Background: De novo thymidylate biosynthesis occurs in the nucleus.
Results: This pathway forms a multienzyme complex that is associated with the nuclear lamina and the DNA replication machinery.
Conclusion: De novo thymidylate biosynthesis occurs at the sites of DNA replication and is enriched at sites of replication initiation.
Significance: The compartmentation of thymidylate biosynthesis at the replication fork may permit regulation of dUTP incorporation into DNA.
Keywords: DNA Replication, DNA Synthesis, Folate Metabolism, Nucleoside Nucleotide Metabolism, Protein-DNA Interaction, DHFR, SHMT1, TYMS, Lamin, Thymidylate, SHMT, DNA Replication, Multienzyme Complex
Abstract
The de novo thymidylate biosynthetic pathway in mammalian cells translocates to the nucleus for DNA replication and repair and consists of the enzymes serine hydroxymethyltransferase 1 and 2α (SHMT1 and SHMT2α), thymidylate synthase, and dihydrofolate reductase. In this study, we demonstrate that this pathway forms a multienzyme complex that is associated with the nuclear lamina. SHMT1 or SHMT2α is required for co-localization of dihydrofolate reductase, SHMT, and thymidylate synthase to the nuclear lamina, indicating that SHMT serves as scaffold protein that is essential for complex formation. The metabolic complex is enriched at sites of DNA replication initiation and associated with proliferating cell nuclear antigen and other components of the DNA replication machinery. These data provide a mechanism for previous studies demonstrating that SHMT expression is rate-limiting for de novo thymidylate synthesis and indicate that de novo thymidylate biosynthesis occurs at replication forks.
Introduction
Regulation of cellular dTTP synthesis is essential for DNA replication and genome stability in the nucleus (1) and mitochondria (2). Depressed de novo thymidylate synthesis resulting from folate deficiency or anti-folate treatment results in deoxyuridine misincorporation into mitochondrial DNA (2) and nuclear DNA leading to genome instability (3). Tetrahydrofolate (THF)2 is a metabolic cofactor that carries and activates single carbons for the synthesis of purine and thymidine nucleotides and for homocysteine remethylation to methionine (4). Folate-mediated one-carbon metabolism is compartmentalized in the mitochondria, nucleus, and cytoplasm of eukaryotic cells (5). The enzymes that constitute the de novo thymidylate pathway include SHMT1, SHMT2α, thymidylate synthase (TYMS), and dihydrofolate reductase (DHFR). Methylene-THF generated by SHMT is the one-carbon donor for the TYMS-catalyzed conversion of dUMP to thymidylate, generating dihydrofolate. DHFR catalyzes the NADPH-dependent reduction of dihydrofolate to regenerate THF for subsequent cycles of de novo thymidylate synthesis (Fig. 1). SHMT1, TYMS, and DHFR have been localized to the nucleus, and their translocation is mediated by post-translational modification with the small ubiquitin-like modifier (SUMO) (6, 7). SHMT1 nuclear translocation is cell cycle-dependent and occurs during the S and G2/M phases and in response to UV damage (7–9). In mice, nuclear localization of the de novo thymidylate synthesis pathway is required to minimize uracil misincorporation into nuclear DNA (10). Intact purified nuclei from mouse liver exhibit de novo thymidylate synthesis activity, whereas nuclei disrupted by sonication lack this activity, indicating that the formation of a multienzyme complex may be required for the pathway to function (6).
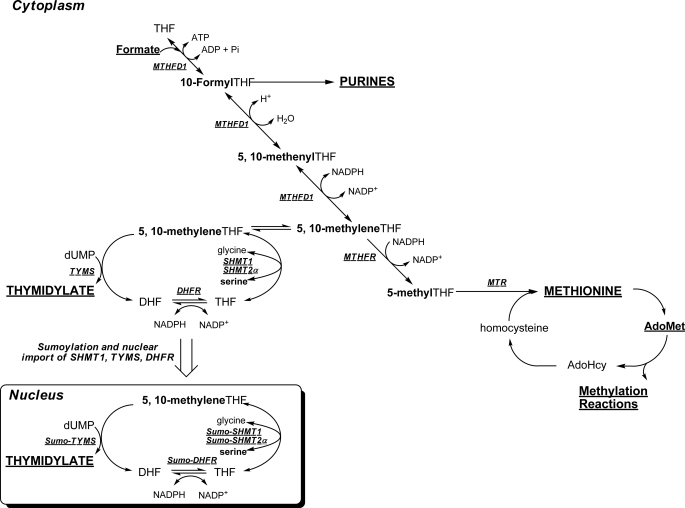
FIGURE 1.
Schematic of folate-mediated one-carbon metabolism in the cytoplasm and nucleus. One-carbon metabolism is required for the de novo synthesis of purines and thymidylate and for the remethylation of homocysteine to methionine. Folate-activated one-carbon units are shown in boldface type. The de novo thymidylate pathway is sumoylated and translocates to the nucleus during S phase. MTHFD1, methylenetetrahydrofolate dehydrogenase; MTR, methionine synthase; MTHFR, methylenetetrahydrofolate reductase; DHF, dihydrofolate.
Previous studies in cell culture and mouse models have shown that SHMT1 expression determines de novo thymidylate synthesis activity, indicating that this enzyme is limiting for de novo thymidylate synthesis in vivo (11, 12). In mammals, there are two SHMT isozymes encoded by distinct genes (13–15). SHMT1 encodes the cytoplasmic/nuclear isozyme (SHMT1), and SHMT2 encodes the mitochondrial (SHMT2) and the cytoplasmic/nuclear (SHMT2α) isoform through alternative promoter use (6, 13–15). This second SHMT2 transcript encodes SHMT2α, which provides functional redundancy with SHMT1 in the de novo thymidylate synthesis pathway. De novo thymidylate biosynthesis activity is reduced by 75% in nuclei isolated from Shmt1−/− mice (6).
Recent evidence indicates that the formation of a multienzyme metabolic complex is required for de novo purine nucleotide biosynthesis in the cytoplasm, referred to as a purinosome (16, 17). Formation of the purinosome is regulated by cell cycle, purine levels, protein kinases (18), microtubule networks (19), and sumoylation (17), with disruption of microtubule networks resulting in the suppression of de novo purine biosynthesis. A nuclear multienzyme complex, termed the replitase, which contained some of the enzyme activities required for the de novo thymidylate cycle, including ribonucleotide reductase, TYMS, and DHFR, as well as DNA polymerase, was identified in mammalian nuclear extracts from S phase cells (20, 21). Other studies have identified SHMT1 as an interacting partner with proliferating cell nuclear antigen (PCNA), indicating that SHMT1 and the de novo thymidylate pathway may localize to sites of DNA synthesis (8, 22). PCNA is a DNA replication and repair processivity factor, which acts as a “sliding clamp” and crucial factor for the localization of proteins to replication forks and repair foci (23, 24).
Shmt1+/− mice accumulate uracil within nuclear DNA (12) and show increased susceptibility to neural tube defects (25) and intestinal cancer (26). In this study, we investigated the presence of a nuclear multienzyme complex that included SHMT and the entire de novo thymidylate pathway, and its association with the DNA replication machinery. The results of this study demonstrate that the de novo thymidylate synthesis pathway is associated with nuclear lamina and that SHMT1 or SHMT2α serve essential roles as scaffold proteins for complex formation. Furthermore, this metabolic complex is enriched at sites of DNA replication initiation, indicating that de novo thymidine nucleotide synthesis occurs at the sites of DNA synthesis.
