Background: KLF16 is the least characterized family member of recently described metabolic regulators.
Results: We extensively characterize mechanisms of DNA binding and chromatin coupling used by KLF16 to regulate metabolic gene expression.
Conclusion: KLF16 is a novel regulator of metabolic genes by regulatable coupling to Sin3-histone deacetylase complexes.
Significance: This knowledge reveals key mechanisms used by KLF16 as a regulator of metabolic gene expression.
Keywords: Endocrinology, Gene Silencing, Kruppel-like Factor (KLF), Metabolism, Sp1, Sin3
Abstract
Krüppel-like factor (KLF) proteins have elicited significant attention due to their emerging key role in metabolic and endocrine diseases. Here, we extend this knowledge through the biochemical characterization of KLF16, unveiling novel mechanisms regulating expression of genes involved in reproductive endocrinology. We found that KLF16 selectively binds three distinct KLF-binding sites (GC, CA, and BTE boxes). KLF16 also regulated the expression of several genes essential for metabolic and endocrine processes in sex steroid-sensitive uterine cells. Mechanistically, we determined that KLF16 possesses an activation domain that couples to histone acetyltransferase-mediated pathways, as well as a repression domain that interacts with the histone deacetylase chromatin-remodeling system via all three Sin3 isoforms, suggesting a higher level of plasticity in chromatin cofactor selection. Molecular modeling combined with molecular dynamic simulations of the Sin3a-KLF16 complex revealed important insights into how this interaction occurs at an atomic resolution level, predicting that phosphorylation of Tyr-10 may modulate KLF16 function. Phosphorylation of KLF16 was confirmed by in vivo 32P incorporation and controlled by a Y10F site-directed mutant. Inhibition of Src-type tyrosine kinase signaling as well as the nonphosphorylatable Y10F mutation disrupted KLF16-mediated gene silencing, demonstrating that its function is regulatable rather than constitutive. Subcellular localization studies revealed that signal-induced nuclear translocation and euchromatic compartmentalization constitute an additional mechanism for regulating KLF16 function. Thus, this study lends insights on key biochemical mechanisms for regulating KLF sites involved in reproductive biology. These data also contribute to the new functional information that is applicable to understanding KLF16 and other highly related KLF proteins.
Introduction
The Sp/KLF3 family contains 24 transcription factors that regulate genes via ubiquitous GC-rich genomic elements (1). The family is defined by extensive (>65%) sequence homology in their C-terminal zinc finger DNA binding domain (2, 3). In contrast, the N-terminal domains are variable, permitting differential cofactor recruitment, thereby defining the function of individual KLF proteins (3). In contrast to the transactivator SP1, KLF proteins activate or repress gene expression (2). As Sp/KLF-binding sites are ubiquitous throughout the genome, Sp/KLF family members likely maintain target gene specificity through diverse mechanisms such as cell type-specific, spatial, and temporal expression patterns as well as competition among family members on regulatory elements. Furthermore, differential coactivator/corepressor recruitment also influences gene regulation (1). Importantly, although ubiquitous GC-rich genomic regulatory regions serve as binding sites for Sp/KLF family members, target specificity may be maintained through distinct post-translational mechanisms (4). Emerging studies on KLF proteins suggest that they may have a wider role in regulation of metabolic and endocrine pathways than previously anticipated. For example, KLF11 regulates the insulin and Pdx1 genes, disruption of which gives rise to diabetes (MODY IV) (5). KLF11 is also involved in cholesterol, glucose, prostaglandin, and neurotransmitter metabolism, further supporting a key regulatory role for this protein in endocrinology (4, 6, 7). Recent studies on KLF9 and -13 suggest a role in steroid metabolism and function in endometrial cells (8), whereas KLF14 has been identified as a key candidate for type II diabetes (9). As KLF9, -13, and -14, along with KLF16, form a structurally related subfamily of KLF proteins, the BTEB-KLF group, they may possess similar functions. However, the precise interrelationship