Background: HNF4α is a key factor regulating hepatocyte differentiation and liver-specific functions.
Results: Acute disruption of HNF4α in the adult liver causes rapid hepatocyte proliferation through direct and indirect control of multiple pathways.
Conclusion: HNF4α is necessary to maintain the differentiated state of hepatocytes in adult liver.
Significance: HNF4α expression maintains normal liver function, and its loss stimulates hepatocytes proliferation, possibly leading to cancer.
Keywords: Cell Proliferation, Gene Knockout, Gene Regulation, General Transcription Factors, Hepatocyte, HNF4α
Abstract
Hepatocyte nuclear factor 4α (HNF4α) regulates genes involved in lipid and bile acid synthesis, gluconeogenesis, amino acid metabolism, and blood coagulation. In addition to its metabolic role, HNF4α is critical for hepatocyte differentiation, and loss of HNF4α is associated with hepatocellular carcinoma. The hepatocyte-specific Hnf4a knock-out mouse develops severe hepatomegaly and steatosis resulting in premature death, thereby limiting studies of the role of this transcription factor in the adult animal. In addition, gene compensation may complicate analysis of the phenotype of these mice. To overcome these issues, an acute Hnf4a knock-out mouse model was generated through use of the tamoxifen-inducible ErT2cre coupled to the serum albumin gene promoter. Microarray expression analysis revealed up-regulation of genes associated with proliferation and cell cycle control only in the acute liver-specific Hnf4α-null mouse. BrdU and ki67 staining confirmed extensive hepatocyte proliferation in this model. Proliferation was associated with induction of the hepatomitogen Bmp7 as well as reduced basal apoptotic activity. The p53/p63 apoptosis effector gene Perp was further identified as a direct HNF4α target gene. These data suggest that HNF4α maintains hepatocyte differentiation in the adult healthy liver, and its loss may directly contribute to hepatocellular carcinoma development, thus indicating this factor as a possible liver tumor suppressor gene.
Introduction
Cancers of the liver and hepatobiliary tract are the seventh most common type of cancer worldwide (1). In the United States, liver cancers have one of the worst 5-year survival rates at 14.4% (2). Along with hepatitis B or C viral infection, the other risk factor for development of hepatocellular carcinoma (HCC)4 is cirrhosis caused by chronic alcohol consumption (3). In addition, it is now recognized that metabolic syndrome, diabetes, and non-alcoholic fatty liver disease may contribute to the development of HCC due to the deleterious effects of these diseases on liver function (4). As a key regulator of liver homeostasis, disruption of hepatocyte nuclear factor 4α (HNF4α, NR2A1) function results in many of these disorders that contribute to HCC.
HNF4α is a member of the nuclear receptor family of transcription factors. Originally designated as an orphan member due to the lack of a known ligand, subsequent studies have identified fatty acids bound in the ligand binding pocket with linoleic acid as the potential endogenous HNF4α ligand (5–8). HNF4α is critical for proper hepatic lipid homeostasis, as evidenced by the development of steatosis in Hnf4α-deficient mouse livers (9). Mutations in the human HNF4A gene result in maturity onset diabetes of the young type 1, a monogenic form of type 2 diabetes mellitus (10). In addition to regulation of multiple metabolic pathways in the liver, HNF4α expression maintains the differentiated state of hepatocytes. Conditional mouse knock-out models demonstrated the requirement of HNF4α expression to maintain proper liver architecture and many epithelial markers in the developing mouse liver (11, 12). Disruption of HNF4α in mature hepatocytes results in epithelial to mesenchymal transition (EMT) (13). The EMT process is associated with tumor progression and metastatic potential (14). Thus, alterations in HNF4α functionality may increase the risk for development of HCC. Indeed, several mouse and human studies have shown that HNF4α expression is diminished in HCC (15–18). Additionally, it was demonstrated that overexpression of HNF4α in rodent HCC models blocks carcinogenesis and metastasis, thus indicating a potential role for HNF4α as a tumor suppressor (16, 19).
Due to its critical role in liver development, it has been difficult to study the role of Hnf4α in the adult mouse. The full-body Hnf4a knock-out mouse is embryonic lethal, and the liver-specific albumin promoter-driven cre recombinase knock-out mice display severe hepatic metabolic disruption and death at 6–8 weeks of age (9, 20). In addition, the possibility exists that gene compensation may occur when Hnf4α is lost during early development, thus obscuring the potential effects of HNF4α in the adult. To overcome these obstacles, a temporal liver-specific knock-out of Hnf4a was generated using the ERT2cre system, which is activated upon tamoxifen exposure (21). Similar to the Alb-Hnf4a knock-out mouse, acute disruption of Hnf4a resulted in hepatomegaly and hepatosteatosis. Interestingly, microarray analysis identified the up-regulation of proliferative genes only in the acute knock-out. Further investigation revealed the impact of acute Hnf4a disruption on the cell cycle, apoptosis, and growth factors, thus suggesting a potential role for HNF4α as a hepatic tumor suppressor.
EXPERIMENTAL PROCEDURES
Animals and Treatments
The Hnf4aF/F and Alb-Hnf4a−/− mouse lines were described previously (9). To generate the conditional temporal Hnf4aF/F;AlbERT2cre mice, Hnf4aF/F mice were crossed with the tamoxifen-inducible hepatocyte-specific Cre recombinase expressing mouse SA+/Cre-ERT2 (21). The mice were on a mixed SvJ129 and FVB background and used at 6–8 weeks of age. For Hnf4aF/F;AlbERT2cre mice, floxed cre-negative or -positive littermates without tamoxifen treatment were used as controls. Mice were fed a diet containing tamoxifen (1 g/kg diet) for up to 5 days (days 1–5) and then returned to regular chow and euthanized at the indicated times (Fig. 1A). The VhlF/F;AlbERT2cre mice were described previously and treated with tamoxifen for 5 days and then returned to a chow diet for 11 days prior to euthanasia (22). Hepatic deletion of VHL results in constitutive activation of HIF2α in the liver, leading to rapid inflammation, lipid accumulation, and fibrosis (22). For BrdU incorporation experiments, miniosmotic pumps containing sterile BrdU were implanted subcutaneously, and mice were euthanized 6 days later. Serum chemistry analysis was performed using the VetScan VS2 comprehensive and liver profiles (Abaxis, Union City, CA). For determination of liver bile acid levels, 20 mg of frozen liver was homogenized in 400 μl of 75% ethanol, incubated at 50 °C for 2 h, and then centrifuged. The supernatant (aqueous fraction) was retained, evaporated, and resuspended in 200 μl of 0.9% saline. Twenty microliters was used for bile acid quantification using the VetSpec Bile Acids kit (Catachem, Oxford, CT). Mice were housed in a temperature- and light-controlled facility and given food and water ad libitum. All animal studies were performed in accordance with the guidelines and approval of the NCI, National Institutes of Health, Animal Care and Use Committee.
FIGURE 1.
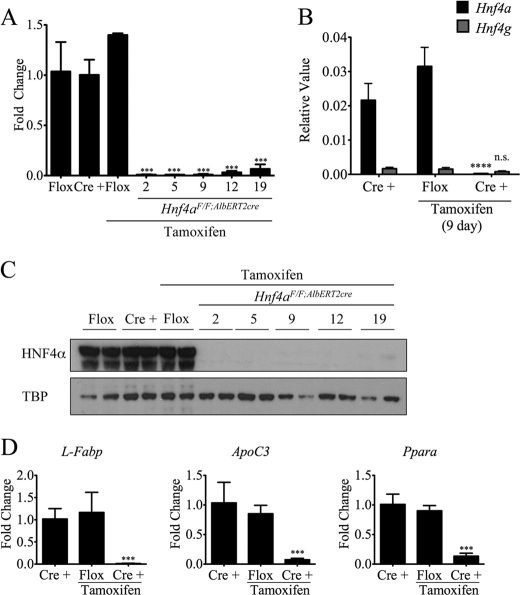
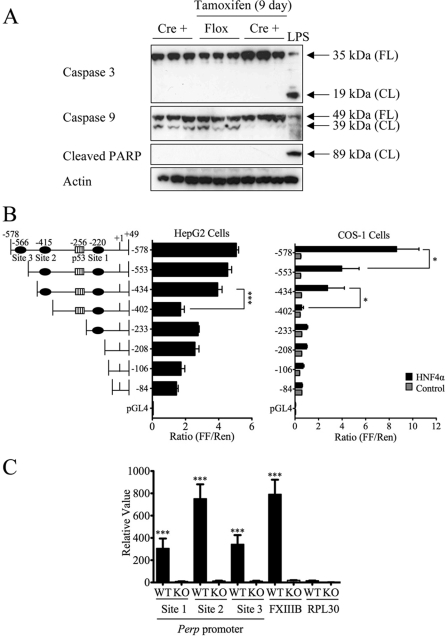
Characterization of the temporal and tissue-specific Hnf4aF/F;AlbERT2cre knock-out mouse. A, Hnf4a mRNA expression as determined by qRT-PCR. Livers (n = 3) from Hnf4aF/F;AlbERT2cre mice (Cre +) were collected at the indicated times after tamoxifen treatment. Nineteen-day Hnf4aF/F mice (Flox) were used as a control for tamoxifen exposure. Values were normalized to actin and represented as -fold change over expression in control Hnf4aF/F mice. Statistically significant changes from Hnf4aF/F control mice as determined by one-way ANOVA are indicated (***, p < 0.0005). B, qRT-PCR expression analysis for Hnf4a and Hnf4g was performed on RNA from 9-day tamoxifen-treated Hnf4aF/F;AlbERT2cre mice. Values shown are relative expression levels. Statistically significant changes from Hnf4aF/F;AlbERT2cre control mice as determined by one-way ANOVA are indicated (****, p < 0.00005). C, Western blot analysis of nuclear protein extracts from two mice per group. Blots were probed with anti-HNF4α antibody or the nuclear loading control TATA-binding protein (TBP). Nineteen-day tamoxifen-treated Hnf4aF/F mice were used as a control for tamoxifen exposure. D, qRT-PCR analysis of known HNF4α target genes in 19-day tamoxifen-treated livers (n = 3). Values were normalized to actin expression and represented as -fold change over control Hnf4aF/F;AlbERT2cre. Statistically significant changes from Hnf4aF/F;AlbERT2cre control mice as determined by one-way ANOVA are indicated (***, p < 0.0005). Error bars, S.D.
