Background: Functional coupling between mitochondrial creatine kinase (MtCK) and adenine nucleotide translocase (ANT) can determine energy transfer pathways in the cell.
Results: We composed a molecular dynamics model of a mitochondrial inner membrane patch, ANT, and MtCK.
Conclusion: Cardiolipin plays an important role in stabilizing MtCK-membrane binding.
Significance: This is a first step in the development of an MtCK, ANT, and membrane interaction model.
Keywords: Bioenergetics, Cardiolipin, Membrane Proteins, Mitochondria, Molecular Dynamics, Adenine Nucleotide Translocase, Creatine Kinase
Abstract
Interaction between mitochondrial creatine kinase (MtCK) and adenine nucleotide translocase (ANT) can play an important role in determining energy transfer pathways in the cell. Although the functional coupling between MtCK and ANT has been demonstrated, the precise mechanism of the coupling is not clear. To study the details of the coupling, we turned to molecular dynamics simulations. We introduce a new coarse-grained molecular dynamics model of a patch of the mitochondrial inner membrane containing a transmembrane ANT and an MtCK above the membrane. The membrane model consists of three major types of lipids (phosphatidylcholine, phosphatidylethanolamine, and cardiolipin) in a roughly 2:1:1 molar ratio. A thermodynamics-based coarse-grained force field, termed MARTINI, has been used together with the GROMACS molecular dynamics package for all simulated systems in this work. Several physical properties of the system are reproduced by the model and are in agreement with known data. This includes membrane thickness, dimension of the proteins, and diffusion constants. We have studied the binding of MtCK to the membrane and demonstrated the effect of cardiolipin on the stabilization of the binding. In addition, our simulations predict which part of the MtCK protein sequence interacts with the membrane. Taken together, the model has been verified by dynamical and structural data and can be used as the basis for further studies.
Introduction
The process of energy transfer between mitochondria and ATPases has been a subject of numerous experimental and theoretical studies (1–8). As a part of a larger energy transfer system, creatine kinase and adenylate kinase shuttles are suggested to play an important role in heart and brain (9–11). Because of the shuttles, mitochondria deliver energy to ATPases in the form of phosphocreatine (PCr)2 and ATP, where highly diffusive PCr provides a temporal and spacial energy buffer. Importance of the shuttles is expected to be large in cells that contain a highly compartmentalized environment with restricted diffusion of metabolites, such as heart muscle cells (12–18). The mitochondrial electron transport chain and ATP synthesis are coupled to phosphorylation of creatine and synthesis of PCr by the mitochondrial isoform of creatine kinase (MtCK) and adenine nucleotides translocase (ANT). Because of the close vicinity of ANT to MtCK, ATP can directly diffuse from ANT to the active site of MtCK and be used for PCr production, leading to the export of PCr from mitochondria as a final product of oxidative phosphorylation. Aspects of such functional coupling between ANT and MtCK have been the subject of kinetic and structural studies (10, 19–22) as well as a theoretical analysis of the coupling kinetics (23). Alternatively to PCr formation from ATP in mitochondria, ATP could leave the mitochondrial intermembrane space via the voltage-dependent anion channel, located on the mitochondrial outer membrane. Importance of the functional coupling between MtCK and ANT has been analyzed by several mathematical models, with some of them predicting a vital role in keeping metabolic stability of the heart (22) and some suggesting a small export of PCr from mitochondria (24). 31P-NMR inversion and saturation analysis on Langendorff-perfused hearts show that the ratio of energy export from mitochondria via PCr and ATP could depend on the workload of the heart and heart conditioning (8, 25). One could speculate that this ratio is critically dependent on the interaction of ANT and MtCK. Although a thermodynamically consistent model of MtCK and ANT interaction exists (23), the proposed kinetic schemes have not been verified by analyzing the interaction between those proteins on a molecular level. Thus, to our knowledge, there is no definite understanding of the kinetic mechanism that explains the functional coupling between MtCK and ANT. As a consequence, this limits the analysis of the importance of the creatine kinase shuttle within a larger energy transfer system.
As a technique to study molecular-level interactions, molecular dynamics (MD) simulations of biomolecular systems have revealed new insight into the molecular-scale phenomena that occur in these complex systems (26, 27). However, many processes in these biomolecular systems occur over length and time scales that are well beyond the current capabilities of atomic-level simulation, i.e. all-atom MD. New MD tools and approaches are being developed that allow modeling of molecular phenomena over longer time and length scales. One such approach is coarse-grained (CG) MD simulation. It is on the basis of atomistic descriptions of microscopic systems and represents these systems on a coarser, mesoscopic level. CG models provide a significant simulation speed-up compared with atomistic models, which is the main reason for their application. CG modeling substantially reduces this computational challenge through a combination of smaller CG degrees of freedom and the fact that in the CG model the effective potential is “smoother” than in the full all-atom model. The first introduction of physics-based CG models in computational biology was carried out by M. Levitt in 1975 (28), and this approach has become increasingly popular in the simulation of complex biological systems (29). Different methodologies have been applied to develop CG models. In structure-based CG models (30–32), the CG interaction parameters are tuned to accurately reproduce structural features of the system (typically, radial distribution functions) generally derived from all-atom MD simulations. Structure-based CG models suffer from a number of limitations. First of all, transferability of interaction parameters to different temperatures is often poor, as thoroughly discussed by Carbone et al. (31). Often, structure-based models requires additional parametrization if one wishes to use them in combination with solvents or other polymer species. Compared with structure-based CG models, thermodynamics-based models rely on a very different coarse-graining strategy. In the thermodynamics-based approach, interaction parameters are chosen to reproduce the selected thermodynamic properties of the system and only a few structural properties. One example of thermodynamics-based CG models is the MARTINI force field (33). The MARTINI model was initially developed for lipid systems (34) and has recently been extended for proteins (35) and carbohydrates (36). The overall aim of the MARTINI coarse-graining approach is to provide a simple model that is computationally fast and easy to use, yet flexible enough to be applicable to a large range of biomolecular systems. On the other hand, because little structural input is taken into account during parametrization, MARTINI can fail to reproduce some of the specific structural characteristics of polymer systems, like spontaneously forming second or higher-order structural rearrangements. However, there are several applications published where the MARTINI force field has been successfully applied to simulate membranes and proteins (37–39).