among BTEB-KLF subfamily members is unclear. For instance, although a targeted Klf9 mutation results in impaired fertility, there is concomitant up-regulation of endometrial Klf13, which may compensate for loss of Klf9 (10). KLF proteins likely provide a local regulatory network in uterine endometrium to maintain hormonal homeostasis through their effects on gene expression. However, evidence of a role for KLF16 in regulating endocrine-metabolic pathways is still lacking. Thus, in this study, our experimental strategy focused first on mechanistically characterizing the function of individual structural domains within KLF16 and subsequently testing the contribution of these mechanisms to the function of the whole protein. We report that KLF16 displays promiscuous selectivity for KLF-binding sites, possesses repression and activation domains that couple to histone deacetylase (HDAC) and histone acetyltransferase (HAT)-mediated pathways, respectively, and interacts with all three isoforms of the corepressor Sin3. KLF16 also regulates the expression of several genes essential for endocrine and metabolic function in a uterine cell model. To better understand these functions, we developed and refined by molecular dynamics the first computational three-dimensional model for the Sin3a PAH2-KLF16 Sin3-interacting domain (SID) complex, which reveals important features contributing to its formation as well as predicted potential mechanisms for its regulation. This prediction, which involves phosphorylation of Tyr-10 and potential disruption between the SID-PAH2, supports that this type of SID is regulated rather than constitutive, as is believed for MAD1 and HBP1. Finally, we experimentally confirmed this signal-induced post-translational mechanism that regulated KLF16 function at the SID level and identified a second signal-induced mechanism to regulate its nuclear translocation. Collectively, these investigations significantly expand our knowledge on the biochemistry of KLF proteins by defining, for the first time, key features that characterize a functional KLF16 protein, which are likely similar in highly related family members. Besides these biochemical discoveries, the characterization of KLF16 as a novel regulated transcription factor in uterine cell biology further underscores the importance of this family of proteins in endocrinology and metabolism.
EXPERIMENTAL PROCEDURES
Reagents and Cell Cultures
Uterine endometrial cell lines were obtained as follows: HEC1A cells (ATCC) and Ishikawa cells (Dr. P. Goodfellow, Washington University, St. Louis). HEC1A and uterine cells were grown in McCoy's 5A and DMEM supplemented with 10% FBS unless otherwise specified. Primary immortalized uterine cells were obtained as a gift from Dr. Hugh S. Taylor (Yale University, New Haven, CT).
KLF16 Plasmids and Constructs
Standard molecular biology techniques were used to clone WT-KLF16 or the KLF16 deletions as follows: N terminus (amino acids 1–124), C terminus (125–252), or C-terminal tail (209–252) into pCMV/Tag 2B (Stratagene) and pM/Gal4 vectors (Invitrogen) (11). Using WT-KLF16 in pM/Gal4 as a template, a library of mutants was generated by mutating serine, threonine, and tyrosine to nonphosphorylatable or phosphomimetic residues using the QuikChange mutagenesis kit (Stratagene). For the generation of nonphosphorylatable residues, serines and threonines were mutated to alanines, whereas tyrosines were mutated to phenylalanines. For phosphomimetic mutations, serines, threonines, and tyrosines were mutated to aspartic acids. The KLF16 Sin3-binding mutant was similarly generated by mutating residues 13 and 14 to prolines (KLF16ADPP). Additionally, WT-KLF16 and N-terminal KLF16 (amino acids 1–124) mutated to A13P/D14P (ADPP); Y10D and Y10F were also generated in pCMV/Tag2B vector. All constructs were verified by sequencing. For BTE reporter assays, we used previously described pBTE0 and pBTE6 reporters (15). CYP1A1 promoters were obtained from Dr. Yoshiaki Fujii-Kuriyama (Sendai, Japan) and cloned into the pGL3-basic luciferase vector (Promega) (16). Ishikawa or primary uterine cells (90% confluent in 6-well plates) were transfected with siRNA against KLF16 (ON-TARGET plus SMART pool siRNA, Dharmacon) or scrambled siRNA using Lipofectamine 2000 (Invitrogen). Each well received 25 nmol of siRNA, 4 μl of Lipofectamine 2000, and 2250 μl of Opti-MEM I (Invitrogen).
