1 counterpart (Fig. 2, model B), whereas in model A these residues would be located at the lower part of helix B, being closer to the potential channel entrance. The protein still showed 55% activity (Fig. 3), and although the mutation of the charged residue in the vicinity of the chromophore generally has a significant impact on the spectrum, the observed λmax shift was only several nm (supplemental Fig. S4). This indicates an important role of these residues for conduction, but not for color tuning, which at a first glance seems to be more consistent with model A than for B. Note, however, that the total charge in model B ChR1 and ChR2 in the active site is conserved, which could account or the missing color shift upon the triple mutation. Nevertheless, model A seems to be favorable in this respect.
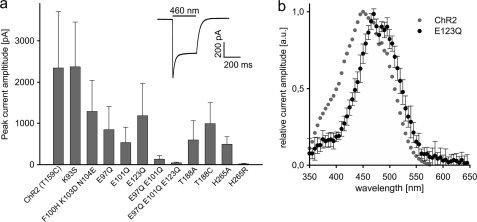
FIGURE 3.
Photocurrents amplitudes (I0) of ChR2-T159C and mutants. a, typical photocurrent is inset. ChR2-T159C was used as standard because of the extra large currents. On the ChR2-T159C background, amino acids that are part of the calculated active site were mutated individually or as doublets or triplets. b shows action spectrum of ChR2 T159C/E123Q at pH 7.2. Action spectra for other mutants are shown in the supplemental Fig. S4.
In separate experiments, we removed the charges from Glu-90, Glu-101, and Glu-123 and replaced them with Gln. The peak current amplitude dropped by 64, 76, and 50% respectively (Fig. 3). In E123Q, the action spectrum was red-shifted by 25 nm (Fig. 3), whereas the other mutants showed only small shifts of a few nm (supplemental Fig. S4). The data suggest that all three residues support function but are individually not of critical importance. However, multiple replacements in the form of E97Q/E101Q or E97Q/E101Q/123Q dropped the activity to 5 or 2% (Fig. 3 and supplemental Table S2).
Fig. 4 shows the cytoplasmic side of the three-dimensional structure based on the alignments A and B. Interestingly a number of titratable residues are present at that location, including Glu-90 and His-134, which have been intensively studied by several groups. They are known to be involved in H+ selectivity and H+ release. Substitution of Glu-90 by Gln or Asp or His-134 by Arg or Asn inhibited H+ conductance and promoted Na+ selectivity (27, 32, 33), suggesting that both residues are responsible for H+ transfer and release into the medium or part of the internal selectivity filter. Ritter et al. (34) and Radu and co-workers (35, 36) monitoring FTIR differences between the dark state and the intermediate P500 reported that ChR2 has two indicative band patterns around 1700 cm−1, implying the existence of two protonated carboxyl residues at least. The first band with higher frequency, 1747(+)/1739(−) cm−1, is assigned to Asp-156 (35, 36), being identical to protonated Asp-115 in BR. The second one, 1729(+)/1739(−) cm−1, is assigned to Glu-90 because it is not observed in the E90Q mutant (34, 35).
FIGURE 4.
The cytoplasmic side of ChR2 focusing on helices A, B, and G for both homology models A and B.
The lower frequency of Glu-90 implies stronger hydrogen bonding, which could be to a hydrophilic exposure of the carboxyl side chain. This would support alignment B, where Glu-90 is located on the edge of helix B. However, it could also mean that Glu-90 is strongly interacting with water molecules supposedly located in the channel. As shown for model A in the supplemental Fig. S6, water distributes in the vicinity of Glu-90 during the MD simulations. Therefore, it is likely that Glu-90 forms hydrogen bonds with the inner water or the backbone atom of helix G, lowering the frequency of this vibration.
