Background: TDP-43 is the principal protein component of cellular inclusion in ALS and FTLD.
Results: Tandem repetitions of TDP-43 residues 331–369 induce cellular aggregates that recruit endogenous TDP-43.
Conclusion: Our results establish a cell-based TDP-43 aggregation model.
Significance: This model will be useful to investigate TDP-43 aggregation and develop strategies/effectors able to prevent/reduce this phenomenon.
Keywords: Neuroscience, RNA, RNA Processing, RNA Splicing, RNA-binding Proteins, Gln/Asn Region, TDP-43, Aggregation Model, hnRNP
Abstract
TDP-43 is one of the major components of the neuronal and glial inclusions observed in several neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal lobar degeneration. These characteristic aggregates are a “landmark” of the disease, but their role in the pathogenesis is still obscure. In previous works, we have shown that the C-terminal Gln/Asn-rich region (residues 321–366) of TDP-43 is involved in the interaction of this protein with other members of the heterogeneous nuclear ribonucleoprotein protein family. Furthermore, we have shown that the interaction through this region is important for TDP-43 splicing inhibition of cystic fibrosis transmembrane regulator exon 9, and there were indications that it was involved in the aggregation process. Our experiments show that in cell lines and primary rat neuronal cultures, the introduction of tandem repeats carrying the 331–369-residue Gln/Asn region from TDP-43 can trigger the formation of phosphorylated and ubiquitinated aggregates that recapitulate many but not all the characteristics observed in patients. These results establish a much needed cell-based TDP-43 aggregation model useful to investigate the mechanisms involved in the formation of inclusions and the gain- and loss-of-function consequences of TDP-43 aggregation within cells. In addition, it will be a powerful tool to test novel therapeutic strategies/effectors aimed at preventing/reducing this phenomenon.
Introduction
In eukaryotic cells, heterogeneous nuclear ribonucleoproteins (hnRNPs)2 play important roles in most gene expression mechanisms such as DNA transcription, DNA replication/repair, pre-mRNA splicing and stability, mRNA export/retention processes, and protein translation (1–5). Up to now, the study of their involvement in human disease has been mostly confined to tumor formation/progression (6, 7). As recently reviewed, however, mislocalization and structural changes of two hnRNPs, TDP-43 and FUS (8–11), have been shown to occur in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). In these two and other related diseases, the identification of TDP-43 protein as a major component of the characteristic ubiquitinated neuronal inclusions (12, 13) has opened up several new avenues of research (reviewed in Refs. 10, 14–19). Most importantly, the discovery that hnRNP proteins may play a fundamental role in causing disease has strongly supported the emerging hypotheses that defects at the mRNA metabolism level may provide a major contribution to development and progression of neurodegenerative disease (20–26).
From a structural point of view, TDP-43 possesses the typical hnRNP architecture. The N-terminal sequence may contribute to the putative interaction of TDP-43 with mutant superoxide dismutase protein (27) and contains a strong nuclear localization signal that keeps TDP-43 predominantly nuclear (28). With regard to RNA binding, the RRM-1 sequence is principally responsible for the binding to the preferred nucleotide sequence represented by (UG)n-repeated sequences (29, 30), whereas RRM-2, which has been recently crystallized (31), has also been suggested to participate in the DNA/RNA binding properties of this protein. The RRM-2 motif has been shown to contain a nuclear export signal that also contributes to regulating its localization (28). Regarding this issue, however, it should be noted that in normal conditions the protein continuously shuttles between the nucleus and the cytoplasm and that the C-terminal sequence and RNA binding activity are also involved in keeping normal cellular distribution, including its intranuclear localization (32). The remaining portion of the molecule is composed of the C-terminal region that makes up almost half of the protein (residues 261–414). In TDP-43, this region contains a glycine-rich motif (33) and a Gln/Asn-rich region that was shown to disrupt hnRNP-A2/TDP-43 complexes (34), possibly causing aggregation. This region was later identified as a potential prion-like domain (35, 36). In general, the C-terminal regions of hnRNP proteins have been generally found to be responsible for mediating protein-protein interactions (37). In this regard, TDP-43 is well known to interact with numerous factors, especially other hnRNP proteins and additional factors that connect it with the general mRNA metabolism (38–40). Finally, in the brains of people affected by ALS/FTLD and additional diseases, now collectively termed as “TDP-43 proteinopathies” (13), the C-terminal region is particularly important because this is where all mutations associated with disease have been found (with just one exception) (41, 42) and its cleavage generates fragments (CTFs) that are associated with cellular toxicity and/or increased aggregate formation potential, as will be further discussed below (43–47). To date, the only minimal sequences required for TDP-43 aggregation have been reported based on peptide studies (48, 49). Another characteristic feature of this region in disease tissues is the specific phosphorylation of several residues particularly serines 409/410 (50), a consistent feature of all sporadic and familiar FTLD-U and ALS cases (51). Phosphorylation has been found to be essential for pathogenicity in a Caenorhabditis elegans animal model (52) and to regulate proteosomally mediated regulation of the CTFs by rendering them resistant to degradation and enhancing their aggregation properties (53). In this respect, understanding the biochemical properties of the C-terminal tail may open the way for the establishment of aggregation models that will be useful to better understand pathogenic processes and might provide a testing benchmark for novel therapeutic options. At the moment, in fact, the only compounds tested for their effect on TDP-43 aggregate formation are represented by methylene blue/dimebon and inorganic zinc (54, 55).
In this work, we have aimed to better elucidate the role of the C-terminal tail of TDP-43 by focusing our attention on the possible relationships between hnRNPs binding to the Gln/Asn region and the interaction of the Gln/Asn region with itself. This has lead to the generation of a TDP-43 aggregation model that recapitulates many but not all features of the aggregates observed in patients.
EXPERIMENTAL PROCEDURES
Plasmid Preparation
The GST-tagged TDP-43 and hnRNP-A2 proteins were generated as mentioned in Buratti et al. (56, 40), respectively.
The generation of GST and EGFP fusion proteins containing the regions from TDP-43(321–366), hnRNPA2(288–322), and hnRNPA2(288–341) was performed using the expression vectors pGEX-4T1 and pEGFP-C2, respectively. The PCR amplification of the TDP-43(321–366) fragment was carried out using the primers TDP-43 forward 321–366 (5′-tatatagaattcgccatggatggct-3′) and TDP-43 reverse 321–366 (5′-tatatagtcgactttaggcctggtttgg-3′) amplified from pGEX3X-TDP-43 (5). The hnRNPA2(288–322) and hnRNPA2(288–341) fragments were amplified using the primers A2 forward EcoRI (5′-tatatagaattctacaatgattttgg-3′), A2 reverse SalI 288–322 (5′-atatatgtcgactttatcctccaccatatg-3′), and A2 reverse SalI 288–341 (5′-atatatatgtcgactttagtatcggctcc-3′), respectively, and using the pGEX3X-hnRNP-A2 (40) as template. All PCR products were cloned in the respective vectors in the EcoRI and SalI restriction sites and checked by sequencing.
The EGFP-Gln/Asn fusion proteins were generated by cloning different numbers of concatemers (see below), codifying each one for the 331–369 residues of TDP-43, in SacI/BamHI restriction sites of the pEGFP-C2 vector. The EGFP-Gln/Asn recombinant proteins, containing a His6 tag epitope at the N terminus, were generated by digesting the different EGFP-Gln/Asn constructs with AgeI and BamHI restriction enzymes. The digested products were cloned in pQE-30 expression vector previously modified by site-directed mutagenesis to add an AgeI restriction site at the 5′ of multicloning sites (MCS). For the creation of TDP-43–12×Gln/Asn, the pFLAG-CMV4-TDP-43WT construct was modified to add an XhoI restriction site at position 1209 of TDP-43. The 12×Gln/Asn repetitions were digested from pEGFP-12×Gln/Asn construct with XhoI/BamHI and subcloned in the same sites of pFLAG-CMV4-TDP-43WT. The constructs pFLAG-TBPH and pFLAG-TBPHΔC(1–332) have been adapted from the construct originally prepared in Ref. 30. The V5-tagged FUS expression plasmid was a generous gift by Tom Ricketts (Harwell, UK), and the plasmid HA-ubiquitin from was Martin Monte (University of Buenos Aires, Buenos Aires, Argentina).
