Background: Apoptotic oligonucleosomal DNA degradation is mediated by DFF40/CAD endonuclease.
Results: Poor DFF40/CAD expression in the cytosol coupled to the caspase-dependent cytosolic processing of ICADL/S impair oligonucleosomal DNA degradation in SK-N-AS cells.
Conclusion: Oligonucleosomal DNA fragmentation during apoptosis is directly correlated with adequate DFF40/CAD cytosolic levels.
Significance: Learning how DFF40/CAD works is crucial for understanding the relevance of apoptosis ending in cancer development.
Keywords: Apoptosis, Caspase, Cell Death, Neuroblastoma, Subcellular Fractionation, DFF40/CAD, Oligonucleosomal DNA Degradation
Abstract
Apoptotic cell death is characterized by nuclear fragmentation and oligonucleosomal DNA degradation, mediated by the caspase-dependent specific activation of DFF40/CAD endonuclease. Here, we describe how, upon apoptotic stimuli, SK-N-AS human neuroblastoma-derived cells show apoptotic nuclear morphology without displaying concomitant internucleosomal DNA fragmentation. Cytotoxicity afforded after staurosporine treatment is comparable with that obtained in SH-SY5Y cells, which exhibit a complete apoptotic phenotype. SK-N-AS cell death is a caspase-dependent process that can be impaired by the pan-caspase inhibitor q-VD-OPh. The endogenous inhibitor of DFF40/CAD, ICAD, is correctly processed, and dff40/cad cDNA sequence does not reveal mutations altering its amino acid composition. Biochemical approaches show that both SH-SY5Y and SK-N-AS resting cells express comparable levels of DFF40/CAD. However, the endonuclease is poorly expressed in the cytosolic fraction of healthy SK-N-AS cells. Despite this differential subcellular distribution of DFF40/CAD, we find no differences in the subcellular localization of both pro-caspase-3 and ICAD between the analyzed cell lines. After staurosporine treatment, the preferential processing of ICAD in the cytosolic fraction allows the translocation of DFF40/CAD from this fraction to a chromatin-enriched one. Therefore, the low levels of cytosolic DFF40/CAD detected in SK-N-AS cells determine the absence of DNA laddering after staurosporine treatment. In these cells DFF40/CAD cytosolic levels can be restored by the overexpression of their own endonuclease, which is sufficient to make them proficient at degrading their chromatin into oligonucleosome-size fragments after staurosporine treatment. Altogether, the cytosolic levels of DFF40/CAD are determinants in achieving a complete apoptotic phenotype, including oligonucleosomal DNA degradation.
Introduction
Apoptotic cell death is critical during embryogenesis and tissue homeostasis in metazoans (1–3). A misbalance in this biological process also plays an important role in the development of different human diseases, including cancer (4, 5). Apoptosis is characterized by a set of morphological and biochemical events, including shrinkage and fragmentation of the nucleus as well as chromatin degradation (3). The execution of apoptotic cell death is governed by caspases, a family of cysteine proteases that, after activation by different pro-apoptotic stimuli, cleave target cell substrates (6–9). DNA degradation is considered one of the hallmarks of apoptotic cell death (1) and usually occurs in a two-step process. First, the chromatin is cleaved into 50–300-kb fragments, termed high molecular weight (HMW)6 DNA fragmentation. Subsequently, DNA is degraded into smaller fragments of oligonucleosomal size, known as low molecular weight (LMW) DNA degradation and visualized as a laddering pattern in an agarose gel electrophoresis (DNA ladder) (2).
The main nuclease responsible for DNA laddering during apoptosis was first identified in humans and mice. The human homologue has been named DFF40 (DNA fragmentation factor, 40-kDa subunit) or caspase-activated nuclease (CPAN), whereas the mouse orthologue is known as caspase-activated DNase (CAD) (10–12). In growing cells DFF40/CAD remains inactive, associated with its chaperone and inhibitor, ICADL (13), also known as DNA fragmentation factor, 45-kDa subunit (DFF45) (12, 14). Two alternatively spliced isoforms of ICAD have been described, the long (ICADL) and the short (ICADS) variants, containing two caspase recognition sites at Asp117 and Asp224 residues. DFF40/CAD activation occurs after caspase-3 processes both ICAD isoforms (12–14). The generation of the knock-out mice for ICAD revealed that this protein acts as a chaperone assisting CAD synthesis (15–17). Therefore, the functional consequences of ICAD deficiency (absence of apoptotic DNA fragmentation) have been attributed to the lack of CAD synthesis.
Upon apoptotic insult, DFF40/CAD needs to be located in the nucleus to degrade the chromatin. It has been described that ICAD-CAD complex is found either in the cytosol (10, 18) or in the nucleus (12, 19–22) of healthy cells. In this work we show that DFF40/CAD protein can be detected in both the cytosolic and the nuclear subcellular compartments of resting cells. However, the levels of the endonuclease in each subcellular compartment are indeed a cell-specific feature. In this sense we have detected that in cells displaying oligonucleosomal DNA degradation upon staurosporine treatment (SH-SY5Y cells), the endonuclease is located in both cytosol and nuclei. However, when DFF40/CAD is not enriched in the cytosolic fraction, cells are defective in the generation of DNA laddering (SK-N-AS cells). To carry out its endonuclease function, DFF40/CAD needs to be located in the same fraction where ICAD is preferentially processed, the cytosolic fraction. Increasing the cytosolic levels of DFF40/CAD in SK-N-AS cells by overexpression of its own endonuclease restores the ability to degrade DNA into LMW fragments. Altogether, our results indicate that despite DFF40/CAD nuclear localization, resting cells require a pool in the cytosol to achieve LMW DNA degradation upon apoptotic insult.
EXPERIMENTAL PROCEDURES
Reagents
All chemicals were obtained from Sigma unless otherwise indicated. The pan-caspase inhibitor q-VD-OPh was from MP Biomedicals Europe (Illkirch, France). Human TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) (CYT-443) and human TNFα (CYT-223) were obtained from AbBcn S. L. (Barcelona, Spain). Anti-Fas (human, activating antibody) (clone CH11) (05-201) was from Millipore Iberica S.A.U. (Madrid, Spain). Cycloheximide was purchased from VWR International Eurolab S.L. (Barcelona, Spain). Nocodazole was from Merck KGaA (Darmstadt, Germany). Antibodies against pro-caspase-3 (9662; 1:2000), pro-caspase-7 (9492; 1:2000), and cleaved poly(ADP-ribose) polymerase (9542; 1:5000) were purchased from Cell Signaling Technology (Beverly, MA). Antibodies against DFF40/CAD (AB16926, 1:500) and α-Fodrin (clone AA6) (MAB1622; 1:40,000) were obtained from Millipore Iberica S.A.U. (Madrid, Spain). Antibodies against pro-caspase-6 (clone 3E8) (M070–3; 1:2,000) and DFF45/ICAD (clone 6B8) (M037–3; 1:40,000) were from MBL (Naka-ku Nagoya, Japan). Anti-PKM2 antibody (H00005315-A01; 1:1000) was from Abnova (Jhongli, Taiwan). Anti-Lamin A/C (clone JOL2) (Ab40567; 1:2000) was from Abcam (Cambridge, UK). Anti-p23 antibody (clone JJ3) (NB300-576; 1:10,000) was obtained from Novus Biological Europe, Inc (Cambridge, UK). Anti-topoisomerase I (2012-4; 1:20,000) was from TopoGEN, Inc. Secondary antibodies against mouse IgG (A9044; 1:10,000) and rabbit IgG (A0545; 1:20,000) were purchased from Sigma.
Cell Lines and Culture Procedures
Human SH-SY5Y, SK-N-AS, SK-N-SH, IMR-5, IMR-32, LAI-5S, LAN-1, and SK-N-BE(2) cell lines were routinely grown in 100-mm culture dishes (BD Biosciences) containing 10 ml of Dulbecco's modified Eagle's medium supplemented with penicillin/streptomycin (100 units/ml and 100 μg/ml, respectively) and 10% of heat-inactivated fetal bovine serum (Invitrogen). Medium was routinely changed every 3 days. Cells were maintained at 37 °C in a saturating humidity atmosphere containing 95% air and 5% CO2. For the different experiments, cells were grown at the adequate cell densities in culture dishes or multiwell plates (BD Biosciences) using the same culture conditions as described above. Some neuroblastoma cell lines employed in this work were generous gifts from Dr. M. Esteller and Dr. M. Berdasco (Institut d'Investigació Biomèdica de Bellvitge (IDIBELL), Barcelona, Spain).
Trypan Blue Exclusion Assays and Chromatin Staining with Hoechst 33258
Trypan blue assays and nuclear morphology by Hoechst 33258 staining were performed as previously established in our laboratory (23). The cell nuclei were visualized with a Nikon ECLIPSE TE2000-E microscope equipped with epifluorescence optics under UV illumination and a Hamamatsu ORCA-ER photographic camera. Uniformly stained nuclei were scored as stage I (partial chromatin condensation in the absence of karyorrhexis) or stage II (nuclear pyknosis and karyorrhexis) morphology.
Oligonucleosomal DNA Degradation Analysis and Quantification
Oligonucleosomal DNA degradation analysis was carried out as previously described (24). Extracted DNA was analyzed in 1.8% agarose gel in 1 mm EDTA, 40 mm Tris acetate, pH 7.6, stained in 0.5 μg/ml of ethidium bromide, and then visualized using a Syngene Gene Genius UV transilluminator equipped with a photographic camera.
