Background: Epithelial-to-mesenchymal transition (EMT) has important biological implications, but mechanisms governing this process are only partially understood.
Results: The nuclear protein “high mobility group A2” regulates expression of the transcription factors Twist and Snail during EMT.
Conclusion: Twist and Snail complement each other in EMT.
Significance: We now understand better how Twist and Snail act interdependently to regulate the EMT.
Keywords: Adherens junction, EMT, Promoters, Tight junctions, Transcription/developmental factors, Transforming growth factor β (TGFβ)
Abstract
Deciphering molecular mechanisms that control epithelial-to-mesenchymal transition (EMT) contributes to our understanding of how tumor cells become invasive and competent for intravasation. We have established that transforming growth factor β activates Smad proteins, which induce expression of the embryonic factor high mobility group A2 (HMGA2), which causes mesenchymal transition. HMGA2 associates with Smad complexes and induces expression of an established regulator of EMT, the zinc finger transcription factor Snail. We now show that HMGA2 can also induce expression of a second regulator of EMT, the basic helix-loop-helix transcription factor Twist. Silencing of endogenous Twist demonstrated that this protein acts in a partially redundant manner together with Snail. Double silencing of Snail and Twist reverts mesenchymal HMGA2-expressing cells to a more epithelial phenotype when compared with single silencing of Snail or Twist. Furthermore, HMGA2 can directly associate with A:T-rich sequences and promote transcription from the Twist promoter. The new evidence proposes a model whereby HMGA2 directly induces multiple transcriptional regulators of the EMT program and, thus, is a potential biomarker for carcinomas displaying EMT during progression to more advanced stages of malignancy.
Introduction
Many successive steps are required for a growing benign hyperplasia to evolve into a fully malignant and metastatic cancer (1). A critical event that enables cancer cells to invade the local tissue, acquire competence for intravasation, and generate progeny with tumor-initiating capacities is the epithelial-to-mesenchymal transition (EMT)3 (2). During EMT, differentiated epithelial cells lose their cell-cell adhesions, become more motile, and exhibit mesenchymal features. For example, loss of E-cadherin expression, a key molecule of the adherens junction and a tumor suppressor gene (CDH1), and induction of vimentin-based intermediate filaments are two of the many established hallmarks of the EMT process (3).
The large numbers of cellular events that characterize the mesenchymal transition are thought to be collectively regulated by a group of transcription factors that coordinate the transcriptional program of EMT. These transcriptional regulators are the zinc finger factors Snail1 (Snail), Snail2 (also known as Slug), ZEB1/δEF1, ZEB2/SIP1, and the basic helix-loop-helix factors E47 and Twist1 (Twist) (4). The complete transcriptomic program that is regulated by these five transcription factors has not yet been fully elucidated. However, clear examples of transcriptional regulation during EMT are the repression of CDH1 by Snail1, Snail2, E47, ZEB1, and ZEB2, a mechanism that is thought to lead to the methylation of the DNA sequences of the CDH1 gene promoter and the terminal silencing of this gene (3). On the other hand, Twist induces expression of genes that promote tumor cell invasiveness (5), and the forkhead transcription factor FoxC2 induces genes of the mesenchymal program (6).
The function and expression of the transcription factors that orchestrate the EMT program is regulated by developmental signal transduction pathways, such as transforming growth factor β (TGFβ), Notch, fibroblast growth factor, and more (2, 7). In vitro studies in immortalized epithelial cells and in carcinoma cell lines, complemented by in vivo studies in transgenic mice, have clearly shown that TGFβ plays a critical role in the control of EMT of tumor cells (8, 9).
The TGFβ pathway includes a versatile network of extracellular signaling factors that regulate important aspects of embryonic development, tissue homeostasis, and progression of disease states, including cancer and tissue fibrosis, where EMT is a process of importance (10). TGFβ ligands signal via cell surface receptor kinases, which activate Smad proteins and additional non-Smad pathways (11), that regulate gene transcription and thus elicit a multitude of physiological responses (12). With respect to its role in cancer, TGFβ suppresses tumorigenesis, because it restricts epithelial cell proliferation and induces apoptosis (9), but also promotes the evolution of carcinomas toward metastasis by promoting EMT, suppressing the beneficial anti-tumoral immune responses and stimulating tumor angiogenesis and cancer-associated fibroblast functions (7, 13).
We previously established a central mechanism that promotes EMT in response to TGFβ and involves two direct target genes of Smad signaling, the high mobility group (HMG) A2 and Snail (14, 15). HMGA2 is a chromatin-binding protein containing three AT-hook domains that enable its binding to the minor groove of DNA and thus organizes protein complexes on enhancers of various genes, leading to regulation of gene expression and cell differentiation (16). HMGA2 is expressed during embryonic development and is much less expressed in adulthood. However, cancers of mesenchymal origin (e.g. fibrosarcomas) and metastatic cancers overexpress HMGA2 (16), which is compatible with a model of transition of tumor cells (via EMT) to phenotypes that reactivate embryonic transcriptional programs (2). We have demonstrated that HMGA2 associates with Smads and together bind to the Snail gene promoter causing Snail expression and EMT (14, 15). Cells that overexpress HMGA2 and undergo mesenchymal transition express high levels of Twist in addition to Snail (14, 15). We hypothesized that Twist and Snail might act in a complementary manner ensuring the robust induction of EMT in cells responding to TGFβ. The present report presents evidence that supports the complementary role of Twist on the side of Snail during EMT driven by HMGA2 overexpression. In addition, we explain how HMGA2 can directly induce Twist transcription by binding to regulatory sequences of this gene. The new work firmly establishes that the EMT program promoted by TGFβ signaling involves a stable crosstalk and interplay of multiple embryonic transcription factors.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
Short hairpin RNA (shRNA) directed against the murine Twist gene (shTwist, 5′-GCAAGATTCAGACCCTCAAAC-3′) was designed using the Invitrogen RNAi Designer online tool. Double-stranded oligonucleotides encoding shTwist or shlacZ were first cloned into a pENTR/U6 vector and then transferred into pBLOCK-iT6-DEST vector (Invitrogen) for stable shRNA expression, according to the manufacturer's instructions. The pENTR/U6 and pENTR/U6/shlacZ control vectors (Invitrogen) were from J. Ericsson (University College Dublin, Conway Institute, Dublin, Ireland). The mammalian expression constructs pcDNA3 encoding HA-tagged hHMGA2 and hHMGA2ΔC, and FLAG-tagged Smad3 and Smad4, have been described previously (14). The murine Twist promoter luciferase plasmid (−1745/+209) was provided by L. R. Howe (Cornell University, New York, NY) (17).
Deletion series of the Twist promoter were obtained by PCR amplification using appropriate primers containing KpnI and HindIII restriction sites, and the corresponding PCR products were cloned into the same restriction sites in a pGL2-Basic vector (Promega, Madison, WI). Primers used for Twist promoter deletion series are listed in supplemental Table S1. The mutants M2 (TTT to CCC) and M3 (TTCTTTT to CCCTCCC) were reconstituted into the Twist −95/+209 promoter by PCR-based site-specific mutagenesis (Stratagene), according to the manufacturer's instructions.