Immunohistochemistry
Liver tissue was fixed in 10% phosphate-buffered formalin for 24 h and then processed in paraffin blocks. Four-micrometer sections were used for H&E staining and immunohistochemistry. A rat anti-BrdU antibody (AbD Serotec, Oxford, UK) and a rabbit anti-human ki67 antibody (ab16667, Abcam (Cambridge, MA)) were used for immunohistochemical detection of proliferation.
Western Blot Analysis
For detection of HNF4α, nuclear protein was prepared using the NE-PER nuclear extraction kit (Thermo Scientific, Rockford, IL). For whole cell extract, 50 mg of liver was homogenized in radioimmune precipitation assay buffer with protease and phosphatase inhibitors and centrifuged at 4 °C for 15 min at 15,000 rpm. Fifty micrograms of protein extract was used for Western blot analysis. Membranes were incubated with antibodies against HNF4α (H1415, Perseus (Tokyo, JP)), TATA-binding protein (Abcam), cyclin D1 (Neomarkers, Fremont, CA), p21 (556431, BD Pharmingen (San Diego, CA)), and actin (ab8227, Abcam). The phospho-ERK1/2, total ERK1/2, phospho-JNK1/2/3, total JNK, phospho-p38, total p38, phospho-AKT, total AKT, phospho-STAT3, total STAT3, phospho-p65, total p65, caspase 3, caspase 9, and cleaved poly(ADP-ribose) polymerase antibodies were all from Cell Signaling Technologies (Beverly, MA).
RNA Analysis
Total RNA was isolated from frozen liver using the RNeasy minikit (Qiagen, Valencia, CA). One microgram of RNA was reverse transcribed and used for qRT-PCR analysis. Primers were designed for gene specificity and to cross exon-exon junctions using Primer-BLAST (NCBI). Results are normalized to actin gene expression. Values given are -fold over control or relative expression value, where appropriate, calculated using the 2ΔCt QPCR calculation method (23).
Microarray Analysis
Dye-coupled cDNA was hybridized to Agilent 44K mouse 60-mer oligonucleotide microarrays (Agilent Technologies, Santa Clara, CA). Three mouse liver samples were independently processed for each genotype analyzed. Microarray data were processed and analyzed using Genespring GX 11.5.1 software (Agilent Technologies). Microarray data were deposited in the NCBI Gene Expression Omnibus under GEO accession number GSE34581. Only those genes changed ≥2.0-fold with a p value of ≤0.05 were considered significant and used for further analysis with Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA). For gene network analysis using IPA, networks were considered highly relevant with a score of ≥30. The network score is the −log of the p value determined by Fisher's exact test result, which takes into account the number of genes in the network, the size of the network, expression values of genes in the network, the number of genes in the data set, and the total number of genes in the IPA database.
Luciferase Assays
The mouse Perp promoter luciferase plasmids were constructed by cloning the upstream region from +49 (relative to the transcription start site) to −578 and successive truncations into the pGL4-basic luciferase vector using primers listed in supplemental Table S1 (Promega, Madison, WI). The luciferase reporters and a Renilla luciferase control vector were cotransfected into HepG2 or COS-1 cells using Fugene 6 transfection reagent (Roche Applied Science). COS-1 cells, which do not endogenously express HNF4α, were cotransfected with a control empty vector or a previously described rat HNF4α expression vector (24). Luciferase assays were performed using the Promega Dual-Luciferase assay kit.
ChIP Assay
Livers of Alb-Hnf4aF/F and Alb-Hn4a−/− mice (n = 3) were perfused with Hanks' balanced salt solution (without Ca2+ and Mg2+) and EDTA followed by Hanks' balanced salt solution with Ca2+ and collagenase. Hepatocyte slurries were washed in DMEM and filtered through a 70-μm strainer and immediately fixed in 1% formaldehyde. Chromatin was prepared using the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technologies). Digested chromatin was incubated overnight with 4 μg of anti-HNF4α (C-19, sc-6556x, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)). One microliter of recovered DNA was used for QPCR analysis. Primers were designed to span an ∼100-bp region of the putative HNF4α binding site (supplemental Table S1). Values were normalized using 2% input control. Values given are -fold over the negative control IgG pull-down using the 2ΔCt QPCR calculation method (23).
Statistical Analysis
All results are expressed as the mean ± S.D. Significance was determined by t test or one-way ANOVA with Bonferroni post-test using Prism 5.0 software (GraphPad Software, La Jolla, CA). A p value of <0.05 was considered significant and is indicated in graphs (*, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.00005).
RESULTS
Generation of Temporal, Hepatocyte-specific Hnf4a Knock-out Mouse
Disruption of the Hnf4a gene in the hepatocyte-specific Alb-Hnf4a−/− mouse results in severe metabolic disturbances in lipid and bile acid handling, which contributes to the early mortality of knock-out mice at 6–8 weeks after birth (9). In order to study the functions of HNF4α in the adult mouse liver without the confounding factors of early mortality and accumulated, severe hepatic malfunction, the Hnf4aF/F mice were crossed with the tamoxifen-inducible hepatocyte-specific Cre recombinase expressing mouse SA+/Cre-ERT2 to generate Hnf4aF/F;AlbERT2cre mice (9, 21). To confirm the inducible recombination of the floxed Hnf4a allele, Hnf4aF/F;AlbERT2cre mice were fed a tamoxifen-containing diet for up to 5 days (day 0 start diet, day 5 remove), at which point the diet was replaced with a normal chow diet. Mice were euthanized on days 2, 5, 9, 12, and 19 after starting the tamoxifen diet. Analysis of Hnf4a mRNA and protein confirmed the complete loss of HNF4α in livers of Hnf4aF/F;AlbERT2cre mice within 48 h of tamoxifen treatment (Fig. 1, A and C). HNF4α expression in normal chow Hnf4aF/F or Hnf4aF/F;AlbERT2cre mice or tamoxifen-treated Hnf4aF/F mice was unchanged, confirming the specificity of the CRE gene activation. A trace amount of HNF4α protein is detectable 19 days after first tamoxifen exposure. The source of this protein may be proliferation and differentiation of hepatic stem/oval cells or loss of Cre recombinase expression in adult hepatocytes after several passages through the cell cycle. Loss of HNF4α transcriptional function was confirmed by the dramatic reduction in mRNA expression of classic HNF4α targets L-Fabp, ApoC3, and Ppara (Fig. 1D) (9, 25, 26). Hnf4g mRNA levels remained low and even decreased slightly after Hnf4a disruption, eliminating the possibility of HNF4γ compensation after HNF4α loss (Fig. 1B).
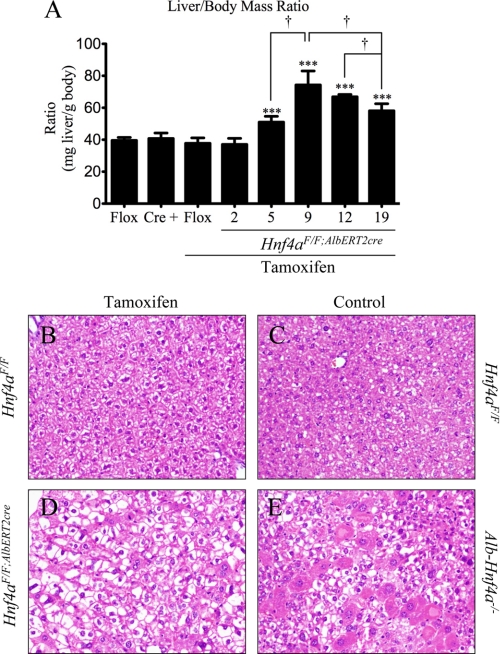
Short term knock-out of Hnf4a recapitulates the physiological changes observed in the developmental Alb-Hnf4a−/− mouse. Serum chemistry analysis of 19-day Hnf4aF/F;AlbERT2cre knock-out mice confirmed liver dysfunction, as revealed by increased levels of alkaline phosphatase, alanine aminotransferase, bile acids, and total bilirubin (Table 1). Similar to the Alb-Hnf4a−/− mouse, the Hnf4aF/F;AlbERT2cre mice also have decreased levels of serum cholesterol, a reflection of the known regulation by HNF4α of genes involved in hepatic cholesterol synthesis and transport (9, 25–27). Hepatomegaly is evident in Hnf4aF/F;AlbERT2cre knock-out mice as early as 5 days post-tamoxifen treatment, peaking at 9 days (Fig. 2A). Although still significantly increased compared with control mice, the liver/body mass ratio decreases slightly by 19 days. This decrease is not due to increased body mass but rather a reduction in liver mass (data not shown). Histological examination of 9-day Hnf4aF/F;AlbERT2cre + tamoxifen mice reveals significant vacuolization of hepatocytes, hypertrophy, and scattered eosinophilic hepatocytes (Fig. 2, B and D). The full Alb-Hnf4a−/− mouse has more extensive eosinophilia and hypertrophy with slightly smaller vacuoles (Fig. 2, C and E). Previously published reports confirmed lipid deposition in the livers of both Hnf4aF/F;AlbERT2cre + tamoxifen and Alb-Hnf4a−/− mice by Oil Red O staining (9, 28). Thus, the temporal hepatocyte-specific Hnf4aF/F;AlbERT2cre mouse model mirrors changes observed in the developmental Alb-Hnf4a−/− mouse with the added advantage of studying early responses to Hnf4α deficiency in healthy adult hepatocytes in vivo.