The aim of this work is to compose a CG MD model of MtCK and ANT interaction on a patch of mitochondrial inner membrane. Here, we describe the model and simulations that predict the physical properties of MtCK and ANT in the membrane and MtCK binding to the membrane. The model is on the basis of the MARTINI coarse-graining approach and simulates a patch of mitochondrial inner membrane, a transmembrane adenine nucleotides translocase in, and a mitochondrial isoform of creatine kinase above the membrane.
MATERIALS AND METHODS
Description of the Modeled System
The model is composed of a membrane, ANT, and MtCK. In all biological membranes, phospholipids are important structural and functional components resulting in spatial separation of the different subcellular compartments. Mitochondrial phospholipids have diverse roles in the regulation of various mitochondrial processes, including apoptosis, electron transport, and protein bindings to membranes (40, 41). Alteration in the content and fatty acid composition of phospholipids within the mitochondria affects many cellular processes, including mitochondrial respiration. Furthermore, various pathological conditions like Barth syndrome are associated with this imbalance (41, 42). Three major types of lipids are found in the mitochondrial inner membrane: phosphatidylcholine (PC), phosphatidylethanolamine (PE), and cardiolipin (CL) in a 2:1:1 molar ratio (43, 44). CL is an unique phospholipid that is almost exclusively localized in the mitochondrial inner membrane where it is synthesized from phosphatidylglycerol and cytidinediphosphate-diacylglycerol (45). It contains three glycerol backbones and four acyl chains. Molar fraction values of CL in the inner mitochondrial membrane vary within the range of 16% to 20% over different species and experimental conditions (46, 47). The CL is asymmetrically distributed between the inner mitochondrial membrane leaflets, showing higher concentration on the matrix side (46). In the mitochondrial membrane, cardiolipin plays an important role in energy metabolism, mainly by providing stability for the individual enzymes and enzyme complexes involved in oxidative phosphorylation (48).
ANT is located in the mitochondrial inner membrane and plays a central role in aerobic cell energetics by moving to the cytosol the ATP molecules generated by oxidative phosphorylation. The ANT consists of about 300 amino acid residues, comprised of three repeat segments of about 100 amino acid residues. The three-dimensional structure of an ANT has been solved by x-ray crystallography at 2.2 Å resolution (49). The size of crystallized ANT is estimated to be approximately 80 × 110 × 90 Å, and a cavity is formed inside ANT with a maximal diameter of 20 Å and a depth of 30 Å. ANT has a basket-like shape formed by six tilted transmembrane segments. Some segments are sharply kinked at the level of proline residues. On the matrix side, three hydrophilic loops contain small amphiphatic helices that are parallel to the membrane surface and tightly surround the carrier. The basket shape opens toward the intermembrane space. During an exchange cycle, ANT undergoes large conformational transitions from a cytosolic open state (c-state), where ADP binds from the cytoplasm, to a matrix open state (m-state), where ATP needs to bind from the mitochondrial matrix (50). The carrier has to recognize the nucleotides, and in contrast to most nucleotide-binding proteins, it only binds adenine nucleotides that are not complexed with magnesium ions.
MtCK (EC 2.7.3.2) is an enzyme that catalyzes the reversible transfer of a phosphoryl group from MgATP to Cr, producing PCr and MgADP. The mitochondrial isoforms generally exist as octamers (51) but can be easily dissociated into dimers (52), i.e. into the basic building block of MtCKs. Dimers are banana-shaped, with the enzymatically active sites of the monomers oriented toward the hollow side. The basis of the dimer/dimer interface is a hydrophobic patch that is strengthened by a number of polar interactions. Dimers assemble into large symmetrical, cube-like octamers with a central channel and have overall dimensions of about 105 × 105 × 86 Å and 422-point group symmetry (51). It exposes two identical faces, top and bottom, perpendicular to their 4-fold axis. MtCK is bound to the outer surface of the inner membrane through electrostatic interactions involving basic residues and negative charges from cardiolipin, which is also associated with the ANT (48). Mostly the octameric form binds rapidly to anionic phospholipids (53). Interaction with the outer membrane may be somewhat weaker because it contains less cardiolipin along with many other anionic phospholipids. Phospholipids with a neutral net charge, like PC or PE, barely interact with MtCK (48).
The Computer Simulation Model
All simulations described in this work were carried out with the GROMACS (GROningen MAchine for Chemical Simulations) molecular dynamics package (version 4.5.4) (54) using the MARTINI coarse-grained force field (FF), version 2.0 (33, 35, 36, 55). This model is on the basis of four-to-one mapping. I.e. on average, four heavy atoms are represented by a single interaction center. As a result, the dynamics is faster because the interactions are much smoother compared with the all-atom type of atomistic interactions. The effective friction caused by the fine-grained degrees of freedom is missing. When interpreting the simulation results, a standard conversion factor of 4 is widely used (34). On the basis of the strategic concern of free energies of hydration and vaporization, the MARTINI model mostly reproduces the correct trends (33). In the MARTINI description of proteins, the secondary structures have to be constructed separately in advance and fixed by the use of a dihedral potential energy function. Processes in which folding and unfolding are playing a substantial role are therefore not suitable for modeling. However, movements of secondary structure elements with respect to each other are possible and were shown to be quite realistic in many applications (56). This work focuses on interactions between proteins and the membrane. A polarized water model was used in all simulations to effectively account for the orientational polarization of water. In the polarized model, a three-bead model is used to represent four water molecules, which is in contrast to the original MARTINI FF. Because of the limitations of the CG model, ion modeling with a MARTINI force field is developed at a qualitative level. More details about the MARTINI CG FF can be found in the literature (33, 35, 36, 55).
All particle pairs, i and j at distance rij interact via a Lennard-Jones potential. The strength of the interaction, determined by the value of the well-depth, ϵij, depends on the types of particles that interact. The well depth ranges from 5.6 kJ/mol for interactions between strongly polar groups to 2.0 kJ/mol for interactions between polar and apolar groups mimicking the hydrophobic effect. The effective size of the particles is governed by the Lennard-Jones parameter, σ = 0.47 nm. In addition to the Lennard-Jones interaction, charged groups (type Q) bearing a charge q interact via a Coulombic energy function with a relative dielectric constant ϵrel = 2.5 (55). The non-bonded Lennard-Jones potential is shifted from rshift = 0.9 nm to 1.2 nm. A real-space cut-off of 1.2 nm and 0.2 nm Fourier grid spacings are used for the particle mesh Ewald type of electrostatic potential. A time step of 20 fs is used, and the neighbor list for non-bonded interactions is updated every 200 fs. These conditions correspond to the standard parameters used in MARTINI FF. All simulation systems are coupled to a Berendsen temperature bath at 309 K (ττ = 0.3 ps), and surface tension is kept constant by using a semi-isotropic Berendsen pressure coupled to a pressure bath at 1 bar (τP = 3 ps) (57). Constant particle number, pressure, and temperature ensembles are simulated assuming periodic boundary conditions.