Recombinant mature TGFβ1 was from BIOSOURCE Inc. Small molecular weight inhibitors: SB505124 against the three type I receptor kinases of the TGFβ family, known as activin receptor-like 4, 5, and 7, was from Sigma; SB202190 against the p38 mitogen-activated protein kinase (MAPK) was from Enzo Life Sciences; LY294002 against the phosphoinositide 3′-kinase was from Cell Signaling Technology; and PD184352 against the MEK1 kinase that activates the MAPKs ERK1 and ERK2 was a kind gift from J. Lennartsson (Ludwig Institute, Uppsala, Sweden).
Cell Culture and Transfections
Parental mouse mammary epithelial NMuMG cells, cell clones overexpressing pMEP4-HA-tagged human HMGA2 (HMGA2-NMuMG) or pMEP4 empty vector (NMuMG-m), clones with stable knockdown of Snail in HMGA2-NMuMG cells (HMGA2-shSnail), and human hepatocarcinoma HepG2 cells have been described previously (14, 15).
HepG2 and NMuMG cells were transiently transfected using the calcium phosphate method and FuGENE HD (Roche Applied Science) or Lipofectamine 2000 (Invitrogen), respectively. Stable transfections of pBLOCK-iT6-DEST vectors (shlacZ and shTwist) in HMGA2-NMuMG and HMGA2-shSnail cells were done using Lipofectamine 2000, and selection was performed with 5 μg/ml blasticidin (Invitrogen). Transient transfections of HMGA2-NMuMG cells with siRNA against mouse Smad4 (Dharmacon ON-TARGETplus SMARTpool l-040687) or non-targeting siRNA control (Dharmacon ON-TARGETplus Non-targeting pool d-001810–10−20) were done using DharmaFECT 1 siRNA transfection reagent (Dharmacon).
Real Time-RT-PCR
Total RNA was isolated from cells using the Qiagen RNeasy mini kit (Qiagen) and digested with DNase I (Qiagen) to remove any contaminating genomic DNA. The reverse transcription reaction was done with 1 μg of RNA and iScript cDNA synthesis kit (Bio-Rad). PCR was performed in a total volume of 10 μl with iQ SYBR Green Supermix (Bio-Rad), 1 μl of cDNA, and 250 nm of each primer. Specific primers were designed according to sequences available in the databases or published by others (supplemental Table S2). Reactions were carried out in triplicates in a CFX96 Real-Time PCR Detection System (Bio-Rad), using conditions as described previously (18). Different controls were used to demonstrate the specificity of the reactions: the reverse transcriptase was omitted (−RT) or the cDNA was replaced with water. These controls were run in every RT-PCR assay, but the figures present only representative and limited numbers of these controls. Gene expression levels were determined with the comparative Ct method using the mouse gapdh mRNA as reference.
Immunoblotting and Immunofluorescence Microscopy
Total protein extracted from cells were subjected to SDS-PAGE and analyzed by immunoblotting, as described (14). Nuclear extracts were isolated with an NXTRACT CelLytic Nuclear Extraction Kit (Sigma-Aldrich), according to the manufacturer's instructions. Rabbit anti-CREB, mouse anti-Twist1, rabbit anti-ZEB1, and rabbit anti-ZEB2/SIP1 were from Santa Cruz Biotechnology; rabbit anti-Snail was from Abcam; rabbit anti-phospho-CREB, rabbit anti-ERK, rabbit anti-phospho-ERK, rabbit anti-p70S6K, and rabbit anti-phospho-p70S6K were from Cell Signaling Technology; mouse anti-E-cadherin, mouse anti-N-cadherin, and mouse anti-PARP1 antibodies were from BD Transduction Laboratories; rabbit anti-fibronectin and mouse anti-α-smooth muscle actin were from Sigma-Aldrich; mouse anti-GAPDH was from Ambion; mouse anti-HA from Roche Applied Science; mouse anti-ZO-1 antibody from Invitrogen; and secondary antibodies, anti-mouse-IgG, and anti-rabbit-IgG coupled to horseradish peroxidase were from GE Healthcare. Chemiluminescence was detected using Immobilon Western chemiluminescent HRP substrate from Millipore.
For immunofluorescence, cells were fixed and stained with phalloidin-FITC (Sigma-Aldrich) or primary antibody and then followed by appropriate Alexa Fluor 488 donkey anti-mouse-IgG or goat anti-rabbit-IgG secondary antibody (Molecular Probes). Photomicrographs were obtained by using a Zeiss Axioplan 2 microscope with a Hammamatsu C4742-95 digital camera, with a Zeiss Plan-neofluar 40×/0.75 objective lens. For phase-contrast microscopy, live cells growing on the culture dish were analyzed on a Zeiss Axiovert 40 CFL microscope with an AxioCam MRc digital camera, using a Zeiss Plan-neofluar 10×/0.3 objective lens. All photography was at ambient temperature in the absence of immersion oil. Primary images were acquired with the Volocity software of the camera. Image memory content was reduced, and brightness-contrast was adjusted using Adobe Photoshop CS3 Extended.
Promoter Reporter Assays
The full-length and deletion constructs of the mouse Twist promoter were co-transfected with reporter plasmid pCMV-βGal for normalization and expression vectors: pcDNA3 (control), pcDNA3-HA-hHMGA2, pcDNA3-HA-hHMGA2ΔC, pcDNA3-FLAG-Smad3, or pcDNA3-FLAG-Smad4 in HepG2 and NMuMG cells. The enhanced luciferase assay kit from BD Pharmingen was used. Normalized promoter activity data are plotted in bar graphs representing mean ± S.D. from triplicate samples. Each independent experiment was repeated at least twice.
DNA Affinity Precipitation Experiments
HepG2 cells were transiently transfected with pcDNA3-HA-hHMGA2. Proteins were extracted from transfected cells in lysis buffer containing protease inhibitors (0.5% Nonidet P-40, 100 mm EDTA, 100 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsulfonyl fluoride, and 1% aprotinin). After preclearing, protein extracts were incubated overnight at 4 °C with biotin-labeled probes described in supplemental Table S3. The biotin-labeled probes were obtained by PCR amplification using AccuPrime Pfx DNA polymerase (Invitrogen). Dynabeads coupled to M-280 Streptavidin (Invitrogen) were then added for 1 h, followed by four washes with 1× B&W buffer (5 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, 1 m NaCl), and resuspended in SDS loading buffer. Bound proteins were resolved by SDS-PAGE and detected by immunoblotting.
Chromatin Immunoprecipitation
ChIP analysis was done as described in a previous study (19). Briefly, HMGA2-NMuMG cells were cultured in 15-cm plates to ∼80% confluence, and one plate was used per ChIP reaction. The cells were cross-linked with 1% formaldehyde and then neutralized with 0.125 m glycine, washed with ice-cold PBS, and resuspended in lysis buffer. Samples were sonicated in a water-bath Diagenode Bioruptor sonicator (output high; 5 cycles of 30 s sonication with 30-s intervals) to yield DNA fragments of 250–500 bp. After removing an aliquot of total cell lysate as input, the supernatant was diluted with ChIP dilution and incubated with 10 μg of rabbit anti-HMGA2 antibody from Santa Cruz Biotechnology or rabbit IgG; the latter was a kind gift from E. Vassilaki (Ludwig Institute, Uppsala, Sweden). The antibodies had been coupled to Protein G Dynabeads (Invitrogen). The beads were washed with ChIP wash buffer and TE buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA). Immunoprecipitated complexes were eluted and reverse cross-linked overnight at 65 °C. DNA was purified and analyzed by real-time PCR with the conditions: 95 °C for 5 min, followed by 39 cycles of 95 °C for 15 s and 60 °C for 1 min. Each independent experiment was repeated at least twice. The specific primers used for the PCR are listed in supplemental Table S3.