Sugiyama et al. (32) substituted the glutamates of helix B individually by alanine and found strong inhibition of photoreceptor currents for Ala-82, Ala-83, Ala-97, and Ala-101 but not for Glu-90. The same finding has been reported by Ruffert et al. (27), proposing a model for helix B that is very similar to our alignment A, because they propose Glu-90 to be located in the middle of helix B. Replacing Glu-90, Glu-97, and Glu-101 with lysines resulted in a large reduction of conductance, highlighting the importance of these residues for channel function. Note that in alignment B, Glu-82/Glu-83 are directed toward the bulk water phase, where they could act as antenna, attracting cations toward the channel. On the other hand, the model building based on the ASR x-ray structure leads to a shorter helix B than when using the BR structure. For this structure, Glu-82/83 could readily be part of helix B. However, to recover the helix during classical MD, longer sampling times would be required, which is beyond the scope of the present manuscript.
Of interest might be K93Q, which does not show Na+ currents nor inhibition of the H+ conductance by Na+ (50). In both alignments, this residue could be part of the potential water channel, with model A being located in the extracellular side, whereas in model B it is located in the cytoplasmic side.
Note that in the present model B, His-265 is located between Glu-90 and His-134, both of which are responsible for cation selectivity (Fig. 4, inset). Therefore we inferred that His-265 might also play an important role. To verify this proposal, we mutated this histidine and measured the photocurrent, which is found to be drastically reduced with regard to the wild type (supplemental Figs. S5 and S6). In model A, His-265 would be still located in the channel but would not interfere directly with Glu-90 and His-134.
Water Distribution
As a result of the extended MD simulations for both models, a large number of water molecules are found in the interior of ChR2 (see supplemental Fig. S7), which is quite different from the situation in other microbial rhodopsins. The solvent molecules are mainly distributed along helix B caused by the polar and charged residues regularly arranged along this helix. This seems to be reasonable because these two helices differ substantially from those in other rhodopsins and therefore are likely responsible for the channel function. It is also noteworthy that solvent molecules are more populated in the extracellular side than the cytoplasmic side in common with other microbial rhodopsins. Although the distribution is separated by retinal in the dark state, the retinal isomerization triggered by light absorption would cause structural changes at the active site and enable cations to pass through.
DC Gate
In agreement with previous studies (35, 36), the present models both locate Cys-128 on helix C and Asp-156 on helix D in the center of the transmembrane region (Fig. 5). Mutation of Cys-128 into Thr/Ser or Ala and mutation of Asp-156 into Ala slows down the photocycle kinetics by 2–4 orders of magnitude (37, 38). According to these original electrical studies and subsequent FTIR experiments for these mutants (36), Asp-156 in ChR2 is probably protonated. Thus it has been implied that Asp-156 forms a hydrogen bond with Cys-128, where Cys-128 might be a donor against a carbonyl of the side chain of Asp-156. Furthermore, this arrangement would explain that the DC hydrogen bond would play a significant role in controlling the channel kinetics, justifying the term “DC gate” (36).
However, the present MD simulations exhibit that Cys-128 is closer to Trp-124 than Asp-156 (see supplemental Fig. S8), implying that Cys-128 is probably a proton donor of a hydrogen bond with the oxygen atom of the backbone of Trp-124, conflicting with the FTIR model. To investigate this in more detail, we also performed a 20-ns MD simulation of the C128T mutant, which shows that the threonine mutant forms an even stronger hydrogen bond with Trp-124 than the wild type (supplemental Fig. S8), which again is in conflict with the FTIR model. Interestingly, the BR x-ray structure supports the present computational model, indicating that Thr-90 (Cys-128 in ChR2) forms two hydrogen bonds with Asp-115 (Asp-156 in ChR2) and Trp-86 (Trp-124 in ChR2). Although the hydrogen atoms are not resolved in the x-ray structures, Thr-90 should obviously be the proton acceptor for Asp-115 and a donor to Trp-86. This hydrogen bond between Thr-90 and Trp-86 is equal to a motif discussed by Ballesteros et al. (39), where the side chain of threonine in the g− conformation (χ1 = gauche−) forms the intrahelical hydrogen bond with the i − 4 carbonyl oxygen. Although there is only a little information about intrahelical hydrogen bonds of cysteines available, we concluded that both the wild type and C128T mutant should form an intrahelical hydrogen bond.