KLF16 Antibody Production, Immunofluorescence, and Confocal Microscopy
A 19-mer KLF16 peptide (AGLDVRAARREAASPGTPC) was synthesized, HPLC-purified, and conjugated to keyhole limpet hemocyanin by the Mayo Clinic Protein Core. Subsequently, a rabbit was immunized with the peptide, and testing and final bleeds were performed by Cocalico Biologicals (Reamstown, PA). Immunofluorescence and confocal microscopy were performed as described (12). KLF16 was localized with a rabbit antiserum (1:1000) and a FITC-AlexaFluor488-conjugated anti-rabbit secondary antibody (1:500; Invitrogen). Sin3a, HDAC1, HDAC2, and dimethyl and trimethyl H3K4 and H3K9 were detected using mouse monoclonal antibodies from Santa Cruz Biotechnology (Sin3a, HDAC1, and -2) or Abcam (di- and trimethyl H3K4 and H3K9) and a rhodamine-AlexaFluor555-conjugated anti-mouse secondary antibody (1:250; Invitrogen). Cellular DNA was stained with DAPI (Vector Laboratories). Fluorescence was observed using an argon-krypton laser on a Zeiss LSM-510 confocal microscope. Antibody specificity for immunostaining was determined using a peptide-blocking assay. KLF16 antibody was preincubated with 100-fold excess of the specific KLF16 peptide (see above) epitope at 4 °C overnight (supplemental Fig. 1). Immunofluorescence was then performed as described above.
CYP1A1 BTE Reporter Constructs and Luciferase Reporter Assay
Uterine cells were grown to 80% confluence in 6-well plates. GAL4 and pBTE reporter assays were performed as described previously (13, 14). Briefly, uterine cells were counted and electroporated. All results were normalized to total protein expression. Additionally, KLF16 protein expression was evaluated by Western blot using anti-FLAG (Sigma). Densitometry (to normalize for transfection efficiency) of protein expression was performed using ImageJ software (National Institutes of Health). Reporter activity was measured using the luciferase kit (Promega). All studies were performed in triplicate in at least three independent experiments with similar results. Error bars indicate S.E. Statistical significance was determined using squared t test analysis.
Random Oligonucleotide Binding Assay
The ROB assay was performed essentially as described (11). A random library of DNA sequences was generated by synthesizing oligonucleotides containing a 12-bp random core sequence flanked on each side by 20 bp (5′-TACAAGATCCGGAATTCCTAC-12N-GACGGATCCGGCGATAAGACA-3′). A forward primer (5′-TACAAGATCCGGAATTCC-3′) and a reverse primer (5′-TGTCTTATCGCCGGATCC-3′) were synthesized to amplify the library. For ROB, we followed a previously published protocol (11).
Cotransfection, Immunoprecipitation, and Western Blot Analysis
Uterine cells were electroporated with 5 μg of either pCMV-Tag2b, pCMVFLAG-KLF16, or pCMVFLAG-KLF16ADPP and either WT-Sin3a, WT-Sin3b-long form, or Sin3b-short form. Forty eight hours later, cell lysate was incubated with anti-FLAG beads (Sigma) overnight at 4 °C. Western blot analysis was performed as described above using anti-Sin3a or anti-Sin3b antibody (Santa Cruz Biotechnology).
Chromatin Immunoprecipitation Assay (ChIP)
Uterine cells were transfected with full-length FLAG-tagged KLF16 expression constructs or control vector as described above. Forty eight hours after transfection, immunoprecipitation was performed using anti-FLAG (Sigma) and ChIP (EZ-Chip kit, Millipore) performed as described previously (17). DNA shearing was performed to produce fragments 200–600 bp in size. A 249-bp PCR product representing the CYP1A1 promoter containing the BTE site was examined on a 2% agarose gel in KLF16-transfected cells compared with pCMV/Tag2b empty vector transfected control cells using following primers: 5′-TCCGCCACCTTTCTCTCCAATC-3′ (forward) and 5′-AAGTCCCCCAGCAACTCACCTGA-3′ (reverse).
Gel Shift Assays
To generate the GST-KLF16-zinc finger fusion protein, BL21 bacteria were induced with isopropyl 1-thio-d-galactopyranoside, and recombinant fusion proteins were then purified using GST-Sepharose beads (Amersham Biosciences) as described previously (11). Gel shift assays were performed as described previously (13, 15). Briefly, a double-stranded DNA probe containing either the BTE (5′-AGCTTGAGAAGGAGGCGTGGCCAACGCATG-3′), typical KLF protein DNA binding domains, the GC box (5′-ATTCGATCGGGGCGGGGCGAGC-3′), or GT box (5′-ATTCGATCGGGGTGGGGCGAGC-3′), highest or lowest consensus ROB sequences (5′-CCGCCCCCCCCC-3′ or 5′-TATATATTGTAT-3′), were end-labeled with [γ-32P]ATP and incubated with 1 μg of GST or GST-KLF16-zinc finger fusion protein, followed by gel electrophoresis. Where indicated, an excess of unlabeled wild-type BTE, GC, GT probe, highest or lowest consensus ROB probes, anti-GST antibody was added.