The simulated bilayers consists of a ternary mixture of palmitoyl-oleoyl-phosphatidylcholine (POPC), palmitoyl-oleoyl-phosphatidylethanolamine (POPE), and diphosphatidyl-sn-glycerol (cardiolipin) lipids in a varying molar ratio. For POPC and POPE lipids, standard MARTINI FF parameters are used (34). Force field parameters for cardiolipin were developed by Dahlberg and Maliniak (58, 59, 60). To set up MARTINI CG FF parameters for general proteins, the two-step protocol is used. First, an atomistic protein structure with all coordinates is converted into a CG model. Then, a suitable MARTINI topology is generated. Both steps are done using publicly available scripts (atom2cg_v2.1.awk and seq2itp.pl) distributed via the MARTINI website. The initial coordinates of the crystallized ANT were taken from the protein data bank (PDB code 2C3E (61)). The crystal structure of human ubiquitous MtCK (51) is used as the MtCK model (PDB code 1QK1). Typically, after a 20-ns membrane relaxation phase, the ANT protein is inserted perpendicularly into the membrane, and then the MtCK protein is positioned on top of the membrane somewhere in the middle of the membrane but not very close to the position of ANT. Then, the simulation box is equilibrated for 50 ns. Finally, the data is collected for about 0.15 μs. The software package VMD (Visual Molecular Dynamics) was used for visualizing molecules (62).
In this work, we consider two types of CG MD models: the large system with simulation box 30 × 30 × 30 nm that contains 2222 phospholipids, 25% of which are CLs, and the small system with simulation box approximately 20 × 20 × 20 nm containing approximately 1230 phospholipids. For small systems, the concentration of CL was variable (0, 4, 8, and 17%). The mitochondrial inner membrane was prepared from a MD simulation of a mixture of randomly mixed MARTINI lipids and polarized water molecule beads. With the twice-higher reference pressure in the direction of the normal to-be-formed membrane, within 5 ns of simulation the mixture spontaneously turned to a flat membrane patch, after which a normal pressure distribution was restored. The size of membranes was roughly the same for all systems. After positioning a pre-equilibrated MtCK protein closely above the membrane (to provide different initial conditions), an additional 20 ns equilibration step was performed. Finally, the ANT transmembrane proteins (three for the large system and one for small systems) were manually inserted into the membrane, with a 20-ns equilibration step following each insertion.
All statistical analysis, including linear regression, analysis of variance, and the calculation of confidence intervals, were performed with SciPy, a set of open source scientific tools for Python.
RESULTS
The Model
A snapshot of the large CG MD model is shown in Fig. 1. The membrane is correctly formed, all hydrophobic tails are inside, and hydrophilic parts are outside, with a thickness of 50 Å. The octameric MtCK is firmly bound to the membrane with its four C-terminal monomers and stays there throughout the 150 ns because of the high CL concentration. Three transmembrane ANTs are shown in different colors, and they all are slightly deformed by the surrounding lipids. Water beads and ions are not shown for clarity. Although all three types of lipids diffused during the simulation from their original place, no apparent grouping was observed, as has been suggested for CL (48). Diffusion coefficients can be estimated from the mean square displacement relation of the Einstein model of Brownian motion. The estimated lateral diffusion coefficients are DCL ≈ 3 × 10−7 cm2/s for cardiolipin and DANT ≈ 6 × 10−8 cm2/s for ANT proteins. The estimates were obtained from averaging diffusion coefficients using simulations of the large system and the two smaller systems with 4 and 17% CL, respectively. The overall density averaged over all systems was approximately 1090 kg/m3.
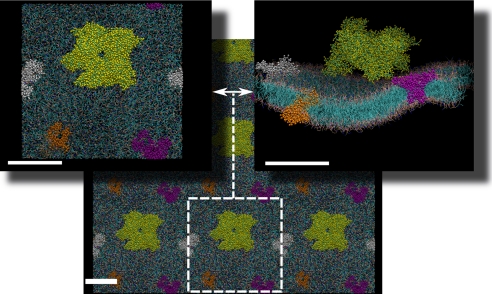
FIGURE 1.
Overall top and side views of a fragment of an inner mitochondrial membrane with one attached MtCK protein (yellow balls and sticks) and three transmembrane ANT proteins (white, purple, and orange). The lower part of the figure shows a larger fragment of the membrane by taking into account the periodicity of the simulation box along the x and y directions. A single simulation box is shown by the white square with dimensions of approximately 300 × 300 Å. The thickness of the membrane is approximately 50 Å. Scale bars = 100 Å.
A snapshot of the small model with 4% CL is shown in Fig. 2. A–C are side views, and D–F are top views. The figure illustrates the roughness and distribution of different types of chemical groups. At shown time moment, the MtCK is bound to the membrane at three locations. In Fig. 2, C and E, the MtCK is cut just above the membrane, and all of its visible residues are in the middle of lipid atoms inside the membrane. Note that one of the binding monomer fragments has penetrated more deeply into the membrane than others. Fig. 2F shows a close view of a binding pocket where charged beads of POPC and POPE have surrounded the part of MtCK, illustrating how lipids can easily reorganize when the environment changes.
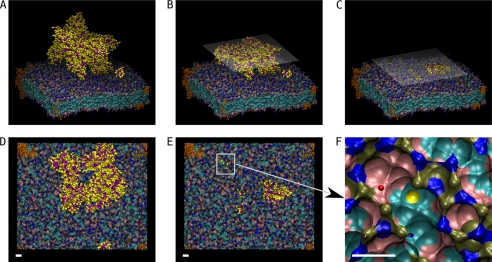
FIGURE 2.