MATERIALS AND METHODS
Cell Lines and Culture Medium
HeLa cells were obtained and cultured as reported previously (6). Cells were cultured in α-MEM or DMEM (Hyclone) supplemented with 10% fetal bovine serum (Hyclone) and penicillin/streptomycin (Mediatech) at 37 °C and 5% CO2. For all experiments, α-MEM (Hyclone) lacking nucleosides was used and supplemented with 10% dialyzed and charcoal treated fetal bovine serum and penicillin/streptomycin (Mediatech) with and without the four canonical deoxyribonucleosides, each at a concentration of 10 mg/liter. Cells were maintained at 37 °C and 5% CO2 for 2 passages over 1 week prior to transfection. The SH-SY5Y human neuroblastoma cell line has been described previously (27). Cells were cultured in α-MEM with 10% dialyzed fetal bovine serum for all experiments. The expression of a dominant negative SHMT1 protein, DN2-SHMT1, was induced in cell lines by the addition of 1 μg/ml tetracycline for a minimum of 4 days prior to experimentation as described previously for SH-SY5Y-FDH cell lines (28).
Generation of Human DN2-SHMT1-expressing Cell Lines
Generation of the pet28a-DN2-SHMT1 cDNA was described previously (29). This DN2-SHMT1 cDNA was cloned into the KpnI and BamHI (New England Biolabs) site of the pcDNA4/TO/myc-His C vector (Invitrogen), generating pcDNA4-DN2-SHMT1. Stable SH-SY5Y cell lines expressing pcDNA6/TR vector (Invitrogen) and pcDNA4-DN2-SHMT1 were generated. SH-SY5Y cells were electroporated with 20 μg of plasmid DNA (pcDNA4-DN2-SHMT1 and pcDNA6/TR vector) at 0.22 kV and 950 microfarads (Bio-Rad Gene-Pulser) and then cultured with α-MEM for 48 h. The medium was replaced with α-MEM containing blasticidin (10 μg/ml) and Zeocin (100 μg/ml; Invitrogen) to select for cells that integrated the plasmid. Individual colonies resistant to Zeocin treatment were selected and expanded using α-MEM containing blasticidin and Zeocin, and clonal lines expressing human DN2-SHMT1 protein were screened by Western blot analyses.
Vectors and Transfection Procedures
The pCMV6-AC-DHFR-GFP, pCMV6-AC-TYMS-GFP, phiYFP-SHMT1, and TagRFP-N-SHMT2α vectors were previously described (6). PmKate2-Lamin B1 was purchased from Evrogen. The pBABE-Puro-SV40 LT vector was deposited in Addgene by Dr. Thomas Roberts and purchased from Addgene (Addgene plasmid 13970). pNTAPB and pCMV-FLAG-MAT-Tag-1 were obtained from Stratagene and Sigma, respectively. PCR was used to create adapters on SHMT1 cDNA for ligation. The forward primers for both pNTAPB-SHMT1 and pCMV-FLAG-SHMT1-MAT-Tag-1 were 5′-ATATAAGCTTATGACGATGCCAGTCAAC-3′, where the underline indicates a HindIII (New England Biolabs Inc.) restriction site. The reverse primer for pNTAPB-SHMT1 was 5′-ATATCTCGAGGAAGTCAGGCAGGCCAGG-3′, where the underline indicates an XhoI (New England Biolabs Inc.) restriction site. The reverse primer for pCMV-FLAG-SHMT1-MAT-Tag-1 was 5′-ATATAGATCTGAAGTCAGGCAGGCCAGG-3′, where the underline indicates a BglII (New England Biolabs Inc.) restriction site. PCRs were conducted as follows: 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 2 min. Following PCR, products were gel-purified using the QIAquick gel extraction kit (Qiagen). Vectors and PCR products were digested with their respective restriction enzymes as per the manufacturer's protocol. Ligation was completed using T4 DNA ligase (Invitrogen) as per the manufacturer's protocol. All transfection procedures were completed using Kit R (Lonza) for a Nucleofector II (Lonza) as per the manufacturer's instructions.
Confocal Microscopy
Following nucleofection, cells were incubated at 37 °C under 5% CO2 for 24 h. For visualization of nuclei in cells not transfected with pmKate2-Lamin B1, DRAQ5 (Alexis Biochemicals) was used per the manufacturer's instructions. Confocal fluorescence microscopy (Leica TCS SP2 system) was used to image all cells at the Cornell Microscope and Imaging Facility.
Cell Cycle Synchronization and Analysis
HeLa cells at 60% confluence were arrested at various cell cycle stages using 30 μm lovastatin (Sigma) (for G1 phase), 2 mm hydroxyurea (Sigma) (for S phase), or 100 ng/ml nocodazole (Sigma) (for G2/M phase) following transfection. Preparation for fluorescence-activated cell sorting (FACS) was described previously (8). FACS analysis was performed at the Biomedical Sciences Flow Cytometry Core Laboratory, Cornell University.
Immunoblotting
SHMT1, SHMT2, TYMS, DHFR, and GAPDH immunoblots were performed as described previously (6, 7). Antibodies directed toward PCNA and the SV40 LT were purchased from Abcam. Mouse anti-PCNA and mouse anti-SV40 large T antigen were diluted in 5% nonfat dry milk (Carnation) containing 1% Nonidet P-40 (U.S. Biologicals) at a concentration of 1 μg/ml and 1:1000, respectively. Goat anti-mouse HRP (Pierce) secondary antibodies were used at a dilution of 1:10,000. SuperSignal West Pico Chemiluminescent substrate was used for visualization (Pierce).
Immunoprecipitation
Immunoprecipitation was conducted using the Dynabeads Protein G immunoprecipitation kit (Invitrogen). For whole cell immunoprecipitation, HeLa cells were lysed using mammalian protein extraction reagent (Pierce) supplemented with 2 mm β-mercaptoethanol (Calbiochem), 0.1 mm EDTA (Fisher), 1 mm PMSF (Alexis Biochemicals), and 1:1000 protease inhibitor mixture (Sigma). For immunoprecipitations from nuclear and cytosolic extracts, nuclei were purified using the Active Motif Nuclear extract kit per the manufacturer's protocol. Antibodies directed toward the following proteins were purchased from Abcam: DNA polymerase δ, uracil DNA glycosylase 2 (UNG2), PCNA, and DHFR. Lamin A/C, Lamin B1, DNA polymerase ϵ, lysine demethylase 1 (KDM1), and replication factor C activator 1 (RFC1) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Non-immune IgG was purchased from Pierce. SHMT1 antibody was obtained as described previously (8). 1 mg of total protein per sample was incubated with 5 μg of antibody overnight at 4 °C. The beads were collected and washed four times with PBS, 0.1% Tween 20 (Fisher). For lamin A/C and lamin B1 immunoprecipitations, washes contained either 150 mm NaCl or 650 mm NaCl in PBS, 0.1% Tween 20. For all immunoprecipitations, 40 μl of SDS-PAGE sample buffer were added to the beads to elute the protein with heating at 100 °C.