In Vivo Phosphorylation Assay
To detect in vivo phosphorylation of KLF16, Ishikawa cells were transfected with EV, FLAG-KLF16, or FLAG-KLF16Y10F. Forty eight hours later, the cells were treated with 5 mCi of [32P]orthophosphate for 4 h. The cells were then lysed in RIPA buffer containing 100 μm sodium orthovanadate. Immunoprecipitation was performed with anti-FLAG M2 beads (Sigma) for 2 h. The beads were washed with RIPA buffer as above. After elution, the denatured protein was run on a 12% gel and autoradiography detected using radiographic film.
RT-PCR
Total RNA was extracted from cells according to the manufacturer's instructions using an RNeasy kit (Qiagen), and 2 μg was used for cDNA synthesis using oligo(dT) primer using SuperScriptTM III first-strand synthesis system for RT-PCR (Invitrogen) per manufacturer's protocol. RT-PCR was performed using Platinum-TaqDNA polymerase (Invitrogen) per manufacturer's protocol. RT-PCR analysis was performed with the following primer sets: FYN, 5′-AATTTCAAATATTGAACAGCTCGGAA-3′(forward), 5′-TTTATAATGTTTGACATGGTCTCCTTT-3′(reverse); LYN, 5′-TTTCCTTATTAGAGAAAGTGAAACAT-3′(forward), 5′-TAATACAAGCCTTCTCCAATCTT-3′(reverse); SRC, 5′-TCCAGATTGTCAACAACACAGA-3′(forward), 5′-TTCTCTGCATTGAGCAGTAA-3′(reverse); YES, 5′-AAACTTGTTCCACTATATGCTGTTGTT-3′(forward), 5′-TTGTCTTCAATTAACCTTGCTAAACCA-3′(reverse); and CYP1A1, 5′-TGATAAGCACGTTGCAGGAGA-3′(forward), 5′-ATAGCACCATCAGGGGTGAG-3′ (reverse). Amplification of human β2-MICROGLOBULIN and GAPDH was performed in the same reaction for all samples as internal controls. Each experiment was done in triplicate. Real time PCR was performed using the IQ-SYBR Green Supermix (Bio-Rad) per the manufacturer's protocol.
CYP1A1 Functional Assays
Uterine cells were electroporated with 10 μg of either FLAG-KLF16 or corresponding empty vector. Thirty six hours later, the cells were treated with either 1 nm TCDD or DMSO. At 48 h, CYP1A1 activity determined using the p450-Glo reporter assay for CYP1A1 (Promega). The cells were treated with Luciferin-CEE, a CYP1A1-specific substrate (1:50), and incubated at 37 °C for 3 h. The medium was then treated with luciferase detection reagent, and readings were obtained using a luminometer. All samples were normalized for protein expression by Western blot using anti-FLAG antibody.
Bioinformatics Analysis
A genome-wide scan for the newly uncovered KLF16 binding consensus (GGGGGGGGCGG) and BTE consensus (GAGGCGTGGCCAAC) within the human genome promoters (−2500 to +1 relative to TSS) was performed using the regulatory sequence analysis tools (RSAT) software with no substitutions and one permitted substitution, respectively. Enrichment of genes for biological processes was analyzed using the gene ontology enrichment analysis software toolkit (GOEAST).
Molecular Modeling and Molecular Dynamic Simulation
The three-dimensional structure of the KLF16 SID complexed with the Sin3a-PAH2 domain was determined using a similar approach to our previously described Sin3a-MAD1 and Sin3a-KLF11 complex (18) as follows: (i) determining the three-dimensional structure of the KLF16 SID by homology modeling using the HBP1 SID taken from the Sin3a-HBP1 complex (Protein Data Bank code 1S5R) as a template (for primary sequences, see Fig. 5a); (ii) obtaining the three-dimensional structure of the Sin3a-PAH2 domain directly obtained from the Sin3a-HBP1 complex; and (iii) determining the three-dimensional complex structure of the KLF16 SID bound with the Sin3a-PAH2 domain (Protein Data Bank code 1G1E) (19) by docking the KLF16 SID to the Sin3a-PAH2 domain to achieve maximal intermolecular interactions between the two partners using AutoDock 3.0.5 (20). The resulting complex was refined by a 2.0-ns (1-fs time step) molecular dynamics (MD) simulation as described previously (21). The model was subjected to structure verification and evaluation using PROCHECK (22). The Ramachandran plot for the model showed 92.5% residues in most favored regions, and hence the model displays appropriate stereochemistry.