The quantification of oligonucleosomal DNA fragments was performed as previously described with some modifications (25). Briefly, after the adequate treatments, 2 × 106 cells were lysed by the addition of 300 μl of DNA fragmentation lysis buffer (0.1% Triton X-100, 20 mm EDTA, 5 mm Tris-HCl, pH 8.5). After the addition of polyethylene glycol (PEG) 8000 and NaCl to a final concentration of 4% and 1 m, respectively, samples were placed on ice for 10 min and then centrifuged at 16,000 × g for 10 min at room temperature. The supernatants were removed and treated with proteinase K and DNase-free RNase A at a final concentration of 200 and 20 μg/ml, respectively. A third part of the supernatants was used to determine the concentration of DNA by adding an equal volume of Hoechst dye solution (0.2 μg/ml Hoechst 33258 in PBS, pH 7.4) to an aliquot (50 μl) of the supernatant. After 20 min at room temperature, fluorescence of the samples was determined at 360-nm excitation, 460-nm emission on a BIO-TEK Synergy HT Fluorometer. The remaining supernatants were used to isolate and precipitate DNA as described for oligonucleosomal DNA degradation analysis.
High Molecular Weight DNA Fragmentation
The procedure employed for these experiments was the same as that described by Barry and Eastman (26) with some modifications. Briefly, 5 × 105 cells were seeded in 12-multiwell plates, and after 24 h they were treated with 1 μm STP for 6 h. Then cells were centrifuged for 5 min at 500 × g and washed once with PBS. Meanwhile, 150 ml of 2% agarose in 1× TBE (89 mm Tris-base, 89 mm boric acid, 2 mm EDTA, pH 8.0) was poured into a horizontal gel support (15 × 15 cm) with the comb at 3.5 cm from one end. Once gelled, the portion of the gel placed 1 mm above the comb was removed by cutting with a scalpel and replaced with 50 ml of 1% agarose, 2% sodium dodecyl sulfate, 64 μg/ml proteinase K in 1× TBE buffer. Before loading, each pellet of cells was resuspended in 15 μl of 1:1 sample buffer (10 mm Tris-HCl, pH 8.8, 50% glycerol, 0.1% bromphenol blue) plus 10 mg/ml RNase A. The gel was electrophoresed at room temperature for 16 h at 45 V. After electrophoresis, the gel was stained in 2 μg/ml ethidium bromide for 2 h and washed twice with distilled water for 30 min. DNA was visualized using a Syngene Gene Genius UV transilluminator coupled with a photographic camera.
Protein Extractions and Western Blotting
Approximately 1 × 106 cells/condition were detached from the 35-mm culture dish, pelleted at 600 × g for 5 min, and washed once with PBS. Then cells were lysed for 15 min on ice with 50 μl of Triton buffer (50 mm Tris-HCl, pH 6.8, 1 mm EDTA, 150 mm NaCl, 1% Triton X-100, 1× protease inhibitor mixture (Roche Applied Science). The supernatants were clarified by centrifuging at 16,000 × g for 5 min at 4 °C. Alternatively, cells were lysed with 100 μl of SET buffer (10 mm Tris-HCl, pH 6.8, 150 mm NaCl, 1 mm EDTA, 1% SDS) and heated at 95 °C for 10 min to obtain total protein extracts. The protein concentration in the supernatants was quantified by a modified Lowry assay (DC protein assay, Bio-Rad), and 15–30 μg of protein were loaded in SDS-polyacrylamide gels. The proteins were electrophoresed and electrotransferred onto polyvinylidene difluoride (PVDF) Immobilon-P membrane (Millipore Ibérica S.A. U) or Protran nitrocellulose transfer membrane (Whatman GmbH, Dassel, Germany). After blocking with Tris-buffered saline, 0.1% Tween 20 containing 5% nonfat dry milk, the membranes were probed with the appropriate specific primary antibodies and incubated with the adequate secondary antibodies conjugated with peroxidase. Finally, immunoblots were developed with the EZ-ECL chemiluminescence detection kit (Biological Industries, Kibbutz Beit-Haemek, Israel).
DEVD-directed Caspase Activity
Quantitative DEVD-like activities in cell lysates were performed as previously described (27). The resulting 96-multiwell microplates were incubated at 35 °C, and caspase activity was measured after 7 h of incubation in a BIO-TEK Synergy HT fluorimeter with an excitation filter of 360 nm (40-nm bandwidth) and emission filter of 530 nm (25-nm bandwidth).
Preparation of Cytosolic Extracts
Cytosolic extracts were obtained by a modified protocol form previously described (27, 28). Cells were grown in 75-cm2 flasks and treated with 1 μm STP for 12 h or left untreated. Cells were collected and centrifuged at 200 × g for 5 min, washed with PBS, resuspended in 5 volumes of cytosolic extraction buffer (20 mm Tris-HCl, pH 7.4, 10 mm KCl, 2 mm MgCl2, 1 mm DTT, 0.5 mm EGTA, 2.5% glycerol, 0.5% Igepal CA-630), and swelled at 4 °C for 15 min. The lysates were centrifuged for 5 min at 600 × g in a microcentrifuge at 4 °C to remove the nuclei, and the supernatants were centrifuged again at 16,000 × g for 15 min and then quantified using the Bio-Rad protein assay Lowry-based method.
Preparation of SH-SY5Y and SK-N-AS Nuclei and Reconstitution of the Cell-free System
Nuclei were obtained as previously described (27). Aliquots of 5 × 107 nuclei/ml were prepared and kept at −80 °C until use.
To reconstitute the system, cytosolic extracts and purified nuclei from the different conditions were adequately combined. Thus, 150 μg of protein from cytosolic extracts treated with 1.5 μg of RNase A for 10 min at 25 °C and 6 × 105 nuclei were incubated at 37 °C for 2 h in a 2× reaction buffer (40 mm Tris-HCl, pH 7.5, 20 mm KCl, 100 mm NaCl, 8 mm MgCl2, 2 mm DTT, 1 mm EGTA, 0.2% Triton X-100, 1× protease inhibitor mixture) in a 100-μl reaction. Reactions were stopped by adding 5 mm EDTA. Nuclei were centrifuged for 15 min at 16,000 × g (4 °C), and the pellet was washed once with cytosolic extraction buffer. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1), and ethanol precipitation and DNA laddering was analyzed in a 1.8% agarose gel.
Chromatin Isolation
The subfractionation protocol was performed as previously described by Méndez and Stillman (29) with some modifications. Cells were seeded in 35-mm culture dishes and treated with 1 μm STP for 6 h. Then cells were collected with their medium, centrifuged at 500 × g for 5 min, washed once with PBS, and resuspended in 3 volumes (60 μl) of buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 0.1% Triton X-100, 1× protease inhibitor mixture). Cells were incubated for 5 min on ice, and nuclei were collected by low speed centrifugation (4 min, 1300 × g, 4 °C). The supernatants were further clarified by high speed centrifugation (20 min, 20,000 × g, 4 °C) obtaining the cytosolic (C) fraction. The resulting pellets were washed twice in buffer A without Triton X-100 and then lysed in 60 μl of Triton buffer (50 mm Tris-HCl, pH 6.8, 1 mm EDTA, 150 mm NaCl, 1% Triton X-100, 1× protease inhibitor mixture). Then, the supernatants were clarified by centrifuging at 16,000 × g for 5 min at 4 °C to obtain the nuclear (N) fraction.
Alternatively, the nuclear fractions were resuspended in 60 μl of buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, supplemented with 1× protease inhibitors) and kept on ice for 15 min. Then samples were clarified by centrifugation (4 min, 1700 × g, 4 °C). Supernatants were considered the nucleoplasmic (N1) fractions. The pellets obtained were washed once in buffer B and centrifuged again under the same conditions. The final resulting pellets were resuspended in 60 μl of SET buffer and heated at 95 °C for 10 min, obtaining the chromatin-enriched (N2) fractions. The protein concentration in the different fractions recovered was determined by the Bio-Rad protein assay Lowry-based method, and 15–20 μg of protein from each fraction were loaded in SDS-polyacrylamide gels.
Sequencing of DFF40/CAD from SK-N-AS Cells
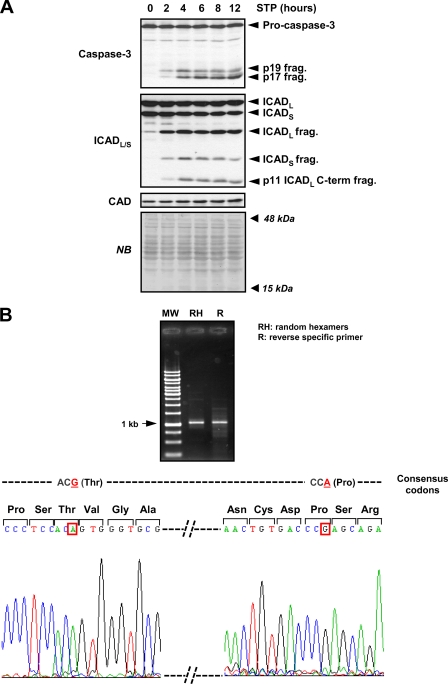
mRNA was isolated from untreated SK-N-AS cells using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, employing for the extraction the RLN buffer (50 mm Tris-HCl, pH 8.0, 140 mm NaCl, 1.5 mm MgCl2, 0.5% Igepal CA-630, 1000 units/ml RNase inhibitor, 1 mm DTT). Total RNA (2 μg) from SK-N-AS was reverse-transcribed (Trasnscriptor First Strand cDNA synthesis kit, Roche Applied Science) using 10 pmol of random hexamer primers or the specific downstream primer (CAD-R; see below) for 30 min at 65 °C. cDNA obtained (2 μl) was amplified by PCR in an Applied Biosystem thermal cycler 2720 with 300 nm concentrations of each primer. The PCR conditions were 95 °C for 20 s, 56 °C for 10 s, and 70 °C for 24 s, repeated 30 cycles, in 1.5 mm MgSO4, 200 nm each dNTP, and 1 unit of KOD Hot Start DNA polymerase (Merck KGaA). Primers used were CAD forward (5′-CAGAGGGCTTGAGGACAT-3′) and CAD reverse (5′-TCAGGCCTCAAACAAAGACCAGGA-3′), amplifying a band of 1080 bp corresponding to the whole ORF of human DFF40/CAD (GenBankTM accession number NM_004402). The PCR products were automatically sequenced in both directions in a 3130XL genetic analyzer (Applied Biosystems).