SHMT1 Tandem Affinity Purification
pNTAPB-SHMT1 was transfected into HeLa cells as described above. For the isolation of TAP-tagged SHMT1 and co-eluting proteins, the Interplay Mammalian TAP System kit (Stratagene) was used following the manufacturer's protocol. For isolation of FLAG-SHMT1-MAT and co-eluting proteins for peptide sequencing, pCMV-FLAG-SHMT1-MAT-Tag-1 was transfected as described above. Approximately 4 × 108 HeLa cells were used per group. The groups were as follows: empty pCMV-FLAG-MAT-Tag-1 control, pCMV-FLAG-SHMT1-MAT-Tag-1 S phase-synchronized cells, and UV-treated cells. UV treatment was accomplished as previously reported (9). 24 h following treatments, cells were washed with 20 ml of PBS, 1 mm PMSF three times. Cells were scraped into 5 ml of PBS, 1 mm PMSF and collected in 50-ml conical tubes (Corning Glass). Cells were pelleted at 1000 rpm for 5 min and suspended in 15 ml of swelling buffer (25 mm HEPES (Fisher), pH 7.8, 1.5 mm MgCl2 (Fisher), 10 mm KCl (Fisher), 0.1% Nonidet P-40 (U.S. Biologicals), 1.0 mm PMSF (Alexis Biochemicals), 1:100 protease inhibitor (Sigma)) and incubated on ice for 10 min. Cells were homogenized with 25 strokes of a B-type Dounce homogenizer, and cell lysis was verified by trypan blue (Fisher) exclusion. The homogenate was then filtered (70-μm mesh; BD Bioscience) and centrifuged at 4000 rpm for 5 min to collect nuclei. Nuclei were washed with 2× swelling buffer and pelleted by centrifugation. Nuclei were suspended in 1 ml of ice-cold sodium phosphate (Fisher), pH 8.0, 75 mm NaCl, 1 mm PMSF, 1:100 protease inhibitor for histidine tag isolation (Sigma). MgCl2 was added to a concentration of 2 mm. Benzonase (Sigma) was added at a concentration of 250 units/ml sample for removal of DNA and incubated for 30 min on ice. Following benzonase treatment, samples were centrifuged at 14,000 rpm for 15 min. The supernatant was collected, and FLAG-SHMT1-MAT and co-eluting proteins were purified using Invitrogen His-TALON magnetic beads per the manufacturer's instructions. Following elution, the eluates were combined and diluted with 1 volume of a solution containing 100 mm Tris, pH 7.4, 2 mm EDTA, and a 1:100 dilution of protease inhibitor mixture. FLAG immunoprecipitation was conducted using FLAG affinity gel (Sigma) per the manufacturer's instructions. The eluate from the FLAG immunoprecipitation was then subjected to protein precipitation using the ND protein precipitation kit (National Diagnostics) per the manufacturer's protocol. The protein pellet was suspended in 20 μl of 20 mm Tris, pH 7.5, and 20 μl of 2× SDS-PAGE loading buffer (Bio-Rad) and heated at 95 °C for 10 min.
Mass Spectrometry
Tandem affinity-purified samples from S phase-blocked and UV-treated cells were run on a 12% polyacrylamide gel and were stained with Novex colloidal blue staining reagent (Invitrogen). The lanes were excised and cut into nine fractions of ∼7 mm by 5-mm areas and sent to the Harvard Microchemistry Facility for peptide digestion, purification, and sequencing. Each excised fraction was reduced, carboxyamidomethylated, and digested with trypsin. Peptide identification of each digestion mixture was performed by microcapillary reversed-phase HPLC nanoelectrospray tandem mass spectrometry on an LTQ-Orbitrap Velos or XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The Orbitrap repetitively surveyed an m/z range from 395 to 1600, whereas data-dependent MS/MS spectra on the 20 (Velos) or 10 (XL) most abundant ions in each survey scan were acquired in the linear ion trap. MS/MS spectra were acquired with relative collision energy of 30%, 2.5-Da isolation width, and recurring ions were dynamically excluded for 60 s. Preliminary sequencing of peptides was facilitated with the SEQUEST algorithm with a 30 ppm mass tolerance against a species-specific (human) subset of the UniProt Knowledgebase. With a custom version of the Harvard Proteomics Browser Suite (Thermo Fisher Scientific), peptide spectrum matches were accepted with mass error of <2.5 ppm and score thresholds to attain an estimated false discovery rate of ∼1%, using a reverse decoy database strategy. Gene ontology term enrichment was performed using DAVID with the total set of proteins with a peptide count of ≥3 and a ratio of experimental group peptide counts to control group peptide counts of >1 (30, 31).
Metabolic Isotope Tracer Studies
Metabolic isotope tracer studies were completed as described previously (32).
Tandem Chromatin Immunoprecipitation
Clones 1, 2, and 3 of pBABE-Puro-SV40 LT-expressing HeLa cells that were selected using puromycin were used for this experiment. Approximately 2 × 108 cells were used per clone. Each plate was washed three times with ice-cold PBS. The bifunctional cross-linker dimethyl 3,3′-dithiobispropionimidate (Pierce) was suspended at a concentration of 5 mm in PBS at pH 8.0, and 20 ml per plate was added. Cells were incubated at 4 °C for 30 min and then washed twice with PBS. 20 ml of ice-cold quenching buffer (100 mm Tris, pH 8.0, 150 mm NaCl) was added per plate, and the cells were incubated for 10 min at 4 °C. Each plate was washed three times with PBS at room temperature. Each plate was treated with a 1% formaldehyde (Fisher) solution in PBS and then incubated at room temperature for 10 min. To quench formaldehyde cross-linking, 3 ml of 1.375 m glycine (Fisher) were added. Plates were washed three times with ice-cold PBS supplemented with 0.5 mm PMSF. Cells were scraped into 5 ml of ice-cold PBS containing 0.5 mm PMSF and centrifuged at 1000 rpm for 5 min at 4 °C. The pelleted cells were resuspended in 10 volumes of swelling buffer and then subjected to Dounce homogenization and centrifugation as reported above. Pelleted nuclei were resuspended in 5 ml of sonication buffer (50 mm Hepes, pH 7.9, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 0.5 mm PMSF, and 1:1000 protease inhibitor mixture). Nuclei were sonicated using a 3-mm microtip probe on a Branson Sonifier 150 at 80% with 10-s pulses, 10 times, with 1-min incubations on ice between pulses. Sonicated nuclei were centrifuged at 14,000 rpm for 15 min at 4 °C, and the supernatant was supplemented with 1 μg/ml sonicated salmon sperm DNA (Invitrogen) and bovine serum albumin (Sigma) at 1 mg/ml. The lysate was precleared using 50 μl of Invitrogen Protein G Dynabeads for 2 h at 4 °C, beads were collected on a magnet, and the supernatant was removed. Samples were divided into 1-ml aliquots for immunoprecipitation. 5 μg of PCNA (Abcam) antibody was incubated for 12 h per 1-ml aliquot at 4 °C. 50 μl of Invitrogen Protein G Dynabeads were added, and the solution was incubated for 1 h at 4 °C. The beads were collected with a magnet and washed twice with 1 ml of sonication buffer. The beads were washed twice with 1 ml of sonication buffer containing 500 mm NaCl and then twice with 1 ml of a solution containing 20 mm Tris, pH 8.0, 1 mm EDTA, 250 mm LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 0.5 mm PMSF, and protease inhibitor mixture. Two more washes were performed with 1 ml of TE buffer. 50 μl of 10 mm DTT was then added to the beads, which were then incubated at 37 °C for 30 min. The eluate was removed, and that step was repeated. The eluates were then combined and diluted 40× with sonication buffer. 10% of the sample was kept for input. The sample was then aliquoted into 1-ml fractions for TYMS, DHFR, SHMT1, and IgG control immunoprecipitations. 5 μg of each antibody was used, and samples were incubated for 12 h at 4 °C. 50 μl of Invitrogen Protein G Dynabeads were added and incubated for 1 h at 4 °C. Washes were performed as above. To elute the proteins, 200 μl of a solution containing 50 mm Tris, pH 8.0, 1 mm EDTA, 1% SDS, and 50 mm NaHCO3 was added, and the beads were heated to 65 °C for 10 min. This step was repeated, and eluates were combined, giving a 400-μl final volume. The input and samples were then treated with 21 μl of 4 m NaCl and incubated for 12 h at 65 °C. 2 μl of RNase A (5 mg/ml) (Rockland Immunochemicals, Inc.) was added to each sample, followed by incubation at 37 °C for 1 h. EDTA was then added to a concentration of 5 mm. 2 μl of proteinase K (10 mg/ml) (Rockland Immunochemicals, Inc.) was then added, and samples were incubated for 2 h at 42 °C. Samples were extracted with phenol/chloroform/isoamyl alcohol (Sigma) once and then with chloroform/isoamyl alcohol (Sigma). 1 μl of glycogen (Sigma) (20 mg/ml) was added, followed by the addition of 40 μl of 3 m sodium acetate (Fisher) and 1 ml of 100% ethanol (Pharmco-AAPER). Samples were vortexed and precipitated for 12 h at −20 °C, followed by centrifugation at 14,000 rpm for 30 min. The pellets were washed with 75% ethanol. Centrifugation was repeated, and the pellet was allowed to dry at room temperature. Samples were then suspended in 100 μl of 10 mm Tris, pH 7.5, for real-time PCR analysis.