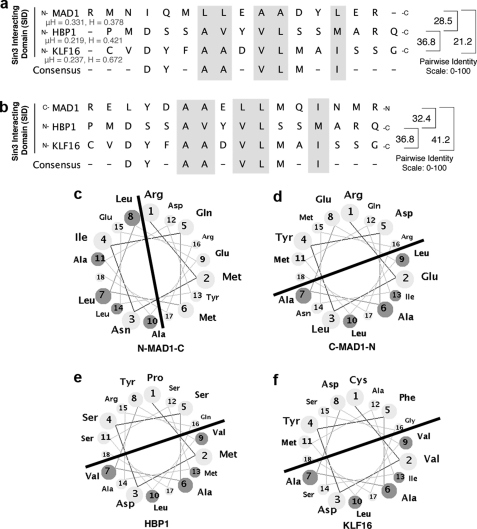
FIGURE 5.
Diagrammatic representation analyses of the MAD1, HBP1, and KLF16 SIDs, multiple SID sequence alignment. Forward (N to C terminus) (a) and reverse (C to N terminus) (b) SID alignments suggest that peptide polarity influences the interaction between the KLF16 SID with the Sin3a PAH2 domains, as the maximal similarity score derived by from pairwise alignments is obtained when the SID of KLF16 and HBP1 adopt the reversed orientation to MAD1. Overall, the high similarity among these peptides in hydrophobicity (H) and hydrophobic moment (μH) along with our previously published circular dichroism studies strongly support that the modeling of KLF16 SID as an amphipathic α-helix is appropriate (18). Comparative helical representation of the MAD1 (c and d), HBP1 (e), and KLF16 (f) SID; similar to MAD1 and HBP1 SID, KLF16 SID is predicted to form an amphipathic α-helix. The line separates the half of the helix containing the grouping of hydrophobic amino acid likely to interact with the Sin3a PHA2 domain. Noteworthy, as with the linear peptide alignment, when these helical wheels are built from the N to C orientation, the hydrophobic part of these helices adopt different orientations, suggesting that the mechanism used by these SID to interact with PAH2 complex are similar in HBP1 and KLF16 but different from MAD1. The main SID residues responsible for these interactions are either identical or conservative substitutions. Based on the sequence analyses of both proteins, these residues within the helix are critical for interacting with PAH2 and form the consensus DXXA(A/V)ϕVLXXIM. Helices were built using the Helical Wheel Applet from Virginia University.
Statistical Analysis
Results are expressed as means ± S.E. Each experiment was repeated in triplicates at least three times. An overall F-test of treatment mean equality and Bonferroni method of multiple comparisons (t tests) was used. Statistical analysis was performed using SAS software (SAS Institute, Cary, NC). All statistical tests were two-sided.