Cloning and Transfection of DFF40/CAD
DFF40/CAD full-length open reading frame cDNA sequence from SK-N-AS cells was subcloned into the pcDNA3 vector (Invitrogen). The construct was named pcDNA3-DFF40/CAD and transfected in SK-N-AS cells using Attractene Transfection Reagent (Qiagen) according to the manufacturer's instructions. Stably transfected cells were selected for a month in the presence of 1 mg/ml G-418 (Geneticin) (Invitrogen) and were used as a pool. DFF40/CAD overexpression was assessed by Western blot analysis.
RESULTS
Human Neuroblastoma-derived Cell Lines Show Distinct Apoptotic Nuclear Phenotypes upon Staurosporine Treatment
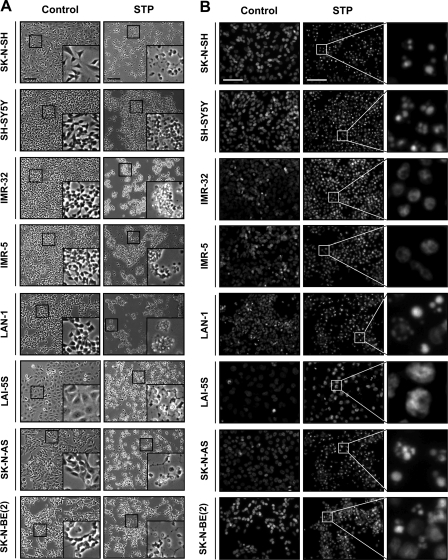
We previously reported the existence of two human neuroblastoma-derived cell lines, IMR-5 and IMR-32, cells that undergo a non-canonical apoptotic cell death upon apoptotic insult (30). This kind of cell death, although caspase-dependent, was characterized by the absence of both nuclear stage II apoptotic morphology (condensation and fragmentation of the cell nuclei) and oligonucleosomal DNA fragmentation (27). In this work we evaluated the morphological appearance of the nuclei and the ability of eight well established human neuroblastoma-derived cells to degrade their DNA into nucleosomal units when challenged with STP. As shown in Figs. 1, A and B, all cell lines tested undergo morphological changes when treated with 1 μm STP for 24 h. Compared with untreated cells, the addition of the alkaloid to the culture media provokes cell shrinkage (Fig. 1A) and an abnormal nuclear morphology (Fig. 1B). However, Hoechst-stained chromatin from STP-treated cells revealed distinct nuclear traits regarding the cell line used. We observed two different types of abnormal nuclei; one of them displayed classical apoptotic nuclei, i.e. highly condensed chromatin and karyorrhexis (in SK-N-SH, SH-SY5Y, LAN-1, SK-N-AS, and SK-N-BE(2) cells), whereas the other presented a partially packaged chromatin (or stage I of apoptotic nuclear morphology) and no evidence of karyorrhexis (in IMR-5, IMR-32, and LAI-5S) (Fig. 1B).
FIGURE 1.
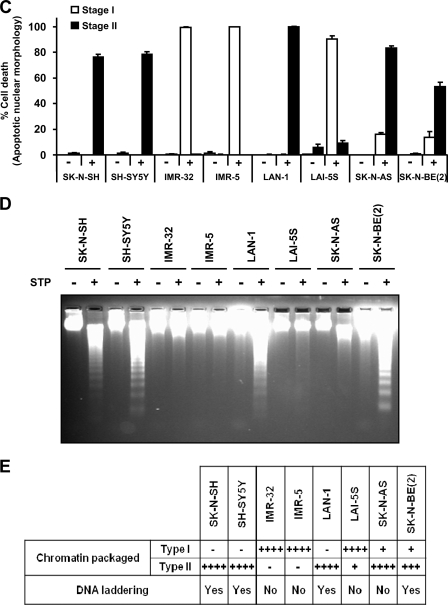
Analysis of apoptotic hallmarks in different human neuroblastoma-derived cell lines upon STP treatment. Eight different neuroblastoma cell lines (SK-N-SH, SH-SY5Y, IMR-32, IMR-5, LAN-1, LAI-5S, SK-N-AS, and SK-N-BE(2)) were left untreated (Control) or treated with 1 μm STP for 24 h. A, microphotographs using a phase contrast microscopy show the morphological appearance of the different cell lines after the indicated treatment. The insets are higher magnifications of the cells framed in the images. The bars indicate 40 μm. B, shown is the morphological appearance of the nuclei stained with Hoechst 33258. The right panels are higher magnifications of the cells framed in the middle panels. The bars indicate 40 μm. C, shown is quantification of cell death by counting stage I or stage II nuclear morphology according to “Experimental Procedures.” Values are expressed as the means ± S.E. D, genomic DNA analysis of the different cell lines by conventional agarose gel electrophoresis and the subsequent ethidium bromide staining is shown. E, shown is a table summarizing the percentage of chromatin-packaged stage observed in the different cell lines and its correlation with their ability to degrade their DNA into LMW size fragments. Note that SK-N-AS cells show stage II nuclear morphology in the absence of a DNA laddering pattern. + means <25%; ++ means 26–50%; +++ means 51–75%, ++++ means >76% of nuclei in stage I and/or stage II apoptotic nuclear morphology.
Despite Presence of Apoptotic Nuclear Morphology and HMW DNA Degradation, SK-N-AS Cells Are Not Able to Hydrolyze Their Chromatin into LMW Fragments upon Apoptotic Stimuli
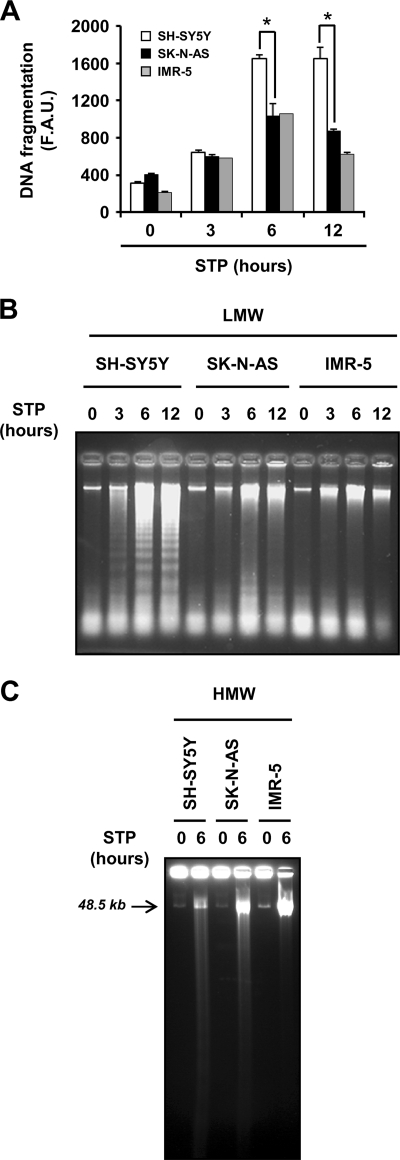
The analysis of the DNA integrity revealed that STP-treated cell lines displaying stage I of nuclear morphology did not degrade their DNA into nucleosomal size fragments (Fig. 1D). As shown in Figs. 1, C and D, most of the cell lines that underwent typical nuclear traits of apoptotic cell death were also able to degrade their DNA into internucleosomal-size fragments. Interestingly, STP did not induce DNA laddering in SK-N-AS cells (Fig. 1D) even though most of them displayed typical apoptotic nuclear morphology (Fig. 1C). Data summarized in Fig. 1E illustrate that apoptotic cells, which preferentially show stage II nuclear morphology, are also able to degrade their chromatin into oligonucleosomal fragments, with the exception of SK-N-AS cells. This peculiarity of SK-N-AS cells was also observed when other apoptotic stimuli were employed, including activators of the intrinsic pathway (e.g. camptothecin, nocodazole, or paclitaxel) (supplemental Fig. S1) as well as inducers of the extrinsic pathway (i.e. CH11 (CD95/Fas agonist antibody), TNF-α, or TRAIL) (supplemental Fig. S2). To further confirm the inability of SK-N-AS cells to generate oligonucleosomal DNA degradation, we used a PEG-based biochemical approach that preferentially allows the recovery of medium-small pieces of nucleotides, such as the internucleosomal-size fragments of DNA (Ref. 25 and Fig. 2B), to be subsequently quantified (Fig. 2A). To control the specificity of this methodology, we took advantage of the intrinsic proprieties characterizing SH-SY5Y and IMR-5 cells, which degrade or not their DNA into LMW size fragments, respectively (27). In this sense, SK-N-AS and IMR-5 cells showed a comparable quantitative profile of DNA degradation after a time-course of STP treatment (Fig. 2A). This is in agreement with the fact that both cell lines, although degrading their chromatin into HMW size fragments, did not display DNA laddering (Ref. 31 and Figs. 2, B and C). Although LMW fragments were not evidenced in both SK-N-AS and IMR-5 cells (Fig. 2B), as depicted in Fig. 2A, STP induced a peak of DNA fragmentation after 6 h, correlating with the presence of HMW DNA degradation (Fig. 2C). However, at this time of treatment, SH-SY5Y cells showed a significant increase in the amount of fragmented DNA compared with SK-N-AS or IMR-5 cells (Fig. 2A). This increase was associated with the presence of LMW DNA fragmentation in SH-SY5Y apoptotic cells (Fig. 2B). These results corroborate a failure of SK-N-AS cells to degrade their DNA into oligonucleosome-size fragments despite the ability to display stage II apoptotic nuclear morphology.