Real-time PCR
Forward and reverse primers surrounding the SV40 origin of replication were 5′-CAGCAGGCAGAAGTATGCAAAGCA-3′ and 5′-TACTTCTGGAATAGCTCAGAGGCCGA-3′, respectively. To amplify the pBABE-Puro vector region, the forward and reverse primers were 5′-ACAGAGTTCTTGAAGTGGTGGCCT-3′ and 5′-TGGTTTGTTTGCCGGATCAAGAGC-3′, respectively. To amplify the large T-antigen insert, the forward and reverse primers were 5′-ACTCCACACAGGCATAGAGTGTCT-3′ and 5′-CCCACCTGGCAAACTTTCCTCAAT-3′, respectively. Real-time PCR analysis was performed using the Quantifast SYBR Green PCR kit (Qiagen). PCR products were quantified using an Applied Biosystems 7500 real-time PCR system.
RESULTS
SHMT1, SHMT2α, DHFR, and TYMS Are Present in Nuclei during S and G2/M Phases
SHMT1, SHMT2α, TYMS, and DHFR have been localized previously to the nucleus and cytoplasm of human and mouse cell lines (6, 8, 10). Nuclear localization of SHMT1 and SHMT2α in mouse embryonic fibroblasts and human cell lines (6, 8) is restricted to the S and G2/M phases of the cell cycle and in response to UV damage (9). In this study, the nuclear localization of TYMS and DHFR was determined as a function of cell cycle. As seen previously for SHMT1, DHFR (supplemental Fig. S1) and TYMS (supplemental Fig. S2) localized to the nucleus during S and G2/M phases but not in G1 phase. These data confirm that all of the enzymes necessary for de novo thymidylate biosynthesis are present within the nucleus during S and G2/M phases during DNA replication and repair but are restricted to the cytoplasm in G1.
Identification of SHMT1-interacting Proteins in Nuclear Extracts
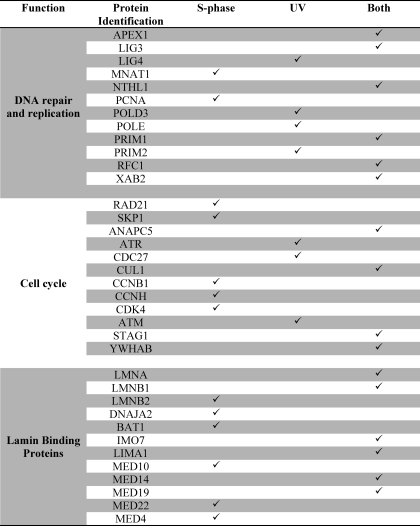
Intact purified nuclei from mouse liver can convert tritium-labeled serine and dUMP to tritium-labeled thymidylate, but this activity is lost in sonicated nuclei, indicating that nuclear integrity and multienzyme complex formation may be necessary for de novo thymidylate synthesis (6). To determine if the de novo thymidylate biosynthesis pathway is present in nuclei within a multienzyme complex, SHMT1 tandem affinity purification and SHMT1 co-precipitation experiments were performed on benzonase-treated nuclear extracts isolated from S phase cells and cells treated with UV (supplemental Fig. S3). Over 833 proteins were identified as SHMT1-interacting proteins. The list was refined by excluding proteins with fewer than three peptides identified, and the remaining proteins were grouped by gene ontology term enrichment using the bioinformatics tool DAVID (Table 1 and supplemental Tables S1–S5). Multiple proteins involved in nucleotide metabolism and DNA replication and repair were identified, as well as 80 lamin-interacting proteins. The majority of the identified proteins known to be involved in DNA replication and repair, as well as lamin-interacting proteins, were found in both S phase and UV-treated samples, although some of these interactions were identified only in one of those two exposures (Table 1).
TABLE 1.
Subsets of proteins identified using tandem affinity purification and MS/MS
A complete listing of proteins identified is available in supplemental Tables S1–S5.
Validation of SHMT1-interacting Partners
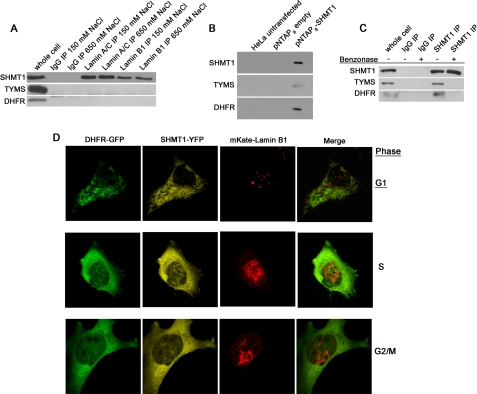
DNA replication occurs on nucleoskeletal structures (34) and disruption of the nuclear lamina results in DNA replication arrests (35, 36). Several proteins involved in DNA replication are lamin-binding proteins, including PCNA, which acts as a processivity factor for DNA replication (37). SHMT1 has been identified previously as a PCNA-interacting partner (8), and this interaction was confirmed in this tandem affinity purification experiment (supplemental Fig. S3 and Table 1). To validate SHMT1 as a lamin-interacting partner, co-immunoprecipitation experiments using benzonase-treated nuclear extracts were performed with antibodies against either lamin A/C or lamin B1. SHMT1 co-precipitated with lamin A/C and lamin B1 antibodies under stringent (650 mm NaCl) and less stringent conditions (150 mm NaCl) (Fig. 2A). Neither TYMS nor DHFR co-precipitated with lamin proteins under these conditions.
FIGURE 2.
SHMT1 is a nuclear lamin-associated protein. A, both lamin A/C and lamin B1 co-immunoprecipitations from benzonase-treated nuclear extracts contained SHMT1 even in high salt conditions (650 mm NaCl), whereas neither contained TYMS or DHFR even under low salt conditions (150 mm NaCl). B, pNTAPb-SHMT1 and pNTAPb empty vectors were transfected into HeLa cells, and tandem affinity purification was performed using streptavidin and calmodulin resins. SHMT1, TYMS, and DHFR were detected in pNTAPb-SHMT1 transfections in samples that were not treated with nucleases. C, the DNA dependence of SHMT1, TYMS, and DHFR interactions were determined by SHMT1 immunoprecipitation in purified nuclear extracts treated with or without benzonase. TYMS and DHFR only immunoprecipitated with SHMT1 in samples lacking benzonase treatment. D, the interaction of SHMT1, lamin B1, and DHFR was investigated by confocal microscopy following transfection of cDNAs encoding GFP-DHFR, SHMT1-YFP, and mKate-lamin B1 fusion proteins in cells blocked in G1 (30 μm lovastatin), S phase (1 mm hydroxyurea), and G2/M (100 ng/ml nocodazole) phases of the cell cycle. Co-localization of DHFR and SHMT1 with lamin B1 is concomitant with nuclear localization of DHFR and SHMT1 during S and G2/M but not during G1 phases of the cell cycle. IP, immunoprecipitation.
Other SHMT1-interacting partners identified in Table 1 were validated by co-immunoprecipitation followed by immunoblotting against the interacting protein. SHMT1, TYMS, and DHFR all co-precipitated with PCNA in nuclear extracts (supplemental Fig. S4, A–C) but not in cytoplasmic extracts (supplemental Fig. S4, A and B). Lysine demethylase 1 (KDM1), which has been recently identified as a folate-binding protein (38), immunoprecipitated with anti-SHMT1 antibodies (supplemental Fig. S4D). Other proteins involved in DNA replication and repair, including DNA polymerases δ and ϵ (POLD and POLE respectively), replication factor C activator 1 (RFC1), and uracil DNA glycosylase 2 (UNG2) (supplemental Fig. S4E) co-immunoprecipitated with SHMT1. In total, eight validations were performed, and no false positives were identified.