RESULTS
Biochemical Studies on DNA Binding Reveal That KLF16 Displays Distinct Selectivity for Different Sp-KLF cis-Regulatory Elements
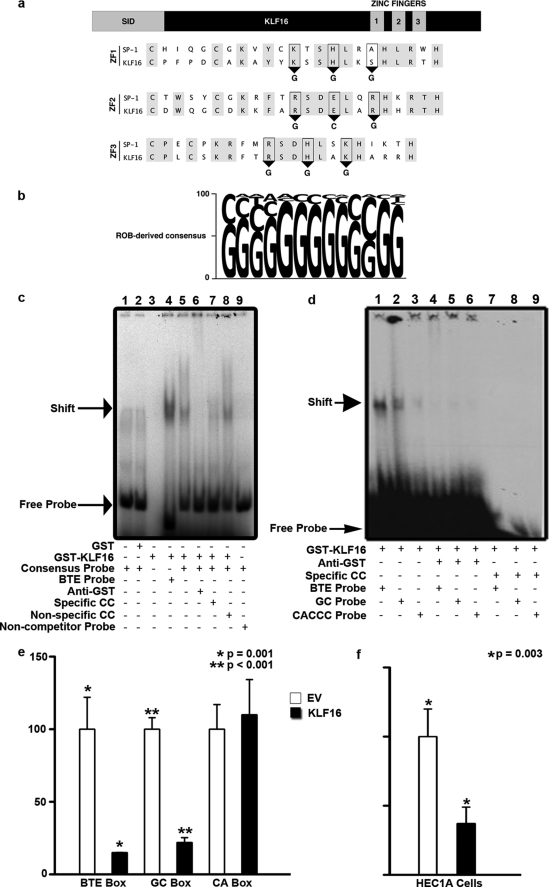
Our studies began by analyzing the DNA binding functions of KLF16, which are key for better understanding this protein and predicting candidate gene targets, as well as the potential competition among KLF family members that may share KLF16 DNA binding activity. Generally, KLF proteins target promoters differentially via three well characterized GC-rich elements as follows: the BTE (GAGGCGTGGCCAAC), GC box (CGGGGCGGGGC), and CA box (CACCC) (11). Interestingly, studies from DNA-bound zinc finger peptides permit prediction of putative DNA sequences that may be recognized by novel zinc finger proteins such as the KLF proteins (11, 23). The amino acid residues within the first (KHA), second (RER), and third (RHK) zinc fingers of KLF16 are identical to corresponding regions within SP1 (Fig. 1a) that bind the sequences GGG (ZF1), GCG (ZF2), and GGG (ZF3), respectively (23). Thus, a priori prediction suggests that KLF16 prefers GC-rich cis-regulatory elements over the CA box KLF sequence. To test this prediction, we performed ROB assays with a library of DNA sequences consisting of 12-bp random cores flanked bilaterally by 16 bp of known sequence, the results of which were aligned to derive a consensus sequence (Fig. 1b). KLF16 binding to this consensus oligonucleotide (GGGGGGGGGCGG) was confirmed by EMSA. Additionally, KLF16 binding specificity was validated by both supershift assay showing complex disruption with anti-GST antibodies and specific site-based competition with cold probes (Fig. 1c). Together, these experiments demonstrated that KLF16 self-selected a GC-rich sequence in vitro that was similar to, yet distinct from, previously described KLF-binding sites. Characterization of this specific DNA element was critical for subsequent identification of candidate KLF16 gene targets by genome-wide analyses of cis-regulatory sequences. Additionally, we comparatively analyzed the binding of KLF16 to the consensus probe with other KLF cis-regulatory sequences, namely the GC, CA. and BTE boxes (Fig. 1, c and d) (1, 2). Densitometric analysis of EMSA data revealed that KLF16 revealed a 5-fold greater preference for the BTE probe compared with the consensus ROB probe (Fig. 1c). In addition, a 17- and 10-fold greater preference for the GC box (very similar to the consensus ROB sequence) and the BTE, respectively, compared with the CA box was observed (Fig. 1d). KLF16 binding to all elements was specific as confirmed by supershift and cold probe competition (Fig. 1, c and d). Thus, the combination of an unbiased approach (ROB), together with three candidate-based studies using known KLF cis-regulatory domains, provided the best comparative information available for any KLF protein, as well as confirmed the a priori prediction (23). KLF16 recognized three different GC-rich sequences with varying affinity, positioning this protein as a candidate to regulate similar sites in gene promoters.
FIGURE 1.