Tissue Culture and Cell Transfection
HeLa, Hep3B, and U2OS cells lines were grown in DMEM/Glutamax-I (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and antibiotic/antimycotic-stabilized suspension (Sigma). The plasmid transfections were carried out with Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. The HEK-293 cells stably transfected with FLAG-TDP-43 have been described elsewhere. These cells were grown in DMEM/Glutamax-I supplemented with 10% fetal bovine serum, antibiotic/antimycotic-stabilized suspension (Sigma), and 100 μg/ml hygromycin (Invivogen). When necessary, the induction of FLAG-TDP-43 was started by adding 1 mg/ml tetracycline at the culture medium. Cultures of trigeminal ganglion neurons were obtained from Wistar rats (7–10 days). Animals were anesthetized with carbonic anhydride and sacrificed by decapitation. The dissociation of trigeminal ganglion neurons was performed in 500 ml of F-12 medium (Invitrogen) containing 0.25 mg/ml trypsin, 1 μg/ml collagenase, and 0.2 mg/ml DNase at room temperature. The samples were centrifuged at 1000 × g for 5 min at RT; the supernatant was discarded, and cells were resuspended in F-12 medium plus 10% fetal bovine serum. Trigeminal ganglion neurons were incubated at 37 °C with 5% CO2 for 3 days in 6-well plates containing F-12 medium plus 10% fetal bovine serum and glasses pre-coated with collagen. Transfection was carried out with Effectene reagent according to manufacturer's instructions, and immunofluorescence (see below) was performed 24 h post-transfection.
Co-immunoprecipitation Assays
For co-immunoprecipitation assays, Hep3B cells (70% of confluence) were transfected with 3 μg of each plasmid. The cells were collected in PBS supplemented with protease inhibitors (Roche Applied Science) and lysed by sonication. The lysates were incubated with 3 μl of anti-GFP antibody (Santa Cruz Biotechnology) for 3 h at 4 °C, and then 30 μl of A/G Plus agarose beads (Santa Cruz Biotechnology) were added. After an overnight incubation at 4 °C, the beads were precipitated and washed three times with PBS. The immunoprecipitates were then analyzed by Western blot using an anti-FLAG antibody (F1804, Sigma).
Protein Expression and Supershift EMSAs
All GST fusion proteins were obtained by transforming Escherichia coli BL21-DE3 with the respective expression vectors, and recombinant protein expression and purification were performed according to the procedure already described by Buratti et al. (56). The His tag recombinant proteins were purified under native conditions according to Qiagen protein purification protocol. Briefly, 5 ml of LB ampicillin/kanamycin pre-cultures (E. coli M15) were grown overnight at 37 °C. The pre-cultures were used to inoculate 400 ml of LB ampicillin/kanamycin until an absorbance of 0.5–0.7. 1 mm isopropyl 1-thio-β-d-galactopyranoside was used to induce the protein expression for 3 h at 37 °C. The cells were centrifuged, and bacterial pellets were resuspended in 30 ml of lysis buffer supplemented with proteases inhibitors (Roche Applied Science mixture tablets). The bacterial solutions were sonicated at maximum power for 10 min on ice and with 10-s pulse intervals. After 40 min of centrifugation at 4000 × g, the supernatants were incubated in agitation at 4 °C with 1 ml of nickel-glutathione beads (Clontech). After three washes with washing buffer, the proteins were eluted at 4 °C with 500 μl of elution buffer in four fractions.
EMSAs were conducted according to procedures already described (29) with minor modifications. The binding buffer contained 20 mm HEPES, pH 7.5, 0.2 mm EDTA, 0.5 mm DTT, and 6% glycerol. The RNA oligonucleotide (UG)6 5′-uguguguguguga-3′ was made by Integrated DNA Technologies. The 5′ end labeling of the oligonucleotide was carried out with polynucleotide kinase according to standard protocols. In all experiments, 100–200 ng of GST-TDP-43 fusion protein and indicated amounts of competitor peptides were incubated for 5 min at room temperature. For supershift analysis, 0.5 μg of GST-hnRNP-A2 and 0.5 ng of labeled oligonucleotide were added, and the mixture was incubated at room temperature for 5 min. Native 5% polyacrylamide gels were run at 100–120 V at 4 °C using 0.5× TBE buffer. Gels were dried and an exposition using Biomax RAR (Eastman Kodak Co.) was performed.
Immunofluorescence Microscopy
For indirect immunofluorescence, U2OS cells were transfected and processed as described previously (32). The anti-FLAG and anti-TDP-43 antibodies were purchased from Sigma and Protein Tech, respectively. The anti-phospho-TDP-43 (Ser(P)-409/410–1) and anti-HA were purchased from COSMO BIO and Santa Cruz Biotechnology, respectively. The secondary antibodies, anti-mouse AlexaFluor, anti-rabbit AlexaFluor. or anti-mouse Far Red, were from Molecular Probes, and the anti-rabbit FITC was from Jackson ImmunoResearch. Cells were analyzed on a Zeiss LSM 510 confocal microscope.
Filter Trap Assay
For filter trap assays, 100–200 ng of GST-TDP-43 were incubated with 0.25–0.5 μm of His-EGFP-Gln/Asn recombinant proteins in reaction buffer (20 mm HEPES, pH 7.5, 2 mm EDTA, 0.5 mm DTT, and 6% glycerol) plus (UG)6 (0.5–1.5 ng). After 20 min at RT, samples were partially solubilized with SDS as final 0.5% (v/v) concentration. The complete reaction mixture was applied on a pre-wetted cellulose acetate 0.2-mm filter (from STERLITECH) using a dot blot chamber. The membrane was washed two times with 500 ml of PBS and immediately used to perform a Western blot.
Cell Lysate Fractionation
U2OS cell were co-transfected with FLAG-TDP-43 WT and EGFP or EGFP-12×Gln/Asn. After 24 h, cells were centrifuged at 1000 × g for 5 min at 4 °C. The cellular pellets were resuspended in 300 μl of lysis buffer (15 mm HEPES, pH 7.5, 250 mm NaCl, 0.5% Nonidet P-40, and 10% glycerol) supplemented with proteases (Roche Applied Science mixture) and phosphatase inhibitors (10 mm NaF, 1 mm β-glycerol phosphate, and 1 mm Na3VO4). Cells were sonicated with 10 pulses (30% of pulse) at 50% of power and centrifuged at 700 × g for 10 min at 4 °C. The pellets were discarded, and total cell lysates were quantified by Bradford. For the fractionation, 450 μg from each total cell lysates were centrifuged at 16,000 × g for 30 min at 4 °C. The supernatants were transferred into a new tube, and the pellets were washed again with lysis buffer, resuspended in 50 μl of urea buffer, and sonicated again. To analyze each fraction by Western blot, 50 μg of total lysates, 50 μg of soluble fraction, and all pellet volume were loaded in a 10% SDS-PAGE.
TAT Peptide Transduction
U2OS cells (3 × 105) were plated in 6-well plates containing coverslips. The next day, the cells were incubated (or not) with 2.5 μm TAT(342–366) and TAT-pControl peptides (see sequences below) for 20 h. Cells were washed three times with PBS, and immunolabeling was carried out using an anti-TDP-43 as mentioned before. The peptide sequences transduced in these experiments are as follows: TAT(342–366) (ggygrkkrrqrrr(TAT)GGsqqnqsgpsgnnqnqgnmqrepnqa) and TAT-pControl (ggygrkkrrqrrr(TAT)GGSHTLRGRRLVFDNQLTIRSPSRREC).
Toxicity Assays
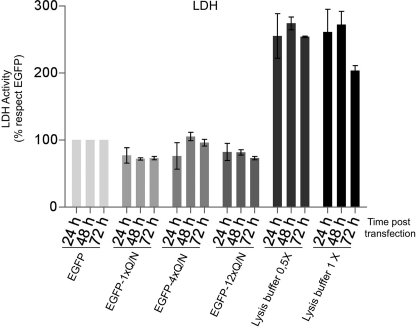
U2OS cells (6 × 104) were plated in 24-well plates and transfected with 0.1 μg of EGFP, EGFP-1×Gln/Asn, EGFP-4×Gln/Asn, or EGFP-12×Gln/Asn constructs (see below). Twenty four, 48, and 72 h after transfection, 50 μl of medium from each condition was used to assay the activity of lactate dehydrogenase enzyme as cell toxicity parameter (G1780, Promega.).