Next, we computed the distance distributions of the sulfur atom of Cys-128 with two oxygen atoms of the side chain of the protonated Asp-156, Oδ1 (C=O), and Oδ2 (COH) (supplemental Fig. S8). In the C128T mutant, Oδ2 is closer to the side chain of the residue 128 than Oδ1, which means that Asp-156 may not be a proton acceptor but a donor in the hydrogen bond. Although it might be also the case for the wild type, the side chain Asp-156 might interact with the backbone of Cys-128 rather than with its side chain, possibly because cysteines are less polar than threonines. In addition, the MD average structure implies that solvent molecules come into the vicinity of Asp-156. In fact, in the BR x-ray structures, a water molecule forms hydrogen bonds with Asp-115 and Leu-87 connecting helix C and D (40). Thus, it is possible that the missing solvent stabilizes the hydrogen bond in the wild type ChR2. However, this issue may be difficult to describe with pure MM methods, because they can fail to reproduce subtle details of complex hydrogen-bonded networks, as reported, for example, for the BR active site (see below). Therefore, more extended QM/MM simulations may be required that are beyond the scope of the present work.
The arrangement with Cys-128 connected to Asp-156 and the Trp-124 backbone would explain why the mutation C128A has a stronger effect than the mutation D156A and why the double mutation C128S/D156A results in a stabilized step function opsin with an open state life time of 30 min (24), which would not be expected for the sole connection of Cys-128 to Asp-156.
Combined QM/MM Simulation
Because classical force field methods can be problematic for complex hydrogen-bonded active site structures (see the supplemental Methods), we performed QM/MM simulations using the approximate DFT method SCC-DFTB (22) in the QM region (41).
The active sites of models A and B are quite different (Fig. 6). In the case of model A, there is a larger similarity to the situation in BR, where a prominent pentagonal hydrogen-bonded structure has been found (42), consisting of retinal, Asp-212, Asp-85, and three water molecules. The important structural detail is the interaction of retinal with a water molecule, although there is no salt bridge with one of the counterions. A QM/MM geometry optimization starting from the MD average structure shows that this is similar for model A, although a partial salt bridge with one of the counterions is formed. The active site in model B is not constituted by a strong pentagonal H-bonded network as in BR, because Lys-103 comes into the active site, which seems to change electrostatic environment significantly. The distance between the carboxyl side chains of the counterions in ChR2 (5.7 Å) is smaller than that in BR structure (6.1 Å; Protein Data Bank 1c3w), which may be caused by the replacement of aspartic acid by glutamic acid that has a longer side chain (supplemental Table S3). Note that the Asp/Glu replacement from BR to ChR also changes the hydrogen bond ASP-85–Thr-89 compared to Glu-123–Thr-127 in ChR2; Thr-127 forms an intrahelical hydrogen bond with the carbonyl oxygen of Glu-123 (Fig. 6) instead of with the Glu-123 side chain.
FIGURE 6.
Active sites optimized by QM/MM method. Left panel, BR from x-ray crystallography. Middle panel, ChR2 from average of MM dynamics simulations for model A. Right panel, model B.
To probe the stability and the flexibility of the active site, we performed a QM/MM MD simulation (for 1 ns). Although the direct interaction with the Schiff base and the closest water molecule are conserved, Glu-123 seems to be more flexible than D85 in BR, whereas Asp-253 appears as similarly rigid as Asp-212 in BR. For Glu-123, we observe a change in the hydrogen bonding pattern during the MD, i.e. the positions of Oϵ1 and Oϵ2 interchange during the simulations highlighting the floppiness of the hydrogen-bonded network.