RESULTS
Mammary Epithelial Cells Overexpressing HMGA2 Strongly Up-regulate Twist
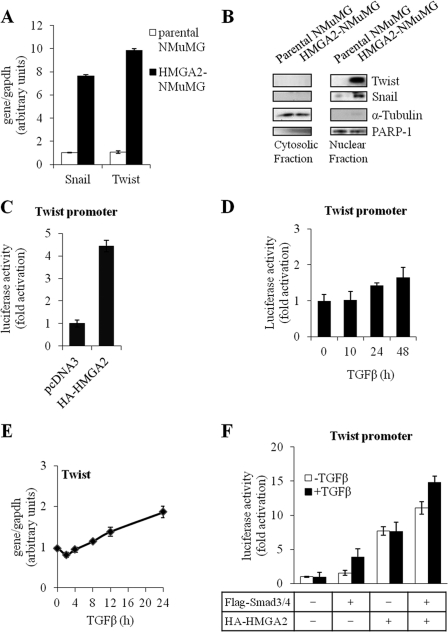
We previously reported that when normal mammary epithelial cells were stably transfected with HMGA2, several transcriptional regulators that promote EMT were up-regulated, and the cells shifted to a mesenchymal phenotype that lacked key features of the parental epithelial phenotype, such as expression of E-cadherin (14, 15). The transcription factors Snail and Twist were highly up-regulated in HMGA2-transfected cells (14). Furthermore, silencing of Snail by short interfering RNA only partially reverted the cells to the epithelial state (14). We therefore hypothesized that Twist, which remains highly expressed in the cells after Snail silencing, could compensate and be responsible for some of the mesenchymal features of these cells. Quantitative RT-PCR analysis of the mRNA from the same two cell types confirmed that cells expressing HMGA2 had dramatically higher levels of Twist mRNA compared with parental NMuMG cells, which paralleled the Snail expression profile (Fig. 1A). Analysis of expression of the corresponding proteins revealed that HMGA2-NMuMG cells expressed high levels of both transcription factors in their nuclear compartment, whereas the parental epithelial NMuMG cells did not express Twist or expressed only minute amounts of Snail protein (Fig. 1B).
FIGURE 1.
HMGA2 induces Twist expression during EMT. A, quantitative RT-PCR analysis of Snail and Twist mRNA levels in parental NMuMG cells and a cell clone of NMuMG expressing constitutively human HMGA2 (HMGA2-NMuMG). Each bar represents mean ± S.D. values from triplicate samples. B, immunoblots of Snail and Twist protein levels in cytosolic and nuclear fractions of cells described in A. PARP-1 and α-tubulin serve as markers for the nuclear and cytosolic fraction respectively. C, D, and F, luciferase reporter assay of Twist promoter constructs in HepG2 cells transiently transfected with pcDNA3 empty vector or HA-HMGA2 (C); stimulated with 5 ng/ml TGFβ1 for the indicated time period (D); transiently transfected with FLAG-Smad3, FLAG-Smad4, and/or HA-HMGA2 plasmids and stimulated or not with 5 ng/ml TGFβ1 (F). Each bar represents mean ± S.D. values of normalized luciferase data from triplicate samples. E, quantitative RT-PCR analysis of Twist mRNA levels in parental NMuMG cells, untreated or treated with 5 ng/ml TGFβ for 2, 4, 8, 12, and 24 h. Each bar represents mean ± S.D. values from triplicate samples.
Induction of endogenous Twist mRNA and protein by HMGA2 was corroborated by promoter activity studies. Using a recombinant construct of the mouse Twist promoter coupled to the luciferase reporter (17) and transient transfection of the promoter construct in the absence or presence of HMGA2, we could demonstrate that HMGA2 induced robust activation of the Twist promoter in the mouse epithelial NMuMG cells (supplemental Fig. S1A) and in human hepatocarcinoma HepG2 cells (Fig. 1C). This suggests that HMGA2 may act directly in regulating Twist gene expression.
Evaluation of TGFβ and Other Signaling Pathways as Regulators of Twist Expression Acting Downstream of HMGA2
We previously established that HMGA2 binds to Smad proteins after TGFβ stimulation and cooperates with them to induce transcription of the Snail promoter (14). We therefore examined if a similar mechanism operated during Twist promoter regulation. Endogenous TGFβ signaling was not sufficient to activate the Twist promoter with early kinetics (Fig. 1, D and E). When a more prolonged time course of TGFβ stimulation was performed, low level Twist promoter activation could be measured 24 and 48 h post-stimulation (Fig. 1D). This is consistent with the weak induction of Twist mRNA and protein by TGFβ alone (14, 15), which only increased after prolonged (12–24 h) stimulation (Fig. 1E). Co-transfection of Smad3 and Smad4, the two major Smads that form complexes in response to TGFβ and are primarily responsible for the EMT phenotype, led to a weak induction of Twist promoter activity, which was enhanced by TGFβ stimulation (Fig. 1F). In contrast to a relatively weak effect shown by Smad3/4 and TGFβ, HMGA2 co-transfection showed a much stronger promoter activation that was not further enhanced by TGFβ (Fig. 1F). The combination of Smad3/4 and HMGA2 exhibited an additive effect both in the absence and presence of TGFβ stimulation (Fig. 1F). These data suggest that, under conditions where the TGFβ pathway is hyperactivated after Smad protein overexpression, weak induction of the Twist promoter can be achieved and this can be further enhanced by co-expressing HMGA2. These studies therefore support the notion that Twist may not be a direct gene target of TGFβ/Smad signaling, whereas HMGA2 can potently induce Twist expression.
It is formally possible that epithelial cells expressing high levels of HMGA2 may express various signaling factors that could then lead to the activation of diverse pathways. We therefore carried out further studies with the aim to test the importance of other signaling mechanisms that could contribute to the induction of Twist expression in cells with high HMGA2 levels. In scrutinizing the TGFβ pathway once again, we observed that silencing by 50% the endogenous Smad4, the central signal transducer of all TGFβ family pathways, had no affect whatsoever on Twist mRNA expression, whereas it suppressed endogenous Snail expression (supplemental Fig. 2A). Likewise, treatment of the cells with SB505124, a low molecular weight inhibitor of the TGFβ family type I receptor serine/threonine kinases, reduced Snail expression by 60%, whereas Twist expression was slightly enhanced in the same cell population (supplemental Fig. S2B). The MAPK pathway inhibitor PD184352, which specifically blocks the activity of the kinase MEK1, showed a small but statistically significant reduction in Twist mRNA levels when tested at two different time points after its addition (supplemental Fig. S2C), suggesting that ERK MAPK signaling partially contributed to the effect of HMGA2 expression on Twist gene induction. The efficacy of the PD184352 inhibitor was verified at both time points using immunoblotting against phosphorylated forms of ERK1 and ERK2 MAPKs, i.e. the MEK1 substrates (supplemental Fig. S2C, right panel). Inhibition of p38 MAPK using the inhibitor SB202190 had no statistically significant effect on Twist mRNA levels, whereas it effectively blocked phosphorylated CREB levels used as controls (supplemental Fig. S2D). Finally, inhibition of the phosphoinositide 3′-kinase (PI3K) using the LY294002 inhibitor, had no impact on Twist mRNA expression levels, but completely blocked the phosphorylation of its downstream kinase target p70S6K (supplemental Fig. S2E).