RESULTS
We have previously identified several members of the hnRNP-A/B protein family as specific interactors of region 321–366 of TDP-43 and have shown that these contacts are very important for maintaining the splicing functionality of TDP-43 (34, 40). Interestingly, half of the residues making up this sequence are represented by Gln/Asn residues, forming a region that is somewhat reminiscent of “prion domains” in which alternating Gln and Asn residues can be arranged in a self-propagating amyloid conformation. In keeping with this possibility, the region spanning residues 320–367 has been shown to be responsible for sequestration of TDP-43 in polyglutamine aggregates and cause loss-of-function effects (36, 57) as predicted by a model previously proposed (14).
To understand the aggregation process, a thorough knowledge of the interactions of TDP-43 and its partners was needed. We have focused on the TDP-43-hnRNP-A2 interaction being one of the most prominent and verified by several proteomic/interaction studies (38–40, 43). We have first extended our investigation of the TDP-43-hnRNP-A2 interactions mapping the minimal sequences needed on both sides. Once this was established and sequences finely mapped, we have looked at the aggregation properties of the minimal TDP-43 region involved in the process and set up a cellular model of aggregation.
hnRNP-A2 Binding Region to TDP-43
As already mentioned, we have previously narrowed down the region of TDP-43 interacting with hnRNP-A2 to residues 321–366 of this protein (34). However, very little data are available with regard to which region of hnRNP-A2 is directly binding to TDP-43. In fact, previous experiments from our laboratory had only loosely identified the C-terminal tail of hnRNP-A2 and -A1 as being responsible for this interaction (40) but not specific short amino acid sequences. Deletion mutants of hnRNP-A2 performed in our laboratory have suggested, using band shift experiments, that the TDP-43-binding site was restricted to residues spanning from 288 to 341 of hnRNP-A2 (data not shown).
To validate this interaction in a more natural setting, we then decided to perform several co-immunoprecipitation (co-IP) analyses using constructs that expressed these hnRNP-A2 and TDP-43 regions fused to EGFP. In the first co-IP experiment, we co-transfected in Hep3B cells constructs EGFP-A2(288–322) or EGFP-A2(288–341) together with a construct that expressed a FLAG-TDP-43 protein. Following IP with an anti-EGFP Ab, the Western blot was probed with an anti-FLAG antibody to detect the eventually associated FLAG-TDP-43 protein. As shown in Fig. 1A, whereas EGFP-A2(288–322) could IP only weakly the FLAG-TDP-43 protein, pulldown efficiency was very much enhanced in the case of the EGFP-A2(288–341) protein. As expected, no signal could be detected using an EGFP protein alone. Moreover, all EGFP constructs were expressed at equal levels in the transfected cells as determined using an anti-EGFP antibody (Fig. 1A, bottom panel).
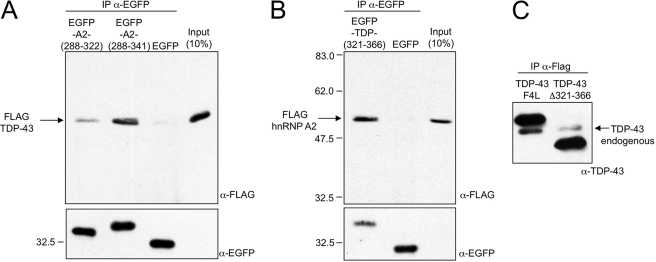
FIGURE 1.
A shows the co-immunoprecipitation between a transfected FLAG-TDP-43 protein and EGFP-hnRNP-A2(288–322), EGFP-hnRNP-A2(288–341), or empty EGFP expressed in Hep3B cells. Following immunoprecipitation with anti-EGFP antibody, the bound FLAG-TDP-43 is detected by Western blot analysis using an anti-FLAG mAb. The expression levels of each of the EGFP constructs is reported in the bottom panel. B shows a similar co-immunoprecipitation experiment but this time between a transfected FLAG-hnRNP-A2 protein and EGFP-TDP-43(321–366) or EGFP alone. Also in this case, following immunoprecipitation with anti-EGFP antibody, the bound FLAG-hnRNP-A2 is detected by Western blot analysis using an anti-FLAG mAb. The expression levels of each EGFP constructs is reported in the bottom panel. C shows a co-immunoprecipitation analysis of endogenous TDP-43 associated with stably transfected cell lines producing a flagged TDP-43 F4L and Δ321–366 TDP-43 mutants. The migration of the endogenous TDP-43 is indicated.
In a second co-IP experiment, we used a similar strategy, differing only in the EGFP molecule utilized, that was fused with the 321–366 residues of TDP-43 (EGFP-TDP-43(321–366)) and co-transfecting this construct with a flagged hnRNP-A2 expression plasmid. As above, IP was performed using an anti-EGFP Ab, and the pulled down flagged hnRNP-A2 molecule was detected using an anti-FLAG antibody. Also in this case, the results reported in Fig. 1B show that EGFP-TDP-43(321–366) could efficiently co-IP the flagged hnRNP-A2 protein and that no signal could be detected for the EGFP protein alone. Finally, we confirmed these results in vivo by immunoprecipitation of TDP-43 from cells that stably expressed the flagged TDP-43 F4L and TDP-43 Δ321–366 mutants previously described from our laboratory (58) (the F4L mutant was chosen as a positive control because it is unable to down-regulate the endogenous TDP-43). Immunoprecipitation using anti-FLAG antibodies shows that the F4L mutant is much more efficient at co-immunoprecipitating endogenous TDP-43 (that is barely detectable in the lane for the Δ321–366 mutant) (Fig. 1C). Taken together, these results show that hnRNP-A2 residues 288–341 and TDP-43 residues 321–366 are sufficient to support the interaction between these two proteins in the natural cellular environment and also without the support of additional protein sequences.
Refining the 321–366 hnRNP-A2 Binding Sequence of TDP-43
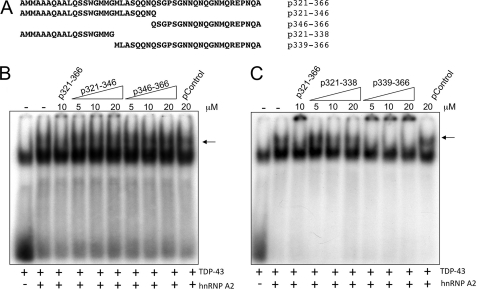
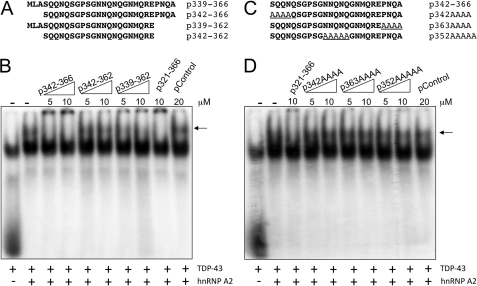
In parallel to these experiments, we were also interested in better defining the 321–366 sequence with the use of synthetic peptides that spanned different portions of this TDP-43 region. As a first approach, we synthesized a number of peptides that divided up this sequence in four different ways (Fig. 2A). Each peptide was used in competition analysis in the presence of TDP-43 bound to a labeled UG6 oligonucleotide and supershifted following the addition of hnRNP-A2 (Fig. 2B, indicated by an arrow). In a first experiment, Fig. 2B shows that neither peptides spanning residues 321–346 nor 346–366 could successfully compete for supershift formation (compare with the 321–366 peptide). In addition, no competition could also be observed for the peptide spanning residues 321–338 (Fig. 2C). However, successful competition could be observed using a peptide spanning the 339–366 region of TDP-43 (Fig. 2C). To see if this region could be reduced even further, we then prepared a second set of peptides where only the extremities of the 339–366 sequence were deleted (Fig. 3A). This analysis showed that efficient competition could be obtained with a peptide spanning residues 342–366 (Fig. 3B). It was noted that within this sequence there were three Gln/Asn-rich stretches (342–345, 352–357, and 363–366), raising the possibility that they were mediating this interaction. Therefore, all these residues were selectively mutated in alanines (Fig. 3C), and the effect of these substitutions was tested in a supershift assay. The results showed that mutating even a single Gln/Asn-rich region within these sequences completely abolished the ability of the peptide to successfully compete in the supershift assay (Fig. 3D). Interestingly, in all these experiments, whenever we used synthetic peptides in EMSA analysis all the successful candidates (p321–366, p339–366, and p342–366) not only disrupted the supershifted complex but also caused the appearance of a highly retarded radioactive signal (composed of TDP-43 bound to radioactive RNA oligonucleotide) that remained within the wells of the polyacrylamide gel (Figs. 2C and 3B). The same phenomenon was also present in our previous work (see Figs. 5 and 8D in Ref. 34). Taking in mind the role of Gln and Asn residues in the protein aggregation process, it was possible that the Gln/Asn residues from this TDP-43 region could be implicated in the aggregation process itself. Consistent with this finding, none of the peptides in which the alanines substituted the Gln/Asn sequences were observed to give rise to aggregate formation in the wells (Fig. 3D). Additionally, the aggregate formation in the wells observed in the presence of the p321–366 peptide was progressively reduced in vitro following addition of increasing amounts of GST-hnRNP-A2(288–341) in the reaction mixture. This experiment suggested that the A2 sequence was displacing or avoiding the interaction of p321–366 peptide with TDP-43 and its aggregation (supplemental Fig. 1).