Excitation Energy Calculation
The optical absorption spectrum of ChR2 exhibits several peaks (λmax) of 447, 470, and 410 nm in the dark-adapted state at neutral pH (34). Because it is difficult to define the exact maximum in the absorption spectrum of ChR2 at neutral pH, instead we adopt the λmax of the action spectrum consisting of a single peak, namely 2.74 eV (452 nm) (Fig. 3 and Table 1) as the reference for the excitation energy calculation (43).
TABLE 1.
Excitation energy (eV) and wavelength in parenthesis (nm)
In SORCI, the QM region consists of the entire chromophore and lysine side chain.
The computed value for the QM/MM optimized structure is 2.64 eV (469 nm) for model A and 2.67 eV (464 nm) for model B. Therefore, both models led to very similar excitation energies, meaning that they cannot be distinguished by such calculations. Because helices C–G are the same in both models, it seems that the different positions of the charged residues in helix B does not have a large impact on the excitation energies, which is a quite reasonable result.
The results have to be compared with the values for BR and SRII as shown in Table 1. QM/MM estimates of excitation energies rely to some degree on an error cancellation, even when using very accurate QM methods like the SORCI method applied in the present study, as we have discussed in detail in our previous work (41, 44–46), leading to some systematic trends in the computed values. In short, we expect excitation energies to be slightly blue-shifted, mostly because of the neglect of protein polarization.
As can be seen from the examples of BR and SRII from Table 1, the computed value of 2.64/2.67 eV seems to be an excellent estimate of the ChR2 λmax. This indicates that our computed structure gives excitation energies in semi-quantitative agreement with the experimental ones, although the calculations cannot be used to distinguish between the two models. In general, proximity of the negative charge to the Schiff base stabilizes the electronic ground state and destabilize the excited state leading to blue shift and vice versa. Therefore, the mutational λmax shifts agree well with the present models where both glutamic acids are supposed to be deprotonated.
DISCUSSION
In this work, we proposed a three-dimensional model of ChR2/ChR1 based on the two alignments in Fig. 1, which result from the observation of the seven-residue periodicity of conserved charged amino acids in the ChR sequences. This pattern is therefore likely to have a functional significance, and indeed, it allows for the formation of a hydrophilic channel, which has been confirmed by our MD simulations (supplemental Fig. S7), because the side chains of Glu-90, Lys-93, Glu-97, Glu-101, and Lys-103 in ChR2 are oriented to inside the protein. Note that these residues have been shown by experiments to be significant for the channel function. However, we were not able to determine the alignment unambiguously; therefore we probed the two possible alignments for helix B, which are similar to the previous suggestion from Ruffert et al. (27) regarding helix B.
The experimentally investigated mutations in this work seem to be consistent with both models A and B as well, leaving the determination of the exact structure for future work. The triple mutant F100H/K103D/N104E has only a small impact on the optical spectrum, a finding in favor of model A at first sight, because model B places these residues prominently into the retinal binding pocket, where such drastic changes are thought to lead to sizable effects. However, the strong H-bonded network may allow for charge shifts upon mutation; therefore this issue is not easy to decide.
A further hint results from the QM/MM MD simulations, which show for model A a much tighter bound binding pocket, whereas for model B larger fluctuations and flip-flop motions occur. From a functional perspective, the effect of isomerization, which leads to channel opening, may be easier to understand on the basis of a tighter bound active site. Model B, on the other hand, has been proposed because of the homology of ASR and DChR, considering their evolutionary relationships. Also the location of Glu-90 at the protein surface in model B more easily explains the red-shifted IR absorption. However, this may also be due to interaction with waters in the channel of model A.