MtCK bound to the membrane by three (of four) C-terminal domains. The fourth C-terminal is above the membrane (not seen in the three-dimensional panels) at this moment. All six subplots show the state of the system in different details, A–C as side views and D–F as top views. The size of the simulation box is 185 × 227 × 191 Å. Because of the periodic boundary condition, one trans-membrane ANT is shared with all four membrane corners, shown in orange. The membrane is shown using isosurface representation. Blue is used for nitrogen groups, brown for phosphate, pink for glycerol groups, and cyan for the rest of carbon atoms in the lipids. A, overall view of a single octameric MtCK protein that is strongly bound to the membrane. Amino acid backbones are shown in purple and side chains in yellow only. B and D, cross-section of the MtCK perpendicular to the normal membrane at 40 Å shown from the side (B) and from the top (D). The half-transparent plane visualizes the cutting face. C and E, a similar cross-section cut by the plane perpendicular to the normal membrane at 7 Å and shown from the side (C) and the top (E). F, enlarged view of the location of the upper left MtCK corner bound to the membrane lipids. The yellow sphere is the closest protein side chain to POPC and POPE. The red sphere represents one Na ion. Scale bar = 10 Å.
The Binding Effect of Cardiolipin
The CL is an important component of the mitochondrial inner membrane. The presence of CL influences protein bindings, including all respiration chain proteins. The measured concentration of CL in the mitochondrial inner membrane varies from 16 to 20% (46, 47). To test the new CG MD model and to show details of the important protein-membrane binding phenomenon, we prepared 12 small systems (three times 0, 4, 8, and 17% of CL in the mixture of POPC and POPE). In the following, we analyzed the dependence of the average binding time for each of the MtCK monomers in the octamer as a function of CL concentration. With the small systems we were able to observe the separation of MtCK during the simulations.
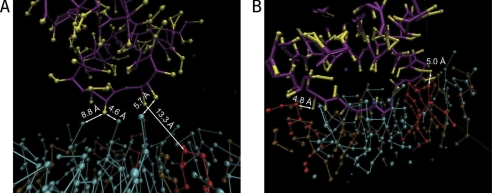
Fig. 3 illustrates the weak and strong binding of the MtCK protein chain and membrane lipids. Fig. 3A shows a moment when nitrogen groups (NC3) of the three POPC lipids (in cyan) is under the strong influence of the monomer of MtCK before detachment. Within the next 20 ps, the monomer moved upward, and the slightly elevated nitrogen groups were dragged back to the average level, increasing the shown distances considerably. In Fig. 3B, the strong binding of MtCK protein chain is illustrated. A fragment of an amino acid chain is deeply penetrated between lipids, and the three CLs are in direct contact with charged residues at distances of approximately 5 Å. This is in contrast with the weakly bound case (Fig. 3A) where the closest CL (in red) is 13.3 Å apart from MtCK. In the strongly bound case of Fig. 3B, the monomer did not detach from the membrane during the simulation, as described below. The distance of 5 Å between MtCK and CL, as in Fig. 3B, is close to the potential energy minimum of non-bonded interactions, leading to stable binding between MtCK and the membrane.
FIGURE 3.
Two different situations of protein chain bindings to membrane lipids. Red, cyan, and brown balls and sticks represent atoms of cardiolipin, POPC, and POPE lipids, respectively. Two monomers of MtCK are shown, one at the time of separation from the membrane (A) and the second corresponding to the residues that remained attached to the membrane throughout the simulation (B). Note that a short fragment of MtCK has penetrated into the lipids in B. Distances to the closest nitrogen groups of POPC and to head groups of cardiolipin are indicated.
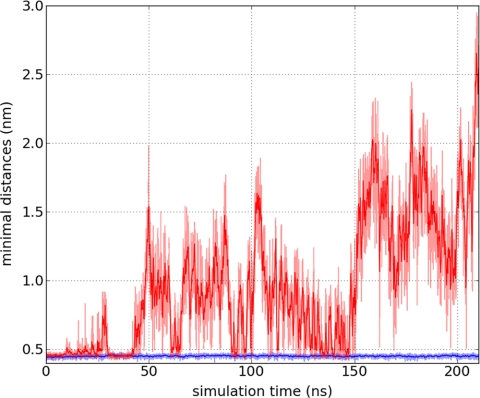
We followed the distance between MtCK monomers and membrane throughout the simulations. Here, the distance is defined as a closest distance between atom groups (a bead in Fig. 3) from MtCK and any lipid in the membrane. As a typical example of the simulations, the distances between the membrane and two monomers are shown in Fig. 4. The first 70 ns is an equilibration time range. In the figure, the weak and strong binding cases are shown in red and blue, respectively. Our results demonstrate that the separation between MtCK and the membrane does not happen instantaneously but through distance oscillations from lower to higher amplitude. The figure shows that in the time range of 70–150 ns, the distance measure fluctuated at some low level, and around the 150 ns time moment, the fluctuations switched to a higher level. We could frequently divide the simulations into several parts according to the average oscillation levels.
FIGURE 4.
Calculated minimal distances between membrane lipids and amino acids of a MtCK protein for weakly (red graph) and strongly bound (blue line) cases during the simulation. At 150 ns, the weakly bound residues are rapidly separating from the membrane. One of the last moments prior to unbinding is shown in Fig. 3A. The strongly bound case is illustrated in Fig. 3B. For clarity, simulated data points (half-transparent lines) are shown with a 10-step moving average (solid lines).
To analyze the duration of the binding between MtCK and the membrane, we introduced the maximal distance between MtCK and the membrane at which MtCK is considered as bound. In our simulations, this distance was taken equal to 10 Å. The binding time for one corner of MtCK is calculated from the moment when at least one residue from the binding corner is closer than 10 Å to the moment when the distances of all residue have exceeded the 10 Å limit. Clearly, at a distance of 10 Å, the MtCK and membrane still interact, but the interaction is considerably weaker. In small systems, because of the use of periodic boundary conditions, some unbound corner monomer of MtCK may approach the membrane from the other side, interact with it, and even bind to it, leading to considerable deformation of MtCK. To exclude this artificial influence from the binding analysis and focus on membrane-MtCK interaction, two additional restrictive limits were used. In the relative binding time calculations, we considered only those moments at which any of the MtCK upper corners were sufficiently far away from the other side of the membrane (distance larger than 20 Å). In addition, to determine the influence of interactions between MtCK and the membrane with regards to binding, we considered only moments at which the distance between the membrane and the corresponding corner of MtCK were closer than 25 Å. This filters out the parts of simulations where MtCK corners are too far from the membrane, as in the case of free diffusion of MtCK.