SHMT1 Interaction with TYMS and DHFR Is DNA-dependent
Neither TYMS nor DHFR were identified as SHMT1- or lamin-interacting proteins by tandem affinity purification. The ability of nuclear SHMT1 to interact with TYMS and DHFR in the absence of DNA digestion was determined by co-precipitation with SHMT1 in small scale tandem affinity purification (Fig. 2B) and SHMT1 immunoprecipitation (Fig. 2C). In the tandem affinity purification, both TYMS and DHFR co-precipitated with SHMT1 in the absence of benzonase treatment (Fig. 2B). However, neither TYMS nor DHFR co-precipitated with SHMT1 in nuclear extracts treated with benzonase, whereas in the absence of benzonase treatment, co-immunoprecipitates contained TYMS and DHFR (Fig. 2C).
Co-localization of de Novo Thymidylate Biosynthesis Pathway with Lamin B1
The co-localization of the de novo thymidylate biosynthesis pathway with the nuclear lamina was investigated by confocal microscopy. HeLa cells transfected with pCMV6-AC-DHFR-GFP, phiYFP-N-SHMT1, and mKate2-lamin B1 expression plasmids exhibited co-localization of lamin B1, SHMT1, and DHFR fusion proteins in filament-like linear structures and clusters in the S and G2/M phases of the cell cycle (Fig. 2D). To control for potential artifacts resulting from lamin B1 fusion protein expression, confocal microscopy was performed on HeLa cells following transfection of pCMV6-AC-DHFR-GFP (supplemental Fig. S5A), pCMV6-AC-TYMS-GFP (supplemental Fig. S5, B and C), or phiYFP-N-SHMT1 (supplemental Fig. S5C) plasmids. All three fusion proteins that constitute the de novo thymidylate biosynthesis cycle co-localized as linear structures within nuclei, demonstrating that the formation of these structures was not of the result of lamin B1 overexpression.
Formation of Thymidylate Biosynthesis Complex Is Nucleotide-independent
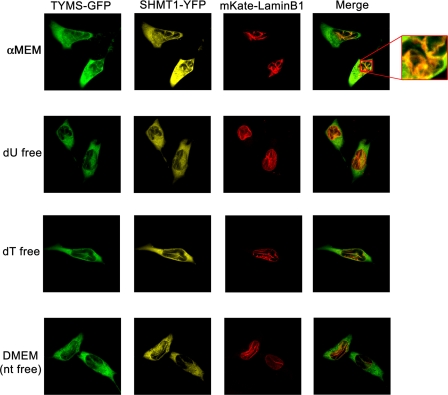
Previous studies have demonstrated that the assembly of the purinosome, a cytoplasmic multienzyme complex comprising the de novo purine biosynthesis enzymes, is regulated by purine levels within the cell. The effect of nucleosides on the formation of the de novo thymidylate biosynthesis pathway complex in nuclei was determined in HeLa cells cultured in medium lacking all nucleosides (DMEM), lacking only deoxyuridine (dU), lacking only thymidine (dT), or replete with all nucleosides (α-MEM) following transfection with the plasmids pCMV6-AC-TYMS-GFP, phiYFP-N-SHMT1, and mKate2-lamin B1 (Fig. 3). The presence or absence of nucleosides in the culture media did not affect the co-localization of SHMT1, TYMS, and lamin B1 as determined by confocal microscopy, demonstrating that formation of the observed linear structures is independent of nucleoside availability.
FIGURE 3.
SHMT1 and TYMS form linear lamin B1 co-localizing structures independent of nucleoside availability. The cDNAs encoding YFP-SHMT1, TYMS-GFP, and mKate-lamin B1 fusion proteins were transfected into cells cultured in medium containing nucleosides (α-MEM), medium without nucleosides (DMEM), and α-MEM that lacks either deoxyuridine (dU) or thymidine (dT) and visualized by confocal microscopy. The co-localization of SHMT1 and TYMS with lamin B1 was independent of nucleosides in the culture medium.
SHMT1 and SHMT2α Are Required for DHFR, TYMS, and Lamin B1 Co-localization
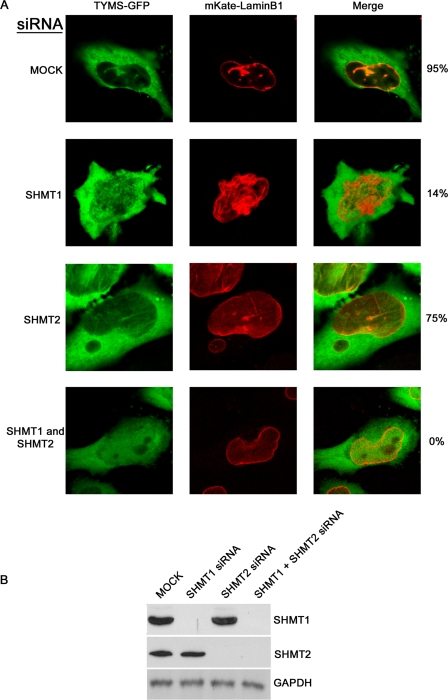
The lamin-binding property of SHMT1 and the lack of interactions among TYMS and DHFR and lamins suggested that SHMT1 and/or SHMT2α may function as scaffold proteins required for DHFR and TYMS localization to the lamina and assembly of the de novo thymidylate synthesis pathway complex. Therefore, the co-localization of TYMS and DHFR with nuclear lamins was investigated in the absence of SHMT. DHFR-GFP and mKate-lamin B1 (supplemental Fig. S6A) or TYMS-GFP and mKate-lamin B1 (Fig. 4A) co-localized in HeLa cells transfected with scrambled siRNA in 95% of the observed cells, but co-localization was reduced to 50 and 75%, respectively, in cells treated with SHMT2 siRNA. Lamin B1 and DHFR or TYMS co-localizing structures were observed in only 5 and 14%, respectively, of cells treated with SHMT1 siRNA and were absent in SHMT1 and SHMT2 siRNA-treated samples. Immunoblotting was performed against SHMT1 and SHMT2 to ensure that the knockdown of those enzymes had occurred (Fig. 4B and supplemental Fig. S6B). These data demonstrate that SHMT1 and SHMT2α function as scaffolds that are required for TYMS and DHFR co-localization with lamin B1.
FIGURE 4.
SHMT1 and SHMT2 are required for TYMS and lamin B1 co-localization. A, cDNAs encoding GFP-TYMS, mKate-lamin B1 fusion proteins, and siRNAs, including scrambled (MOCK), SHMT1, SHMT2, or both SHMT1 and SHMT2, were transfected into HeLa cells. Cells were blocked in S phase using hydroxyurea (2 mm) and visualized with confocal microscopy. For each siRNA treatment, 100 cells were counted. The presence of DHFR and lamin B1 co-localizing structures occurred in 95 ± 4, 14 ± 4.5, 75 ± 5.7, and 0 ± 0.6% of cells transfected with scrambled, SHMT1, SHMT2, and both SHMT1 and SHMT2 siRNAs, respectively. Error is expressed in S.D. (n = 3). B, immunoblotting was performed on siRNA-treated samples to ensure knockdown of SHMT1 and SHMT2. GAPDH was visualized as a loading control.
TYMS, DHFR, and SHMT2 Do Not Affect SHMT1 and Lamin B1 Co-localization
The effect of TYMS, DHFR, and SHMT2 expression on SHMT1 co-localization with lamin B1 was investigated in HeLa cells expressing SHMT1-YFP and mKate-lamin B1 (supplemental Fig. S7A). Lamin B1 and SHMT1 co-localized independent of reduced TYMS, DHFR, or SHMT2 expression, which was achieved by siRNA treatment. Immunoblotting was performed against TYMS, DHFR, and SHMT2 to ensure that the knockdown of those enzymes had occurred (supplemental Fig. S7B). These data demonstrate that SHMT1 is essential for the de novo thymidylate synthesis pathway complex formation and that SHMT1 anchors the de novo thymidylate pathway to lamin B1.