Helices C–G are the same in model A and B; therefore, the discussion of the so called DC gate may be independent of a particular alignment. Our MD model leads to different conclusions regarding the interaction of Cys-128 and Asp-156 than the proposed model from FTIR measurements, in particular regarding the C128T mutant. The interactions of threonines at site i with backbone carbonyl oxygens at site i − 4 are of particular interest, because they may distort the helical structure (39). This is also found with the side chain of Thr-127 on helix C (Fig. 1), which also assumes a g− conformation forming the intrahelical hydrogen bond with Glu-123 according to our MD simulations, thereby distorting the helical structure leading to lower helicities at residues 126 and 127 in the wild type (supplemental Fig. S9). Furthermore, we also found that the C128T mutation additionally reduces the helicity at residue 126. Because proline generally disrupts a helical structure, Pro-129 also destabilizes helix C (supplemental Fig. S9). As a result, helix C is bent and flexible around the structural motif consisting of Thr-127, Cys-128, and Pro-129.
It should be also emphasized that Thr-127 has an intrahelical hydrogen bond with the backbone of Glu-123 in the active site (Fig. 6). Although Thr-89 in BR (Thr-127 in ChR2) interacts with the Asp-85 side chain (Glu-123 in ChR2) in the dark state (Fig. 6), it has been proposed that Thr-89 forms an intrahelical hydrogen bond with Asp-85 in the O-intermediate state (47). Therefore, it is also possible that the corresponding hydrogen in ChRs may alter the bonding pattern during the photocycle.
Taking into account the flexibility and the connection to the active site, the intrahelical hydrogen bonds may be important for the channel on and off gating during the ChR photocycle. Note that the three residues 127–129 are highly conserved among ChRs (Fig. 1), supporting the present model. In addition, it has been reported that C128X mutations dramatically slow down the photocycle (37, 38).
Coming to Thr-159 on helix D, it has been recognized that the mutation T159C causes 10 times larger photocurrents compared with the ChR2 wild type in Xenopus oocytes (31). Moreover, the T159M mutation alters the ion selectivity of the channel in favor of K+ with strong consequences on the reversal potential in living cells (31). In the present model, we could not find any polar side chains that could interact with Thr-159. Instead, the side chain of Thr-159 in the g− conformation forms the intrahelical hydrogen bond with Ser-155, causing a distortion of helix D as shown in Fig. 5. Because Thr-159 is located close to the DC gate, it is likely that Thr-159 and its hydrogen bond act cooperatively with the DC gate. This is in line with experimental findings that modification of the DC gate has a reduced influence in VChR1 or the C1V1 hybrid (24), which naturally contain a cysteine at the 159 position. Furthermore, we can see intrahelical hydrogen bonds between Ser-155–Gly-151 on helix D and Thr-188–Tyr-184 on helix E in the vicinity of the retinal binding pocket (Fig. 5). Therefore, Ser-155 and Thr-188 may contribute to the function in the same way that the DC gate and Thr-159 do. To prove this, we measured the photocurrent of ChR2 mutants T188A and T188C. Both constructs showed normal targeting and expression levels as evaluated from enhanced cyan fluorescent protein fluorescence. The mutations result in a drastic reduction of the photocurrent showing the importance of this structural motif for the ChR function (supplemental Figs. S5 and S6). T188A showed hardly any photocurrents, whereas photocurrents of T188C were approximately five times smaller than that of wild type. This might be related to the fact that cysteine can still form an intrahelical hydrogen bond, different from alanine, although it can expected to be weaker than that of threonine.
Based on the electron-microscopy structure (10), it has been proposed that the ChR2 channel is located at the dimer interface on the 2-fold axis, lined by transmembrane helices C and D. To investigate this issue in more detail, we performed 30-ns simulations of ChR2 using the dimeric displacement based on the cryoelectron microscopy result.
With the 2-fold axis between helices C and D, as suggested by cryoelectron microscopy, the simulations do not show any steric hindrance and do exhibit the densely packed hydrophobic residues between the proteins. Note that the alignment about the two helices is uniquely determined. Therefore it is less likely that the channel pore is located at the dimer interface.