TABLE 1.
Serum chemistry analysis
Statistical differences were determined by one-way ANOVA. Significant difference between control and tamoxifen-treated mice of the same genotype is indicated.
| Control |
Tamoxifen (19-day) |
|||
|---|---|---|---|---|
| Flox | Cre+ | Flox | Cre+ | |
| Alkaline phosphatase (units/liter) | 80.3 ± 21.5 | 98.3 ± 10.4 | 103.0 ± 13.5 | 884 ± 360a |
| Alanine aminotransferase (units/liter) | 89.7 ± 15.4 | 93.5 ± 38.1 | 97.8 ± 37.3 | 163.3 ± 36.2b |
| Bile acids (μmol/liter) | 9.0 ± 7.8 | 7.3 ± 2.6 | 5.5 ± 3.7 | 70.8 ± 30.1c |
| Total bilirubin (mg/dl) | 0.20 ± 0.00 | 0.15 ± 0.06 | 0.18 ± 0.05 | 0.38 ± 0.05a |
| Cholesterol (mg/dl) | 89.7 ± 3.5 | 87.8 ± 9.3 | 66.5 ± 1.3c | 21.3 ± 4.6a |
a p < 0.0005.
b p < 0.05.
c p < 0.005.
FIGURE 2.
Acute hepatic disruption of Hnf4a results in hepatomegaly and hepatocyte hypertrophy and vacuolization. A, liver and body mass (n = 6) was assessed at the indicated time points. Nineteen-day Hnf4aF/F mice were used as a control for tamoxifen exposure. Significant differences from Hnf4aF/F + tamoxifen (19-day) are indicated (***, p < 0.0005). Significant differences between tamoxifen-treated Hnf4aF/F;AlbERT2cre mice at different time points are indicated (†, p < 0.05). B–E, representative H&E staining of 9-day tamoxifen-treated Hnf4aF/F (B) and Hnf4aF/F;AlbERT2cre mice (D) as well as Hnf4aF/F (C) and Alb-Hnf4a−/− mice (E). Error bars, S.D.
Hepatocyte Proliferation in Response to Acute HNF4α Deficiency
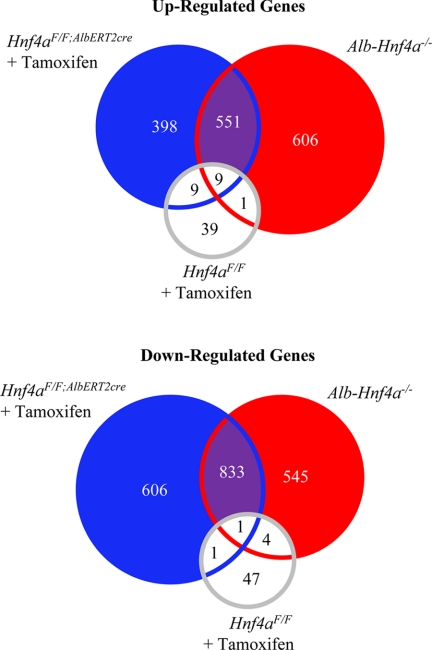
To determine the pathways influenced by HNF4α, gene expression analysis was performed on livers from Hnf4aF/F;AlbERT2cre + tamoxifen (19 day) and Alb-Hnf4a−/− mice and their respective wild-type controls (Fig. 3). The overall number of genes significantly altered in the two different knockouts was similar: 2388 genes in Hnf4aF/F;AlbERT2cre + tamoxifen mice and 2535 in Alb-Hnf4a−/− mice. Those genes commonly regulated in both the Hnf4aF/F;AlbERT2cre + tamoxifen (19 day) and Alb-Hnf4a−/− mice may be direct HNF4α transcriptional targets. The 1384 commonly regulated genes identified were further subjected to IPA gene network analysis with a score of 30 or greater considered highly relevant within the data set. As expected, the highest scoring network identified was lipid metabolism (Table 2). Interestingly, gene networks involved in cell cycle, cellular growth, proliferation, and the inflammatory response were also significantly affected by HNF4α deficiency. When microarray data were analyzed separately for the genotypes, cell growth and proliferation pathways were highly significant only in the Hnf4aF/F;AlbERT2cre + tamoxifen-treated mice. Although gene network analysis indicated significant changes in cell growth and proliferation for both knock-out genotypes, the highest scoring proliferation network in the Alb-Hnf4a−/− mouse data set was 14, indicating that the Hnf4aF/F;AlbERT2cre + tamoxifen data set was driving the identification of proliferation gene networks in the combined analysis (data not shown). The microarray data suggest that the increase in liver/body mass ratio in acutely HNF4α-deficient livers may be due in part to hepatocyte proliferation and not solely to lipid accumulation.
FIGURE 3.
Microarray analysis to determine HNF4α-regulated genes. RNA was extracted from livers (n = 3) of control Hnf4aF/F, Hnf4aF/F;AlbERT2cre, and Alb-Hnf4a−/− mice as well as 19-day tamoxifen-treated Hnf4aF/F and Hnf4aF/F;AlbERT2cre mice. Those genes significantly changed from the respective control genotype were used to identify the common and uniquely regulated genes in Alb-Hnf4a−/− and Hnf4aF/F;AlbERT2cre + tamoxifen mice.
TABLE 2.
Gene network analysis
Microarray results were processed through Ingenuity Pathway Analysis software to identify networks of genes most significantly affected by the phenotype and assigned a score based on the degree of relevance of the network to the genes in the data set. Networks with a score higher than 30 were considered highly relevant.
| Score | |
|---|---|
| Up-regulated in bothHnf4aF/F;AlbERT2cre + tamoxifen and Alb-Hnf4a−/− | |
| Cellular function and maintenance, dermatological diseases and conditions, genetic disorder | 38 |
| Antimicrobial response, inflammatory response, dermatological diseases and conditions | 36 |
| Cellular development, cell cycle, cellular growth and proliferation | 32 |
| Down-regulated in bothHnf4aF/F;AlbERT2cre + tamoxifen and Alb-Hnf4a−/− | |
| Lipid metabolism, small molecule biochemistry, vitamin and mineral metabolism | 48 |
| Tumor morphology, inflammatory response, cell cycle | 32 |
| Lipid metabolism, molecular transport, small molecule biochemistry | 32 |
| Up-regulated inHnf4aF/F;AlbERT2cre + tamoxifen | |
| Dermatological diseases and conditions, genetic disorder, antimicrobial response | 34 |
| Cell death, cellular development, cellular growth and proliferation | 34 |
| Skeletal and muscular system development and function, cellular growth and proliferation, cellular movement | 32 |
| Nervous system development and function, organ morphology, inflammatory disease | 32 |
| Up-regulated inAlb-Hnf4a−/− | |
| Antimicrobial response, inflammatory response, dermatological diseases and conditions | 34 |
| Cellular movement, immune cell trafficking, cell-to-cell signaling and interaction | 34 |
| Small molecular biochemistry, organ development, reproductive system development and function | 32 |
| Cancer, reproductive system disease, cellular development | 30 |
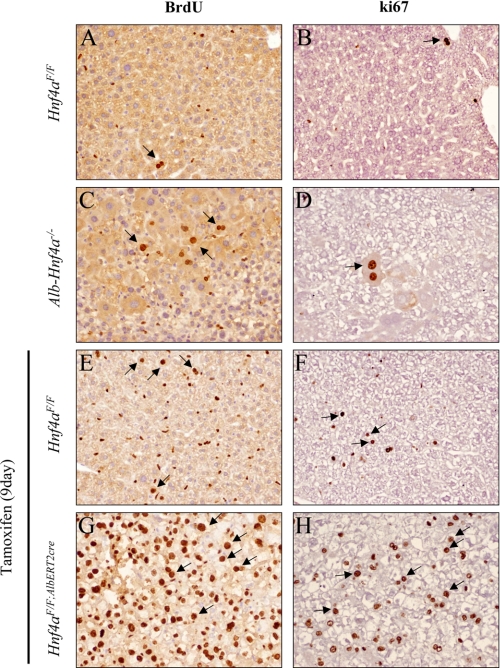
Hepatocyte proliferation was assessed by BrdU incorporation in the livers of both Hnf4aF/F;AlbERT2cre and Alb-Hnf4a−/− mice. Tamoxifen treatment resulted in a higher degree of BrdU labeling in Hnf4aF/F + tamoxifen (9 day) livers compared with normal diet Hnf4aF/F mice (Fig. 4, A and E). The small, oblong nuclei labeled after tamoxifen treatment are characteristic of non-parenchymal cells, such as Kupffer or stellate cells. Acute disruption of Hnf4α expression in the Hnf4aF/F;AlbERT2cre + tamoxifen mice resulted in widespread hepatic nuclei labeling (Fig. 4G), whereas limited foci of proliferation were evident in Alb-Hnf4a−/− mice (Fig. 4C). Proliferation in the livers of Hnf4aF/F;AlbERT2cre + tamoxifen mice was further confirmed using the endogenous proliferation marker ki67 (Fig. 4H). Similar results were obtained with proliferating cell nuclear antigen (PCNA) staining (data not shown). Because the proliferation signal initiated after HNF4α deficiency is more robust in the acute knock-out model, the expression of cell cycle regulators in 9-day tamoxifen-treated Hnf4aF/F;AlbERT2cre mice was examined.