SHMT2α Functions as Scaffold Protein Independent of SHMT1
Previous studies have demonstrated that SHMT1 and SHMT2α are functionally redundant in nuclear de novo thymidylate synthesis (6). The dependence of the SHMT2α-lamin interaction on SHMT1, TYMS, and DHFR expression was investigated in HeLa cells expressing SHMT2α-RFP and mKate-lamin B1 fusion proteins (supplemental Fig. S8A). SHMT2α and lamin B1 co-localized independent of TYMS, DHFR, and SHMT1 expression. Immunoblotting was performed against TYMS, DHFR, and SHMT1 to ensure that the knockdown of those enzymes had occurred (supplemental Fig. S8B). These data demonstrate that SHMT2α co-localizes with lamin B1 even in the absence of SHMT1, further supporting the functional redundancy of SHMT2α and SHMT1 in nuclear de novo thymidylate synthesis.
SHMT1 Scaffold Function Can Determine de Novo Thymidylate Synthesis Capacity
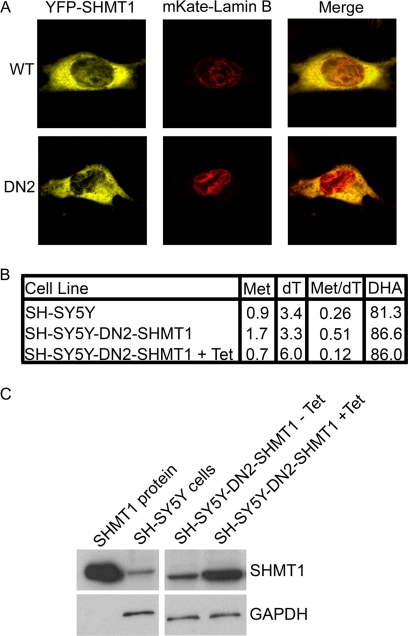
Previous studies of SHMT1-deficient mice (12) and cell culture models (11, 32) have demonstrated strong correlations between SHMT expression and de novo thymidylate synthesis capacity, although enzyme kinetic studies do not indicate that SHMT1 is catalytically rate-limiting in the de novo thymidylate biosynthesis pathway (39). To distinguish between the catalytic and scaffold functions of SHMT1 in nuclear de novo thymidylate biosynthesis, the effect of expressing a dominant negative SHMT1 protein in SH-SY5Y cells that express endogenous SHMT1 on de novo thymidylate synthesis was determined. SHMT is a tetramer, best described as a dimer of obligate dimers, in which amino acid residues from each monomer in the obligate dimer contribute to both active sites in the dimer (29, 40). Previously, we have described the generation of a dominant negative SHMT1 protein, termed DN2-SHMT1(Y82A/Y83F/K257Q), that dimerizes with wild type SHMT1, creating a stable protein that lacks catalytic activity in both active sites of the dimer (29). Tyr-82 is required for hydrophobic stacking with the p-aminobenzoylglutamate moiety of tetrahydrofolate, and the Y82A mutation eliminates folate binding. The side chains of Tyr-83 and Lys-257 are required for SHMT1 catalytic activity. Expression of DN2-SHMT1 in cells expressing endogenous SHMT1 results in the formation of inactive SHMT1 tetramers and a decrease in the specific activity of SHMT1 in the cell. Co-expression of YFP-DN2-SHMT1 and mKate-lamin B fusion proteins resulted in the formation of SHMT1-lamin B linear structures that were indistinguishable from structures resulting from coexpression of wild-type YFP-SHMT1 and mKATE-lamin B1 (Fig. 5A), indicating that the mutations in DN2-SHMT1 did not impair lamin binding activity.
FIGURE 5.
Dominant negative SHMT1 (DN2-SHMT1) localizes with lamin B1 and enhances SHMT1 activity for de novo thymidylate biosynthesis. A, cDNAs encoding mKate-lamin B1 and either YFP-SHMT1 or YFP-DN2-SHMT1 fusion proteins were expressed in HeLa cells, and co-localization of the proteins was determined by confocal microscopy. Both SHMT1 and DN2-SHMT1 colocalize with lamin B1. B, the effect of tetracycline (Tet)-inducible expression of DN2-SHMT1 on SHMT1 activity in de novo thymidylate biosynthesis was determined in SH-SY5Y cells. Cellular protein and DNA were isolated from cells cultured for 8 days in defined medium containing l-[2,3,3-2H3]serine. Isotopic enrichment of the one carbon derived from l-[2,3,3-2H3]serine into cellular protein pools was determined by the detection of labeled Met and dehydroalanine (DHA). Dehydroalanine is formed from serine during the derivatization procedure. Enrichment of [2,3,3-2H3]serine in thymidine (dT) was determined by analysis of nuclear DNA. All values are expressed as the percentage of l-[2,3,3-2H3]serine-derived carbons that contain two deuterium atoms (CD2) in the target compound (the ratio of carbons containing two deuterium atoms in the target compound divided by the total number of carbons that contain one or two deuterium atoms × 100). Two independent experiments were performed with duplicate measurements for each sample, and identical values were obtained within each experiment. The results from one experiment are shown. The data demonstrate that tet-inducible expression of DN2-SHMT1 decreased SHMT1 specific activity in homocysteine remethylation to methionine in the cytoplasm by 60%, whereas DN2-SHMT1 expression enhances SHMT1 specific activity in de novo thymidylate biosynthesis by 45% C, immunoblotting was performed on SH-SY5Y-DN2-SHMT1 cells in a tet-inducible system. The SH-SY5Y-DN2-SHMT1 cells exhibited increased levels of total SHMT1 in response to tetracycline. These cells were used for isotopic tracer studies. GAPDH was used as a loading control.
To determine the effect of DN2-SHMT1 expression on SHMT1 activity in cells with respect to de novo thymidine biosynthesis and homocysteine remethylation, metabolic isotope tracer experiments were performed using l-[2H3]serine and SH-SY5Y cells that express DN2-SHMT1 from a tet-inducible promoter (28). In this assay, the methylene-THF that is generated by SHMT from l-[2H3]serine is incorporated into methionine or thymidine with the donated one-carbon unit retaining two deuterium atoms (CD2) that were present on the hydroxymethyl group of serine (Fig. 1). Alternatively, if [l-2H3]serine enters the mitochondria, the hydroxymethyl group will be released from mitochondria as formate containing a single deuterium atom (CD1) and enter the methylene-THF pool with one deuterium atom (CD1), which can be incorporated into methionine or thymidylate (Fig. 1). CD2-methylene-THF can also be converted to CD1 methylene-THF through its reversible conversion to methenyl-THF through the activity of MTHFD1, which was identified to interact with SHMT1 in the nucleus both during S phase and in response to UV (supplemental Table S2).
The effect of DN2-SHMT1 expression on CD2 enrichment in thymidine in DNA and on serine and methionine in cellular protein was determined following the culture of SH-SY5Y with l-[2H3]serine for 8 days. Fig. 5B shows that 81.3–86.6% of the isotopically labeled serine used for protein synthesis was unmetabolized because it retained both deuteriums on the C3 carbon in SH-SY5Y cells and SH-SY5Y-DN2-SHMT1 cells in the presence and absence of tetracycline. If the methionine and thymidylate one-carbon units were only derived from SHMT1, the mass +2 species of these metabolites should be ∼80% of the labeled species. Expression of DN2-SHMT1 suppressed CD2 incorporation into methionine by 60%, consistent with a loss of SHMT1 specific activity in the cytoplasm. Interestingly, CD2 incorporation into thymidine within nuclear DNA was increased by 82%, indicating that DN2-SHMT1 expression increased the flux of SHMT1-derived one-carbon units into thymidylate, presumably by recruiting wild type SHMT1 subunits to the lamina within the SHMT1 tetramer. These results demonstrate that the lamin-binding activity and scaffold function of SHMT1 contribute more than its catalytic activity to de novo thymidylate biosynthesis in this cell line (Fig. 5B).