FIGURE 2.
SK-N-AS cells degrade their DNA into HMW but not into LMW fragments. A, quantification of LMW-fragmented DNA in SK-N-AS, SH-SY5Y, and IMR-5 cells in a time-course experiment (3, 6, and 12 h) with 1 μm STP. Quantification was carried out as described under “Experimental Procedures.” The fluorescence values acquired were expressed as arbitrary units (F.A.U., fluorescence arbitrary units). All experiments were repeated at least three times. Values are expressed as the means ± S.E. Student's t tests were used to determine the statistical significance. p < 0.01 was considered to be significant. B, genomic DNA analysis of the samples obtained in A is shown. C, HMW fragmentation of the DNA in SH-SY5Y, SK-N-AS, and IMR-5 cells left untreated (0) or treated with 1 μm STP for 6 h is shown.
SK-N-AS Cells Undergo Caspase-dependent Cell Death after STP Treatment with Proper Activation of Caspase-3 and ICAD Processing
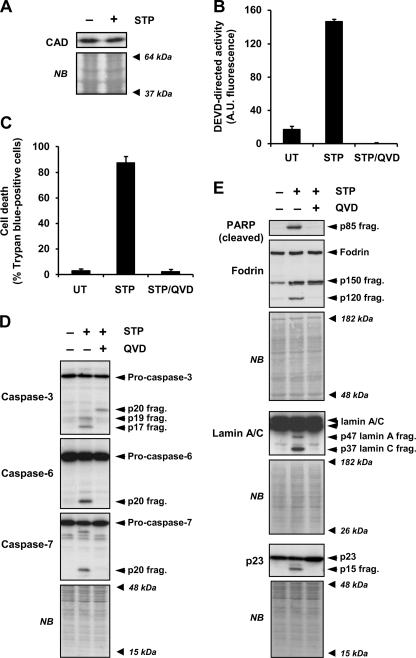
It is well accepted that DFF40/CAD is the main endonuclease responsible for the DNA hydrolysis into LMW size fragments during apoptosis (10, 12, 32). As shown in Fig. 3A, SK-N-AS cells expressed DFF40/CAD, maintaining the protein levels after 24 h in the presence of STP. To look in greater depth at the molecular mechanisms involved in the cell death process induced by STP in SK-N-AS cells, we analyzed the activation status of caspases. A failure in the activation of these proteases could explain the defectiveness of SK-N-AS cells in degrading their DNA into LMW fragments. In this regard, STP induced a caspase-dependent cell death as the addition of q-VD-OPh, a broad pan-caspase inhibitor, completely abrogated caspase activity (Fig. 3B) and, therefore, cell death (Fig. 3C). In addition, executioner pro-caspases, i.e. pro-caspase-3, -6, and -7, were processed into their respective subunits (Fig. 3D). This processing was avoided by the addition of q-VD-OPh to the culture media (Fig. 3D). Moreover, STP induced the proteolysis of general, e.g. poly(ADP-ribose) polymerase, as well as specific caspase substrates (Fig. 3E). Hence, caspase-3 cleaved α-fodrin (α-spectrin), giving a 120-kDa fragment (27), caspase-6 processed lamin A/C into 47- and 37-kDa fragments, respectively (33), and caspase-7 proteolyzed p23 co-chaperone, generating a 17-kDa fragment (34) (Fig. 3E). Finally, and according to the results obtained, q-VD-OPh impaired the proteolysis of all caspases substrates tested (Fig. 3E). Having discarded a general defect in the activation of the executioner caspases, we focused on the minimal components required for oligonucleosomal DNA fragmentation, the caspase-3-ICAD-DFF40/CAD axis (14). As shown in Fig. 4A, we observed the activation of caspase-3 during a time-course of STP treatment. This strictly correlated with the processing of ICAD, DFF45/ICADL (long), and DFF35/ICADS (short) isoforms into their respective fragments. Moreover, the p11 C-terminal fragment, resulting from caspase-3-mediated cleavage of DFF45/ICADL at the residue Asp224, was soon observed at 2 h of treatment (Fig. 4A). The generation of this p11 fragment has been proposed as the determinant for the proper release of DFF40/CAD from its chaperone (35). Of note, p11 detection was evidenced throughout the time of STP treatment in a period in which oligonucleosomal DNA hydrolysis should take place (Figs. 2B and 4A). In addition to that, DFF40/CAD protein levels remained unaltered during the time of STP treatment (Fig. 4A). To exclude a potential mutation in the dff40/cad gene of SK-N-AS cells affecting its function, the cDNA corresponding to the ORF of the endonuclease was obtained and entirely sequenced in both directions. The results showed a wild-type sequence with no mutations affecting the amino acid composition. However, compared with the human DFF40/CAD nucleotide consensus sequence from the NCBI nucleotide data base (NCBI reference sequence NM_004402.2), we observed two silent polymorphic variants corresponding to the 531 and 954 nucleotides of the DFF40/CAD coding sequence (Fig. 4B). This polymorphic combination has been previously reported by Wang and co-workers (12) after cloning and sequencing the dff40 gene from HeLa cells.
FIGURE 3.
STP-induced apoptosis in SK-N-AS cells is a caspase-dependent process. A, SK-N-AS cells were left untreated (−) or treated (+) for 24 h with 1 μm STP. Total extracts were obtained, and DFF40/CAD protein levels were analyzed by Western blot. B and C, SK-N-AS cells were left untreated (UT) or treated with 1 μm STP alone or in the presence of the pan-caspase inhibitor q-VD-OPh (20 μm) (STP/QVD). DEVD-like activity (B) or cell viability by trypan blue exclusion assay (C) was performed after 24 h of treatment. D and E, SK-N-AS cells were left untreated (−) or treated (+) with 1 μm STP alone or plus q-VD-OPh (20 μm) for 6 h. Pro-caspase-3, -6, and -7 processing in their active fragments (D) and cleavage of poly(ADP-ribose) polymerase (PARP (cleaved)), α-Fodrin, lamin A/C, and p23 co-chaperone (E) were analyzed by Western blot. The apparent molecular mass of the specific fragments is indicated on the right of the panels. The membranes were stained with naphthol blue (NB) to assess equal loading. A.U., fluorescence arbitrary units.
FIGURE 4.
Caspase-3/ICAD/CAD axis is properly activated in STP-treated SK-N-AS cells. A, protein extracts from SK-N-AS cells treated with 1 μm STP for the indicated times were analyzed by Western blotting to assess caspase-3 activation (upper panel). The membrane was re-probed with a monoclonal antibody to analyze the cleavage of ICAD (middle panel) and subsequently with an antibody against DFF40/CAD (lower panel). The bands corresponding to the unprocessed proteins or the specific fragments are indicated on the right of the panel. Protein loading was assessed by staining the membranes with naphthol blue (NB). B, shown are PCR products of dff40/cad from SK-N-AS cells amplified by either random hexamers (RH) or a specific reverse primer (R) (see “Experimental Procedures”) (upper panel) and cDNA sequencing (lower scheme). The data obtained reveal two silent polymorphisms compared with the nucleotide consensus sequence for human dff40/cad gene (NCBI reference sequence NM_004402.2) at 531 (G to A) and 954 (A to G) nucleotides of the coding sequence (framed in red boxes in the lower scheme). MW, molecular weight marker.