All the above data suggest that HMGA2 minimally activates other signal transduction pathways that contribute to Twist gene regulation. It is therefore likely that HMGA2 regulates Twist gene expression by acting on its promoter, possibly via direct binding to its DNA sequences.
HMGA2 Regulates Twist Promoter Expression by Direct Binding to Its DNA
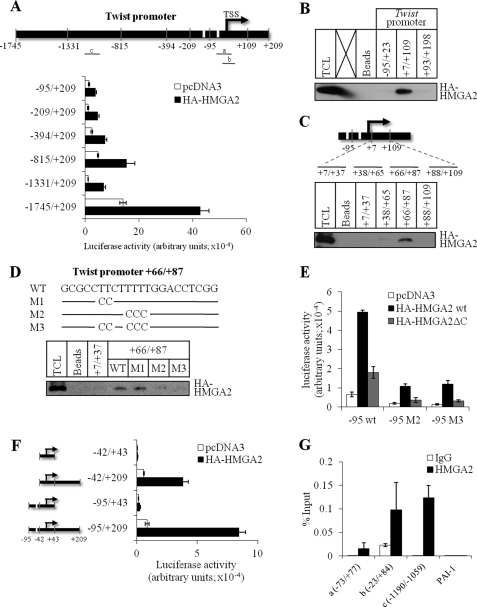
We first mapped putative regions of the mouse Twist promoter that are responsible for regulation by HMGA2, using a panel of deletion mutants of the promoter (Fig. 2A and supplemental Fig. S1A). All promoter constructs tested, spanning from −1745 to −95 bp relative to the transcription start site (TSS) of the mouse Twist gene, exhibited significant activation by co-transfected HMGA2 (Fig. 2A and supplemental Fig. S1A). This result suggested that HMGA2 may bind at multiple sites along the Twist promoter; however, an important and necessary site may lie very proximal to the TSS of the Twist gene. To experimentally identify binding of HMGA2 to the mouse Twist promoter, we synthesized three oligonucleotides spanning the region between −95 and +198 bp relative to the TSS and performed DNA affinity precipitation (DNAP) assays (Fig. 2B). HMGA2 bound with high specific affinity to oligonucleotide +7/+109 relative to the TSS, whereas it failed to bind the upstream −95/+23 or the downstream +93/+198 sequences (Fig. 2B). Further fine mapping of this region of the mouse Twist gene using shorter oligonucleotides in the DNAP assay, revealed binding of HMGA2 to the +66/+87 bp segment relative to the TSS (Fig. 2C). This 22-bp DNA segment spans a stretch of A:T base pairs (Fig. 2D). Mutagenesis of the T-rich stretch to C (mutants M1–M3) in the +66/+87 bp oligonucleotide was employed to test whether HMGA2 binding was affected. Although mutant M1 had no effect, mutants M2 and M3 almost completely abolished binding of HMGA2 to the +66/+87 bp oligonucleotides, suggesting that the last three nucleotides in the TTCTTTT motif are important for binding (Fig. 2D).
FIGURE 2.
HMGA2 binds directly to the Twist promoter. A, schematic diagram of the murine Twist promoter (top panel). Nucleotide numbers were assigned relative to the TSS, and two putative TATA boxes are denoted by white bars. Fragments a, b, and c indicate PCR amplicons analyzed in panel G with ChIP. Luciferase reporter assays of a deletion series of Twist promoter constructs in HepG2 cells transiently transfected with pcDNA3 or HA-HMGA2 plasmid (bottom panel). B and C, biotin-labeled probes spanning −95 to +23, +7 to +109, and +93 to +198 (B), and probes spanning +7 to +37, +38 to +65, +66 to +87, and +88 to +109 (C), were used in DNAP experiments using extracts of HepG2 cells transiently transfected with HA-HMGA2 plasmid. D, nucleotide sequences of wild-type (WT) and mutant (M1–3) probes corresponding to the +66 to +87 region of the Twist promoter. A line indicates unaltered sequences. Binding of HMGA2 to WT or mutant probes was assessed by DNAP experiments using extracts of HepG2 cells as described in B. Probe +7/+37 is not bound by HMGA2 and serves as an additional negative control. TCL, total cell lysates in B–D. E, luciferase reporter assays of wild type or mutants (M2 and M3) −95/+209 Twist promoter constructs in HepG2 cells transiently transfected with pcDNA3, HA-HMGA2 wt, or HA-HMGA2ΔC plasmids. F, luciferase reporter assays of a deletion series of Twist promoter constructs based on the −95/+209 Twist promoter in HepG2 cells transiently transfected with pcDNA3 or HA-HMGA2 plasmid. Each bar represents mean ± S.D. values of normalized luciferase data from triplicate samples. G, ChIP assays in HMGA2-NMuMG cells were used to analyze binding of HMGA2 protein along the Twist promoter using anti-HMGA2 or control IgG antibodies. Quantitative PCR was performed, and values are expressed as percentage of input DNA. Each bar represents mean ± S.D. values from triplicate samples. The PCR amplicons, −73/+77, −23/+84, and −1190/−1059, used are denoted as a, b, and c in panel A.
We then introduced these two mutations, M2 and M3, in the context of the −95 to +209 mouse Twist promoter and tested for transcriptional activation by HMGA2 in HepG2 and NMuMG cells (Fig. 2E and supplemental Fig. S1B). The basal levels of promoter activity of the M2 and M3 mutants were significantly reduced compared with the wild-type promoter (Fig. 2E and supplemental Fig. S1B). HMGA2 could further enhance the promoter activity of the M2 and M3 mutants, however, the levels of activity never reached the high levels of wild-type promoter activity induced by HMGA2 (Fig. 2E and supplemental Fig. S1B). The ability of HMGA2 to induce activity by promoters carrying the M2 and M3 mutations that abolish HMGA2 binding to the promoter suggested that the short C-terminal regulatory domain of HMGA2, which becomes phosphorylated and controls HMGA2 function (20, 21), may contribute to the transcriptional activation of the Twist promoter. We therefore tested Twist promoter activation by using a mutant HMGA2 construct lacking its C-terminal acidic domain (ΔC); this mutant activated partially the wild-type short Twist promoter, however, it was completely unable to mediate activation of the two mutant Twist promoters, M2 and M3 (Fig. 2E and supplemental Fig. S1B). These data demonstrate that, in the absence of the regulatory C-terminal domain of HMGA2, the binding of HMGA2 to the TTCTTTT motif of the Twist promoter is absolutely critical for regulation of the promoter. However, in the wild-type context of HMGA2, the TTCTTT motif is not absolutely critical and HMGA2 may bind to additional sequences between the −95 and +198 positions relative to the TSS of the mouse Twist gene. However, this sequence lacks any obvious A:T-rich motif excluding the putative TATA-box (see the schematic illustration of the promoter in Fig. 2A).