De Novo Thymidylate Pathway Is Associated with Episomal DNA
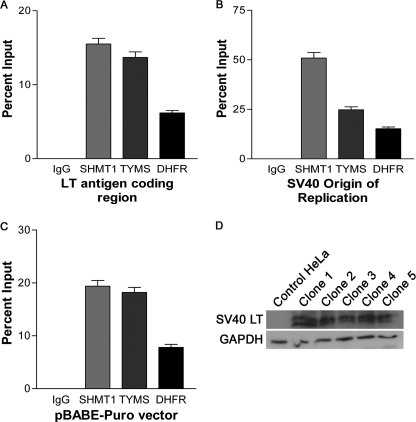
The interaction of SHMT1 and the de novo thymidylate cycle with proteins involved in DNA replication and repair, including UNG2, RFC1, PCNA, and DNA polymerases δ and ϵ (Table 1 and supplemental Fig. S4E) indicates that the thymidylate biosynthesis pathway may function at the replication fork. During DNA replication initiation, RFC interacts with DNA at the primer-template junction and loads PCNA onto the DNA template, which allows for the processive replication of DNA by DNA polymerases δ and ϵ (23). Mammalian origins of replication are not well characterized; therefore, a model system of episomally replicating DNA was employed to determine if the de novo thymidylate biosynthesis pathway is present at the replication fork (Fig. 6). HeLa cells were transfected with the pBABE-Puro-SV40 LT plasmid, which contains both the SV40 LT cDNA as well as the SV40 origin of replication. The SV40 LT is sufficient for replication initiation at the SV40 origin of replication (41) and has also been shown to induce S phase (42). Tandem ChIP experiments using antibodies directed against PCNA followed by IgG, SHMT1, TYMS, or DHFR showed that all three enzymes required for de novo thymidylate synthesis are present on DNA, with the greatest occupancy observed at the SV40 origin of replication (Fig. 6B). Because of the bidirectional nature of DNA replication, regions either upstream or downstream of the SV40 origin of replication were also probed. The required enzymes for de novo thymidylate biosynthesis were present within the SV40 LT coding region (Fig. 6A) and also within a randomly chosen area of the vector referred to as pBABE-Puro vector (Fig. 6C). These data demonstrate that de novo thymidylate biosynthesis localizes to replication forks and suggests the processive synthesis and incorporation of dTTP into DNA during DNA replication.
FIGURE 6.
The de novo dTMP synthesis pathway is associated with episomal DNA in tandem chromatin immunoprecipitations. HeLa cells were transfected with pBABE-Puro-SV40 LT vector and selected for using puromycin. Clones were isolated and immunoblots performed against SV40 LT to ensure that clones were expressing SV40 LT (D). Tandem ChIP assays were performed as described under “Materials and Methods” using antibodies directed against PCNA and then antibodies directed against non-immune IgG, SHMT1, TYMS, or DHFR. Three separate SV40 LT-expressing clones were subjected to tandem ChIP in duplicate. Error is expressed as S.E. (error bars). Regions of the pBABE-Puro-SV40 LT vector were probed for the presence of SHMT1, TYMS, or DHFR using real-time PCR (A–C). All three enzymes precipitated regions of the vector, including the large T antigen coding region (A), SV40 origin of replication (B), and downstream region of the SV40 origin (C). The highest occupancy was observed around the SV40 origin.
DISCUSSION
This study demonstrates that the de novo thymidylate synthesis pathway is present in the nucleus as a metabolic complex associated with the nuclear lamina and that SHMT1 and SHMT2α anchor the de novo thymidylate synthesis metabolic complex to the nuclear lamina. Previous studies have shown that de novo thymidylate synthesis activity in isolated nuclei is impaired following sonication, indicating a role for nuclear architecture in thymidylate synthesis (6). The results of this study provide a mechanism for this observation by demonstrating that SHMT1 and SHMT2α are lamin-binding proteins that serve as scaffold proteins that are essential for assembly of the de novo thymidylate synthesis pathway into a multienzyme complex in the nucleus.
The SHMT1 scaffold function probably accounts for its role as a limiting factor for de novo thymidylate biosynthesis in cell culture and mouse models (11, 12, 32). Uracil levels in nuclear DNA, which are a proxy for thymidylate synthesis capacity, are elevated in Shmt1+/− mice compared with wild-type littermates (12), demonstrating a key role of this enzyme in determining thymidylate synthesis capacity in vivo. The results from this study indicate that the scaffold function of SHMT may be a more important determinant of de novo thymidylate biosynthesis capacity than its catalytic activity. Expression of a dominant negative SHMT1 that lacked catalytic activity but retained lamin binding activity in neuroblastoma reduced the incorporation of SHMT1-derived one-carbon units into methionine synthesis, which occurs in the cytoplasm, but enhanced the incorporation of SHMT1-derived one-carbon units into de novo thymidylate synthesis, which occurs in the nucleus. Furthermore, the identification of MTHFD1 as a component of the nuclear thymidylate biosynthesis complex indicates that formate may be an important source of nuclear methylene-THF for de novo thymidylate biosynthesis.
These data also support a role for de novo thymidylate biosynthesis at the sites of nuclear DNA synthesis. TYMS and DHFR, like SHMT1, were shown to be present in the nucleus only during S and G2/M phases and thereby enable nuclear de novo thymidylate synthesis during DNA replication and repair. The nuclear lamina has been observed to play an important role in tethering DNA to the nuclear periphery during replication and repair, and in this study, it is shown to support the formation of the de novo thymidylate synthesis multienzyme complex. The results of this study also indicate that de novo thymidylate biosynthesis occurs at the sites of DNA synthesis and is associated with the DNA replication machinery. SHMT1 was shown to interact with PCNA, RFC1, and the DNA polymerases δ and ϵ, and PCNA was shown to interact with SHMT1, TYMS, and DHFR. SHMT1, TYMS, and DHFR proteins are associated with replicating DNA and are enriched at sites of DNA replication initiation.
Thymidine nucleotides are unique from the other nucleotides required for DNA replication. They are not essential during DNA replication, because dUTP can be incorporated into DNA in lieu of dTTP. Furthermore, although all other nucleotide synthesis occurs in the cytoplasm, we have shown that the de novo thymidylate biosynthesis pathway localizes to the nucleus and that its presence in the nucleus is essential to prevent uracil accumulation in nuclear DNA (10). The finding that SHMT1 is an essential scaffold for the formation of the de novo thymidylate biosynthesis multicomplex accounts for the sensitivity of Shmt+/− mice to increased uracil misincorporation in DNA, increased risk of intestinal cancer (26), and neural tube defects (25). Furthermore, the nuclear compartmentation of de novo thymidylate biosynthesis at the replication fork may permit the regulation of dUTP incorporation into DNA, as opposed to its misincorporation in DNA, for the regulation of transcriptional networks and biological pathways, as has been demonstrated in Drosophila (33, 43).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK58144 and HD059120 (to P. J. S.).

This article contains supplemental Figs. S1–S8 and Tables S1–S5.
- THF
- tetrahydrofolate
- SHMT
- serine hydroxymethyltransferase
- TYMS
- thymidylate synthase
- DHFR
- dihydrofolate reductase
- SUMO
- small ubiquitin-like modifier
- PCNA
- proliferating cell nuclear antigen
- SV40 LT
- SV40 large T antigen
- MEM
- minimum essential medium
- TAP
- tandem affinity purification.