FIGURE 2.
Competition analysis of interaction between TDP-43 and hnRNP-A2 (measured using a band shift assay) using different peptides based on the 321–366 sequence is shown. A scheme of the peptides used in this experiment is reported in A. B and C show the band shift assays in which increasing quantities (as indicated) of the different synthetic peptides were used as competitors. A nonrelated peptide (pControl) was also used as control. In each experiment, the supershifted (UG)6-TDP-43-hnRNPA2 interaction is indicated by an arrow.
FIGURE 3.
A and B show competition analysis of the interaction between TDP-43 and hnRNP-A2 (measured using a band shift assay) using different peptides based on the 339–366-residue sequence. In this experiment, increasing quantities (as indicated) of the different synthetic peptides were used as competitors, and a nonrelated peptide (pControl) was used as control. The supershifted (UG)6-TDP-43-hnRNPA2 interaction is indicated by an arrow. C and D show the effect on (UG)6-TDP-32-hnRNPA2 supershift formation (indicated by an arrow) using peptides based on the 342–366 sequence in which the various Gln/Asn-rich regions were mutated to alanines. Also in this case, increasing quantities (as indicated) of the different synthetic peptides were used as competitors and a nonrelated peptide (pControl) was used as control.
FIGURE 5.
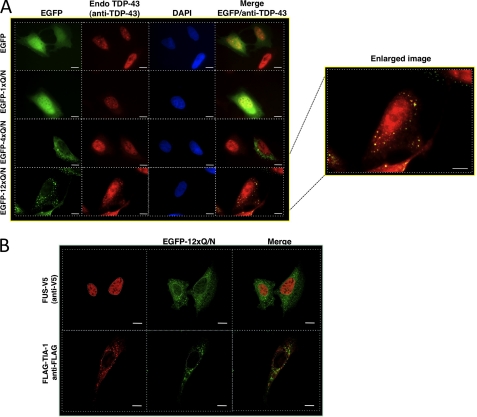
A shows an immunofluorescence of U2OS cells transfected with EGFP, EGFP-1×Gln/Asn, EGFP-4×Gln/Asn, or EGFP-12×Gln/Asn (green). The cellular distribution of endogenous TDP-43 was assayed using an anti-TDP-43 Ab (red). Cells nuclei were visualized with DAPI. Co-localization of endogenous TDP-43 aggregates with those from EGFP-12×Gln/Asn was observed (yellow) (scale bars, 10 μm). An enlarged image of the co-localization is shown (enlarged image). B shows an immunofluorescence of the cellular distribution of V5-FUS and FLAG-TIA-1 (red) proteins in an EGFP-12×Gln/Asn (green) background transfection. Nuclei are highlighted with white dotted lines. Scale bars, 10 μm.
FIGURE 8.
As an indicator of cellular toxicity, the release of lactate dehydrogenase (LDH) into the media was used following transfection of the 1×Gln/Asn, 4×Gln/Asn, and 12×Gln/Asn constructs in U2OS cells. Lactate dehydrogenase levels were measured at 24, 48, and 72 h after cells were transfected with the indicated constructs. Data from three separate experiments were analyzed, and the results are reported with standard deviation.
Taken together, all these in vitro data demonstrated that the residues 342–366 from TDP-43 conform to the minimal sequence necessary to interact with hnRNPA2. Additionally, the rich composition in Gln/Asn residues of this sequence and the observation that all the competing peptides caused the production of pre-aggregates make this region a good target to study its role in TDP-43 aggregation.
Use of Tandem Repeats Carrying the 331–369 Gln/Asn-rich Region Alone Can Promote Aggregation in Vitro and in Transfected U2OS Cells
According to the results observed previously, it was then decided to determine whether this Gln/Asn region was capable of inducing a similar kind of aggregate formation in vivo. Initially, we used synthetic hybrid peptides, including the 321–366 and 342–366 sequences linked to a fluorophore and fused to an HIV sequence, to facilitate their introduction into the cell. It was observed that the peptides entered all cells causing a limited cytoplasmic TPD-43 aggregation phenomenon (supplemental Fig. 2). At this point, it was hypothesized that for the formation of inclusions, there may be a need of an aggregation focus, and it was decided to try with plasmids carrying multiple copies of the peptide sequence. To achieve this, we chose the 331–369 region rather than the minimal 342–366 sequence studied in vitro to provide some additional degree of structural integrity. To achieve this aim, we engineered several plasmids that carried 1, 4, or 12 different copies of the 331–369 region fused to EGFP (Fig. 4, A and B).
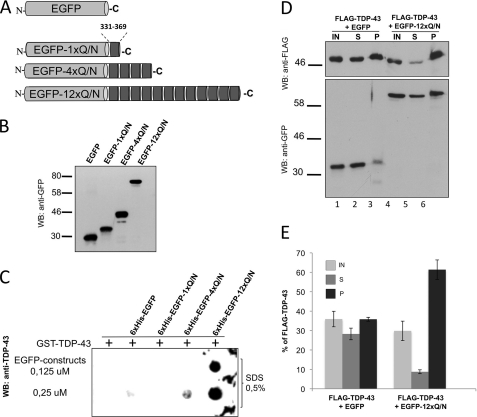
FIGURE 4.
A shows a schematic representation of constructs generated by cloning different repetitions (1, 4, or 12) of the Gln/Asn region from TDP-43 (residues 331–369) in the C terminus of the EGFP protein. The same constructs were subcloned to include an N-terminal His6 tag (see “Experimental Procedures”). B shows a Western blot of purified His6-EGFP-Gln/Asn constructs using an anti-GFP antibody. In a filter-binding assay (C), different concentrations of these proteins were incubated in vitro with recombinant GST-TDP-43 to test the aggregate formation. D, U2OS cells were co-transfected with FLAG-TDP-43 and EGFP-12×Gln/Asn constructs. Cell lysates were fractioned in soluble (S) and insoluble (pellet (P)) fractions, and the presence of FLAG-TDP-43 was detected by Western blot using an anti-FLAG Ab (IN, inputs). The correct expression of EGFP and EGFP-12×Gln/Asn was detected using an anti-GFP Ab. Quantification from three independent experiments is shown in E.
First of all, these constructs were cloned in a bacterial expression vector to include an N-terminal His6 tag. Following their expression and purification, the purified proteins were then mixed with GST-TDP-43 wild type and an unlabeled UG oligonucleotide. Aggregate formation was then measured using a filter binding assay (Fig. 4C). This experiment showed that with respect to the EGFP protein or the EGFP proteins carrying one and four repeats (EGFP-1×Gln/Asn and EGFP 4×Gln/Asn), only the EGFP protein carrying 12 repeats (EGFP-12×Gln/Asn) was capable of efficiently promoting TDP-43 aggregation.
This ability of EGFP-12×Gln/Asn to sequester the wild type TDP-43 protein to form aggregates in vitro was then assayed in a more natural setting following its transfection in U2OS cells together with FLAG-TDP-43 (co-transfection with EGFP was used as a control). The transfected samples were then centrifuged, and the soluble and pelleted fractions were analyzed by Western blot both for FLAG-TDP-43 expression and EGFP/EGFP-12×Gln/Asn expression. As shown in Fig. 4D, co-transfection with EGFP-12×Gln/Asn substantially increased the amount of FLAG-TDP-43 in the P fraction (lane 6) as opposed to co-transfection with EGFP (lanes 1–3). These results were confirmed by three independent experiments. Their quantification is shown in Fig. 4E. All these results indicated that the interaction of TDP-43 with multiple repeats composed by the 331–369 TDP-43 sequence can induce in vitro and in vivo TDP-43 aggregation.