We have presented two models of the helix A and B arrangements in ChR and have discussed the expected properties on the bases of experimental data and QM/MM calculations. In both models, the key characteristics are the charged residues in helix B that define in conjunction with residues of helix C and G the ion conducting pore that goes all the way through from the extracellular phase to the intracellular bulk phase. This is the fundamental distinction from the pumping rhodopsins where the retinal is never simultaneously connected to the extra- and intracellular phases by a water column and hydrogen bond networks. A combination of modeling and electrical studies of mutants revealed residues that are important for conductance and ion selectivity. The light switch is conserved in ChR from other microbial rhodopsin, but details about early retinal isomerization and gating of the channel remain unknown and will be addressed in a more extended project already in progress. Strictly speaking, we are unable to prefer any of those two models, and we could even imagine that both arrangements may be realized in different ChRs or even in one ChR under variable ionic or voltage conditions.
Supplementary Material
This work was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (to H. C. W.).

This article contains supplemental text, references, Tables S1–S4, and Figs. S1–S10.
P. Hegemann, F. Zhang, and K. Deisseroth, unpublished observation.
- ChR
- channelrhodopsin
- VChR
- Volvox ChR
- DChR
- D. salina ChR
- ASR
- Anabaena sensory rhodopsin
- BR
- bacteriorhodopsin
- MD
- molecular dynamics
- QM
- quantum mechanics
- MM
- molecular mechanics
- SORCI
- spectroscopy-oriented configuration interaction
- SCC-DFTB
- self-consistent charge density functional tight binding.
REFERENCES
- 1. Nagel G., Ollig D., Fuhrmann M., Kateriya S., Musti A. M., Bamberg E., Hegemann P. (2002) Channelrhodopsin-1. A light-gated proton channel in green algae. Science 296, 2395–2398 [DOI] [PubMed] [Google Scholar]
- 2. Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., Ollig D., Hegemann P., Bamberg E. (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sineshchekov O. A., Jung K. H., Spudich J. L. (2002) Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 99, 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suzuki T., Yamasaki K., Fujita S., Oda K., Iseki M., Yoshida K., Watanabe M., Daiyasu H., Toh H., Asamizu E., Tabata S., Miura K., Fukuzawa H., Nakamura S., Takahashi T. (2003) Archaeal-type rhodopsins in Chlamydomonas. Model structure and intracellular localization. Biochem. Biophys. Res. Commun. 301, 711–717 [DOI] [PubMed] [Google Scholar]
- 5. Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 [DOI] [PubMed] [Google Scholar]
- 6. Ishizuka T., Kakuda M., Araki R., Yawo H. (2006) Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res. 54, 85–94 [DOI] [PubMed] [Google Scholar]
- 7. Li X., Gutierrez D. V., Hanson M. G., Han J., Mark M. D., Chiel H., Hegemann P., Landmesser L. T., Herlitze S. (2005) Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U.S.A. 102, 17816–17821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adamantidis A. R., Zhang F., Aravanis A. M., Deisseroth K., de Lecea L. (2007) Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450, 420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fenno L., Yizhar O., Deisseroth K. (2011) The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller M., Bamann C., Bamberg E., Kühlbrandt W. (2011) Projection structure of channelrhodopsin-2 at 6 Å resolution by electron crystallography. J. Mol. Biol. 414, 86–95 [DOI] [PubMed] [Google Scholar]
- 11. Mitaku S., Hirokawa T., Tsuji T. (2002) Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18, 608–616 [DOI] [PubMed] [Google Scholar]
- 12. Wolf S., Böckmann M., Höweler U., Schlitter J., Gerwert K. (2008) Simulations of a G protein-coupled receptor homology model predict dynamic features and a ligand binding site. FEBS Lett. 582, 3335–3342 [DOI] [PubMed] [Google Scholar]