We investigated the possibility that HMGA2 could function via additional promoter sequences using a small panel of deletion mutants and HMGA2 co-transfection in the same promoter activation assays (Fig. 2F and supplemental Fig. S1C). Deletion of the +44 to +209 segment (−95/+43) of the promoter that also spans the TTCTTTT motif essentially neutralized promoter activation by HMGA2, despite the presence of the upstream TATA-box (Fig. 2F and supplemental Fig. S1C). In agreement with a minor role of the TATA-box as a sequence that mediates activation of the promoter by HMGA2, complete deletion of the TATA-box in the −42 to +209 promoter construct (−42/+209) showed decreased (by 2-fold) but still strong activation by HMGA2 (Fig. 2F and supplemental Fig. S1C). Finally, a very small promoter fragment spanning the TSS and devoid of TATA-box or HMGA2-binding motif (−42/+43) was completely inactive and resulted in background promoter activity (Fig. 2F and supplemental Fig. S1C).
Finally, we confirmed the in vitro DNA binding assays of Fig. 2B using ChIP assays on the endogenous Twist promoter chromatin in HMGA2-NMuMG cells (Fig. 2G). HMGA2 showed robust association (over that measured by a nonspecific antibody) with the Twist promoter chromatin as represented by three specific genomic fragments (fragments a–c). In addition, binding of HMGA2 with the Twist promoter chromatin was specific, because HMGA2 failed to associate to an unrelated gene promoter with relatively high A:T sequence content, the plasminogen activator inhibitor 1 (Fig. 2G). Fragments a and b on the Twist promoter overlap with the proximal TTCTTTT motif (Fig. 2A and supplemental Table S3), which corroborates the importance of the in vitro DNAP assays at the endogenous gene level. Interestingly, fragment c maps further upstream at positions −1190 to −1059 relative to the TSS of the mouse Twist gene (Fig. 2A and supplemental Table S3). This fragment was selected due to its very long stretch of A-rich sequence, and as shown in Fig. 2G, it scored highly positive, suggesting that HMGA2 can associate with this sequence as well. However, deletion of this sequence in the context of the promoter construct −815/−209 allowed HMGA2 to activate promoter activity (Fig. 2A and supplemental Fig. S1A), suggesting that the upstream fragment c is not by itself the only critical regulatory element of the Twist promoter that responds to HMGA2. These data therefore support the model in which HMGA2 activates Twist promoter activity by directly binding to DNA sequences of this gene and highlight an importance for the C-terminal regulatory domain of HMGA2 in mediating promoter activation.
Contribution of Twist to the Mesenchymal Phenotype Induced by HMGA2
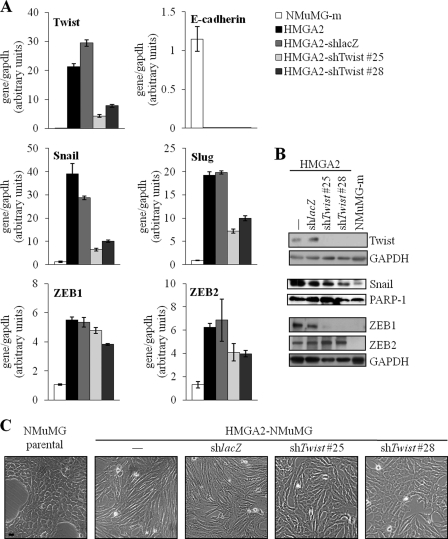
To address functionally the role of Twist downstream of HMGA2, we silenced endogenous Twist mRNA in the mesenchymal HMGA2-NMuMG cells using shRNA vectors (Fig. 3A). QRT-PCR analysis confirmed up to 6-fold knockdown in several clones.4 We selected two individual clones (#25 and #28) and a single control clone expressing an shRNA targeting the bacterial LacZ mRNA (HMGA2-shlacZ) for further analysis (Fig. 3A).
FIGURE 3.
Twist depletion in HMGA2-NMuMG cells causes a mild re-epithelialization. A, quantitative RT-PCR analysis of Twist, Snail, Slug, ZEB1, ZEB2, and E-cadherin mRNA levels; and B, immunoblot of Twist, Snail, ZEB1, and ZEB2 proteins in NMuMG-m, HMGA2-NMuMG cells (HMGA2, −), and its derivatives constitutively expressing shRNA against Twist (HMGA2-shTwist #25 and #28) or control lacZ (HMGA2-shlacZ). NMuMG-m is a sub-clone of parental NMuMG, which exhibits a highly polarized epithelial morphology, and serves as a mock NMuMG-transfected clone for HMGA2-NMuMG cells (14, 15). Each bar represents mean ± S.D. values from triplicate samples. C, cellular morphology of parental NMuMG cells, HMGA2-NMuMG cells, and its derivatives described in A. Bar, 10 μm.
Molecular analysis of the EMT transcription factors at the mRNA level confirmed that silencing of endogenous Twist led to a concomitant down-regulation of Snail and Slug (Fig. 3A). In contrast, in the same cells, the level of ZEB1 was weakly down-regulated, and the level of ZEB2 mRNA was reduced by 25% (Fig. 3A). Analysis of the corresponding protein levels (Fig. 3B) revealed that the strongest effect of Twist silencing was on ZEB1 expression, Snail was down-regulated but to intermediate levels, and ZEB2 was not affected at all. During these analyses we failed to identify a Slug antibody that provided reproducible and reliable protein data (not shown). We therefore conclude that Twist significantly contributes to the sustained expression of Snail and ZEB1 and possibly to the expression of Slug. Conversely, Snail can also weakly contribute to the sustained induction of Twist by HMGA2.