Cellular Distribution of the EGFP-12×Gln/Asn Aggregates
We performed a preliminary characterization of this aggregation process in connection with endogenous TDP-43 distribution by transfecting EGFP, EGFP-1×Gln/Asn, EGFP-4×Gln/Asn, and EGFP-12×Gln/Asn in U2OS cells and HEK-293 (data not shown). This transfection experiment (Fig. 5A) showed that aggregate formation of the EGFP carrying Gln/Asn repeats could not be observed at all for the control EGFP and EGFP-1×Gln/Asn constructs. However, aggregate formation was weakly apparent for the EGFP-4×Gln/Asn construct (diffuse green spots in the cytoplasm) and much more prominent for EGFP-12×Gln/Asn expression. In this respect, it should be noted that transfecting three times the amount of EGFP-4×Gln/Asn with respect to EGFP-12×Gln/Asn did not result in similar aggregate formation, suggesting that this phenomenon is not just a consequence of the absolute amount of Gln/Asn repeats introduced into cells (data not shown). Most importantly, the aggregates formed by the EGFP-12×Gln/Asn constructs were observed to co-localize with the endogenous TDP-43 (Fig. 5A, enlarged image). The observation that neither FUS protein (fused to a V5 epitope) nor TIA-1 (a typical stress granules marker expressed as a FLAG-tagged protein) co-localized with these aggregates shows both the specificity of the EGFP-12×Gln/Asn for sequestering endogenous TDP-43 and that these aggregates are different from stress granules. In the case of TIA-1, in fact, it is known that just the overexpression of this protein can induce stress granule formation rendering useful this phenomenon to differentiate stress granules from other kind of aggregates (59). It should be kept in mind, however, that Liu-Yesucevitz et al. (60) reported the presence of TIA-1 in spinal cord aggregates of ALS patients. Finally, cell fractionation studies performed following the co-transfection of V5-tagged FUS and EGFP-12×Gln/Asn construct showed that in contrast with the TDP-43 case (Fig. 4D) no change in solubility could be observed for FUS confirming its absence in EGFP-12×Gln/Asn aggregates (data not shown).
Role of TDP-43 Functional Domains on Aggregate Co-localization
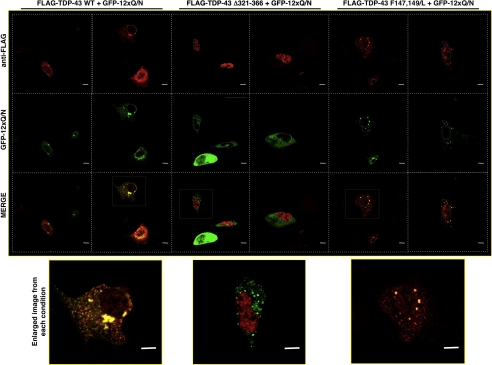
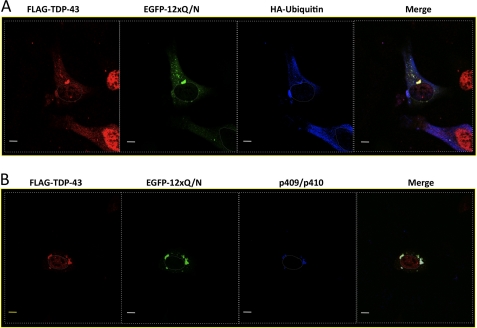
To eventually understand the triggering factors for aggregation, it is essential to know the functional domains/properties of TDP-43 that are important for EGFP-12×Gln/Asn-TDP-43 co-localization. Fig. 6 shows co-transfection of the EGFP-12×Gln/Asn construct with FLAG-TDP-43 wild type and two mutants, one carrying a deletion in the 321–366 region (FLAG-TDP-43-Δ321–366) and the other carrying two Phe/Leu substitutions in RRM-1 that render it unable to bind RNA (FLAG-TDP-43 F147L,F149L). To avoid any interference from endogenous TDP-43, U2OS cells were treated with TDP-43 siRNA before transfecting these siRNA-resistant mutants. The results of this experiment clearly showed that co-localization with EGFP-12×Gln/Asn aggregates occurs with FLAG-TDP-43 wild type and with the mutant lacking RNA binding capacity (FLAG-TDP-43 F147L,F149L). Interestingly, in the case of FLAG-TDP-43 wild type, it was observed occasionally that the nuclei of the cells containing extensive aggregation were almost devoid of TDP-43 (see enlarged image at Fig. 6, bottom left panel). Conversely, the co-localization with the EGFP-12×Gln/Asn aggregates was completely abolished in the case of FLAG-TDP-43-Δ321–366, the mutant lacking the Gln/Asn region. No cytoplasmic co-localization could be observed when any of these mutants were co-transfected with EGFP alone (supplemental Fig. 3). These experiments demonstrate that the EGFP-12×Gln/Asn aggregates can sequester TDP-43 only if it carries the 321–366 sequence. Moreover, once recruited in these aggregates the FLAG-TDP-43 wild type was observed to become both ubiquitinated (Fig. 7A) and phosphorylated in the two 409 and 410 serine residues (Fig. 7B) that represent two distinguishing features of the inclusions observed in the brain of ALS and FTLD patients. These results were confirmed by Western blot analysis (supplemental Fig. 4). It should also be noted that in several instances we observed aggregates of EGFP-12×Gln/Asn that lack TDP-43 and also lack ubiquitination and phosphorylation, suggesting that EGFP-12×Gln/Asn can aggregate in the absence of much full-length TDP-43. We have not been able to detect cleaved fragments, but they could be a late event in the development of the disease that is not easily reproducible in cell culture.
FIGURE 6.
Immunofluorescence of cellular distribution of FLAG-TDP-43WT, FLAG-TDP-43 Δ321–366, and FLAG-TDP-43 F147L,F149L (red) co-transfected with EGFP-12×Gln/Asn (green) in U2OS cells is shown. Enlarged images from selected co-localization events are shown (bottom panels). Nuclei are highlighted with white dotted lines. Scale bars, 10 μm.
FIGURE 7.
Immunofluorescence experiment shows that aggregates are positive for ubiquitination and abnormal phosphorylation in TDP-43 residues Ser-409/410. A, FLAG-TDP-43WT (red) was co-transfected with EGFP-12×Gln/Asn (green) plus HA-ubiquitin (blue). The co-localization of aggregates with HA-ubiquitin is observed as white. B, cells were co-transfected with FLAG-TDP-43WT (red) and EGFP-12×Gln/Asn (green), and the phosphorylation of Ser-409/410 TDP-43 residues was detected using a specific anti-Ser(P)-409/410 Ab (blue). The co-localization of the aggregates with phosphorylated Ser-409/410 residues is observed as white. Nuclei are highlighted with white dotted lines. Scale bars, 10 μm.
It is not really known if the aggregates in the brains of patients are neurotoxic (see the review by Baloh (61) for a discussion on this subject). It was then of interest to examine whether transfection with the Gln/Asn repeat constructs was associated with increased cellular toxicity as determined by lactate dehydrogenase release. The results of this assay failed to detect any increased release of lactate dehydrogenase levels in the presence of the 1×Gln/Asn, 4×Gln/Asn, and 12×Gln/Asn constructs with respect to controls (Fig. 8). In this respect, it should be noted that preliminary immunohistochemical studies of our inclusions with anti-p62 and anti-LC3 antibodies were suggestive of an initial autophagic response (data not shown); this observation may provide an explanation for the lack of toxicity and deserves further studies.
We have previously shown that human TDP-43 could replace the Drosophila TBPH in the development of the neuromuscular junction (34, 60) and that they shared also their functional properties in splicing assays (30). Interestingly we observe aggregate co-localization when the EGFP-12×Gln/Asn construct was co-transfected with Drosophila TBPH showing once more the considerable functional homology of human TDP-43 (supplemental Fig. 5). In keeping with this similarity, especially with regard to the role played by the C-terminal tail, deleting the C-terminal tail of TBPH also abolished co-localization with EGFP-12×Gln/Asn (supplemental Fig. 5).