KLF16 DNA binding domain displayed selectivity for distinct cis-regulatory elements. a, sequence alignment of the zinc fingers of SP1 and KLF16 revealing critical conserved residues (gray). Predicted KLF16 binding to GGG, GCG, and GGG bases via zinc fingers 1–3, respectively (ZF 1, 2, 3), is shown. b, ROB assay for KLF16 in uterine cells. 32P-Labeled ROB oligonucleotides were incubated with GST-KLF16 and separated by nondenaturing gel electrophoresis. The consensus sequence derived from 45 sequenced oligonucleotides after seven rounds of ROB is shown. The font size of each nucleotide corresponds to frequency of occurrence. c, EMSA using either 32P-labeled consensus KLF16-ROB (lanes 1, 2 and 5–8), BTE (lane 4), or noncompetitor probe (lane 9) with either 1 μg of GST protein (lane 2), GST-KLF16-zinc finger (ZF) (lanes 3–9), or probe alone (lane 1). Where indicated, the following were also added: 1 μg of anti-GST (lane 6), 500 m excess of unlabeled KLF16-ROB consensus probe (cold competitor, lane 7), or 500 m excess unlabeled noncompetitor probe (lane 8). Specific complexes formed between GST-KLF16 and either labeled BTE or KLF16-ROB probe (lanes 5 and 8) are indicated by an arrow. Anti-GST disrupted the GST-KLF16-ZF/consensus probe complex (lane 6). Addition of excess unlabeled consensus probe competed for the binding (lane 7), whereas an unrelated noncompetitor probe did not (lane 8). GST-KLF16-ZF did not shift the labeled noncompetitor probe (lane 9). Lane 1 contained only labeled KLF16-ROB consensus probe and no protein, whereas lane 3 contained 1 μg of GST-KLF16-ZF fusion protein alone. d, EMSA comparing GST-KLF16-ZF (1 μg per lane; lanes 1–9) protein binding to either 32P-labeled CYP1A1 BTE (lanes 1, 4, and 7), KLF consensus element (GC box) (lanes 2, 5, and 8), and CA box (lanes 3, 6, and 9). GST-KLF16-ZF protein (lanes 1–3) specifically bound all three KLF-binding elements (lanes 1–3, arrow indicates the shift), the BTE box with highest affinity (lane 1), robust binding to the GC box (lane 2), and the CA box with least affinity (lane 3). KLF16 binding to all elements was specifically disrupted by 1 μg of anti-GST (lanes 4–6: lane 4, BTE; lane 5, GC; lane 6, CA). Binding was lost on addition of 500 m fold excess unlabeled specific cold competitor (lanes 7–9: lane 7, BTE; lane 8, GC; lane 9, CA). e, uterine cells were cotransfected with 7.5 μg of FLAG-KLF16 or corresponding empty vector and either a 6×-BTE-, 6×-GC-, or 6×-CA-luciferase reporter vector (2.5 μg). Luciferase levels normalized to total protein levels and binding showed significant repression compared with empty vector, when KLF16 was cotransfected with either the 6×BTE (*, p = 0.001) or 6×GC-luciferase reporter (**, p < 0.001). There was no repression of 6×-CA-luciferase (f). HEC1A uterine cells were cotransfected for 48 h with either 7.5 μg of FLAG-KLF16 or corresponding parent pCMV/FLAG vector and 2.5 μg of luciferase reporter containing 6×-tandem BTEs. Luciferase activity normalized to protein concentrations show that compared with EV, KLF16 decreased luciferase expression by 63% (*, p = 0.003).
To correlate the EMSA results with transcriptional activity, we performed reporter assays using luciferase constructs containing six tandem repeats of the BTE, GC, or CA box in uterine cells. Results from these experiments were congruent with relative selectivity defined by EMSA (Fig. 1e). KLF16 exerted negligible silencing effects on the 6×-CA box compared with empty vector. In contrast, KLF16 inhibited luciferase activity driven by both the 6×-GC box (80%; p < 0.001) and 6×-BTE (85%; p = 0.001) compared with empty parental vector (Fig. 1e). KLF16 was also a repressor of BTE-luciferase in another uterine cell line, HEC1A (63%; p = 0.003) (Fig. 1f). KLF16 therefore displayed sequence selectivity for an SP1-like GC-rich binding site, distinguishing it from others like KLF1 that bind CA boxes, which has been the paradigm for all KLF transcription factors.
To complement these experiments, we performed genome-wide bioinformatics analysis using the newly uncovered KLF16 and BTE consensus sequences as probes, which revealed ∼354 and 28 matches, respectively, the majority of which map to characterized genes. Ontological analysis of these genes revealed significant enrichment for targets involved in metabolic, endocrine, and reproductive functions (p < 0.05) (supplemental Table 1). These data represent the most detailed characterization to date of the functional properties of the KLF16 DNA binding domain, critical for achieving promoter recognition. In addition, the presence of the preferred KLF16-binding site in a significant number of genes involved in metabolic and endocrine functions suggests that KLF16 may play a role in their regulation.