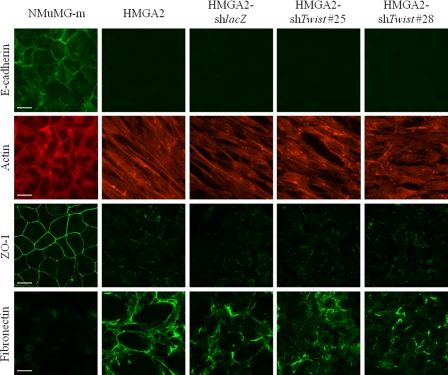
Morphological analysis of these stably transfected clones demonstrated partial reversion of the mesenchymal phenotype to an epithelial one, because the cells with reduced Twist levels maintained features of the spindle-like morphology and regained only partial cell-cell adhesion (Fig. 3C). To further confirm the microscopic observations, we performed immunofluorescence analysis of the same stable cell clones and compared the pattern of adhesion and extracellular matrix markers to the pattern of NMuMG-m cells or HMGA2-NMuMG cells (Fig. 4). HMGA2-NMuMG and HMGA2-shlacZ control cells exhibited the characteristic mesenchymal phenotype with complete loss of E-cadherin (adherens junction marker) and ZO-1 (tight junction marker), enhanced fibronectin (extracellular matrix marker) deposition, and characteristic actin stress fiber organization (Fig. 4). Interestingly, the two clones that exhibited significantly reduced levels of Twist expression (Fig. 3A) showed no evidence of E-cadherin-positive adherens junctions, whereas some ZO-1-positive tight junctions could be observed in one of the clones (shTwist #28) (Fig. 4). The same cell clones exhibited an actin cytoskeleton that resembled more that of mesenchymal cells with strong stress fibers (Fig. 4). However, clones shTwist #25 and #28 also showed scattered cells with cortical actin assembly (Fig. 4). In addition, in one of the clones (shTwist #28), the marker that showed the strongest response to Twist silencing was fibronectin, whose extracellular deposition was significantly reduced (Fig. 4). The immunofluorescence data were confirmed by immunoblotting of total cell lysates from the same cell clones (supplemental Fig. S3A). Although no detectable levels of E-cadherin or ZO-1 could be measured after Twist silencing, there was a reduction in expression of mesenchymal protein markers: N-cadherin, in the case of clone shTwist #25; and fibronectin and α-smooth muscle actin, in the case of clone shTwist #28. As expected, Twist silencing had no impact on the robust expression of the HMGA2 protein that is overexpressed in these clones (supplemental Fig. S3A). The marker analysis at the mRNA, protein, and microscopic level allows us to conclude that silencing of endogenous Twist expression in NMuMG cells that overexpress HMGA2 was insufficient to drive the reversion of the cell phenotype toward epithelial differentiation.
FIGURE 4.
Protein marker analysis in HMGA2-NMuMG cells after Twist depletion. Immunostaining for E-cadherin, ZO-1, and fibronectin and phalloidin staining (actin) in HMGA2-shTwist clones, as described in Fig. 3A. Bar, 20 μm.
Twist and Snail Together Contribute to the Mesenchymal Phenotype Induced by HMGA2
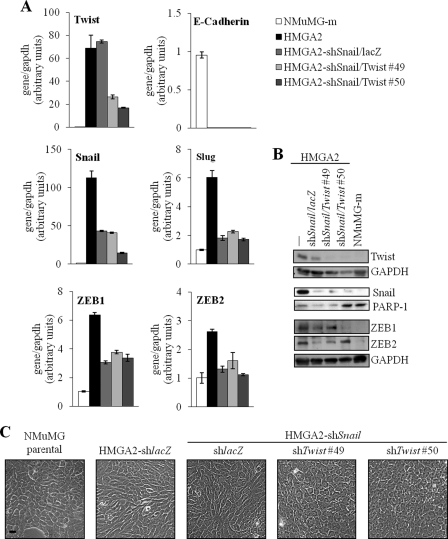
Because silencing Twist (Figs. 3 and 4) or Snail (14) alone partially reverted HMGA2-NMuMG cells to epithelial cells, we decided to combine silencing of Snail and Twist in the same mesenchymal cells that overexpress HMGA2 (Fig. 5). This was achieved by stable transfection of the previously established HMGA2-shSnail clones (Fig. 5) (14), with the same vectors expressing shRNAs targeting Twist as those used for single Twist silencing in the experiments of Figs. 3 and 4. Stable transfection with a nonspecific (shlacZ) vector confirmed continuous robust Twist mRNA and only small reduction in protein expression in the selected clones (Fig. 5, A and B). In contrast, several stable clones exhibiting significant (50–70%) silencing of endogenous Twist were obtained after transfection of the shTwist vector, and two of them (clones #49 and #50) were selected for detailed analysis (Fig. 5, A and B). Molecular analysis of additional transcriptional regulators of the EMT program at the mRNA and protein levels showed an interconnected pattern of regulation (Fig. 5, A and B). At the mRNA level, Slug, ZEB1, and ZEB2 expression were significantly reduced upon silencing of both Snail and Twist, as was the case in cells where only Snail was silenced (Fig. 5A). At the protein level, as explained above, we failed to analyze Slug levels. However, ZEB1 and ZEB2 showed reciprocal patterns (Fig. 5B). When both Snail and Twist were significantly reduced, then some cells exhibited strong ZEB2 down-regulation with relatively high, albeit reduced ZEB1 levels (clone #49), and some cells showed strong ZEB1 down-regulation with relatively intact and high levels of ZEB2 (clone #50). Overall these data confirm the intimate interconnection between the various EMT transcription factors and suggest that silencing of both Snail and Twist leads to a more profound down-regulation of the levels of all five EMT transcription factors (at mRNA and protein levels combined), which was anticipated to cause a more efficient epithelial reversion of the cell phenotype.
FIGURE 5.
Twist and Snail depletion in HMGA2-NMuMG cells causes stronger epithelial reversion. A, quantitative RT-PCR analysis of Twist, Snail, Slug, ZEB1, ZEB2, and E-cadherin mRNA levels; and B, immunoblots of Twist, Snail, ZEB1, and ZEB2 proteins in NMuMG-m and HMGA2-NMuMG cells (HMGA2, −), and cell clones of HMGA2-shSnail constitutively expressing shRNA against Twist (HMGA2-shSnail/Twist #49 and #50) or its control HMGA2-shSnail/lacZ. Each bar represents mean ± S.D. values from triplicate samples. C, cellular morphology of parental NMuMG, HMGA2-shlacZ, and HMGA2-shSnail/Twist clones described in A. Bar, 10 μm.
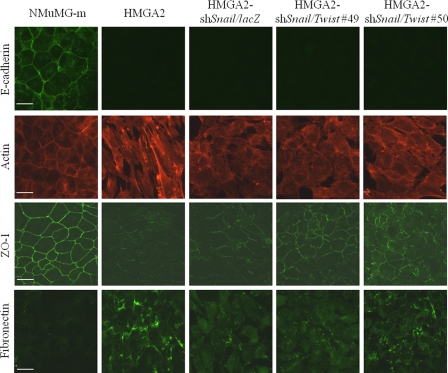
Consistent with this notion, microscopic analysis of the clones that exhibit knockdown of both Snail and Twist revealed rather homogeneous epithelial morphology (Fig. 5C), which was significantly more differentiated than the phenotype of cells with single Snail or single Twist knockdown (Figs. 3C and 5C). To further extend these observations, we studied various protein markers using immunofluorescence microscopy (Fig. 6). The epithelial tight junction marker ZO-1 provided clear evidence that clones exhibiting both low Snail and Twist expression had the best organized tight junctions, with more robust and contiguous assembly of the ZO-1-positive junctional complexes, whereas the single Snail knockdown exhibited interrupted, punctate-like tight junction assemblies (Fig. 6, ZO-1 immunofluorescence). The ZO-1 profile was recapitulated by the actin cytoskeleton pattern, as clones with both Snail and Twist silenced showed the strongest cortical arrangement of their cytoskeleton that resembled to a large extent the cytoskeleton of mock-transfected NMuMG cells (Fig. 6, actin fluorescence). Fibronectin levels were also suppressed after double Snail and Twist knockdown, supporting the epithelial reversion of these stable cell clones (Fig. 6, fibronectin immunofluorescence). The immunofluorescence data were confirmed by immunoblotting of total cell lysates from the same cell clones (supplemental Fig. S3B). Although silencing of Snail and Twist had no impact on HMGA2 transgene expression, strong ZO-1 protein levels were evident, accompanied by much weaker fibronectin protein expression and even lower levels of the other two mesenchymal protein markers, N-cadherin and α-smooth muscle actin.