FIGURE 4.
Extensive hepatocyte proliferation after acute hepatic disruption of Hnf4a. BrdU pumps were implanted subcutaneously for 6 days prior to euthanasia of Hnf4aF/F (A and B) and Alb-Hnf4a−/− mice (C and D) on a normal chow diet. On day 3 of tamoxifen treatment, a BrdU pump was implanted subcutaneously in Hnf4aF/F (E and F) and Hnf4aF/F;AlbERT2cre (G and H) mice. Tamoxifen was removed on day 5, and mice were returned to a normal chow diet until euthanasia on day 9. Slides were stained for BrdU incorporation (A, C, E, and G, dark brown) or ki67 (B, D, F, and H, dark brown) and counterstained with hematoxylin for visualization of nuclei (light purple). Experiments were performed on four animals per group, and representative images are shown. Examples of hepatocyte nuclei staining are indicated by arrows.
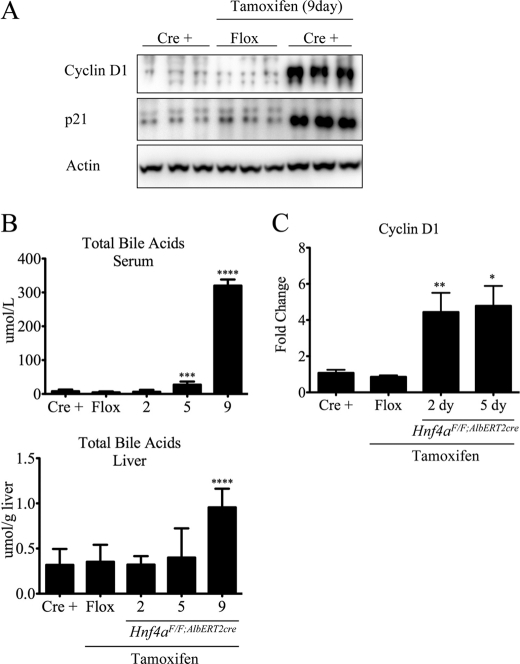
Consistent with the microarray data, several genes important in proliferation were up-regulated in Hnf4aF/F;AlbERT2cre knock-out mice. Notably, the cyclins A2, B1, D1, and E2 (Ccna2, Ccnb1, Ccnd1, and Ccne2, respectively) as well as cyclin-dependent kinase 1 (Cdk1), myelocytomatosis oncogene (c-Myc), and Pcna were up-regulated (Table 3). Stem cell marker prominin 1 (Prom1/CD133) expression was up-regulated 72-fold after Hnf4a disruption, suggesting a loss of hepatocyte differentiation. The apoptosis-, senescence-, and cell cycle-related gene p21 (Cdkn1a) was induced 18-fold in Hnf4aF/F;AlbERT2cre knock-out mice. Gene expression of another member of the Cip/Kip family of cyclin-dependent kinase inhibitors, p27 (Cdkn1b), was reduced 40% in tamoxifen-treated Hnf4aF/F;AlbERT2cre mice. Expression of the p53/p63-regulated gene Perp was dramatically suppressed in tamoxifen-treated Hnf4aF/F;AlbERT2cre mice. Up-regulation of cyclin D1 and p21 at the protein level was confirmed in Hnf4aF/F;AlbERT2cre knock-out mice by Western blot analysis (Fig. 5A). Together, these data indicate that HNF4α expression suppresses adult hepatocyte proliferation.
TABLE 3.
Changes in gene expression after 9 days of Hnf4a knockout
Data shown as -fold change over control Hnf4aF/F mice. Significant differences from control as determined by one-way ANOVA are indicated.
| Control, Hnf4aF/F;AlbERT2cre | Tamoxifen (9-day) |
||
|---|---|---|---|
| Hnf4aF/F | Hnf4aF/F;AlbERT2cre | ||
| Ccna2 | 1.01 ± 0.21 | 1.20 ± 0.55 | 4.55 ± 0.34a |
| Ccnb1 | 1.01 ± 0.15 | 2.15 ± 2.55 | 19.0 ± 1.991a |
| Ccnd1 | 1.23 ± 0.75 | 1.67 ± 0.84 | 22.75 ± 9.47b |
| Ccnd2 | 1.02 ± 0.28 | 0.68 ± 0.16 | 0.95 ± 0.13 |
| Ccnd3 | 1.05 ± 0.41 | 1.11 ± 0.10 | 1.61 ± 0.70 |
| Ccne2 | 1.02 ± 0.25 | 0.69 ± 0.09 | 2.72 ± 1.97b |
| Cdk1 | 1.12 ± 0.26 | 2.62 ± 1.31 | 16.79 ± 7.66b |
| Cdk2 | 1.02 ± 0.23 | 1.14 ± 0.48 | 2.06 ± 0.88 |
| Cdk4 | 1.03 ± 0.29 | 0.88 ± 0.11 | 1.33 ± 0.15 |
| Cdk6 | 1.00 ± 0.01 | 0.64 ± 0.08** | 0.47 ± 0.09a |
| p53 | 1.62 ± 0.70 | 0.44 ± 0.06 | 1.68 ± 1.78 |
| p63 | 1.12 ± 0.64 | 0.52 ± 0.14 | 0.58 ± 0.22 |
| Perp | 1.04 ± 0.34 | 1.03 ± 0.01 | 0.07 ± 0.02c |
| Prom1 | 1.03 ± 0.29 | 0.81 ± 0.13 | 71.8 ± 32.4b |
| Pcna | 1.00 ± 0.05 | 0.93 ± 0.07 | 2.61 ± 0.96b |
| cMyc | 1.02 ± 0.26 | 1.46 ± 1.05 | 5.85 ± 0.28c |
| p21 | 1.17 ± 0.83 | 2.16 ± 0.42 | 18.5 ± 1.37a |
| p27 | 1.01 ± 0.15 | 1.37 ± 0.64 | 0.56 ± 0.14b |
| Xiap | 1.05 ± 0.37 | 0.37 ± 0.02b | 0.20 ± 0.02c |
| cIap1 | 1.00 ± 0.09 | 1.22 ± 0.09 | 1.29 ± 0.33 |
| cIap2 | 1.01 ± 0.18 | 1.19 ± 0.20 | 1.86 ± 1.27 |
| Puma | 1.01 ± 0.19 | 0.87 ± 0.22 | 0.85 ± 0.32 |
| Bcl2 | 1.02 ± 0.24 | 0.76 ± 0.32 | 0.88 ± 0.33 |
| Bcl-lx | 1.03 ± 0.31 | 1.51 ± 0.31 | 1.66 ± 0.98 |
a p < 0.0005.
b p < 0.05.
c p < 0.005.
FIGURE 5.
Induction of cyclin D1 precedes elevated serum bile acids. A, Western blot analysis of liver whole cell extracts (n = 3) from 9-day tamoxifen-treated Hnf4aF/F;AlbERT2cre mice. Actin was used as a loading control. B, serum and liver total bile acid concentration of Hnf4aF/F;AlbERT2cre mice (n = 3) after the indicated tamoxifen treatment. Five-day Hnf4aF/F mice were used as a control for tamoxifen exposure. Significant differences from tamoxifen-treated Hnf4aF/F mice as determined by Student's t test are indicated (***, p < 0.0005; ****, p < 0.00005). C, qRT-PCR analysis of mRNA from livers of Hnf4aF/F;AlbERT2cre mice (n = 3) at the indicated time points. Data are expressed as a relative value compared with actin expression. Five-day tamoxifen-treated Hnf4aF/F mice were used as a control for tamoxifen exposure. Statistically significant changes from Hnf4aF/F + tamoxifen mice as determined by Student's t test are indicated (*, p < 0.05; **, p < 0.005). Error bars, S.D.
Lack of Inflammatory Signal after Acute Deletion of HNF4α in Liver
Hepatocyte proliferation in the context of partial hepatectomy or carbon tetrachloride exposure occurs in a two-step process: priming of the hepatocyte by inflammatory cytokines that in turn stimulates growth factor production and release (29, 30). It was proposed that increases in bile acids after such liver injury is one mechanism to initiate the regenerative process via a nuclear receptor-dependent mechanism (31). Liver bile acid levels were maintained at control levels until 9 days post-tamoxifen in Hnf4aF/F;AlbERT2cre mice (Fig. 5B). Serum bile acid levels were statistically increased by 5 days of tamoxifen treatment in Hnf4aF/F;AlbERT2cre mice, reaching a maximum at 9 days of treatment (Fig. 5B). However, increased cyclin D1 gene expression precedes serum and, more importantly, liver bile acid elevation, and thus it is unlikely to be the initiating proliferative signal in Hnf4aF/F;AlbERT2cre mice (Fig. 5C).