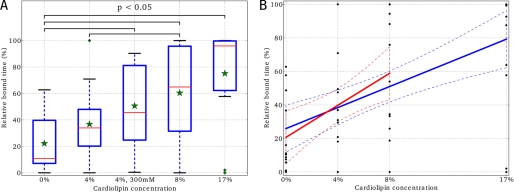
The results of the binding time analysis are summarized in Fig. 5. Fig. 5 shows the dependence of relative bound time to CL concentration. The relative bound time of a MtCK corner monomer is defined as a percentage of binding duration to the time when the conditions described above are satisfied for the particular corner. For example, when one of the MtCK monomers is bound throughout the simulation, as corresponding to the blue trace in Fig. 4, the relative bound time is 100%. When the monomer binds and detaches several times during the simulation, the relative bound time will be smaller. In Fig. 5A, the green stars correspond to averaged relative bound times taken over all four membrane-sided corners of MtCK and the three independent simulations. Clearly, the relative bound time increases with the increase of CL concentration. The results of analysis of variance-based statistical analysis are shown with black clamps that indicate when the average relative bound times of two systems are different at a confidence level of p < 0.05. Clearly, the relative bound time is very small in the absence of CL, in contrast to the case with 17% CL. This demonstrates the importance of CL in the stabilization of MtCK binding to the membrane.
FIGURE 5.
Influence of cardiolipin on MtCK binding to the membrane. Here, relative bound time for MtCK is shown for simulations with a different amount of cardiolipin in the membranes. As explained in the text, the MtCK monomer was considered to be bound to the membrane if it was closer than 10 Å from the membrane. Two different statistical methods both show (A and B) that the increase of cardiolipin content in the membrane results in stronger binding of MtCK. A, on the basis of analysis of variance and Fisher's least significant difference (LSD) statistical methods (71), it can be concluded that at a p < 0.05 confidence level the average relative bound times (showed as green stars) are mostly different and that cardiolipin content is at least one of the factors influencing MtCK binding to the membrane. Here, bound times are graphically summarized as box plots for every system. Because of the relatively short simulation time, there are some data variances and some outliers (green diamonds). The bottom and top of the blue rectangles are the lower and upper quartiles (the 25th and 75th percentile, respectively). The red lines are the 50th percentile, i.e. the medians. The ends of the whiskers are mostly the minimum and maximum of all the data in a system. A green diamond in the system of 4% is an outlier. I.e. the data beyond the 1.5 interquartile range of the upper quartile. In the simulated membranes, the concentration of cardiolipin was changed from 0% up to a molar ratio of 17%. The simulated system (4%, 300 mm) refers to the simulations with elevated NaCl salt concentration. B, the relationship between CL content and the relative bound time was analyzed by linear regression for all simulations, except the elevated NaCl salt case. Two linear regression lines (solid lines) together with their 95% confidence intervals (dashed lines) are calculated over the entire set of data points (blue) and over smaller subset 0, 4, and 8% CL (red).
In addition to comparing the binding times between simulations, we performed a linear regression analysis of the relationship between the binding time and CL concentration (Fig. 5B). We excluded the high salt concentration case from the analysis to focus on the role of CL. Because the larger separation step between 8 and 17% of CL can bias the regression analysis, the results were calculated both over the CL concentration subrange (0 to 8%) and full range (0 to 17%). As both fits demonstrate (r2 equal to 0.23 and 0.29 for the subrange and full range, respectively), higher CL concentrations resulted in longer binding durations. The slope of both regression lines, 4.8 and 3.1, were found to be significantly different from zero (the corresponding p values are 3 × 10−3 and 9 × 10−5, respectively). The 95% confidence interval of the slope, calculated over the full range of CL concentrations, is 3.1 ± 1.5.
High Salt Concentration Effect
It has been reported that the binding of MtCK to the mitochondrial inner membrane is reduced with high salt concentration (63, 64). Already, 100 mm of KCl solution affects the ionic composition of a medium that is suggested to be important for solubility of MtCK (63). In our CG-MD models, ion modeling was applied only at a qualitative level because of principal limitations of MARTINI FF. Because MARTINI FF provides parameters only for Na+ ions and by itself does not differentiate in between K+ and Na+ ions, we used Na+ ions in our simulations as representatives of salt ions. In an attempt to observe a salt effect, we chose the highest salt strength value available from the literature (64): 300 mm. The small system with 4% of CL was modified with an elevated salt concentration. However, as Fig. 5A shows, we could not distinguish between the two systems with normal and elevated physiological conditions at a confidence level of p < 0.05. This could be because of the limitation of MARTINI FF in modeling ions in general or because of the limited simulation time we used.
DISCUSSION
In this work, we composed a molecular dynamics model of MtCK and ANT interaction on a patch of mitochondrial inner membrane. To our knowledge, this is the first computer model to simulate MtCK binding to the membrane and movement of MtCK and ANT in the membrane. In addition to estimating diffusion coefficients for ANT and CL, we demonstrated the importance of CL-MtCK interaction to the binding of MtCK to the mitochondrial membrane. The simulations also predicted the sequence of MtCK residues interacting with the membrane during the binding.
To compare the simulation results with experimental data, we estimated the diffusion coefficients for CL and ANT within the membrane patch. Unfortunately, to our knowledge, the amount of experimental data available for lateral diffusion coefficients of ANT and cardiolipin is rather limited. Gupte et al. (65) reported a lateral diffusion coefficient of ANT in the isolated inner membranes of rat liver mitochondria with a diffusion coefficient of DANT = 1.7 × 10−9 cm2/s at 273 K. The temperature coefficient, Q10, is also unknown for ANT in mitochondrial membrane. We used the Q10 determined for a lateral diffusion of rhodopsin: Q10 ≈ 1.7 (66). Using this temperature coefficient, the diffusion coefficient of ANT is DANT ≈ 1.1 × 10−8 cm2/s at 309 K, the temperature used in the simulations. Considering the rough speed-up factor of 4 in MARTINI CG MD simulations (34), the calculated diffusion coefficient for ANT in the mitochondrial inner membrane is estimated to be DANT–calc = 6.0 × 10−8/4 = 1.5 × 10−8 cm2/s, which is in excellent agreement with the estimate on the basis of the experimental value of Gupte et al. (65). Of course, the speed-up factor 4 varies in different simulated systems, and to map the dynamics more accurately between CG and AA, a long AA MD type of simulation is necessary.