FIGURE 6.
Twist and Snail depletion in HMGA2-NMuMG cells causes tight junction reassembly. Immunostaining for E-cadherin, ZO-1, and fibronectin and phalloidin staining (actin) in HMGA2-shSnail/Twist clones, as described in Fig. 5A. Bar, 20 μm.
On the other hand, analysis for E-cadherin-positive adherens junctions clearly demonstrated that none of the double-silenced clones re-expressed this important epithelial marker (Fig. 6, E-cadherin immunofluorescence). This result was quantified using total cell lysate immunoblotting and mRNA analysis, both showing undetectable levels of E-cadherin in clones with both Snail and Twist silenced (Fig. 5A and supplemental Fig. S3B). The latter suggests that, although mesenchymal HMGA2-expressing cells revert into epithelial cells, which build rather robust tight junction assemblies upon silencing of both Snail and Twist (Fig. 6), these cells are not bona fide epithelial cells because they failed to assemble adherens junctions due to the dominant suppression of E-cadherin by HMGA2 (Figs. 5A and 6 and supplemental Fig. S3B). We therefore conclude that, although Snail and Twist mediate a fair number of changes in the EMT caused by HMGA2 overexpression, these transcriptional repressors obviously are not solely responsible for the suppression of E-cadherin expression.
DISCUSSION
In this investigation, we provide four mechanistic messages that are of importance for the process of EMT: (a) Twist and Snail play partially redundant roles during EMT of mammary epithelial cells; (b) Twist contributes, although weakly, to Snail expression and, conversely, Snail contributes to Twist expression in the cascade of transcriptional events that lead to EMT downstream of the TGFβ/HMGA2 signaling module; (c) HMGA2 suppresses expression of E-cadherin by yet unknown mechanisms that do not seem to incorporate the action of Snail and Twist; and (d) HMGA2 binds directly to promoter sequences of the Twist gene and activates this promoter without activating downstream TGFβ/Smad or MAPK, PI3K signaling.
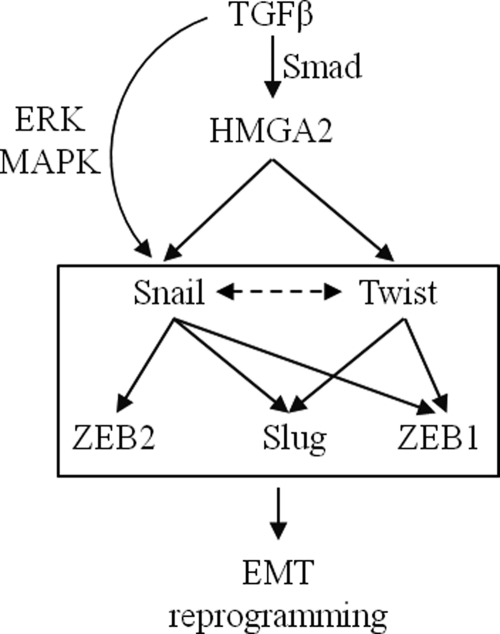
Based on the model proposed by our previous investigations (14, 15), TGFβ induces HMGA2 expression, which then organizes transcriptional induction of two major EMT regulators, Snail and Twist (Fig. 7). These two transcription factors might act in a complementary manner that ensures the robust and stable transition of epithelial cells to the mesenchymal phenotype. The evidence provided by silencing experiments of Twist alone or of both Snail and Twist in mesenchymal cells that overexpress HMGA2 (Figs. 3–6) provides a more complicated but probably more realistic model of regulation. Twist weakly but reproducibly contributes to Snail expression, because silencing of Twist perturbs the expression of Snail and silencing of Snail weakly affects the expression of Twist (Figs. 3A, 5A, and 7, dotted arrow). Furthermore, ZEB2 seems to take input mainly from Snail, whereas ZEB1 and Slug seem to take inputs from both Snail and Twist (Figs. 3A, 5A, and 7).
FIGURE 7.
Diagram of the molecular pathway that leads to EMT downstream of TGFβ and HMGA2, as analyzed in this study. TGFβ, via Smads induces expression of HMGA2. HMGA2 then binds to the promoters of Snail and Twist and induces their expression. The Snail gene also receives additional inputs from TGFβ, such as ERK MAPK signaling. The five pro-EMT transcription factors (boxed) positively cross-regulate each other to elicit nuclear reprogramming that leads to EMT. Snail and Twist cross-regulate expression of each other albeit weakly (dotted arrow). Snail regulates expression of Slug, ZEB1, and ZEB2, whereas Twist regulates expression of Slug and ZEB1. The possible cross-regulation between ZEB1, ZEB2, and Slug was not examined in this study.
We previously demonstrated a direct interaction of HMGA2 with Smads and with the Snail promoter (14). In mammary epithelial cells responding to TGFβ and undergoing EMT, Snail is an immediate-early gene, whereas Twist is a late gene (Fig. 1, D and E) (15). It is possible that long term silencing of endogenous genes by stable shRNA constructs bypasses the dynamic regulation of gene expression that operates in the EMT program. In other words, HMGA2 rapidly induces Snail expression in the absence of any contribution by Twist; later on, Twist expression is induced. However, once Twist is induced, it contributes to the maintenance of Snail levels. Thus, when Twist is silenced stably, it also perturbs the long term induction of Snail downstream of HMGA2 (Fig. 3, A and B). This mechanism requires further investigation to directly demonstrate that Twist protein binds to regulatory sequences of the Snail gene and directly induces expression of Snail. Overall, this model of a tight functional interconnection between Snail and Twist downstream of HMGA2 is in good agreement with a recent report on the cooperation of Snail and Twist in inducing expression of ZEB1 in NMuMG cells after stimulation with TGFβ (22), which is further validated by the data herein (Figs. 3B, 5B, and 7).
It is also evident from the analysis of epithelial markers after silencing of Twist (Figs. 4 and 6), that when epithelial cells turn into mesenchymal, under the influence of high HMGA2 expression, Snail and Twist clearly contribute to this transition, and a hallmark of their action is the lack of tight junctions. For this reason, silencing of Snail and Twist restores tight junction assembly despite the presence of high levels of HMGA2 in the cells (Fig. 6). However, under the same conditions, the adherens junctions (Fig. 6) and the total E-cadherin levels (Figs. 3A and 5A) do not revert. This observation opens the exciting possibility that HMGA2 may directly suppress expression of E-cadherin. If this stands true, it will represent a new aspect of the multiple functions HMGA2 mediates, because this nuclear protein often associates with the assembly of enhanceosomes and the positive regulation of transcription (16, 23). The preliminary evidence we provide about regulation of E-cadherin expression by HMGA2 may shed new light about how HMGA2 may also repress transcription of target genes. Alternatively, HMGA2 may act on E-cadherin indirectly via another transcriptional repressor, whose levels are induced by high HMGA2 expression. Experiments are underway to investigate this possibility.