Lack of DNA LMW Fragmentation in SK-N-AS Cells Is Not Due Either to Non-nuclear DFF40/CAD Sublocalization or Inherent Refractoriness of Their Chromatin to Be Degraded
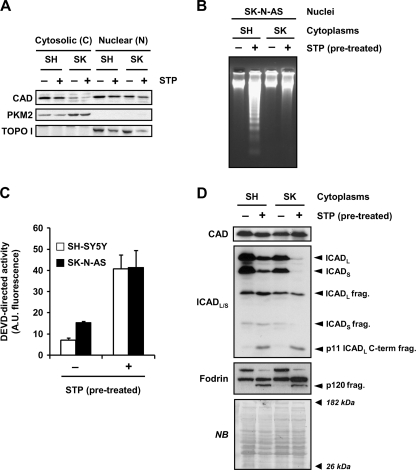
Given that the apoptotic function of DFF40/CAD is nuclear, one possible explanation of the defect described for SK-N-AS cells could rely on a non-nuclear sublocalization of the endonuclease in dying cells. For this reason, we isolated cytosolic and nuclear fractions from SH-SY5Y or SK-N-AS cells in the presence or absence of STP. Fig. 5A shows that, despite a differential distribution of DFF40/CAD in the cytosolic fractions, the endonuclease was found in the nuclear fractions of both SK-N-AS and SH-SY5Y cells either in untreated or STP-treated cells. At this point we thought the refractoriness of SK-N-AS cells to degrade their DNA into oligonucleosomal fragments could rely on the chromatin structure, affecting the endonuclease accessibility to the internucleosomal sites (36). For this reason we performed cell-free assays employing non-treated SK-N-AS isolated nuclei that were incubated with cytoplasmic extracts from either untreated or STP-treated SK-N-AS or SH-SY5Y cells. As shown in Fig. 5B, chromatin of isolated SK-N-AS nuclei was digested into oligonucleosomal DNA fragments when incubated with cytoplasmic extracts from STP-treated SH-SY5Y cells. Nevertheless, the cytoplasm from STP-treated SK-N-AS cells, displaying a DEVDase activity comparable with that obtained in STP-treated SH-SY5Y cytoplasmic extracts (Fig. 5C), failed to generate a DNA laddering pattern (Fig. 5B). The inability of STP-treated SK-N-AS cytoplasm to degrade DNA was due neither to an absence of DFF40/CAD protein (Fig. 5D, upper panel) nor to an impairment in ICAD processing (Fig. 5D, middle panel). Moreover, the detection of the 120-kDa fragment of α-fodrin in the cytoplasmic fractions from STP-treated SK-N-AS cells by Western blot indicated that caspase-3 was properly activated (Fig. 5D, lower panel).
FIGURE 5.
Despite DFF40/CAD nuclear protein levels being comparable between SH-SY5Y and SK-N-AS cells, only the cytoplasm from STP-treated SH-SY5Y cells generate DNA laddering in isolated nuclei of SK-N-AS cells. A, SK-N-AS (SK) and SH-SY5Y (SH) cells treated (+) or not (−) with 1 μm STP for 6 h were subjected to a biochemical subfractionation procedure to separate the cytosolic (C) from the nuclear (N) fractions. Protein extracts were quantified, loaded onto a polyacrylamide gel, electrotransferred to PVDF membranes, and incubated with an anti-DFF40/CAD antibody (upper panel). Membranes were re-probed with an anti-PKM2 (middle panel) and an anti-topoisomerase I (TOPO I, lower panel) antibody to verify the purity of the cytosolic and nuclear fractions, respectively. B, C, and D, shown is the cell-free system assay. Cytoplasmic cell lysates were obtained from SH-SY5Y (SH) or SK-N-AS (SK) cells treated (+) or not (−) with 1 μm STP for 12 h. Isolated nuclei from untreated SK-N-AS cells were incubated for 2 h at 37 °C with 150 μg of the different cytoplasmic extracts. Then, nuclei were collected, and DNA was purified and analyzed by conventional agarose gel electrophoresis and ethidium bromide staining (B). After the cell-free system reaction, aliquots were taken, and quantitative DEVDase assay (C) or DFF40/CAD, ICAD, and α-Fodrin detection by Western blot (D) were performed. In C, DEVDase activity was carried out by incubating 25 μg of extracts with Ac-DEVD- aminofluoromethylcoumarin for 7 h in the fluorimeter. The data obtained are expressed as arbitrary units. Protein loading in D was assessed by staining the membrane with naphthol blue (NB). A.U., fluorescence arbitrary units.
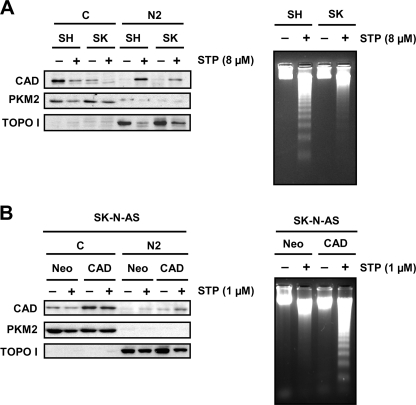
Cytosolic Levels of DFF40/CAD Determine Refractoriness of SK-N-AS Cells to Degrade DNA into LMW Size Fragments
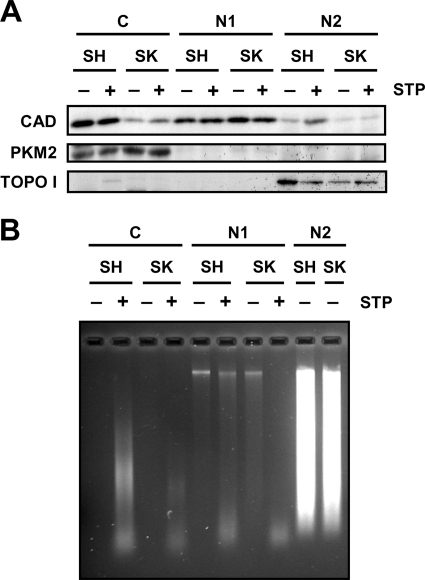
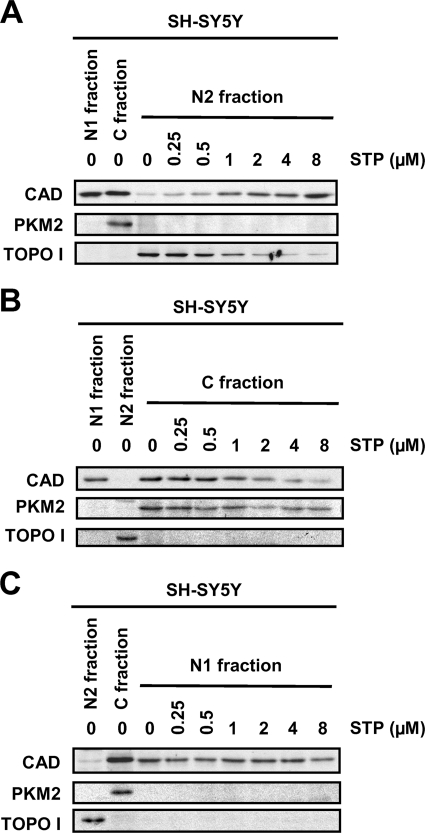
At this point, DFF40/CAD was found in the nuclear fraction of both cell lines (Fig. 5A). However, SK-N-AS chromatin was degraded into LMW fragments in a cell-free assay only when extracts from STP-treated SH-SY5Y cells were employed (Figs. 5B). We then sought to analyze whether DFF40/CAD of SK-N-AS cells associates with the chromatin upon STP treatment. For this purpose we proceeded with a modified protocol from a chromatin binding assay previously developed by Méndez and Stillman (29). We obtained a chromatin-nuclear matrix fraction (N2 in Fig. 6). As expected, DFF40/CAD was evidenced in the chromatin-enriched fraction of SH-SY5Y cells treated with STP (Figs. 6, A and B). However, the endonuclease was not recovered in the chromatin-enriched fraction of SK-N-AS cells upon apoptotic stimulus (Fig. 6A). Taking advantage of the subfractionation method employed, besides the C and N2, we obtained one additional fraction corresponding to the nucleoplasmic (N1) fraction. As can be observed in Figs. 5A (for the cytosolic (C) fractions) and 6A (for both, cytosolic (C) and nucleoplasmic (N1) fractions), we found evidence that DFF40/CAD was recovered in both fractions in untreated SH-SY5Y cells. However, in non-treated SK-N-AS cells, DFF40/CAD was barely detected in the cytosolic fraction (Figs. 5A and 6A) but enriched in the nucleoplasmic fraction (Fig. 6A). To verify the purity of cytosolic and chromatin-enriched fractions, we analyzed the distribution of PKM2, a cytosolic protein kinase involved in cell metabolism, and topoisomerase I, respectively (Fig. 6A, middle and lower panels). As shown in Fig. 6B, we corroborated that DNA is mainly purified in the chromatin-enriched fraction after extracting from the different fractions obtained. Some traces of DNA, probably fragmented DNA, were only detected in the STP-treated cytosolic (C) fraction of SH-SH5Y cells. As stated earlier, although DFF40/CAD levels were comparable between nucleoplasmic (N1) fractions of SH-SY5Y- and SK-N-AS-growing cells, we found a differential recovery of the endonuclease in their respective cytosolic (C) fractions (Figs. 5A and 6A). We, therefore, sought to determine whether DFF40/CAD recovered in the chromatin-enriched fraction from STP-treated SH-SY5Y cells actually comes from its cytosolic pool. As shown in Fig. 7A, the levels of DFF40/CAD detected in the chromatin-enriched (N2) fraction of STP-treated SH-SY5Y cells increased in a dose-dependent manner. This correlated with a decrease in the amounts of DFF40/CAD observed in the cytosolic (C) fraction (Fig. 7B), whereas the endonuclease levels remained unaltered in the nucleoplasmic (N1) fraction (Fig. 7C).
FIGURE 6.
DFF40/CAD is not detected in the chromatin-enriched fraction of SK-N-AS cells upon STP treatment. A, SK-N-AS (SK) and SH-SY5Y (SH) cells were treated (+) or not (−) with 1 μm STP for 6 h. After that, we performed the biochemical fractionations as described in Fig. 5A. From the nuclear (N in Fig. 5A) fraction obtained, we further separated the chromatin-enriched (N2) fraction from the nucleoplasmic (N1) fraction, and DFF40/CAD protein was analyzed by Western blot (upper panel). PKM2 (middle panel) and topoisomerase I (TOPO I, lower panel) were run as cytosolic and nuclear protein markers, respectively. B, genomic DNA analysis of the different fractions obtained in A was achieved by conventional agarose gel electrophoresis and subsequent ethidium bromide staining. Note that DNA is mainly observed in the N2 chromatin-enriched fraction in both cell lines.