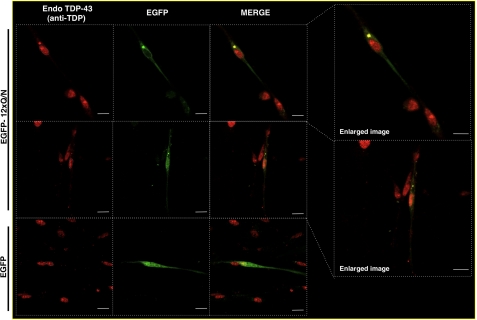
Finally, all these experiments were performed in non-neuronal cell lines, and thus we considered it important to check if the EGFP-12×Gln/Asn construct was capable of inducing aggregate formation also in neuronal cells. To perform this, we prepared primary neuronal cultures from rat trigeminal dorsal ganglia and transfected the EGFP-12×Gln/Asn construct into them. As shown in Fig. 9, transfection of EGFP-12×Gln/Asn caused the appearance of one or few more round cytoplasmic inclusions that are highly similar to what is commonly observed in the neurons of affected patients.
FIGURE 9.
Immunofluorescence assay shows the formation of aggregates in primary neuronal cultures. Trigeminal ganglion neurons were transfected with EGFP or EGFP-12×Gln/Asn (green), and the cellular distribution of endogenous TDP-43 (red) was analyzed using an anti-TDP-43 Ab. An enlarged image from co-localization is shown (right panels). Nuclei are highlighted with white dotted lines. Scale bars, 10 μm.
DISCUSSION
TDP-43 is a typical hnRNP protein whose involvement in several neurodegenerative diseases has recently received a lot of attention by the research community (14, 15, 33). A detailed knowledge of its biochemical properties and hence its basic role in biological processes such as transcription (62, 63), pre-mRNA splicing (56, 64–66), microRNA processing (67), and mRNA stability (68, 69) is essential to develop novel diagnostic and therapeutic approaches. As usual for most hnRNP proteins, these functional properties are mediated by its characteristic domains that include an N-terminal sequence, two RNA recognition motif domains, and a Gly-rich C-terminal region.
The C-terminal region is particularly significant as it has been found that TDP-43 mutations associated with disease (41, 42, 70) are mostly localized within the C-terminal region and are generally thought to enhance inclusion formation (46, 71, 72). Indeed, on the basis of sequence homology, an intriguing hypothesis has also been recently made regarding the presence in this region of a potentially infectious “prion domain” from residue 277 to 414 (35), a characteristic that may be shared with other proteins involved in ALS/FTLD (73). With regard to the C-terminal tail, our previous work on TDP-43-hnRNP interactions suggested that the interaction of TDP-43 with its 321–366 Gln/Asn-rich sequence was stronger than the interaction with hnRNP-A2 as the 321–366-residue peptide was able to disrupt the TDP-43-hnRNP-A2 complex and produce a material that is retained at the origin of the gel (34). Consistent with this observation, it was recently shown that the presence of a Gln/Asn-rich region spanning residues 320–367 is responsible for sequestration of TDP-43 in polyglutamine aggregates and causes loss-of-function effects (36, 57).
At the moment, in fact, the simple conclusion that TDP-43 aggregates are just toxic by themselves and can thus explain the neurodegeneration has not been upheld by several studies, as recently reviewed (61). Regarding this issue, a large series of reports have focused on the potential toxicity of overexpression of the whole TDP-43 (certainly toxic with or without aggregation) and/or several large CTFs similar to those found in the brain extracts from patients. It is certain, in fact, that overexpression of TDP-43 and/or its fragments can promote aggregation in the cellular cytoplasm, as discussed in many recent reviews (61, 74). How they may do this is still not clear, and a “two-hit” model has been recently proposed (75). However, the significance of cleavage in the pathological pathway is not well established, and hence the significance of CTFs aggregation should be cautiously interpreted.
Our results are in keeping with this view, as cytotoxicity measurements failed to detect a significant effect following transfection with our 1×, 4×, and 12×Gln/Asn constructs into U2OS cells. Moreover, this lack of toxicity of the aggregations per se has been also reported by Johnson et al. (76) using a yeast aggregation model in which toxicity was associated with aggregation only when the RRM motifs and glycine-rich domains were included in the expressed constructs (either singly or in pairs). This finding further strengthens the hypothesis that the pathological significance of the aggregates probably resides in their ability to function as a “TDP-sink” in the cytoplasm, resulting in various loss-of-function effects within the nucleus. An alternative explanation that cannot be excluded is that aggregates may be toxic by themselves.
We show here that a repeated TDP-43 amino acid sequence 331–369 is capable of inducing TDP-43 aggregate formation both in non-neuronal and neuronal cells that mimic many but not all of the characteristics of the inclusions found in patients (see Table 1 for a comparison between the characteristics of our inclusions and patient observations). In particular, this system is capable of recapitulating some of the morphological features of TDP-43 aggregates found in ALS and FTLD brains. In fact it produces round and large formations, as we observed in rat dorsal ganglion neurons or granular dot-like inclusions, as we observed in U2OS and HEK-293 cells (data not shown). Their localization is predominantly cytoplasmic with occasional nuclear occurrence. Our data demonstrate that the key interaction for aggregate formation is TDP-43 with itself through the 331–369 region. In fact, TDP-43 147/149 Phe → Leu is not able to bind RNA, but it is efficient in forming aggregates, whereas TDP-43 Δ321–366 does not induce aggregation (Fig. 6). It is also clear from the results shown in Fig. 7 that ubiquitination and phosphorylation follow the aggregation. It is also worth noting that in some cells, where there is substantial cytoplasmic aggregation, there is a scant presence of TDP-43 in the nucleus, strengthening the hypothesis that sequestering TDP-43 in the aggregates essentially results in a situation equivalent to a nuclear knock-out of TDP-43 with its well known consequences (77–81).
TABLE 1.
Comparison between the 12×Gln/Asn-induced aggregates and patient aggregates
| Aggregate characteristics | 12×Gln/Asn aggregates | Pathological aggregates |
|---|---|---|
| Origin | Expression of 339–369-residue Gln/Asn repeats | Unknown |
| Localization | Preferentially cytoplasmic | Preferentially cytoplasmic |
| Time of generation | 24 h | Unknown (probably late in lifetime) |
| Stress granules | Apparently not | Uncertain |
| TIA-1 co-localization | No | Uncertaina |
| Ubiquitination | Yes | Yes |
| Hyperphosphorylation | Yes | Yes |
| Cleavage | No | Yes |
| Empty nuclei | In some cells following transient transfection | Yes in some neurons |
| Overexpression of endogenous TDP-43 | No | Uncertain |
a There is some controversy regarding this issue as initial reports did not find any co-localization in the aggregates from the spinal cords of three sporadic ALS patients (83). However, a later and more detailed study reported that co-localization between TIA-1 and TDP-43 aggregates could be observed in four ALS patients (spinal cord) and five FTLD-U patients (brains) (60).
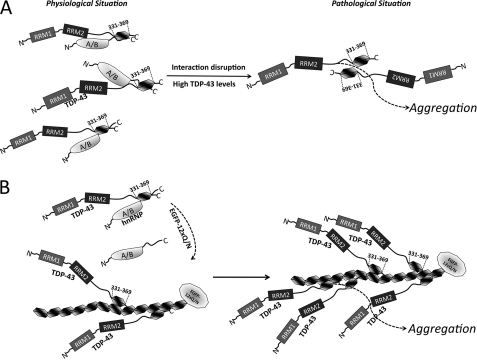
The observation that the FLAG-TDP-43-Δ321–366 construct does not co-localize with the EGFP-12×Gln/Asn aggregates in U2OS cells suggests that the normal interactions of TDP-43 with other hnRNPs might keep TDP-43 soluble. This is consistent with the substantial overlap of the 321–366 region with the site that interacts with hnRNP-A2 and with the result observed in supplemental Fig. 1. Moreover, the irrelevance for aggregation of the TDP-43 RNA binding domain mutant suggests that protein-protein contact may modulate the aggregation process and that the trigger may be a defect in the TDP-43 normal interactions. This would also be consistent with the observation that TDP-43 aggregates in patients do not contain hnRNP proteins (82). All these considerations are summarized in the schematic model reported in Fig. 10. Indeed, in our case the aggregation seems to be a process that involves mainly TDP-43 self-interactions and that leads to phosphorylation, ubiquitination, and possibly nuclear TDP-43 depletion.