- 13. Watanabe H. C., Mori Y., Tada T., Yokoyama S., Yamato T. (2010) Molecular mechanism of long-range synergetic color tuning between multiple amino acid residues in conger rhodopsin. Biophysics 6, 67–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vogeley L., Sineshchekov O. A., Trivedi V. D., Sasaki J., Spudich J. L., Luecke H. (2004) Anabaena sensory rhodopsin. A photochromic color sensor at 2.0 A. Science 306, 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu Z., Gunner M. R. (2005) Energetics of quinone-dependent electron and proton transfers in Rhodobacter sphaeroides photosynthetic reaction centers. Biochemistry 44, 82–96 [DOI] [PubMed] [Google Scholar]
- 17. Olsson M. H. M., Sondergaard C. R., Rostkowski M., Jensen J. H. (2011) PROPKA3: Consistent treatment of internal and surface residues in empirical pK(a) predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]
- 18. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) GROMACS. Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 19. Feller S. E., Gawrisch K., MacKerell A. D. (2002) Polyunsaturated fatty acids in lipid bilayers. Intrinsic and environmental contributions to their unique physical properties. J. Am. Chem. Soc. 124, 318–326 [DOI] [PubMed] [Google Scholar]
- 20. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM. The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elstner M., Porezag D., Jungnickel G., Elsner J., Haugk M., Frauenheim T., Suhai S., Seifert G. (1998) Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 58, 7260–7268 [Google Scholar]
- 22. Dinner A. R., Lopez X., Karplus M. (2003) A charge-scaling method to treat solvent in QM/MM simulations. Theor. Chem. Acc. 109, 118–124 [Google Scholar]
- 23. Neese F. (2003) A spectroscopy oriented configuration interaction procedure. J. Chem. Phys. 119, 9428–9443 [Google Scholar]
- 24. Yizhar O., Fenno L. E., Prigge M., Schneider F., Davidson T. J., O'Shea D. J., Sohal V. S., Goshen I., Finkelstein J., Paz J. T., Stehfest K., Fudim R., Ramakrishnan C., Huguenard J. R., Hegemann P., Deisseroth K. (2011) Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berthold P., Tsunoda S. P., Ernst O. P., Mages W., Gradmann D., Hegemann P. (2008) Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell 20, 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ernst O. P., SánchezMurcia P. A., Daldrop P., Tsunoda S. P., Kateriya S., Hegemann P. (2008) Photoactivation of channelrhodopsin. J. Biol. Chem. 283, 1637–1643 [DOI] [PubMed] [Google Scholar]
- 27. Ruffert K., Himmel B., Lall D., Bamann C., Bamberg E., Betz H., Eulenburg V. (2011) Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem. Biophys. Res. Commun. 410, 737–743 [DOI] [PubMed] [Google Scholar]
- 28. Govorunova E. G., Spudich E. N., Lane C. E., Sineshchekov O. A., Spudich J. L. (2011) New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. mBio. 2, e00115–00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stegemann S., Bock R. (2006) Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell 18, 2869–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gunaydin L. A., Yizhar O., Berndt A., Sohal V. S., Deisseroth K., Hegemann P. (2010) Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392 [DOI] [PubMed] [Google Scholar]
- 31. Berndt A., Schoenenberger P., Mattis J., Tye K. M., Deisseroth K., Hegemann P., Oertner T. G. (2011) High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. U.S.A. 108, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sugiyama Y., Wang H., Hikima T., Sato M., Kuroda J., Takahashi T., Ishizuka T., Yawo H. (2009) Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem. Photobiol. Sci. 8, 328–336 [DOI] [PubMed] [Google Scholar]