Two different binding situations were introduced in this study in detail, and are also shown in Fig. 3. The crystallographic study has shown that one cubic octameric MtCK enzyme consists of eight monomers and 379 residues in each (51). It has been suggested that this binding is stabilized by electrostatic interactions between anionic lipids, CL in this case, and several C-terminal patches of basic residues (Lys, Arg, or His, depending on the MtCK isoform) at the end of monomers (44, 48). It is also known that phospholipids with a neutral net charge like PC or PE barely interact with the kinases. Figs. 3 and 5 are in agreement with these views, as the monomer of MtCK mainly interacts with CL. According to our simulations in both the weak and strongly bound cases (Fig. 3, A and B), corners of one MtCK are bound to the membrane lipids by the end of their C-terminal monomers. Looking at the exact sequence of one MtCK monomer in the PDB (PDB code 1QK1), for example, the residues that form cardiolipin anchors are typically in the range of Arg-360 to His-379. The residues that strongly interact with CL are mostly Arg (Arg-369, Arg-364, Arg-361) and, in some cases, Lys-378. Often, residues from Gln-366 up to the end form, approximately, a line segment slightly bent toward the membrane, as seen in Fig. 3B. The last turn at the end of the monomer, consisting of Glu-363, Arg-364, and Gly-365, finishes the straight part and turns into the α-helix, which interacts with CL less frequently. It is interesting that the last residue, His-379, does not interact with CL often. Rather, it is pointed up toward MtCK. In the weakly bound case (Fig. 3A), the monomer that interacts with lipids includes mostly the last turn range, i.e. from Arg-361 to Ile-368.
Model Simplifications
When composing the CG MD model, we had to make several simplifications to reduce the computational demands. The main simplification was to reduce the size of the simulation box as much as possible. The simulations presented in this work took several months on the available computer cluster.
On the basis of experimental results, it is known that MtCK is localized in both the peripheral intermembrane space and the cristae space (67). With a height of about 9 nm, the MtCK octamer is found to be bound on both sides by the inner and outer membrane in the intermembrane space, although generally there is more space between cristae membranes and MtCKs. MtCK can be bound either to both sides or only one side. In this work, the model of MtCK is bound to only one side of the phospholipid membrane for two reasons. It is computationally less expensive to develop and test the main components of the model, and secondly, the coupling between MtCK and ANT has been reported on mitoplasts that lack a mitochondrial outer membrane (68), suggesting that the binding of MtCK does not critically depend on the existence of the outer membrane.
Because of a relatively small simulation box, we observed in some simulations that the MtCK octamer gets attracted by monomers on the “top” (Fig. 1) to the other side of the membrane. We used periodic boundary conditions in the simulation of the membrane, ANT, and MtCK. Thus, because of periodicity, the top of the simulation box is connected to the bottom. As a result, when MtCK moves too far away from the membrane, the upper part of the octamer could be influenced by the other side of the membrane. In simulations that were exposed to this influence, we observed that the MtCK octamer was dragged to a diagonal position between two sides of the same membrane and often gets deformed during this process. In our analysis, we discarded the moments when MtCK octamer upper corners were too close to the bottom side of the membrane. One way to avoid this effect is to increase the simulation box size. However, that approach leads to a rapid increase in computational demand. Another solution is to use elastic networks around proteins. The elastic network conserves tertiary and quartenary structures more faithfully without sacrificing realistic dynamics of proteins. Although the use of elastic networks together with MARTINI FF is still an active field of research, there are a number of realistic simulations published (69, 70).
As shown in Fig. 4, the oscillation with the larger distances near the end of the simulations is a common phenomenon in many simulated systems. This is a strong argument to extend the duration of simulations in the future to study the interaction between MtCK and ANT. However, the duration of the current simulations was long enough to see the influence of CL to MtCK binding. Using the newly developed model in a somewhat longer simulation, it might be possible to observe other important phenomena, such as the induced cluster formation of CL close to MtCK binding pockets (43), and coupling of two ANTs into a single working unit through CL interaction (61), and reveal details of these mechanisms. Also, the relatively short simulation duration could be the reason why high salt concentrations were not observed to affect the binding to anionic phospholipids.
In conclusion, we developed a CG MD model that is a part of the creatine kinase shuttle, including mitochondrial isoform MtCKs, transmembrane ANTs, and cardiolipin in the membrane as the important factor for coupling these proteins. The model is validated against many structural and dynamical experimental properties and can be used as the base model for further developments.
Acknowledgments
We thank David W. Schryer (Institute of Cybernetics at Tallinn University of Technology) for help with the text.
This work was supported by Wellcome Trust Fellowship WT081755 (to M. V. and P. P.) and Estonian Science Foundation MOBILITAS Postdoctoral Research Grant MJD80.
- PCr
- phosphocreatine
- MtCK
- mitochondrial creatine kinase
- ANT
- adenine nucleotide translocase
- MD
- molecular dynamics
- CG
- coarse-grained
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- CL
- cardiolipin
- FF
- force field
- POPC
- palmitoyl-oleoyl-phosphatidylcholine
- POPE
- palmitoyl-oleoyl-phosphatidylethanolamine.