FIGURE 10.
In a physiological situation, TDP-43 is interacting with its partners (i.e. mainly other hnRNP proteins of the A/B family) through its 331–369 region to carry out the normal biological functions (A, left side). When this normal interaction becomes disrupted, maybe through an increase in its expression levels, TDP-43 molecules are more prone to produce aggregates and generate a pathological situation (A, right side). This hypothesis is supported by our data where the addition of multiple repetitions of 331–369 TDP-43 sequence (EGFP-12×Gln/Asn) can successfully compete the TDP-43-hnRNP interaction (B, left side), inducing the recruitment of TDP-43 molecules and generating an aggregation condition that mimics the pathological situation (B, right side).
Finally, it is important to highlight that because all the inclusions reported in this paper were induced using the same basic construct (EGFP-12×Gln/Asn), it is clear that this system may be a formidable research tool as it represents a much needed cell-based TDP-43 aggregation model. Such a model will be fundamental for the identification of the factors involved in the formation of the original aggregate core and its impact in cellular metabolism and to follow the modifications that are seen in the final stage of the aggregates observed in the brains of patients. The aggregation model described in this study is the subject of the patent application RM2011A000582.
Supplementary Material
Acknowledgments
We thank Dr. Maurizio Romano for the FLAG-TBPH plasmids and Dr. Marco Baralle for helping with trigeminal dorsal ganglion neuron extraction.
This work was supported by AriSLA.

This article contains supplemental Figs. 1–5.
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- FTLD
- frontotemporal lobar degeneration
- ALS
- amyotrophic lateral sclerosis
- CTF
- C-terminal fragment
- EGFP
- enhanced green fluorescent protein
- IP
- immunoprecipitation
- TBPH
- TAR-binding protein homolog.
REFERENCES
- 1. Krecic A. M., Swanson M. S. (1999) hnRNP complexes. Composition, structure, and function. Curr. Opin. Cell Biol. 11, 363–371 [DOI] [PubMed] [Google Scholar]
- 2. Martinez-Contreras R., Cloutier P., Shkreta L., Fisette J. F., Revil T., Chabot B. (2007) hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 623, 123–147 [DOI] [PubMed] [Google Scholar]
- 3. He Y., Smith R. (2009) Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell. Mol. Life Sci. 66, 1239–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prasanth K. V., Prasanth S. G., Xuan Z., Hearn S., Freier S. M., Bennett C. F., Zhang M. Q., Spector D. L. (2005) Regulating gene expression through RNA nuclear retention. Cell 123, 249–263 [DOI] [PubMed] [Google Scholar]
- 5. Dreyfuss G., Kim V. N., Kataoka N. (2002) Messenger RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205 [DOI] [PubMed] [Google Scholar]
- 6. Carpenter B., MacKay C., Alnabulsi A., MacKay M., Telfer C., Melvin W. T., Murray G. I. (2006) The roles of heterogeneous nuclear ribonucleoproteins in tumor development and progression. Biochim. Biophys. Acta 1765, 85–100 [DOI] [PubMed] [Google Scholar]
- 7. Chen M., David C. J., Manley J. L. (2010) Tumor metabolism: hnRNP proteins get in on the act. Cell Cycle 9, 1863–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Budini M., Baralle F. E., Buratti E. (2011) Regulation of gene expression by TDP-43 and FUS/TLS in frontotemporal lobar degeneration. Curr. Alzheimer Res. 8, 237–245 [DOI] [PubMed] [Google Scholar]
- 9. Warraich S. T., Yang S., Nicholson G. A., Blair I. P. (2010) TDP-43. A DNA and RNA-binding protein with roles in neurodegenerative diseases. Int. J. Biochem. Cell Biol. 42, 1606–1609 [DOI] [PubMed] [Google Scholar]
- 10. Lagier-Tourenne C., Polymenidou M., Cleveland D. W. (2010) TDP-43 and FUS/TLS. Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fiesel F. C., Kahle P. J. (2011) TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration FEBS J. 278, 3550–3568 [DOI] [PubMed] [Google Scholar]
- 12. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 13. Geser F., Martinez-Lage M., Kwong L. K., Lee V. M., Trojanowski J. Q. (2009) Amyotrophic lateral sclerosis, frontotemporal dementia, and beyond. The TDP-43 diseases. J. Neurol. 256, 1205–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buratti E., Baralle F. E. (2009) The molecular links between TDP-43 dysfunction and neurodegeneration. Adv. Genet. 66, 1–34 [DOI] [PubMed] [Google Scholar]
- 15. Chen-Plotkin A. S., Lee V. M., Trojanowski J. Q. (2010) TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Rev. Neurol. 6, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gendron T. F., Josephs K. A., Petrucelli L. (2010) Review. Transactive response DNA-binding protein 43 (TDP-43). Mechanisms of neurodegeneration. Neuropathol. Appl. Neurobiol. 36, 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Colombrita C., Onesto E., Tiloca C., Ticozzi N., Silani V., Ratti A. (2011) RNA-binding proteins and RNA metabolism. A new scenario in the pathogenesis of amyotrophic lateral sclerosis. Arch. Ital. Biol. 149, 83–99 [DOI] [PubMed] [Google Scholar]
- 18. Mackenzie I. R., Rademakers R., Neumann M. (2010) TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9, 995–1007 [DOI] [PubMed] [Google Scholar]
- 19. Dormann D., Haass C. (2011) TDP-43 and FUS: a nuclear affair. Trends Neurosci. 34, 339–348 [DOI] [PubMed] [Google Scholar]
- 20. Strong M. J. (2010) The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS). J. Neurol. Sci. 288, 1–12 [DOI] [PubMed] [Google Scholar]
- 21. van Blitterswijk M., Landers J. E. (2010) RNA processing pathways in amyotrophic lateral sclerosis. Neurogenetics 11, 275–290 [DOI] [PubMed] [Google Scholar]
- 22. Anthony K., Gallo J. M. (2010) Aberrant RNA processing events in neurological disorders. Brain Res. 1338, 67–77 [DOI] [PubMed] [Google Scholar]
- 23. Ticozzi N., Ratti A., Silani V. (2010) Protein aggregation and defective RNA metabolism as mechanisms for motor neuron damage. CNS Neurol. Disord. Drug Targets 9, 285–296 [DOI] [PubMed] [Google Scholar]
- 24. Rabin S. J., Kim J. M., Baughn M., Libby R. T., Kim Y. J., Fan Y., Libby R. T., La Spada A., Stone B., Ravits J. (2010) Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum. Mol. Genet. 19, 313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ule J. (2008) Ribonucleoprotein complexes in neurologic diseases. Curr. Opin. Neurobiol. 18, 516–523 [DOI] [PubMed] [Google Scholar]
- 26. Bäumer D., Ansorge O., Almeida M., Talbot K. (2010) The role of RNA processing in the pathogenesis of motor neuron degeneration. Expert Rev. Mol. Med. 12, e21. [DOI] [PubMed] [Google Scholar]
- 27. Higashi S., Tsuchiya Y., Araki T., Wada K., Kabuta T. (2010) TDP-43 physically interacts with amyotrophic lateral sclerosis-linked mutant CuZn superoxide dismutase. Neurochem. Int. 57, 906–913 [DOI] [PubMed] [Google Scholar]
- 28. Winton M. J., Igaz L. M., Wong M. M., Kwong L. K., Trojanowski J. Q., Lee V. M. (2008) Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 283, 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buratti E., Baralle F. E. (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 30. Ayala Y. M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F. E. (2005) Human, Drosophila, and C.elegans TDP43. Nucleic acid binding properties and splicing regulatory function. J. Mol. Biol. 348, 575–588 [DOI] [PubMed] [Google Scholar]
- 31. Kuo P. H., Doudeva L. G., Wang Y. T., Shen C. K., Yuan H. S. (2009) Structural insights into TDP-43 in nucleic acid binding and domain interactions. Nucleic Acids Res. 37, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ayala Y. M., Zago P., D'Ambrogio A., Xu Y. F., Petrucelli L., Buratti E., Baralle F. E. (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785 [DOI] [PubMed] [Google Scholar]
- 33. Buratti E., Baralle F. E. (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 13, 867–878 [DOI] [PubMed] [Google Scholar]
- 34. D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y. M., Baralle F. E. (2009) Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 37, 4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cushman M., Johnson B. S., King O. D., Gitler A. D., Shorter J. (2010) Prion-like disorders. Blurring the divide between transmissibility and infectivity. J. Cell Sci. 123, 1191–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Udan M., Baloh R. H. (2011) Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion 5, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cartegni L., Maconi M., Morandi E., Cobianchi F., Riva S., Biamonti G. (1996) hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259, 337–348 [DOI] [PubMed] [Google Scholar]