FIGURE 7.
DFF40/CAD detected in the chromatin-enriched fraction of STP-treated SH-SY5Y cells comes from the cytosolic pool of the endonuclease. A, SH-SY5Y cells were treated with different concentrations of STP (0, 0.25, 0.5, 1, 2, 4, 8 μm) for 6 h, and then cells were fractionated to obtain the chromatin-enriched or N2 (A), the cytosolic or C (B), and the nucleoplasmic or N1 (C) fractions. DFF40/CAD protein detection was performed by Western blot in the different fractions obtained. PKM2 (middle panel) and topoisomerase I (TOPO I, lower panel) were run as cytosolic and nuclear protein markers, respectively.
Taken together, the results indicate that DFF40/CAD that accumulates into the chromatin-enriched fraction upon STP treatment is derived from the cytosolic rather than the nucleoplasmic pool in SH-SY5Y cells. In STP-treated SK-N-AS cells, the lack of DFF40/CAD in the chromatin-enriched fraction could be explained by a problem in the chromatin binding properties of the endonuclease. However, after 6 h of treatment employing a higher concentration of the alkaloid (8 μm), DFF40/CAD was detected in the chromatin-enriched (N2) fraction (Fig. 8A). This enrichment correlated with the disappearance of DFF40/CAD from the cytosolic fraction (Fig. 8A). Interestingly, even if DFF40/CAD was located at the chromatin-enriched fraction after 6 h with 8 μm STP, SK-N-AS cells were still defective at degrading their DNA into oligonucleosome-size fragments (Fig. 8A). We then sought to corroborate that DFF40/CAD cytosolic levels in untreated cells were determinant in promoting oligonucleosomal DNA fragmentation after STP treatment. For this purpose we augmented the DFF40/CAD levels in SK-N-AS cells by overexpressing their own endonuclease. As shown in Fig. 8B, the protein levels of DFF40/CAD were efficiently increased in the cytosolic fraction recovered from SK-N-AS cells that were stably transfected with the endonuclease (SK-N-AS-DFF40/CAD) compared with cells that were stably transfected with the empty vector (SK-N-AS-Neo). After 1 μm STP treatment, DFF40/CAD was only detected in the chromatin-enriched (N2) fraction from SK-N-AS-DFF40/CAD cells correlating with the presence of oligonucleosomal DNA fragments (Fig. 8B).
FIGURE 8.
The lack of DNA laddering in STP-treated SK-N-AS cells is not due to a defect in the translocation of DFF40/CAD to the chromatin-enriched fraction but determined by the cytosolic levels of the endonuclease. A, SK-N-AS (SK) and SH-SY5Y (SH) cells were treated (+) or not (−) with 8 μm STP for 6 h. Then cells were fractionated to obtain the cytosolic (C) and the chromatin-enriched (N2) fractions. DFF40/CAD protein was assessed in the cytosolic (C) and the chromatin-enriched fraction (N2). Otherwise, DNA was extracted and analyzed by conventional agarose gel electrophoresis to verify the oligonucleosomal DNA degradation. B, SK-N-AS cells stably transfected with empty pcDNA3 (Neo) or pcDNA3-DFF40/CAD (CAD) plasmids were treated with 1 μm STP (+) or left untreated (−) for 6 h. Cells were fractionated to obtain the cytosolic (C) and the chromatin-enriched (N2) fractions, and DFF40/CAD protein was assessed by Western blot. Alternatively, DNA was extracted and analyzed by conventional agarose gel electrophoresis to analyze the DNA laddering. Note that only DFF40/CAD overexpressing cells display a DNA laddering pattern upon STP treatment. PKM2 (middle panel) and topoisomerase I (TOPO I, lower panel) were run as cytosolic and nuclear protein markers, respectively.
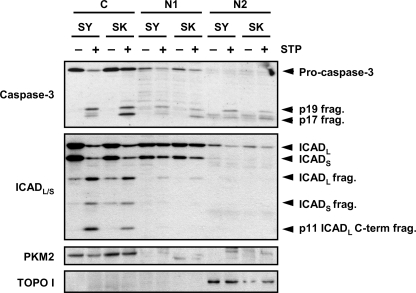
To explain the need for a specific pool of DFF40/CAD to degrade DNA into LMW fragments, we analyzed the activation of caspase-3 and the processing of ICAD in the different biochemical fractions obtained. As depicted in Fig. 9, pro-caspase-3 and ICADL/S showed a preferential localization in the cytosolic (C) fraction in both untreated SH-SY5Y and SK-N-AS cells. After STP treatment, the caspase-mediated processing of ICADL/S was observed in the cytosolic fraction in both cell lines, correlating with the detection of the active p17 subunit of caspase-3 (Fig. 9).
FIGURE 9.
The translocation of DFF40/CAD from the cytosolic to the chromatin-enriched fraction is due to a preferential processing of ICADL/S in the cytosol. SK-N-AS (SK) and SH-SY5Y (SH) cells were treated (+) or not (−) with 1 μm STP for 6 h. Then cells were fractionated to obtain the cytosolic (C), the nucleoplasmic (N1), and the chromatin-enriched (N2) fractions, and caspase-3 and ICAD cleavage were analyzed by Western blot. PKM2 and topoisomerase I (TOPO I) were run as cytosolic and nuclear protein markers, respectively.
Altogether, our results show that cells require certain levels of DFF40/CAD in the chromatin fraction to promote oligonucleosomal DNA degradation during apoptotic cell death. The caspase-dependent accumulation of the endonuclease in this fraction relies on a cytosolic pool that is translocated to the nucleus upon apoptotic stimuli. DFF40/CAD is required to be present in the cytosol because of the preferential processing of DFF45/ICADL in this subcellular compartment.
DISCUSSION
It is well accepted that apoptosis is a biological consequence of a series of complex biochemical and cellular events. Regardless of the specific intracellular pathway activated, apoptosis is depicted morphologically by condensation (highly packaged chromatin, or pyknosis) and fragmentation of the nuclei (karyorrhexis) (also known as stage II) and biochemically by oligonucleosomal DNA degradation (also known as “DNA laddering”) (37, 38). These phenomena are accepted as the hallmarks of apoptotic cell death. These two events have been traditionally considered to occur concomitantly (1, 2, 30, 39). In this work we establish that STP provokes different cell death features, clearly distinguishable among eight different human neuroblastoma-derived cell lines. We describe a set of cell lines displaying a non-classical type of apoptotic nuclear morphology upon STP treatment that corresponds with that previously described as stage I (partial condensation of the chromatin around the nuclear membrane) (40). The presence of this apoptotic nuclear phenotype always correlates with an absence of a DNA laddering pattern, which has been previously reported for IMR-5 and IMR-32 cells (27, 30), and is now extended to LAI-5S cells. We also show that the presence of stage II apoptotic nuclear morphology normally correlates with the ability of the cell to achieve oligonucleosomal DNA degradation. However, despite sharing stage II nuclear morphology, SK-N-AS was the only cell line unable to disintegrate its chromatin into LMW size fragments. Although examples have already been reported of cells displaying apoptotic nuclear morphology in the absence of oligonucleosomal DNA degradation during cell death (36, 41, 42), the role of DFF40/CAD in this cellular paradigm has not been addressed yet.
The existence of a such cellular model could be a valuable tool to discern how cells control the final steps of apoptotic cell death. Indeed, we have taken advantage of SK-N-AS cell innate behavior to understand why endogenous expression of DFF40/CAD as well as ICAD processing are not enough to generate DNA laddering in an intracellular environment in which stage II apoptotic nuclear morphology occurs. Comparative biochemical approaches between SK-N-AS cells and a cellular model that undergoes a complete apoptotic phenotype upon STP treatment, such as SH-SY5Y cells (27, 30, 31), allow us to observe a dissimilar subcellular distribution of DFF40/CAD. In fact, although SH-SY5Y cells show both cytosolic and nuclear pools, in SK-N-AS cells the endonuclease is barely detected in the cytosolic fraction. The subcellular localization of DFF40/CAD remains controversial as data reported in the bibliography seem to be contradictory. On the one hand, several laboratories establish that DFF40/CAD is exclusively located in the cytosol of the cells, translocating to the nucleus after caspase-mediated ICAD processing (10, 18). On the other hand, other authors indicate a nuclear localization of DFF40/CAD in proliferating healthy cells, and therefore, further translocation to the nucleus is not needed during apoptosis (12, 19–22). These data, which seem to be contradictory, can be explained by the results presented in this work, establishing that the subcellular localization of endogenous DFF40/CAD happens in a cell-specific manner. In addition, we establish for the first time that the DFF40/CAD subcellular distribution will condition the capability of a cell to die while degrading or not its chromatin into oligonucleosomal fragments. We show here that this biochemical apoptotic feature seems to be directly correlated with the amount of DFF40/CAD protein content present in the cytosol. Accordingly, the analysis of both total and cytosolic DFF40/CAD protein content across the different human-derived neuroblastoma cell lines shows a direct correlation between the cytosolic, but not the total, levels and the ability of the cell to generate DNA laddering after STP treatment (supplemental Fig. S3). In this regard SK-N-AS cells are able to degrade their chromatin into LMW fragments once the cytosolic pool is enhanced after exogenous expression of its own endonuclease. The preferential processing of ICADL/S in the cytosol explains the requirement for a cell to express the adequate amounts of DFF40/CAD in the same subcellular compartment, leading to DNA degradation into LMW fragments during apoptosis. Along the same line, the activation of, at least, caspase-3 is necessary for the proper cleavage of ICADL/S. Indeed, the release of a caspase-3-mediated p11 C-terminal fragment from ICADL is required for apoptotic nuclease activity (35). In this regard the main processing of pro-caspase-3 takes place in the cytosol of both cells lines analyzed even if caspase-3 active fragments can also be evidenced in the nuclear fractions after STP treatment. However, nuclear ICAD-CAD complex does not seem to be affected, maybe because it is not reached by the limited amounts of active caspase-3 detected in the nuclear fractions. In the case of SK-N-AS cells, the cleaved ICAD in the cytoplasm only liberates a small pool of CAD, due to the low cytosolic levels.