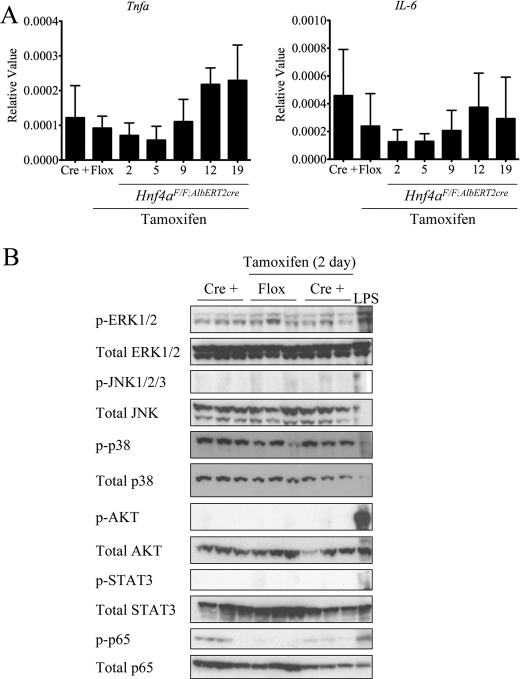
It was recently shown that inhibition of HNF4α leads to a miRNA-mediated pathway resulting in stimulation of the IL-6-STAT3 inflammatory axis in the context of HCC (32). Expression of the two major inflammatory cytokines, Il-6 and Tnfa, was assessed by qRT-PCR. Basal expression levels were near the lower limit of detection and did not significantly increase at any time point after tamoxifen exposure in the Hnf4aF/F;AlbERT2cre mice (Fig. 6A). Consistent with the lack of cytokine induction, none of the major cellular signaling pathways were activated in Hnf4aF/F;AlbERT2cre mice after 2 days of tamoxifen treatment (Fig. 6B). This suggests that loss of HNF4α up-regulates a hepatotrophic factor independent of Kupffer cell activation to drive hepatocyte proliferation.
FIGURE 6.
No activation of cytokine signaling pathways after temporal Hnf4a deletion. A, qRT-PCR analysis of mRNA from livers of Hnf4aF/F;AlbERT2cre mice (n = 3) at the indicated time points. Nineteen-day Hnf4aF/F mice were used as a control for tamoxifen exposure. B, Western blot analysis of liver whole cell extracts (n = 3) from 2-day tamoxifen-treated Hnf4aF/F;AlbERT2cre mice. Total unphosphorylated protein as well as actin was used as a loading control. Liver extract from a 6-h LPS-treated mouse was used as a positive control. Error bars, S.D.
Loss of Hnf4α Induces Hepatotrophic Factor Bone Morphogenetic Protein 7 (Bmp7)
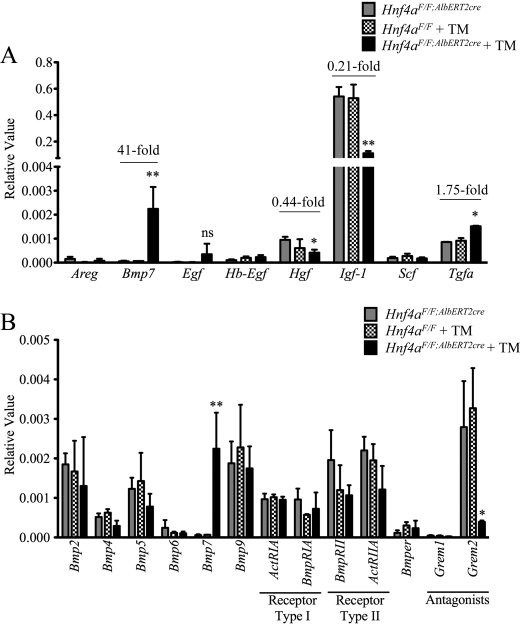
There are a wide variety of growth factors capable of stimulating hepatocyte proliferation, including hepatocyte growth factor (HGF), stem cell factor/kit-ligand (SCF/KITL), BMP7, insulin-like growth factor 1 (IGF-1), and the epidermal growth factor (EGF) receptor ligands EGF heparin-binding EGF-like growth factor, transforming growth factor α (TGFα), and amphiregulin (reviewed in Refs. 29 and 30). The expression of these growth factors was assessed in livers from 9-day tamoxifen-treated Hnf4aF/F;AlbERT2cre mice. Hgf, Igf-1, and Tgfa are the only mitogens studied that are normally produced at low levels in quiescent hepatocytes, as evidenced by the higher basal transcript levels in control Hnf4aF/F;AlbERT2cre mice (Fig. 7A). Tgfa was induced 1.75-fold in tamoxifen-treated Hnf4aF/F;AlbERT2cre mice, whereas Hgf and Igf-1 were reduced to 0.44 and 0.21, respectively, compared with control Hnf4aF/F;AlbERT2cre mice. The reduction in Igf-1 expression would be expected as it is regulated by the transcription factor hepatocyte nuclear factor 1α (HNF1α), whose expression is dependent on HNF4α (33–35). Egf expression was too variable in tamoxifen-treated Hnf4aF/F;AlbERT2cre mice to conclude a significant up-regulation. The most striking change was the induction of Bmp7, over 40-fold in Hnf4a-deficient mice.
FIGURE 7.
The growth factor Bmp7 is specifically induced by acute hepatic Hnf4a disruption. qRT-PCR analysis of livers from Hnf4aF/F;AlbERT2cre mice (n = 3) 9 days after tamoxifen treatment. A, gene expression analysis of major growth factors involved in liver regeneration expressed as relative value normalized to actin expression. -Fold change from Hnf4aF/F + tamoxifen is indicated for statistically significant changes (*, p < 0.05; **, p < 0.005). B, gene expression analysis of Bmp-related family members expressed as relative value normalized to actin expression. Statistically significant changes from Hnf4aF/F + tamoxifen mice as determined by one-way ANOVA are indicated (*, p < 0.05; **, p < 0.005). ns, not significant; Error bars, S.D.
To determine if this was a specific up-regulation of Bmp7 or a general activation of Bmps, expression of several members of the Bmp family, their receptors, and regulators was evaluated. Among the Bmps with detectable hepatic expression, only Bmp7 was significantly altered in tamoxifen-treated Hnf4aF/F;AlbERT2cre mice (Fig. 7B). Interestingly, expression of the Bmp antagonist Gremlin2 (Grem2/Prdc) was significantly reduced in tamoxifen-treated Hnf4aF/F;AlbERT2cre mice.
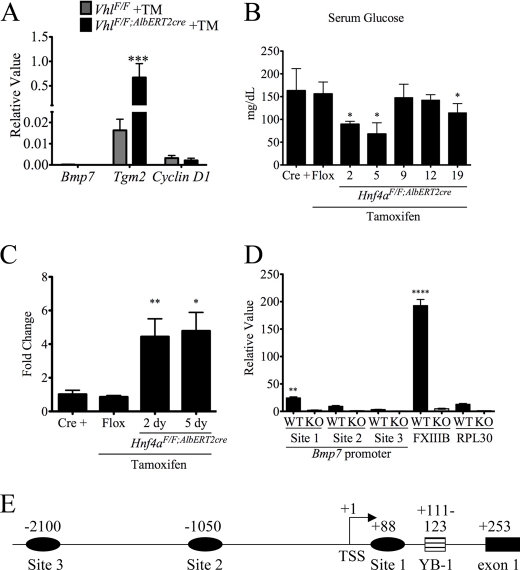
A recent study found elevated BMP7 levels in a hepatitis B mouse model of HCC and further identified elevated BMP7 in human cirrhotic liver and HCC samples (36). To assess whether the induction of Bmp7 in Hnf4aF/F;AlbERT2cre + tamoxifen mice was associated with steatosis and liver damage or cellular proliferation, Bmp7 levels were measured in the VhlF/F;AlbERT2cre mouse, which displays hepatic steatosis, inflammation, and fibrosis but not cellular proliferation (22). Transglutaminase 2 (Tgm2), a marker of fibrosis, is significantly up-regulated in VhlF/F;AlbERT2cre + tamoxifen mice, but Bmp7 levels remain nearly undetectable (Fig. 8A). Cyclin D1 mRNA, one marker for cellular proliferation, was slightly decreased from basal levels. This suggests that Bmp7 is not up-regulated by steatosis or fibrosis alone in mouse liver.
FIGURE 8.
Regulation of the Bmp7 promoter. A, qRT-PCR analysis of livers of VhlF/F + tamoxifen (TM) and VhlF/F;AlbERT2cre + tamoxifen mice. Results are expressed as a relative value, and statistically significant difference as determined by Student's t test from VhlF/F + tamoxifen is indicated (***, p < 0.0005). B, fed serum glucose concentrations (mg/dl). Statistically significant changes from Hnf4aF/F;AlbERT2cre control mice as determined by one-way ANOVA are indicated (*, p < 0.05). C, qRT-PCR analysis of Bmp7 mRNA from livers of Hnf4aF/F;AlbERT2cre treated with tamoxifen for 2 and 5 days. Significant differences from control Hnf4aF/F mice are indicated (*, p < 0.05; **, p < 0.005). D, ChIP assay with HNF4α antibody performed on liver extracts from Hnf4aF/F (WT) and Alb-Hnf4a−/− (KO) mice. QPCR values for immunoprecipitated DNA were normalized to input DNA and expressed as a relative value. Statistically significant changes from WT RPL30 as determined by Student's t test are indicated (**, p < 0.005; ****, p < 0.00005). E, schematic of the mouse Bmp7 promoter indicating potential HNF4α binding sites as well as the previously characterized Y-box protein binding site (YB-1). The position of the central nucleotide of the putative HNF4α binding site is given relative to the transcription start site (TSS). Error bars, S.D.