- 33. Gradmann D., Berndt A., Schneider F., Hegemann P. (2011) Rectification of the channelrhodopsin early conductance. Biophys. J. 101, 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ritter E., Stehfest K., Berndt A., Hegemann P., Bartl F. J. (2008) Monitoring light-induced structural changes of Channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J. Biol. Chem. 283, 35033–35041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Radu I., Bamann C., Nack M., Nagel G., Bamberg E., Heberle J. (2009) Conformational changes of channelrhodopsin-2. J. Am. Chem. Soc. 131, 7313–7319 [DOI] [PubMed] [Google Scholar]
- 36. Nack M., Radu I., Gossing M., Bamann C., Bamberg E., von Mollard G. F., Heberle J. (2010) The DC gate in Channelrhodopsin-2. Crucial hydrogen bonding interaction between C128 and D156. Photochem. Photobiol. Sci. 9, 194–198 [DOI] [PubMed] [Google Scholar]
- 37. Berndt A., Yizhar O., Gunaydin L. A., Hegemann P., Deisseroth K. (2009) Bi-stable neural state switches. Nat. Neurosci. 12, 229–234 [DOI] [PubMed] [Google Scholar]
- 38. Bamann C., Gueta R., Kleinlogel S., Nagel G., Bamberg E. (2010) Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry 49, 267–278 [DOI] [PubMed] [Google Scholar]
- 39. Ballesteros J. A., Deupi X., Olivella M., Haaksma E. E., Pardo L. (2000) Serine and threonine residues bend α-helices in the χ1 = g− conformation. Biophys. J. 79, 2754–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luecke H., Schobert B., Richter H. T., Cartailler J. P., Lanyi J. K. (1999) Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 291, 899–911 [DOI] [PubMed] [Google Scholar]
- 41. Wanko M., Hoffmann M., Strodel P., Koslowski A., Thiel W., Neese F., Frauenheim T., Elstner M. (2005) Calculating absorption shifts for retinal proteins. Computational challenges. J. Phys. Chem. B 109, 3606–3615 [DOI] [PubMed] [Google Scholar]
- 42. Shibata M., Tanimoto T., Kandori H. (2003) Water molecules in the Schiff base region of bacteriorhodopsin. J. Am. Chem. Soc. 125, 13312–13313 [DOI] [PubMed] [Google Scholar]
- 43. Tsunoda S. P., Hegemann P. (2009) Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem. Photobiol. 85, 564–569 [DOI] [PubMed] [Google Scholar]
- 44. Hoffmann M., Wanko M., Strodel P., König P. H., Frauenheim T., Schulten K., Thiel W., Tajkhorshid E., Elstner M. (2006) Color tuning in rhodopsins. The mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. J. Am. Chem. Soc. 128, 10808–10818 [DOI] [PubMed] [Google Scholar]
- 45. Wanko M., Hoffmann M., Frauenheim T., Elstner M. (2008) Effect of polarization on the opsin shift in rhodopsins. 1. A combined QM/QM/MM model for bacteriorhodopsin and pharaonis sensory rhodopsin II. J. Phys. Chem. B 112, 11462–11467 [DOI] [PubMed] [Google Scholar]
- 46. Wanko M., Hoffmann M., Frähmcke J., Frauenheim T., Elstner M. (2008) Effect of polarization on the opsin shift in rhodopsins. 2. Empirical polarization models for proteins. J. Phys. Chem. B 112, 11468–11478 [DOI] [PubMed] [Google Scholar]
- 47. Phatak P., Frähmcke J. S., Wanko M., Hoffmann M., Strodel P., Smith J. C., Suhai S., Bondar A. N., Elstner M. (2009) Long-distance proton transfer with a break in the bacteriorhodopsin active site. J. Am. Chem. Soc. 131, 7064–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Birge R. R., Zhang C. F. (1990) 2-Photon double resonance spectroscopy of bacteriorhodopsin. Assignment of the electronic and dipolar properties of the low-lying 1AG-star(-)-like and 1B-U-star+-like pi,pi-star states. J. Chem. Phys. 92, 7179–7195 [Google Scholar]
- 49. Chizhov I., Schmies G., Seidel R., Sydor J. R., Lüttenberg B., Engelhard M. (1998) The photophobic receptor from Natronobacterium pharaonis. Temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 75, 999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berndt A. (2011) Mechanismus und anwendungsbezogene Optimierung von Channelrhodopsin-2. Ph.D. Dissertation, Humboldt University zu Berlin, Berlin, Germany [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.