Despite our findings, we cannot rule out the possibility of other cellular paradigms in which the activation of the caspase-3/ICAD/CAD axis occurs in other subcellular compartments than in the cytosol. In these cases we can hypothesize that if CAD protein levels are adequate, the cell should be able to degrade its chromatin into oligonucleosome-size fragments upon apoptotic insult. Therefore, the limiting aspect for the occurrence of internucleosomal apoptotic DNA degradation should be directly related to the levels of mature caspase-3, rather than pro-caspase-3, afforded at the same subcellular compartment in which ICAD-CAD complex resides. Then, in healthy cells, the subcellular localization of pro-caspase-3 and ICAD-CAD complex could be different without affecting the capacity to degrade their chromatin into LMW fragments during apoptosis. In this regard, it has been recently stated that nuclear localization of active caspase-3, but not pro-caspase-3, is closely related to the occupancy of its p3 recognition site, located in the large subunit, possibly by an as yet unknown non-degradable substrate (43).
Based on the last recommendations formulated by the Nomenclature Committee on Cell Death (44), STP-induced cell death in SK-N-AS cells as well as in SH-SY5Y cells can be biochemically grouped into the “caspase-dependent intrinsic apoptosis” category. However, from a classical point of view, whereas SH-SY5Y cells undergo typical apoptosis, SK-N-AS cells show an atypical apoptotic cell death characterized by an incomplete degradation of their chromatin. There are many examples in the literature of cells displaying atypical apoptotic cell death that can be classified as “caspase-dependent intrinsic apoptosis.” These cell types include the human acute lymphoblastic leukemia cell line MOLT-4 (42), the human breast carcinoma-derived cell line MCF-7 (45), the human androgen-independent prostatic cancer cell DU-145 (41), the human neuronal-like cell NT2 (46), and some human neuroblastoma-derived cell lines such as IMR-5 and IMR-32 (27, 30). Therefore, a cell committed to death can die despite occasional and non-critical alterations in the intracellular pathway triggered. For example, when caspase-3-defective MCF-7 cells die, they activate the canonical intracellular apoptotic pathway bypassing the absence of caspase-3 (45). Hence, the presence of caspase-3 should be dispensable for cell death regardless of the apoptotic inducer employed. However, the biological implications concerning caspase-3 activation are far from irrelevant when considering an entire cellular population. Indeed, the activation of caspase-3 during anti-cancer radiotherapy prompts tumor cell repopulation (47). Similar to the caspase-3 example, oligonucleosomal DNA degradation is also considered a dispensable biochemical feature for the cell death of a single cell or even for a well limited group of cells (48). However, the ability of a certain pool of cells to degrade their chromatin into LMW-size fragments or not could entail disparate biological consequences for their neighbors. In this regard, although the information is limited concerning the cell death process in the absence of oligonucleosomal DNA fragmentation, it is considered that chromatin degradation into LMW fragments confers some advantages to the organism. For example, DNA degradation may minimize the risk of transfer of non-degraded cellular or viral DNA carrying potential oncogenes from a doomed to an adjacent healthy cell (49–51). Moreover, it has also been proposed that intact circulating nucleosomes in blood from non- or partially degraded nuclear chromatin could trigger some autoimmune diseases, such as systemic lupus erythematosus, in which autoantibodies against heterochromatin are generated (52). Finally, it is thought that phagocytic activity of healthy neighboring cells can be facilitated if the chromatin of apoptotic bodies is already fragmented (53). So the importance of revealing the molecular mechanisms regulating the final apoptotic steps, such as DNA degradation into oligonucleosomal pieces, resides in the biological consequences for neighboring cells. The identification of other cellular models, including primary tumors as well as non-tumor cells, emulating SK-N-AS cell behavior might be a valuable tool to detect new critical points affecting downstream apoptotic signals, i.e. oligonucleosomal DNA fragmentation. Therefore, knowledge of the molecular mechanisms governing the full activation of DFF40/CAD might be crucial for improving current therapeutic approaches in diseases concerning DNA degradation, e.g. cancer or autoimmune disorders.
Supplementary Material
Acknowledgments
We are indebted to the other members from both laboratories for helpful criticisms. We are especially grateful to Joaquin López and Patricia Ortega for technical support.
This work was supported by Ministerio de Ciencia e Innovación/Fondo Europeo de Desarrollo Regional (FEDER) Grants BFU2008-01328 and TRA2009-0185 (to V. J. Y.) and SAF2010-19953 (to J. X. C.), Centro de Investigacion Biomedica en Red sobre Enfermedades Neurodegenerativas Grant CB06/05/0042, and Generalitat de Catalunya Grant SGR2009-346.

This article contains supplemental Figs. 1–3.
- HMW
- high molecular weight
- LMW
- low molecular weight
- DFF40/CAD
- DNA fragmentation factor, 40-kDa subunit/caspase-activated DNase (CAD)
- ICAD
- inhibitor of CAD
- DFF45
- DNA fragmentation factor, 45-kDa subunit
- STP
- staurosporine
- q-VD-OPh
- N-(2-Quinolyl)valyl-aspartyl-(2,6-difluorophenoxy)methyl ketone.
REFERENCES
- 1. Steller H. (1995) Mechanisms and genes of cellular suicide. Science 267, 1445–1449 [DOI] [PubMed] [Google Scholar]
- 2. Jacobson M. D., Weil M., Raff M. C. (1997) Programmed cell death in animal development. Cell 88, 347–354 [DOI] [PubMed] [Google Scholar]
- 3. Hipfner D. R., Cohen S. M. (2004) Connecting proliferation and apoptosis in development and disease. Nat. Rev. Mol. Cell Biol. 5, 805–815 [DOI] [PubMed] [Google Scholar]
- 4. de Bruin E. C., Medema J. P. (2008) Apoptosis and non-apoptotic deaths in cancer development and treatment response. Cancer Treat. Rev. 34, 737–749 [DOI] [PubMed] [Google Scholar]
- 5. Baguley B. C. (2011) The paradox of cancer cell apoptosis. Front. Biosci. 16, 1759–1767 [DOI] [PubMed] [Google Scholar]
- 6. Timmer J. C., Salvesen G. S. (2007) Caspase substrates. Cell Death Differ. 14, 66–72 [DOI] [PubMed] [Google Scholar]
- 7. Lüthi A. U., Martin S. J. (2007) The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 14, 641–650 [DOI] [PubMed] [Google Scholar]
- 8. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dix M. M., Simon G. M., Cravatt B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. (1998) A caspase-activated DNase that degrades DNA during apoptosis and its inhibitor ICAD. Nature 391, 43–50 [DOI] [PubMed] [Google Scholar]
- 11. Halenbeck R., MacDonald H., Roulston A., Chen T. T., Conroy L., Williams L. T. (1998) CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45. Curr. Biol. 8, 537–540 [DOI] [PubMed] [Google Scholar]
- 12. Liu X., Li P., Widlak P., Zou H., Luo X., Garrard W. T., Wang X. (1998) The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. U. S. A. 95, 8461–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sakahira H., Enari M., Nagata S. (1998) Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391, 96–99 [DOI] [PubMed] [Google Scholar]
- 14. Liu X., Zou H., Slaughter C., Wang X. (1997) DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 89, 175–184 [DOI] [PubMed] [Google Scholar]
- 15. Zhang J., Liu X., Scherer D. C., van Kaer L., Wang X., Xu M. (1998) Resistance to DNA fragmentation and chromatin condensation in mice lacking the DNA fragmentation factor 45. Proc. Natl. Acad. Sci. U. S. A. 95, 12480–12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J., Lee H., Lou D. W., Bovin G. P., Xu M. (2000) Lack of obvious 50-kilobase pair DNA fragments in DNA fragmentation factor 45-deficient thymocytes upon activation of apoptosis. Biochem. Biophys. Res. Commun. 274, 225–229 [DOI] [PubMed] [Google Scholar]
- 17. Nagase H., Fukuyama H., Tanaka M., Kawane K., Nagata S. (2003) Mutually regulated expression of caspase-activated DNase and its inhibitor for apoptotic DNA fragmentation. Cell Death Differ. 10, 142–143 [DOI] [PubMed] [Google Scholar]
- 18. Sabol S. L., Li R., Lee T. Y., Abdul-Khalek R. (1998) Inhibition of apoptosis-associated DNA fragmentation activity in nonapoptotic cells. The role of DNA fragmentation factor-45 (DFF45/ICAD). Biochem. Biophys. Res. Commun. 253, 151–158 [DOI] [PubMed] [Google Scholar]