REFERENCES
- 1. Klingenberg M. (2010) Wanderings in bioenergetics and biomembranes. Biochim. Biophys. Acta 1797, 579–594 [DOI] [PubMed] [Google Scholar]
- 2. Scheffler I. (2007) Mitochondria, 2nd Ed., J. Wiley and Sons, Inc., Hoboken, NJ [Google Scholar]
- 3. Vendelin M., Kongas O., Saks V. (2000) Regulation of mitochondrial respiration in heart cells analyzed by reaction-diffusion model of energy transfer. Am. J. Physiol. Cell Physiol. 278, C747–764 [DOI] [PubMed] [Google Scholar]
- 4. Saks V. A., Kongas O., Vendelin M., Kay L. (2000) Role of the creatine/phosphocreatine system in the regulation of mitochondrial respiration. Acta Physiol. Scand. 168, 635–641 [DOI] [PubMed] [Google Scholar]
- 5. Wu F., Jeneson J. L., Beard D. (2007) Oxidative ATP synthesis in skeletal muscle is controlled by substrate feedback. Am. J. Physiol. Cell. Physiol. 292, C115–124 [DOI] [PubMed] [Google Scholar]
- 6. Wu F., Zhang J., Beard D. (2009) Experimentally observed phenomena on cardiac energetics in heart failure emerge from simulations of cardiac metabolism. Proc. Natl. Acad. Sci. 106, 7143–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu F., Zhang E. Y., Zhang J., Bache R. J., Beard D. A. (2008) Phosphate metabolite concentrations and ATP hydrolysis potential in normal and ischaemic hearts. J. Physiol. 586, 4193–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vendelin M., Hoerter J. A., Mateo P., Soboll S., Gillet B., Mazet J. L. (2010) Modulation of energy transfer pathways between mitochondria and myofibrils by changes in performance of perfused heart. J. Biol. Chem. 285, 37240–37250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bessman S. P., Geiger P. J. (1981) Transport of energy in muscle. The phosphorylcreatine shuttle. Science 211, 448–452 [DOI] [PubMed] [Google Scholar]
- 10. Wallimann T., Wyss M., Brdiczka D., Nicolay K., Eppenberger H. (1992) Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands. The “phosphocreatine circuit” for cellular energy homeostasis. Biochem. J. 281, 21–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dzeja P. P., Terzic A. (2003) Phosphotransfer networks and cellular energetics. J. Exp. Biol. 206, 2039–2047 [DOI] [PubMed] [Google Scholar]
- 12. Kaasik A., Veksler V., Boehm E., Novotova M., Minajeva A., Ventura-Clapier R. (2001) Energetic crosstalk between organelles. Architectural integration of energy production and utilization. Circ. Res. 89, 153–159 [DOI] [PubMed] [Google Scholar]
- 13. Abraham M. R., Selivanov V. A., Hodgson D. M., Pucar D., Zingman L. V., Wieringa B., Dzeja P. P., Alekseev A. E., Terzic A. (2002) Coupling of cell energetics with membrane metabolic sensing. Integrative signaling through creatine kinase phosphotransfer disrupted by M-CK gene knock-out. J. Biol. Chem. 277, 24427–24434 [DOI] [PubMed] [Google Scholar]
- 14. Vendelin M., Birkedal R. (2008) Anisotropic diffusion of fluorescently labeled ATP in rat cardiomyocytes determined by raster image correlation spectroscopy. Am. J. Physiol. Cell Physiol. 295, C1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sokolova N., Vendelin M., Birkedal R. (2009) Intracellular diffusion restrictions in isolated cardiomyocytes from rainbow trout. BMC Cell Biol. 10, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramay H. R., Vendelin M. (2009) Diffusion restrictions surrounding mitochondria. A mathematical model of heart muscle fibers. Biophys. J. 97, 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sepp M., Vendelin M., Vija H., Birkedal R. (2010) ADP compartmentation analysis reveals coupling between pyruvate kinase and ATPases in heart muscle. Biophys. J. 98, 2785–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jepihhina N., Beraud N., Sepp M., Birkedal R., Vendelin M. (2011) Permeabilized rat cardiomyocyte response demonstrates intracellular origin of diffusion obstacles. Biophys. J. 101, 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacobus W. E., Lehninger A. L. (1973) Creatine kinase of rat heart mitochondria. Coupling of creatine phosphorylation to electron transport. J. Biol. Chem. 248, 4803–4810 [PubMed] [Google Scholar]
- 20. Saks V. A., Chernousova G. B., Gukovsky D. E., Smirnov V. N., Chazov E. I. (1975) Studies of energy transport in heart cells. Mitochondrial isoenzyme of creatine phosphokinase. Kinetic properties and regulatory action of Mg2+ ions. Eur. J. Biochem. 57, 273–290 [DOI] [PubMed] [Google Scholar]
- 21. Barbour R. L., Ribaudo J., Chan S. H. (1984) Effect of creatine kinase activity on mitochondrial ADP/ATP transport. Evidence for a functional interaction. J. Biol. Chem. 259, 8246–8251 [PubMed] [Google Scholar]
- 22. Saks V. A., Kuznetsov A. V., Vendelin M., Guerrero K., Kay L., Seppet E. K. (2004) Mol. Cell. Biochem. 256–257, 185–199 [DOI] [PubMed] [Google Scholar]
- 23. Vendelin M., Lemba M., Saks V. (2004) Analysis of functional coupling: mitochondrial creatine kinase and adenine nucleotide translocase. Biophys. J. 87, 696–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hettling H., van Beek J. (2011) Analyzing the functional properties of the creatine kinase system with multiscale “sloppy” modeling. PLoS Comput. Biol. 7, e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vendelin M., Mateo P., Soboll S., Gillet B., Mazet J., Hoerter J. (2010) J. Biol. Chem. 285, le22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khalili-Araghi F., Gumbart J., Wen P. C., Sotomayor M., Tajkhorshid E., Schulten K. (2009) Molecular dynamics simulations of membrane channels and transporters. Curr. Opin. Struct. Biol. 19, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durrant J. D., McCammon J. A. (2011) Molecular dynamics simulations and drug discovery. BMC Biol. 9, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levitt M., Warshel A. (1975) Computer simulation of protein folding. Nature 253, 694–698 [DOI] [PubMed] [Google Scholar]
- 29. Voth G. (2008) Coarse-graining of Condensed Phase and Biomolecular Systems, pp. 1–40, CRC Press, Boca Raton, FL [Google Scholar]
- 30. Milano G., Müller-Plathe F. (2005) Mapping atomistic simulations to mesoscopic models. A systematic coarse-graining procedure for vinyl polymer chains. J. Phys. Chem. B 109, 18609–18619 [DOI] [PubMed] [Google Scholar]
- 31. Carbone P., Varzaneh H. A., Chen X., Müller-Plathe F. (2008) Transferability of coarse-grained force fields. The polymer case. J. Chem. Phys. 128, 064904 [DOI] [PubMed] [Google Scholar]
- 32. Sun Q., Faller R. (2005) Comput. Chem. Eng. 29, 2380–2385 [Google Scholar]
- 33. Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., de Vries A. H. (2007) The MARTINI force field. Coarse-grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 [DOI] [PubMed] [Google Scholar]