Little is known regarding the transcriptional regulation of Bmp7, and no studies have been conducted on its regulation in liver. A mouse Bmp7 promoter luciferase construct was induced by high glucose levels in cultured kidney cells (37). Fed serum glucose levels were measured at various time points after tamoxifen treatment in Hnf4aF/F;AlbERT2cre mice (Fig. 8B). Serum glucose levels do not exceed control levels at any point after Hnf4a disruption. The initial drop in glucose levels may be due to the tamoxifen diet. The Bmp7 induction, similar to cyclin D1, precedes the onset of cholestasis at day 5 (Fig. 8C). HNF4α has been shown to interact with the nuclear receptor corepressor 2 (SMRT/NCoR) complex, resulting in histone deacetylase recruitment and inhibition of transcription (38). The mouse Bmp7 promoter was assessed using Genomatix MatInspector software for HNF4α binding sites. Three putative binding sites were found within the first 3 kb of the promoter (Fig. 8E). To determine if HNF4α binds to the Bmp7 promoter in vivo, extracts were prepared from liver of Hnf4aF/F and Alb-Hnf4a−/− mice for the ChIP assay. The Alb-Hnf4a−/− liver extracts serve as a control for antibody specificity. The HNF4α binding site in the promoter of the coagulation factor 13B (FXIIIB) was used as a positive control (39). HNF4α binding was significantly enriched on Bmp7 site 1 in Hnf4aF/F liver compared with the negative control RPL30 intron 2 and liver extracts from Alb-Hnf4a−/− mice (Fig. 8D). However, this enrichment was minimal compared with the enrichment on the known HNF4α binding site in the FXIIIB promoter. Thus, it is unlikely that HNF4α recruits histone deacetylase complexes to actively repress Bmp7 transcription in normal adult liver.
Regulation of p53/63-dependent Desmosomal Gene Perp by HNF4α
The potential repression of apoptosis in HNF4α-deficient livers was assessed by Western blot analysis of caspase 3 or 9 activation. No significant caspase activation was detected, as indicated by caspase or poly(ADP-ribose) polymerase cleavage (Fig. 9A). There is a slight increase in full-length caspase 3 and 9 in the tamoxifen-treated Hnf4aF/F;AlbERT2cre mice that may indicate defective programmed cell death in these livers. Microarray and gene expression analysis revealed a significant down-regulation of the gene Perp (Table 2). PERP was first identified as an effector of p53-dependent apoptosis and is up-regulated during apoptosis but not cell cycle arrest (40, 41).
FIGURE 9.
Identification of Perp as a novel HNF4α target gene. A, Western blot analysis of whole cell extracts blotted with antibodies that recognize full-length (FL) and/or cleaved (CL) proteins. B, luciferase assay with the indicated Perp promoter firefly luciferase constructs transfected into HepG2 and COS-1 cells. For COS-1 cells, luciferase plasmids were cotransfected with a control (gray bars) or HNF4α (black bars) expression vector. Firefly luciferase values are normalized to a Renilla luciferase vector. Significant differences as determined by Student's t test are indicated (*, p < 0.05; ***, p < 0.0005). C, HNF4α ChIP assay performed with liver extracts from Hnf4aF/F (WT) and Alb-Hnf4a−/− (KO) mice. QPCR values for immunoprecipitated DNA were normalized to input DNA and expressed as a relative value. Statistically significant changes from WT RPL30 as determined by Student's t test are indicated (***, p < 0.0005). Error bars, S.D.
Because the decrease in Perp expression in the HNF4α-deficient liver is of a similar magnitude as verified direct HNF4α targets, the first 2 kb of the Perp promoter were analyzed for potential HNF4α binding sites. Three putative binding sites (−220, −415, and −566 bp from the transcription start site) are located within the first 1 kb of the Perp proximal promoter (Fig. 9B). To confirm the presence of a functional HNF4α binding site, promoter luciferase constructs were tested in HepG2 cells that basally express HNF4α. Luciferase expression in HepG2 cells was reduced when putative HNF4α binding site 2 (−415 bp) was deleted (Fig. 9B). In a parallel experiment, rat HNF4α was overexpressed in COS-1 cells that do not basally express HNF4α. The full-length Perp promoter luciferase activity was significantly induced in COS-1 cells when HNF4α was co-transfected. This induction was completely lost when site 2 was deleted. Deletion of site 3 resulted in incomplete loss of luciferase activity, suggesting that this site cooperates with site 2 to achieve full transactivation of the Perp promoter. The ChIP assay from Hnf4aF/F mouse liver extracts confirmed recruitment of HNF4α to site 2 in the Perp promoter (Fig. 9C). The reduced binding signals at HNF4α site 1 and site 3 may be due to the resolution of the ChIP assay (DNA average length 200 bp) and close proximity of the sites. The specificity of the immunoprecipitation was confirmed by the absence of enrichment in Alb-Hnf4a−/− mouse liver extracts or the nonspecific gene RPL30.
DISCUSSION
The present study demonstrated that acute loss of HNF4α in the adult mouse liver initiates a robust proliferative response in normal hepatocytes, suggesting that HNF4α may be a true tumor suppressor. This response is transitory in nature because microarray analysis indicated that changes in cell cycle control and other genes involved in hepatocyte proliferation were associated with the Hnf4aF/F;AlbERT2cre knock-out and not with the Alb-Hnf4a−/− mice in which only small regenerating foci of hepatic periportal oval cells were observed previously (42). This latter finding was confirmed in the present study using BrdU incorporation. However, the widespread BrdU staining observed in the Hnf4aF/F;AlbERT2cre knock-out mice indicates that loss of HNF4α stimulates proliferation in both hepatocytes and oval cells. Although loss of HNF4α can initiate proliferation, the severe metabolic disruption in the hepatocytes may prevent continuous cell division. In order to progress to cancer, other genetic changes are probably necessary in conjunction with the loss of HNF4α.
The findings in the Hnf4aF/F;AlbERT2cre knock-out mouse are in agreement with previous in vitro genomic and transcriptomic analysis identifying cell cycle-, apoptosis-, and other cancer-related genes as direct HNF4α targets (43). Among the genes identified in that study were the apoptosis-associated genes Cideb and Sec16b that were strongly down-regulated in the Hnf4aF/F;AlbERT2cre knock-out mouse model as well (data not shown). In the current study, Perp was revealed as an HNF4α target gene in the liver by microarray, ChIP, and luciferase promoter assays. PERP is induced by p53 in response to apoptosis but not cell cycle arrest and is capable of inducing cell death (40, 41). As well as being an important effector of apoptosis, PERP is a desmosomal protein essential for epithelial integrity, particularly in the skin (44). The regulation of Perp expression by HNF4α is consistent with the broad number of cell adhesion-related genes previously identified as HNF4α targets, including E-cadherin, claudins 1 and 3, and desmocollin 2 (11). Disruption of Perp promoted UVB-induced squamous cell carcinoma that was attributed to loss of PERP apoptotic functions as well as its desmosomal role (45). The cumulative effect of the loss of several apoptosis genes in Hnf4aF/F;AlbERT2cre knock-out mice may shift the steady-state equilibrium between apoptosis and proliferation. Indeed, Western blot analysis revealed a small increase in basal, full-length caspase 3 and 9 protein expression, which suggests that HNF4α-deficient mice may have a defect in response to mitochondrial damage.
It is difficult to interpret the significant increase in p21 expression observed in Hnf4aF/F;AlbERT2cre knock-out mice given the context-specific ability of p21 to both promote and inhibit apoptosis and the cell cycle (Fig. 5A) (reviewed in Ref. 46). The lack of activation of apoptosis and the increase in proliferation markers in Hnf4aF/F;AlbERT2cre knock-out mice suggest that p21 may be functioning in its antiapoptotic/proproliferation role by preventing caspase 3 cleavage and assisting with cyclin D-CDK complex assembly (reviewed in Refs. 46 and 47). p21 was previously identified in HepG2 and HCT116 cells as being positively regulated by HNF4α (48). The induction of p21 in the context of HNF4α depletion suggests that HNF4α does not significantly contribute to the induction of p21 in the whole liver.
The proliferative signal in Hnf4aF/F;AlbERT2cre knock-out mice is probably due to alteration of several pathways due to the pleiotropic nature of HNF4α target genes in hepatocytes. Although current models of hepatocyte regeneration suggest a two-hit hypothesis of inflammation and then proliferation, no changes in hepatic cytokine levels or activation of inflammation-associated signaling pathways were detected. However, it is possible that activation of these pathways occurred very rapidly and transiently and was thus missed during the present analysis. A recent report demonstrated increased proliferation and invasiveness and decreased apoptosis in HepG2 cells and mouse models of HCC in which HNF4α was knocked down (32). The proliferative phenotype was linked to HNF4α regulation of the IL-6/STAT3 signaling pathway through a miRNA feedback loop. However, no activation of STAT3 or Il-6 expression was observed in Hnf4aF/F;AlbERT2cre knock-out mice. This result suggests that the miRNA-mediated IL-6/STAT3 signaling pathway only functions in the context of active HCC and not in the healthy liver. HNF4α was previously shown to regulate the expression of miR-122, a highly expressed liver-specific miRNA important in hepatocyte differentiation and proliferation (49). Thus, HNF4α may regulate the expression of a large set of miRNA and RNAs in order to maintain the differentiated hepatocyte phenotype.