- 19. Samejima K., Earnshaw W. C. (1998) ICAD/DFF regulator of apoptotic nuclease is nuclear. Exp. Cell Res. 243, 453–459 [DOI] [PubMed] [Google Scholar]
- 20. Samejima K., Earnshaw W. C. (2000) Differential localization of ICAD-L and ICAD-S in cells due to removal of a C-terminal NLS from ICAD-L by alternative splicing. Exp. Cell Res. 255, 314–320 [DOI] [PubMed] [Google Scholar]
- 21. Chen D., Stetler R. A., Cao G., Pei W., O'Horo C., Yin X. M., Chen J. (2000) Characterization of the rat DNA fragmentation factor 35/Inhibitor of caspase-activated DNase (short form). The endogenous inhibitor of caspase-dependent DNA fragmentation in neuronal apoptosis. J. Biol. Chem. 275, 38508–38517 [DOI] [PubMed] [Google Scholar]
- 22. Lechardeur D., Drzymala L., Sharma M., Zylka D., Kinach R., Pacia J., Hicks C., Usmani N., Rommens J. M., Lukacs G. L. (2000) Determinants of the nuclear localization of the heterodimeric DNA fragmentation factor (ICAD/CAD). J. Cell Biol. 150, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gozzelino R., Sole C., Llecha N., Segura M. F., Moubarak R. S., Iglesias-Guimarais V., Perez-Garcia M. J., Reix S., Zhang J., Badiola N., Sanchis D., Rodriguez-Alvarez J., Trullas R., Yuste V. J., Comella J. X. (2008) BCL-XL regulates TNF-α-mediated cell death independently of NF-κB, FLIP, and IAPs. Cell Res. 18, 1020–1036 [DOI] [PubMed] [Google Scholar]
- 24. Ribas J., Yuste V. J., Garrofé-Ochoa X., Meijer L., Esquerda J. E., Boix J. (2008) 7-Bromoindirubin-3′-oxime uncovers a serine protease-mediated paradigm of necrotic cell death. Biochem. Pharmacol. 76, 39–52 [DOI] [PubMed] [Google Scholar]
- 25. Ioannou Y. A., Chen F. W. (1996) Quantitation of DNA fragmentation in apoptosis. Nucleic Acids Res. 24, 992–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barry M. A., Eastman A. (1993) Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch. Biochem. Biophys. 300, 440–450 [DOI] [PubMed] [Google Scholar]
- 27. Yuste V. J., Bayascas J. R., Llecha N., Sánchez-López I., Boix J., Comella J. X. (2001) The absence of oligonucleosomal DNA fragmentation during apoptosis of IMR-5 neuroblastoma cells. Disappearance of the caspase-activated DNase. J. Biol. Chem. 276, 22323–22331 [DOI] [PubMed] [Google Scholar]
- 28. Widlak P., Garrard W. T. (2006) The apoptotic endonuclease DFF40/CAD is inhibited by RNA, heparin, and other polyanions. Apoptosis 11, 1331–1337 [DOI] [PubMed] [Google Scholar]
- 29. Méndez J., Stillman B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle. Assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boix J., Llecha N., Yuste V. J., Comella J. X. (1997) Characterization of the cell death process induced by staurosporine in human neuroblastoma cell lines. Neuropharmacology 36, 811–821 [DOI] [PubMed] [Google Scholar]
- 31. Yuste V. J., Sánchez-López I., Solé C., Moubarak R. S., Bayascas J. R., Dolcet X., Encinas M., Susin S. A., Comella J. X. (2005) The contribution of apoptosis-inducing factor, caspase-activated DNase, and inhibitor of caspase-activated DNase to the nuclear phenotype and DNA degradation during apoptosis. J. Biol. Chem. 280, 35670–35683 [DOI] [PubMed] [Google Scholar]
- 32. Kawane K., Fukuyama H., Yoshida H., Nagase H., Ohsawa Y., Uchiyama Y., Okada K., Iida T., Nagata S. (2003) Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat. Immunol. 4, 138–144 [DOI] [PubMed] [Google Scholar]
- 33. Ruchaud S., Korfali N., Villa P., Kottke T. J., Dingwall C., Kaufmann S. H., Earnshaw W. C. (2002) Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 21, 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walsh J. G., Cullen S. P., Sheridan C., Lüthi A. U., Gerner C., Martin S. J. (2008) Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl. Acad. Sci. U. S. A. 105, 12815–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang D., Kidd V. J. (1998) Cleavage of DFF-45/ICAD by multiple caspases is essential for its function during apoptosis. J. Biol. Chem. 273, 28549–28552 [DOI] [PubMed] [Google Scholar]
- 36. Kuribayashi N., Sakagami H., Iida M., Takeda M. (1996) Chromatin structure and endonuclease sensitivity in human leukemic cell lines. Anticancer Res. 16, 1225–1230 [PubMed] [Google Scholar]
- 37. Wyllie A. H., Morris R. G., Smith A. L., Dunlop D. (1984) Chromatin cleavage in apoptosis. Association with condensed chromatin morphology and dependence on macromolecular synthesis. J. Pathol. 142, 67–77 [DOI] [PubMed] [Google Scholar]
- 38. Zamzami N., Kroemer G. (1999) Condensed matter in cell death. Nature 401, 127–128 [DOI] [PubMed] [Google Scholar]
- 39. Samejima K., Tone S., Earnshaw W. C. (2001) CAD/DFF40 nuclease is dispensable for high molecular weight DNA cleavage and stage I chromatin condensation in apoptosis. J. Biol. Chem. 276, 45427–45432 [DOI] [PubMed] [Google Scholar]
- 40. Susin S. A., Daugas E., Ravagnan L., Samejima K., Zamzami N., Loeffler M., Costantini P., Ferri K. F., Irinopoulou T., Prévost M. C., Brothers G., Mak T. W., Penninger J., Earnshaw W. C., Kroemer G. (2000) Two distinct pathways leading to nuclear apoptosis. J. Exp. Med. 192, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oberhammer F., Wilson J. W., Dive C., Morris I. D., Hickman J. A., Wakeling A. E., Walker P. R., Sikorska M. (1993) Apoptotic death in epithelial cells. Cleavage of DNA to 300- and/or 50-kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 12, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Falcieri E., Martelli A. M., Bareggi R., Cataldi A., Cocco L. (1993) The protein kinase inhibitor staurosporine induces morphological changes typical of apoptosis in MOLT-4 cells without concomitant DNA fragmentation. Biochem. Biophys. Res. Commun. 193, 19–25 [DOI] [PubMed] [Google Scholar]
- 43. Luo M., Lu Z., Sun H., Yuan K., Zhang Q., Meng S., Wang F., Guo H., Ju X., Liu Y., Ye T., Lu Z., Zhai Z. (2010) Nuclear entry of active caspase-3 is facilitated by its p3-recognition-based specific cleavage activity. Cell Res. 20, 211–222 [DOI] [PubMed] [Google Scholar]
- 44. Galluzzi L., Vitale I., Abrams J. M., Alnemri E. S., Baehrecke E. H., Blagosklonny M. V., Dawson T. M., Dawson V. L., El-Deiry W. S., Fulda S., Gottlieb E., Green D. R., Hengartner M. O., Kepp O., Knight R. A., Kumar S., Lipton S. A., Lu X., Madeo F., Malorni W., Mehlen P., Nuñez G., Peter M. E., Piacentini M., Rubinsztein D. C., Shi Y., Simon H. U., Vandenabeele P., White E., Yuan J., Zhivotovsky B., Melino G., Kroemer G. (2012) Molecular definitions of cell death subroutines. Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 19, 107–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jänicke R. U., Sprengart M. L., Wati M. R., Porter A. G. (1998) Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 273, 9357–9360 [DOI] [PubMed] [Google Scholar]
- 46. Walker P. R., Leblanc J., Carson C., Ribecco M., Sikorska M. (1999) Neither caspase-3 nor DNA fragmentation factor is required for high molecular weight DNA degradation in apoptosis. Ann. N.Y. Acad. Sci. 887, 48–59 [DOI] [PubMed] [Google Scholar]
- 47. Huang Q., Li F., Liu X., Li W., Shi W., Liu F. F., O'Sullivan B., He Z., Peng Y., Tan A. C., Zhou L., Shen J., Han G., Wang X. J., Thorburn J., Thorburn A., Jimeno A., Raben D., Bedford J. S., Li C. Y. (2011) Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 17, 860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schulze-Osthoff K., Walczak H., Dröge W., Krammer P. H. (1994) Cell nucleus and DNA fragmentation are not required for apoptosis. J. Cell Biol. 127, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de la Taille A., Chen M. W., Burchardt M., Chopin D. K., Buttyan R. (1999) Apoptotic conversion. Evidence for exchange of genetic information between prostate cancer cells mediated by apoptosis. Cancer Res. 59, 5461–5463 [PubMed] [Google Scholar]
- 50. Yan B., Wang H., Li F., Li C. Y. (2006) Regulation of mammalian horizontal gene transfer by apoptotic DNA fragmentation. Br. J. Cancer 95, 1696–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yan B., Wang H., Xie D., Wakamatsu N., Anscher M. S., Dewhirst M. W., Mitchel R. E., Chen B. J., Li C. Y. (2009) Increased skin carcinogenesis in caspase-activated DNase knockout mice. Carcinogenesis 30, 1776–1780 [DOI] [PubMed] [Google Scholar]
- 52. Herrmann M., Voll R. E., Zoller O. M., Hagenhofer M., Ponner B. B., Kalden J. R. (1998) Impaired phagocytosis of apoptotic cell material by monocyte-derived macrophages from patients with systemic lupus erythematosus. Arthritis Rheum. 41, 1241–1250 [DOI] [PubMed] [Google Scholar]
- 53. Peitsch M. C., Mannherz H. G., Tschopp J. (1994) The apoptosis endonucleases. Cleaning up after cell death? Trends Cell Biol. 4, 37–41 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.