The mechanism by which HMGA2 induces expression of Twist seems to involve direct association with A:T base pairs in the proximal promoter of Twist, immediately downstream of the TSS of this gene (Fig. 2 and supplemental Fig. S1). The motif identified here is in agreement with the motif identified in the Snail promoter (14), and more generally, with the affinity that HMGA2 shows for the minor groove of DNA, usually associated with A:T-rich sequences (16). The present ChIP analysis also implicates additional upstream sites on the Twist promoter where HMGA2 associates (Fig. 2G). However, the long A-stretch of this upstream sequence does not conform to the same sequence motif as that identified in the proximal Twist promoter or other genes, and deletion experiments indicate that this sequence may not be as critical as the proximal site for promoter activation in transfected cells (Fig. 2 and supplemental Fig. S1). Further analysis is needed to understand better the contribution of HMGA2 on the regulation of the extended Twist gene locus and the modifications of its chromatin.
We also found that Smads together with HMGA2 can regulate the Twist promoter (Fig. 1F). However, the magnitude of effects we measured is drastically smaller from the regulation of the Snail promoter by HMGA2 and Smads (14). This is in agreement with the evidence that, although HMGA2 appears to induce robust Twist mRNA and protein levels (Fig. 1, A and B), which correlates with the robust activation of the Twist promoter (Fig. 1C), TGFβ induces Twist mRNA (Fig. 1D) and Twist promoter activity (Fig. 1E) weakly and only after prolonged stimulation. Thus, protein complexes of Smads with HMGA2 may not play as important a role in transcriptional induction of Twist compared with their role in induction of Snail. Within a TGFβ-enriched environment, the activated Smad complex can obviously also participate in the up-regulation of Twist; however, it cannot play the primary role. This suggests that the genomic organization of the Twist gene may require the productive cooperation between HMGA2 and another transcriptional partner to facilitate robust transcriptional induction of Twist. Such a cofactor remains to be identified.
In summary, the current study provides deep and more comprehensive analysis of the relative roles Snail and Twist play, under the instructions of the nuclear factor HMGA2, in promoting the mesenchymal transition.
Supplementary Material
Acknowledgments
We thank Johan Ericsson (University College Dublin Conway Institute, Dublin, Ireland), Louise R. Howe (Cornell University, New York, NY), Johan Lennartsson and Eleftheria Vasilaki (Ludwig Institute for Cancer Research, Uppsala Branch, Sweden), and Robert A. Weinberg (Whitehead Institute for Biomedical Research, Cambridge, MA) for reagents and advice during the course of this work and members of our laboratory for suggestions and useful discussions.
This work was supported in part by the Ludwig Institute for Cancer Research and the Swedish Cancer Society (project numbers: 4855-B03-01XAC and CAN 2006/1078 to A. M.).

This article contains supplemental Figs. S1–S3 and Tables S1–S3.
E.-J. Tan, S. Thuault, L. Caja, T. Carletti, C.-H. Heldin, and A. Moustakas, unpublished data.
- EMT
- epithelial-to-mesenchymal transition
- DNAP
- DNA affinity precipitation
- HMG
- high mobility group
- TSS
- transcription start site
- ZEB1/2
- Zinc finger E-box binding homeobox 1/2
- NMuMG
- Namru murine mammary gland
- TGFβ
- transforming growth factor β.
REFERENCES
- 1. Nguyen D. X., Bos P. D., Massagué J. (2009) Metastasis. From dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284 [DOI] [PubMed] [Google Scholar]
- 2. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 3. Berx G., van Roy F. (2009) Involvement of members of the cadherin superfamily in cancer. Cold Spring Harbor Perspect. Biol. 1, a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peinado H., Olmeda D., Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 5. Eckert M. A., Lwin T. M., Chang A. T., Kim J., Danis E., Ohno-Machado L., Yang J. (2011) Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell 19, 372–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mani S. A., Yang J., Brooks M., Schwaninger G., Zhou A., Miura N., Kutok J. L., Hartwell K., Richardson A. L., Weinberg R. A. (2007) Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc. Natl. Acad. Sci. U.S.A. 104, 10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moustakas A., Heldin C. H. (2007) Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 98, 1512–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akhurst R. J., Derynck R. (2001) TGF-β signaling in cancer. A double-edged sword. Trends Cell Biol. 11, S44–51 [DOI] [PubMed] [Google Scholar]
- 9. Pardali K., Moustakas A. (2007) Actions of TGF-β as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta 1775, 21–62 [DOI] [PubMed] [Google Scholar]
- 10. Moustakas A., Heldin C. H. (2009) The regulation of TGFβ signal transduction. Development 136, 3699–3714 [DOI] [PubMed] [Google Scholar]
- 11. Kang J. S., Liu C., Derynck R. (2009) New regulatory mechanisms of TGF-β receptor function. Trends Cell Biol. 19, 385–394 [DOI] [PubMed] [Google Scholar]
- 12. Heldin C. H., Landström M., Moustakas A. (2009) Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 21, 166–176 [DOI] [PubMed] [Google Scholar]
- 13. Bierie B., Moses H. L. (2010) Transforming growth factor β (TGF-β) and inflammation in cancer. Cytokine Growth Factor Rev. 21, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thuault S., Tan E. J., Peinado H., Cano A., Heldin C. H., Moustakas A. (2008) HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J. Biol. Chem. 283, 33437–33446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thuault S., Valcourt U., Petersen M., Manfioletti G., Heldin C. H., Moustakas A. (2006) Transforming growth factor-β employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 174, 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fusco A., Fedele M. (2007) Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7, 899–910 [DOI] [PubMed] [Google Scholar]
- 17. Howe L. R., Watanabe O., Leonard J., Brown A. M. (2003) Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 63, 1906–1913 [PubMed] [Google Scholar]
- 18. Valcourt U., Kowanetz M., Niimi H., Heldin C. H., Moustakas A. (2005) TGF-β and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell 16, 1987–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koinuma D., Tsutsumi S., Kamimura N., Taniguchi H., Miyazawa K., Sunamura M., Imamura T., Miyazono K., Aburatani H. (2009) Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol. Cell. Biol. 29, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noro B., Licheri B., Sgarra R., Rustighi A., Tessari M. A., Chau K. Y., Ono S. J., Giancotti V., Manfioletti G. (2003) Molecular dissection of the architectural transcription factor HMGA2. Biochemistry 42, 4569–4577 [DOI] [PubMed] [Google Scholar]
- 21. Sgarra R., Maurizio E., Zammitti S., Lo Sardo A., Giancotti V., Manfioletti G. (2009) Macroscopic differences in HMGA oncoproteins post-translational modifications. C-terminal phosphorylation of HMGA2 affects its DNA binding properties. J. Proteome Res. 8, 2978–2989 [DOI] [PubMed] [Google Scholar]
- 22. Dave N., Guaita-Esteruelas S., Gutarra S., Frias À., Beltran M., Peiró S., de Herreros A. G. (2011) Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J. Biol. Chem. 286, 12024–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merika M., Thanos D. (2001) Enhanceosomes. Curr. Opin. Genet. Dev. 11, 205–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.