In adult mammals, BMP7 is a circulating cytokine produced primarily by the kidney and bone, but elevated expression has been detected in several tumor types, including breast, melanoma, and prostate (50–53). Although the liver does not normally produce BMP7, BMP7 receptors are found on hepatocytes, and neutralization of circulating BMP7 in a mouse partial hepatectomy model inhibited hepatocyte regeneration (54). In the reverse experiment, these authors demonstrated increased liver proliferation after partial hepatectomy when recombinant BMP7 was infused into the mice. BMP7 was also able to antagonize TGFβ and inhibit fibrosis in a mouse CCl4-induced liver fibrosis model (55). In contrast to the beneficial effects observed in the rodent liver fibrosis model, BMP7 expression is elevated in patients with chronic liver diseases, raising the question of species-specific effects of BMP7 (56, 57). A recent study using a hepatitis B model of HCC in mice as well as human HCC samples confirmed up-regulation of BMP7 in cirrhotic and cancerous liver tissues (36). The same study also demonstrated increased cell viability, migration, and invasiveness in the hepatoma cell line Hep3B.
The mechanism by which loss of HNF4α induces Bmp7 remains elusive. It is likely that a transcriptional regulator of Bmp7 is up-regulated after HNF4α disruption. A recent report identified BMP7 as a target of MYC (myelocytomatosis oncogene) in medulloblastoma-derived cell lines, binding an unspecified location near the transcription start site (58). Indeed, a small 5.85-fold increase in c-myc mRNA was observed in 9-day Hnf4aF/F;AlbERT2cre knock-out mice. A transcription factor binding site search of the mouse Bmp7 promoter identified a putative E-box/c-MYC binding site −130 bp from the transcription start site. Additionally, several putative binding sites for the c-MYC-regulated protein E2-F1 are located within the proximal promoter of Bmp7. E2-F1 mRNA was previously shown to be up-regulated in the Hnf4aF/F;AlbERT2cre knock-out mice (28). It is interesting to note that Bmp7 was not induced in the VhlF/F;AlbERT2cre mouse model, which displays significant steatosis, cholestasis, and fibrosis but no proliferation, suggesting that the mechanism of induction is linked to proliferation factors. Elevated Bmp7 expression in the livers of Hnf4aF/F;AlbERT2cre knock-out mice is consistent with reports of increased circulating BMP7 in patients with liver disease. Thus, BMP7 expression may be a marker of liver disease, and the Hnf4aF/F;AlbERT2cre knock-out mice may be a model for studying the role of BMP7 in liver function.
This study establishes HNF4α not only as a key regulator of hepatic differentiation, maintenance of the liver phenotype, and metabolism but also as a potential tumor suppressor. In an otherwise healthy liver, acute disruption of HNF4α in hepatocytes initiates a robust proliferative response. The combined effects of HNF4α regulation on cellular adhesion, EMT, growth factors, apoptosis, and proliferation position it as an important factor in hepatic tumorigenesis. The new temporal and spatial modulating Hnf4a knock-out mouse model generated in this study circumvents the early mortality observed in the developmental Alb-Hnfa−/− mouse, thus allowing for future studies on the role of HNF4α in fibrosis and tumorigenesis.
Supplementary Material
Acknowledgments
The SA+/Cre-ERT2 mouse was provided by Daniel Metzger and Pierre Chambon (Department of Physiological Genetics, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France).
This work was funded by the intramural research program at the National Cancer Institute, National Institutes of Health (NIH).

This article contains supplemental Table S1.
- HCC
- hepatocellular carcinoma
- EMT
- epithelial to mesenchymal transition
- ANOVA
- analysis of variance
- miRNA
- microRNA
- IPA
- Ingenuity Pathway Analysis
- QPCR
- quantitative PCR
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Ferlay J., Shin H. R., Bray F., Forman D., Mathers C., Parkin D. M. (2010) GLOBOCAN 2008, Version 1.2. Cancer incidence and mortality worldwide. IARC CancerBase No. 10. International Agency for Research on Cancer, Lyon, France [Google Scholar]
- 2. Howlader N., Noone A. M., Krapcho M., Neyman N., Aminou R., Waldron W., Altekruse S. F., Kosary C. L., Ruhl J., Tatalovich Z., Cho H., Mariotto A., Eisner M. P., Lewis D. R., Chen H. S., Feuer E. J., Cronin K. A., Edwards B. K. (eds) (2011) SEER Cancer Statistics Review, 1975–2008, NCI, National Institutes of Health, Bethesda, MD, http://seer.cancer.gov/csr/1975_2008 [Google Scholar]
- 3. Llovet J. M., Burroughs A., Bruix J. (2003) Hepatocellular carcinoma. Lancet 362, 1907–1917 [DOI] [PubMed] [Google Scholar]
- 4. Shaw J. J., Shah S. A. (2011) Rising incidence and demographics of hepatocellular carcinoma in the USA. What does it mean? Expert Rev. Gastroenterol. Hepatol. 5, 365–370 [DOI] [PubMed] [Google Scholar]
- 5. Wisely G. B., Miller A. B., Davis R. G., Thornquest A. D., Jr., Johnson R., Spitzer T., Sefler A., Shearer B., Moore J. T., Miller A. B., Willson T. M., Williams S. P. (2002) Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure 10, 1225–1234 [DOI] [PubMed] [Google Scholar]
- 6. Dhe-Paganon S., Duda K., Iwamoto M., Chi Y. I., Shoelson S. E. (2002) Crystal structure of the HNF4 α ligand binding domain in complex with endogenous fatty acid ligand. J. Biol. Chem. 277, 37973–37976 [DOI] [PubMed] [Google Scholar]
- 7. Sladek F. M., Zhong W. M., Lai E., Darnell J. E. (1990) Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 4, 2353–2365 [DOI] [PubMed] [Google Scholar]
- 8. Yuan X., Ta T. C., Lin M., Evans J. R., Dong Y., Bolotin E., Sherman M. A., Forman B. M., Sladek F. M. (2009) Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS ONE 4, e5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayhurst G. P., Lee Y. H., Lambert G., Ward J. M., Gonzalez F. J. (2001) Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamagata K., Furuta H., Oda N., Kaisaki P. J., Menzel S., Cox N. J., Fajans S. S., Signorini S., Stoffel M., Bell G. I. (1996) Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature 384, 458–460 [DOI] [PubMed] [Google Scholar]
- 11. Battle M. A., Konopka G., Parviz F., Gaggl A. L., Yang C., Sladek F. M., Duncan S. A. (2006) Hepatocyte nuclear factor 4α orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc. Natl. Acad. Sci. U.S.A. 103, 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parviz F., Matullo C., Garrison W. D., Savatski L., Adamson J. W., Ning G., Kaestner K. H., Rossi J. M., Zaret K. S., Duncan S. A. (2003) Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 34, 292–296 [DOI] [PubMed] [Google Scholar]
- 13. Santangelo L., Marchetti A., Cicchini C., Conigliaro A., Conti B., Mancone C., Bonzo J. A., Gonzalez F. J., Alonzi T., Amicone L., Tripodi M. (2011) The stable repression of mesenchymal program is required for hepatocyte identity. A novel role for hepatocyte nuclear factor 4α. Hepatology 53, 2063–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thiery J. P., Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 15. Lazarevich N. L., Shavochkina D. A., Fleishman D. I., Kustova I. F., Morozova O. V., Chuchuev E. S., Patyutko Y. I. (2010) Deregulation of hepatocyte nuclear factor 4 (HNF4) as a marker of epithelial tumor progression. Exp. Oncol. 32, 167–171 [PubMed] [Google Scholar]
- 16. Ning B. F., Ding J., Yin C., Zhong W., Wu K., Zeng X., Yang W., Chen Y. X., Zhang J. P., Zhang X., Wang H. Y., Xie W. F. (2010) Hepatocyte nuclear factor 4 α suppresses the development of hepatocellular carcinoma. Cancer Res. 70, 7640–7651 [DOI] [PubMed] [Google Scholar]
- 17. Lazarevich N. L., Cheremnova O. A., Varga E. V., Ovchinnikov D. A., Kudrjavtseva E. I., Morozova O. V., Fleishman D. I., Engelhardt N. V., Duncan S. A. (2004) Progression of HCC in mice is associated with a down-regulation in the expression of hepatocyte nuclear factors. Hepatology 39, 1038–1047 [DOI] [PubMed] [Google Scholar]
- 18. Kalkuhl A., Kaestner K., Buchmann A., Schwarz M. (1996) Expression of hepatocyte-enriched nuclear transcription factors in mouse liver tumors. Carcinogenesis 17, 609–612 [DOI] [PubMed] [Google Scholar]
- 19. Yin C., Lin Y., Zhang X., Chen Y. X., Zeng X., Yue H. Y., Hou J. L., Deng X., Zhang J. P., Han Z. G., Xie W. F. (2008) Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4α gene. Hepatology 48, 1528–1539 [DOI] [PubMed] [Google Scholar]
- 20. Chen W. S., Manova K., Weinstein D. C., Duncan S. A., Plump A. S., Prezioso V. R., Bachvarova R. F., Darnell J. E. (1994) Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8, 2466–2477 [DOI] [PubMed] [Google Scholar]
- 21. Schuler M., Dierich A., Chambon P., Metzger D. (2004) Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis 39, 167–172 [DOI] [PubMed] [Google Scholar]