REFERENCES
- 1. Samsonoff W. A., Reston J., McKee M., O'Connor B., Galivan J., Maley G., Maley F. (1997) Intracellular location of thymidylate synthase and its state of phosphorylation. J. Biol. Chem. 272, 13281–13285 [DOI] [PubMed] [Google Scholar]
- 2. Anderson D. D., Quintero C. M., Stover P. J. (2011) Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blount B. C., Mack M. M., Wehr C. M., MacGregor J. T., Hiatt R. A., Wang G., Wickramasinghe S. N., Everson R. B., Ames B. N. (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage. Implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. U.S.A. 94, 3290–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fox J. T., Stover P. J. (2008) Folate-mediated one-carbon metabolism. Vitam. Horm. 79, 1–44 [DOI] [PubMed] [Google Scholar]
- 5. Tibbetts A. S., Appling D. R. (2010) Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 30, 57–81 [DOI] [PubMed] [Google Scholar]
- 6. Anderson D. D., Stover P. J. (2009) SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS One 4, e5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson D. D., Woeller C. F., Stover P. J. (2007) Small ubiquitin-like modifier-1 (SUMO-1) modification of thymidylate synthase and dihydrofolate reductase. Clin. Chem. Lab. Med. 45, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 8. Woeller C. F., Anderson D. D., Szebenyi D. M., Stover P. J. (2007) Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J. Biol. Chem. 282, 17623–17631 [DOI] [PubMed] [Google Scholar]
- 9. Fox J. T., Shin W. K., Caudill M. A., Stover P. J. (2009) A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair. J. Biol. Chem. 284, 31097–31108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macfarlane A. J., Anderson D. D., Flodby P., Perry C. A., Allen R. H., Stabler S. P., Stover P. J. (2011) Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J. Biol. Chem. 286, 44015–44022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oppenheim E. W., Adelman C., Liu X., Stover P. J. (2001) Heavy chain ferritin enhances serine hydroxymethyltransferase expression and de novo thymidine biosynthesis. J. Biol. Chem. 276, 19855–19861 [DOI] [PubMed] [Google Scholar]
- 12. MacFarlane A. J., Liu X., Perry C. A., Flodby P., Allen R. H., Stabler S. P., Stover P. J. (2008) Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J. Biol. Chem. 283, 25846–25853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garrow T. A., Brenner A. A., Whitehead V. M., Chen X. N., Duncan R. G., Korenberg J. R., Shane B. (1993) Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J. Biol. Chem. 268, 11910–11916 [PubMed] [Google Scholar]
- 14. Stover P. J., Chen L. H., Suh J. R., Stover D. M., Keyomarsi K., Shane B. (1997) Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J. Biol. Chem. 272, 1842–1848 [DOI] [PubMed] [Google Scholar]
- 15. Girgis S., Nasrallah I. M., Suh J. R., Oppenheim E., Zanetti K. A., Mastri M. G., Stover P. J. (1998) Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene 210, 315–324 [DOI] [PubMed] [Google Scholar]
- 16. An S., Kumar R., Sheets E. D., Benkovic S. J. (2008) Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106 [DOI] [PubMed] [Google Scholar]
- 17. Field M. S., Anderson D. D., Stover P. J. (2011) Mthfs is an essential gene in mice and a component of the purinosome. Front. Genet. 2, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. An S., Kyoung M., Allen J. J., Shokat K. M., Benkovic S. J. (2010) Dynamic regulation of a metabolic multienzyme complex by protein kinase CK2. J. Biol. Chem. 285, 11093–11099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. An S., Deng Y., Tomsho J. W., Kyoung M., Benkovic S. J. (2010) Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc. Natl. Acad. Sci. U.S.A. 107, 12872–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prem veer Reddy G., Pardee A. B. (1980) Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc. Natl. Acad. Sci. U.S.A. 77, 3312–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noguchi H., Prem veer Reddy G., Pardee A. B. (1983) Rapid incorporation of label from ribonucleoside disphosphates into DNA by a cell-free high molecular weight fraction from animal cell nuclei. Cell 32, 443–451 [DOI] [PubMed] [Google Scholar]
- 22. Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., Goldberg D. S., Li N., Martinez M., Rual J. F., Lamesch P., Xu L., Tewari M., Wong S. L., Zhang L. V., Berriz G. F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H. W., Elewa A., Baumgartner B., Rose D. J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S. E., Saxton W. M., Strome S., Van Den Heuvel S., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K. C., Harper J. W., Cusick M. E., Roth F. P., Hill D. E., Vidal M. (2004) A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 24. Ulrich H. D. (2009) Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair 8, 461–469 [DOI] [PubMed] [Google Scholar]
- 25. Beaudin A. E., Abarinov E. V., Noden D. M., Perry C. A., Chu S., Stabler S. P., Allen R. H., Stover P. J. (2011) Shmt1 and de novo thymidylate biosynthesis underlie folate-responsive neural tube defects in mice. Am. J. Clin. Nutr. 93, 789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macfarlane A. J., Perry C. A., McEntee M. F., Lin D. M., Stover P. J. (2011) Shmt1 heterozygosity impairs folate-dependent thymidylate synthesis capacity and modifies risk of Apc(min)-mediated intestinal cancer risk. Cancer Res. 71, 2098–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Girgis S., Suh J. R., Jolivet J., Stover P. J. (1997) 5-Formyltetrahydrofolate regulates homocysteine remethylation in human neuroblastoma. J. Biol. Chem. 272, 4729–4734 [DOI] [PubMed] [Google Scholar]
- 28. Anguera M. C., Field M. S., Perry C., Ghandour H., Chiang E. P., Selhub J., Shane B., Stover P. J. (2006) Regulation of folate-mediated one-carbon metabolism by 10-formyltetrahydrofolate dehydrogenase. J. Biol. Chem. 281, 18335–18342 [DOI] [PubMed] [Google Scholar]
- 29. Zanetti K. A., Stover P. J. (2003) Pyridoxal phosphate inhibits dynamic subunit interchange among serine hydroxymethyltransferase tetramers. J. Biol. Chem. 278, 10142–10149 [DOI] [PubMed] [Google Scholar]
- 30. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 31. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools. Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herbig K., Chiang E. P., Lee L. R., Hills J., Shane B., Stover P. J. (2002) Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J. Biol. Chem. 277, 38381–38389 [DOI] [PubMed] [Google Scholar]
- 33. Deutsch W. A., Spiering A. L. (1982) A new pathway expressed during a distinct stage of Drosophila development for the removal of dUMP residues in DNA. J. Biol. Chem. 257, 3366–3368 [PubMed] [Google Scholar]
- 34. Hozák P., Hassan A. B., Jackson D. A., Cook P. R. (1993) Visualization of replication factories attached to nucleoskeleton. Cell 73, 361–373 [DOI] [PubMed] [Google Scholar]
- 35. Spann T. P., Moir R. D., Goldman A. E., Stick R., Goldman R. D. (1997) Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 136, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moir R. D., Spann T. P., Herrmann H., Goldman R. D. (2000) Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 149, 1179–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shumaker D. K., Solimando L., Sengupta K., Shimi T., Adam S. A., Grunwald A., Strelkov S. V., Aebi U., Cardoso M. C., Goldman R. D. (2008) The highly conserved nuclear lamin Ig-fold binds to PCNA. Its role in DNA replication. J. Cell Biol. 181, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luka Z., Moss F., Loukachevitch L. V., Bornhop D. J., Wagner C. (2011) Histone demethylase LSD1 is a folate-binding protein. Biochemistry 50, 4750–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reed M. C., Nijhout H. F., Neuhouser M. L., Gregory J. F., 3rd, Shane B., James S. J., Boynton A., Ulrich C. M. (2006) A mathematical model gives insights into nutritional and genetic aspects of folate-mediated one-carbon metabolism. J. Nutr. 136, 2653–2661 [DOI] [PubMed] [Google Scholar]
- 40. Szebenyi D. M., Liu X., Kriksunov I. A., Stover P. J., Thiel D. J. (2000) Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex. Evidence for asymmetric obligate dimers. Biochemistry 39, 13313–13323 [DOI] [PubMed] [Google Scholar]
- 41. Abdel-Aziz W., Malkas L. H., Wills P. W., Hickey R. J. (2003) The DNA synthesome. Its potential as a novel in vitro model system for studying S phase-specific anticancer agents. Crit. Rev. Oncol. Hematol. 48, 19–33 [DOI] [PubMed] [Google Scholar]
- 42. Ahuja D., Sáenz-Robles M. T., Pipas J. M. (2005) SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 24, 7729–7745 [DOI] [PubMed] [Google Scholar]
- 43. Békési A., Pukáncsik M., Muha V., Zagyva I., Leveles I., Hunyadi-Gulyás E., Klement E., Medzihradszky K. F., Kele Z., Erdei A., Felföldi F., Kónya E., Vértessy B. G. (2007) A novel fruitfly protein under developmental control degrades uracil-DNA. Biochem. Biophys. Res. Commun. 355, 643–648 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.