- 34. Marrink S., de Vries A., Mark A. (2004) J. Phys. Chem. B 108, 750–760 [Google Scholar]
- 35. Monticelli L., Kandasamy S., Periole X., Larson R., Tieleman D. P., Marrink S. (2008) J. Chem. Theory Comput. 4, 819–834 [DOI] [PubMed] [Google Scholar]
- 36. López C., Rzepiela A., de Vries A., Dijkhuizen L., Hünenberger P., Marrink S. (2009) J. Chem. Theory Comput. 5, 3195–3210 [DOI] [PubMed] [Google Scholar]
- 37. Song B., Yuan H., Jameson C., Murad S. (2011) Mol. Phys. 109, 1511–1526 [Google Scholar]
- 38. Samuli Ollila O. H., Louhivuori M., Marrink S. J., Vattulainen I. (2011) Protein shape change has a major effect on the gating energy of a mechanosensitive channel. Biophys. J. 100, 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murtola T., Vuorela T., Hyvönen M., Marrink S., Karttunen M., Vattulainen I. (2011) Soft Matter 7, 8135–8141 [Google Scholar]
- 40. Palsdottir H., Hunte C. (2004) Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 [DOI] [PubMed] [Google Scholar]
- 41. Paradies G., Petrosillo G., Paradies V., Ruggiero F. (2010) Oxidative stress, mitochondrial bioenergetics, and cardiolipin in aging. Free Radic. Biol. Med. 48, 1286–1295 [DOI] [PubMed] [Google Scholar]
- 42. Haines T. (2009) A new look at cardiolipin. Biochim. Biophys. Acta 1788, 1997–2002 [DOI] [PubMed] [Google Scholar]
- 43. Maniti O., Lecompte M. F., Marcillat O., Desbat B., Buchet R., Vial C., Granjon T. (2009) Mitochondrial creatine kinase binding to phospholipid monolayers induces cardiolipin segregation. Biophys. J. 96, 2428–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schlattner U., Gehring F., Vernoux N., Tokarska-Schlattner M., Neumann D., Marcillat O., Vial C., Wallimann T. (2004) C-terminal lysines determine phospholipid interaction of sarcomeric mitochondrial creatine kinase. J. Biol. Chem. 279, 24334–24342 [DOI] [PubMed] [Google Scholar]
- 45. Houtkooper R. H., Vaz F. M. (2008) Cardiolipin, the heart of mitochondrial metabolism. Cell. Mol. Life Sci. 65, 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krebs J. J., Hauser H., Carafoli E. (1979) Asymmetric distribution of phospholipids in the inner membrane of beef heart mitochondria. J. Biol. Chem. 254, 5308–5316 [PubMed] [Google Scholar]
- 47. Hoch F. (1992) Cardiolipins and biomembrane function. Biochim. Biophys. Acta 1113, 71–133 [DOI] [PubMed] [Google Scholar]
- 48. Schlattner U., Tokarska-Schlattner M., Ramirez S., Brückner A., Kay L., Polge C., Epand R. F., Lee R. M., Lacombe M. L., Epand R. M. (2009) Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim. Biophys. Acta 1788, 2032–2047 [DOI] [PubMed] [Google Scholar]
- 49. Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G. J., Brandolin G. (2003) Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 [DOI] [PubMed] [Google Scholar]
- 50. Robinson A. J., Overy C., Kunji E. R. (2008) The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. 105, 17766–17771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eder M., Fritz-Wolf K., Kabsch W., Wallimann T., Schlattner U. (2000) Crystal structure of human ubiquitous mitochondrial creatine kinase. Proteins 39, 216–225 [DOI] [PubMed] [Google Scholar]
- 52. Wyss M., Smeitink J., Wevers R. A., Wallimann T. (1992) Mitochondrial creatine kinase: a key enzyme of aerobic energy metabolism. Biochim. Biophys. Acta 1102, 119–166 [DOI] [PubMed] [Google Scholar]
- 53. Rojo M., Hovius R., Demel R., Wallimann T., Eppenberger H. M., Nicolay K. (1991) Interaction of mitochondrial creatine kinase with model membranes. A monolayer study. FEBS Lett. 281, 123–129 [DOI] [PubMed] [Google Scholar]
- 54. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 55. Yesylevskyy S. O., Schäfer L. V., Sengupta D., Marrink S. J. (2010) Polarizable water model for the coarse-grained MARTINI force field. PLoS Comput. Biol. 6, e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yefimov S., van der Giessen E., Onck P. R., Marrink S. J. (2008) Mechanosensitive membrane channels in action. Biophys. J. 94, 2994–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Dinola A., Haak J. R. (1984) J. Chem. Phys. 81, 3684 [Google Scholar]
- 58. Dahlberg M., Maliniak A. (2010) J. Chem. Theory Comput. 6, 1638–1649 [DOI] [PubMed] [Google Scholar]
- 59. Dahlberg M., Maliniak A. (2008) Molecular dynamics simulations of cardiolipin bilayers. J. Phys. Chem. B 112, 11655–11663 [DOI] [PubMed] [Google Scholar]
- 60. Dahlberg M. (2007) Polymorphic phase behavior of cardiolipin derivatives studied by coarse-grained molecular dynamics. J. Phys. Chem. B 111, 7194–7200 [DOI] [PubMed] [Google Scholar]
- 61. Nury H., Dahout-Gonzalez C., Trézéguet V., Lauquin G., Brandolin G., Pebay-Peyroula E. (2005) Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 579, 6031–6036 [DOI] [PubMed] [Google Scholar]
- 62. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 63. Saks V. A., Khuchua Z. A., Kuznetsov A. V., Veksler V. I., Sharov V. G. (1986) Heart mitochondria in physiological salt solution. Not ionic strength but salt composition is important for association of creatine kinase with the inner membrane surface. Biochem. Biophys. Res. Commun. 139, 1262–1271 [DOI] [PubMed] [Google Scholar]
- 64. Lipskaya T. Y. (2001) Biochem. 66, 1098–1111 [DOI] [PubMed] [Google Scholar]
- 65. Gupte S. S., Chazotte B., Leesnitzer M. A., Hackenbrock C. R. (1991) Two-dimensional diffusion of F1F0-ATP synthase and ADP/ATP translocator. Testing a hypothesis for ATP synthesis in the mitochondrial inner membrane. Biochim. Biophys. Acta 1069, 131–138 [DOI] [PubMed] [Google Scholar]
- 66. Wey C. L., Cone R. A., Edidin M. A. (1981) Lateral diffusion of rhodopsin in photoreceptor cells measured by fluorescence photobleaching and recovery. Biophys. J. 33, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schlattner U., Tokarska-Schlattner M., Wallimann T. (2006) Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta 1762, 164–180 [DOI] [PubMed] [Google Scholar]
- 68. Saks V. A., Kuznetsov A. V., Kupriyanov V. V., Miceli M. V., Jacobus W. E. (1985) Creatine kinase of rat heart mitochondria. The demonstration of functional coupling to oxidative phosphorylation in an inner membrane-matrix preparation. J. Biol. Chem. 260, 7757–7764 [PubMed] [Google Scholar]
- 69. Wohlert J., Berglund L. (2011) J. Chem. Theory Comput. 7, 753–760 [Google Scholar]
- 70. Wee C. L., Gavaghan D., Sansom M. S. (2010) Interactions between a voltage sensor and a toxin via multiscale simulations. Biophys. J. 98, 1558–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Montgomery D., Runger G. (2010) Applied Statistics and Probability for Engineers, pp. 524–526, Wiley, New York [Google Scholar]