- 22. Qu A., Taylor M., Xue X., Matsubara T., Metzger D., Chambon P., Gonzalez F. J., Shah Y. M. (2011) Hypoxia-inducible transcription factor 2α promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology 54, 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Inoue Y., Hayhurst G. P., Inoue J., Mori M., Gonzalez F. J. (2002) Defective ureagenesis in mice carrying a liver-specific disruption of hepatocyte nuclear factor 4α (HNF4α). HNF4α regulates ornithine transcarbamylase in vivo. J. Biol. Chem. 277, 25257–25265 [DOI] [PubMed] [Google Scholar]
- 25. Ladias J. A., Hadzopoulou-Cladaras M., Kardassis D., Cardot P., Cheng J., Zannis V., Cladaras C. (1992) Transcriptional regulation of human apolipoprotein genes ApoB, ApoCIII, and ApoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2, and EAR-3. J. Biol. Chem. 267, 15849–15860 [PubMed] [Google Scholar]
- 26. Mietus-Snyder M., Sladek F. M., Ginsburg G. S., Kuo C. F., Ladias J. A., Darnell J. E., Jr., Karathanasis S. K. (1992) Antagonism between apolipoprotein AI regulatory protein 1, Ear3/COUP-TF, and hepatocyte nuclear factor 4 modulates apolipoprotein CIII gene expression in liver and intestinal cells. Mol. Cell. Biol. 12, 1708–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stroup D., Chiang J. Y. (2000) HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7α-hydroxylase gene (CYP7A1). J. Lipid Res. 41, 1–11 [PubMed] [Google Scholar]
- 28. Zhang Y., Bonzo J. A., Gonzalez F. J., Wang L. (2011) Diurnal regulation of the early growth response 1 (Egr-1) protein expression by hepatocyte nuclear factor 4α (HNF4α) and small heterodimer partner (SHP) cross-talk in liver fibrosis. J. Biol. Chem. 286, 29635–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alison M. R., Islam S., Lim S. (2009) Stem cells in liver regeneration, fibrosis and cancer. The good, the bad, and the ugly. J. Pathol. 217, 282–298 [DOI] [PubMed] [Google Scholar]
- 30. Riehle K. J., Dan Y. Y., Campbell J. S., Fausto N. (2011) New concepts in liver regeneration. J. Gastroenterol. Hepatol. 26, 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang W., Ma K., Zhang J., Qatanani M., Cuvillier J., Liu J., Dong B., Huang X., Moore D. D. (2006) Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 312, 233–236 [DOI] [PubMed] [Google Scholar]
- 32. Hatziapostolou M., Polytarchou C., Aggelidou E., Drakaki A., Poultsides G. A., Jaeger S. A., Ogata H., Karin M., Struhl K., Hadzopoulou-Cladaras M., Iliopoulos D. (2011) An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147, 1233–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nolten L. A., Steenbergh P. H., Sussenbach J. S. (1995) Hepatocyte nuclear factor 1 α activates promoter 1 of the human insulin-like growth factor I gene via two distinct binding sites. Mol. Endocrinol. 9, 1488–1499 [DOI] [PubMed] [Google Scholar]
- 34. Kuo C. J., Conley P. B., Chen L., Sladek F. M., Darnell J. E., Jr., Crabtree G. R. (1992) A transcriptional hierarchy involved in mammalian cell-type specification. Nature 355, 457–461 [DOI] [PubMed] [Google Scholar]
- 35. Lee Y. H., Sauer B., Gonzalez F. J. (1998) Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1α knockout mouse. Mol. Cell. Biol. 18, 3059–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu J. W., Hsia Y., Yang W. Y., Lin Y. I., Li C. C., Tsai T. F., Chang K. W., Shieh G. S., Tsai S. F., Wang H. D., Yuh C. H. (2012) Identification of the common regulators for hepatocellular carcinoma induced by hepatitis B virus X antigen in a mouse model. Carcinogenesis 33, 209–219 [DOI] [PubMed] [Google Scholar]
- 37. Wang S., Hirschberg R. (2011) Y-box protein-1 is a transcriptional regulator of BMP7. J. Cell. Biochem. 112, 1130–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Torres-Padilla M. E., Sladek F. M., Weiss M. C. (2002) Developmentally regulated N-terminal variants of the nuclear receptor hepatocyte nuclear factor 4α mediate multiple interactions through coactivator and corepressor-histone deacetylase complexes. J. Biol. Chem. 277, 44677–44687 [DOI] [PubMed] [Google Scholar]
- 39. Inoue Y., Peters L. L., Yim S. H., Inoue J., Gonzalez F. J. (2006) Role of hepatocyte nuclear factor 4α in control of blood coagulation factor gene expression. J. Mol. Med. 84, 334–344 [DOI] [PubMed] [Google Scholar]
- 40. Attardi L. D., Reczek E. E., Cosmas C., Demicco E. G., McCurrach M. E., Lowe S. W., Jacks T. (2000) PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 14, 704–718 [PMC free article] [PubMed] [Google Scholar]
- 41. Reczek E. E., Flores E. R., Tsay A. S., Attardi L. D., Jacks T. (2003) Multiple response elements and differential p53 binding control Perp expression during apoptosis. Mol. Cancer Res. 1, 1048–1057 [PubMed] [Google Scholar]
- 42. Stanulovi V. S., Kyrmizi I., Kruithof-de Julio M., Hoogenkamp M., Vermeulen J. L., Ruijter J. M., Talianidis I., Hakvoort T. B., Lamers W. H. (2007) Hepatic HNF4α deficiency induces periportal expression of glutamine synthetase and other pericentral enzymes. Hepatology 45, 433–444 [DOI] [PubMed] [Google Scholar]
- 43. Bolotin E., Liao H., Ta T. C., Yang C., Hwang-Verslues W., Evans J. R., Jiang T., Sladek F. M. (2010) Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology 51, 642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ihrie R. A., Marques M. R., Nguyen B. T., Horner J. S., Papazoglu C., Bronson R. T., Mills A. A., Attardi L. D. (2005) Perp is a p63-regulated gene essential for epithelial integrity. Cell 120, 843–856 [DOI] [PubMed] [Google Scholar]
- 45. Beaudry V. G., Jiang D., Dusek R. L., Park E. J., Knezevich S., Ridd K., Vogel H., Bastian B. C., Attardi L. D. (2010) Loss of the p53/p63 regulated desmosomal protein Perp promotes tumorigenesis. PLoS Genet. 6, e1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cazzalini O., Scovassi A. I., Savio M., Stivala L. A., Prosperi E. (2010) Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat. Res. 704, 12–20 [DOI] [PubMed] [Google Scholar]
- 47. Sherr C. J., Roberts J. M. (1999) CDK inhibitors. Positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 48. Hwang-Verslues W. W., Sladek F. M. (2008) Nuclear receptor hepatocyte nuclear factor 4α1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol. Endocrinol. 22, 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu H., He J. H., Xiao Z. D., Zhang Q. Q., Chen Y. Q., Zhou H., Qu L. H. (2010) Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 52, 1431–1442 [DOI] [PubMed] [Google Scholar]
- 50. Morrissey C., Brown L. G., Pitts T. E., Vessella R. L., Corey E. (2010) Bone morphogenetic protein 7 is expressed in prostate cancer metastases, and its effects on prostate tumor cells depend on cell phenotype and the tumor microenvironment. Neoplasia 12, 192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alarmo E. L., Rauta J., Kauraniemi P., Karhu R., Kuukasjärvi T., Kallioniemi A. (2006) Bone morphogenetic protein 7 is widely overexpressed in primary breast cancer. Genes Chromosomes Cancer 45, 411–419 [DOI] [PubMed] [Google Scholar]
- 52. Rothhammer T., Poser I., Soncin F., Bataille F., Moser M., Bosserhoff A. K. (2005) Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res. 65, 448–456 [PubMed] [Google Scholar]
- 53. Masuda H., Fukabori Y., Nakano K., Takezawa Y., Suzuki T., Yamanaka H. (2003) Increased expression of bone morphogenetic protein-7 in bone metastatic prostate cancer. Prostate 54, 268–274 [DOI] [PubMed] [Google Scholar]
- 54. Sugimoto H., Yang C., LeBleu V. S., Soubasakos M. A., Giraldo M., Zeisberg M., Kalluri R. (2007) BMP-7 functions as a novel hormone to facilitate liver regeneration. FASEB J. 21, 256–264 [DOI] [PubMed] [Google Scholar]
- 55. Zeisberg M., Yang C., Martino M., Duncan M. B., Rieder F., Tanjore H., Kalluri R. (2007) Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J. Biol. Chem. 282, 23337–23347 [DOI] [PubMed] [Google Scholar]
- 56. Tacke F., Gäbele E., Bataille F., Schwabe R. F., Hellerbrand C., Klebl F., Straub R. H., Luedde T., Manns M. P., Trautwein C., Brenner D. A., Schölmerich J., Schnabl B. (2007) Bone morphogenetic protein 7 is elevated in patients with chronic liver disease and exerts fibrogenic effects on human hepatic stellate cells. Dig. Dis. Sci. 52, 3404–3415 [DOI] [PubMed] [Google Scholar]
- 57. Chayanupatkul M., Honsawek S., Vejchapipat P., Chongsrisawat V., Poovorawan Y. (2009) Elevated serum bone morphogenetic protein 7 levels and clinical outcome in children with biliary atresia. Eur. J. Pediatr. Surg. 19, 246–250 [DOI] [PubMed] [Google Scholar]
- 58. Fiaschetti G., Castelletti D., Zoller S., Schramm A., Schroeder C., Nagaishi M., Stearns D., Mittelbronn M., Eggert A., Westermann F., Ohgaki H., Shalaby T., Pruschy M., Arcaro A., Grotzer M. A. (2011) Bone morphogenetic protein-7 is a MYC target with prosurvival functions in childhood medulloblastoma. Oncogene 30, 2823–2835 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.