- 38. Ling S. C., Albuquerque C. P., Han J. S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D. W. (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. U.S.A. 107, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Freibaum B. D., Chitta R. K., High A. A., Taylor J. P. (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail. An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 41. Barmada S. J., Finkbeiner S. (2010) Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Disease-associated pathways. Rev. Neurosci. 21, 251–272 [DOI] [PubMed] [Google Scholar]
- 42. Pesiridis G. S., Lee V. M., Trojanowski J. Q. (2009) Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum. Mol. Genet. 18, R156–R162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dormann D., Capell A., Carlson A. M., Shankaran S. S., Rodde R., Neumann M., Kremmer E., Matsuwaki T., Yamanouchi K., Nishihara M., Haass C. (2009) Proteolytic processing of TAR DNA binding protein-43 by caspases produces C-terminal fragments with disease defining properties independent of progranulin. J. Neurochem. 110, 1082–1094 [DOI] [PubMed] [Google Scholar]
- 45. Igaz L. M., Kwong L. K., Chen-Plotkin A., Winton M. J., Unger T. L., Xu Y., Neumann M., Trojanowski J. Q., Lee V. M. (2009) Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 284, 8516–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. (2009) Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 18, 3353–3364 [DOI] [PubMed] [Google Scholar]
- 47. Yang C., Tan W., Whittle C., Qiu L., Cao L., Akbarian S., Xu Z. (2010) The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One 5, e15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen A. K., Lin R. Y., Hsieh E. Z., Tu P. H., Chen R. P., Liao T. Y., Chen W., Wang C. H., Huang J. J. (2010) Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J. Am Chem. Soc. 132, 1186–1187 [DOI] [PubMed] [Google Scholar]
- 49. Saini A., Chauhan V. S. (2011) Delineation of the core aggregation sequences of TDP-43 C-terminal fragment. ChemBioChem 12, 2495–2450 [DOI] [PubMed] [Google Scholar]
- 50. Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., Akiyama H. (2008) Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neumann M., Kwong L. K., Lee E. B., Kremmer E., Flatley A., Xu Y., Forman M. S., Troost D., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2009) Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liachko N. F., Guthrie C. R., Kraemer B. C. (2010) Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J. Neurosci. 30, 16208–16219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y. J., Gendron T. F., Xu Y. F., Ko L. W., Yen S. H., Petrucelli L. (2010) Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol. Neurodegener. 5, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamashita M., Nonaka T., Arai T., Kametani F., Buchman V. L., Ninkina N., Bachurin S. O., Akiyama H., Goedert M., Hasegawa M. (2009) Methylene blue and dimebon inhibit aggregation of TDP-43 in cellular models. FEBS Lett. 583, 2419–2424 [DOI] [PubMed] [Google Scholar]
- 55. Caragounis A., Price K. A., Soon C. P., Filiz G., Masters C. L., Li Q. X., Crouch P. J., White A. R. (2010) Zinc induces depletion and aggregation of endogenous TDP-43. Free Radic. Biol. Med. 48, 1152–1161 [DOI] [PubMed] [Google Scholar]
- 56. Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., Baralle F. E. (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fuentealba R. A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M. I., Weihl C. C., Baloh R. H. (2010) Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 285, 26304–26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ayala Y. M., De Conti L., Avendaño-Vázquez S. E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., Baralle F. E. (2011) TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 30, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu-Yesucevitz L., Bilgutay A., Zhang Y. J., Vanderweyde T., Vanderwyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., Petrucelli L., Wolozin B. (2010) Tar DNA-binding protein-43 (TDP-43) associates with stress granules. Analysis of cultured cells and pathological brain tissue. PLoS One 5, e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baloh R. H. (2011) TDP-43. The relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. FEBS J. 278, 3539–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ou S. H., Wu F., Harrich D., García-Martínez L. F., Gaynor R. B. (1995) Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Abhyankar M. M., Urekar C., Reddi P. P. (2007) A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues. Role for TDP-43 in insulator function. J. Biol. Chem. 282, 36143–36154 [DOI] [PubMed] [Google Scholar]
- 64. Mercado P. A., Ayala Y. M., Romano M., Buratti E., Baralle F. E. (2005) Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 33, 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bose J. K., Wang I. F., Hung L., Tarn W. Y., Shen C. K. (2008) TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J. Biol. Chem. 283, 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dreumont N., Hardy S., Behm-Ansmant I., Kister L., Branlant C., Stévenin J., Bourgeois C. F. (2010) Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 38, 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Buratti E., De Conti L., Stuani C., Romano M., Baralle M., Baralle F. (2010) Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 277, 2268–2281 [DOI] [PubMed] [Google Scholar]
- 68. Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., Baekelandt V., Alunni-Fabbroni M., Schulz J. B., Kahle P. J. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) down-regulates histone deacetylase 6. EMBO J. 29, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Volkening K., Leystra-Lantz C., Yang W., Jaffee H., Strong M. J. (2009) Tar DNA-binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res. 1305, 168–182 [DOI] [PubMed] [Google Scholar]
- 70. Banks G. T., Kuta A., Isaacs A. M., Fisher E. M. (2008) TDP-43 is a culprit in human neurodegeneration, and not just an innocent bystander. Mamm. Genome 19, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., Gitler A. D. (2009) TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Arai T., Hasegawa M., Nonoka T., Kametani F., Yamashita M., Hosokawa M., Niizato K., Tsuchiya K., Kobayashi Z., Ikeda K., Yoshida M., Onaya M., Fujishiro H., Akiyama H. (2010) Phosphorylated and cleaved TDP-43 in ALS, FTLD and other neurodegenerative disorders and in cellular models of TDP-43 proteinopathy. Neuropathology 30, 170–178 [DOI] [PubMed] [Google Scholar]
- 73. Gitler A. D., Shorter J. (2011) RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion 5, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cohen T. J., Lee V. M., Trojanowski J. Q. (2011) TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol. Med. 17, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pesiridis G. S., Tripathy K., Tanik S., Trojanowski J. Q., Lee V. M. (2011) A “two-hit” hypothesis for inclusion formation by carboxyl-terminal fragments of TDP-43 protein linked to RNA depletion and impaired microtubule-dependent transport. J. Biol. Chem. 286, 18845–18855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Johnson B. S., McCaffery J. M., Lindquist S., Gitler A. D. (2008) A yeast TDP-43 proteinopathy model. Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 105, 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ayala Y. M., Misteli T., Baralle F. E. (2008) TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc. Natl. Acad. Sci. U.S.A. 105, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Feiguin F., Godena V. K., Romano G., D'Ambrogio A., Klima R., Baralle F. E. (2009) Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 583, 1586–1592 [DOI] [PubMed] [Google Scholar]
- 79. Kraemer B. C., Schuck T., Wheeler J. M., Robinson L. C., Trojanowski J. Q., Lee V. M., Schellenberg G. D. (2010) Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 119, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Polymenidou M., Lagier-Tourenne C., Hutt K. R., Huelga S. C., Moran J., Liang T. Y., Ling S. C., Sun E., Wancewicz E., Mazur C., Kordasiewicz H., Sedaghat Y., Donohue J. P., Shiue L., Bennett C. F., Yeo G. W., Cleveland D. W. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tollervey J. R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A. L., Zupunski V., Patani R., Chandran S., Rot G., Zupan B., Shaw C. E., Ule J. (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Neumann M., Igaz L. M., Kwong L. K., Nakashima-Yasuda H., Kolb S. J., Dreyfuss G., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2007) Absence of heterogeneous nuclear ribonucleoproteins and survival motor neuron protein in TDP-43 positive inclusions in frontotemporal lobar degeneration. Acta Neuropathol. 113, 543–548 [DOI] [PubMed] [Google Scholar]
- 83. Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.