KLF16 Regulates Endogenous Target Genes for Metabolic and Endocrine Pathways through Binding to Distinct Sp-KLF Sites
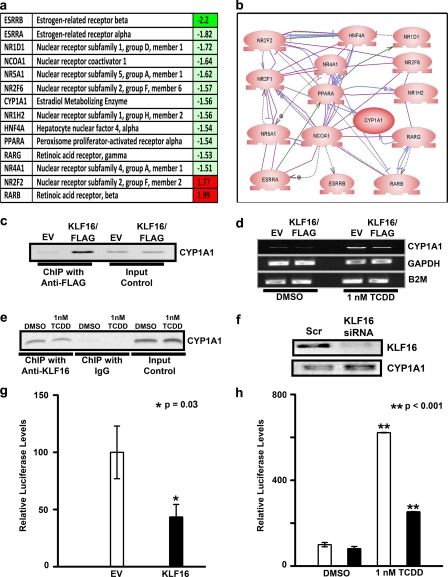
To identify bona fide KLF16 gene targets that participate in metabolic and endocrine functions in endometrial cells, we utilized several complementary approaches as follows: pathway-specific microarray analysis with pathway reconstruction, ChIP, promoter assays, and EMSA. Overexpression of KLF16 in uterine epithelial cells combined with microarray analysis revealed that KLF16 regulates genes comprising key nodes within the metabolic-endocrine biosynthetic pathways such as NRR2 and RARB, nuclear hormone receptors ESRRA and ESRRB, and the estrogen-metabolizing enzyme CYP1A1 (Fig. 2, a and b). Therefore, these results demonstrate that KLF16 is at least as important as KLF9 and -13 in regulating endometrial genes, most notably those implicated in metabolic and endocrine homeostasis.
FIGURE 2.
KLF16 functions as transcription factor involved in the regulation of metabolic and endocrine gene expression in uterine cells. a, selected identifiers for KLF16-regulated genes. Most of these genes (80%) are down-regulated (green) by this transcription factor, which is congruent with the predominant gene-silencing activity of KLF16. Notably, from these genes, 22% participate in estrogen metabolism. b, semantic-based pathway reconstruction algorithms integrate most of the genes found regulated by KLF16 in the PCR microarray into a seamless pathway, showing active interaction among its nodes. KLF16 regulated the expression of several genes connected with metabolism of sex steroid as well as environmental contaminant toxins in endometrial cells. c, chromatin immunoprecipitation was performed on uterine cells transfected with either pCMV/FLAG- or pcDNA3HIS-KLF16 or corresponding empty vectors. Anti-FLAG or anti-HIS antibodies were used for immunoprecipitation, respectively. Shown is a representative sample where FLAG-KLF16 but not empty vector bound a 249-bp CYP1A1 genomic element containing the BTE (−55 to −41) in uterine cells. d, uterine cells were transfected with either FLAG-KLF16 or corresponding empty vector. The cells were treated with 1 nm dioxin to induce CYP1A1 expression. Total RNA was extracted at 48 h, and RT-PCR was done. KLF16 repressed TCDD induction of CYP1A1 mRNA expression by 50% (p < 0.05) compared with the corresponding empty vector. Housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β2 microglobulin (B2M) were used as loading controls. e, ChIP was performed for endogenous KLF16 in the presence or absence of 1 nm dioxin. Positive amplification of the CYP1A1 promoter demonstrates that KLF16 binds this genomic element described above both in the presence and absence of dioxin. f, uterine cells were transfected with KLF16 siRNA to knock down endogenous KLF16 levels, which was confirmed by Western blot using anti-KLF16 (upper panel). CYP1A1 mRNA levels were increased 2-fold in cells treated with KLF16 siRNA (lower panel). g, uterine cells were transfected with 7.5 μg of FLAG-KLF16 or corresponding empty vector and 2.5 μg of CYP1A1 promoter-luciferase constructs containing the BTE (−55 to −41). Results of luciferase-reporter assays normalized to protein expression revealed that KLF16 significantly repressed CYP1A1-luciferase activity compared with empty vector control (46%; *, p = 0.03). h, CYP1A1 enzymatic activity was measured using a well characterized CYP substrate-reporter assay. Uterine cells were transfected with either KLF16 (black bars) or empty vector control (white bars) and subsequently treated with either 1 nm TCDD or DMSO control. A luciferin-derived CYP1A1 substrate was added to the medium and enzyme activity determined by reporter activity. CYP1A1 enzymatic activity was significantly inhibited by KLF16 (60%; **, p < 0.001) in TCDD treated cells.
Given our microarray results and recognition by KLF16 of the BTE site (present in several cytochrome P450 enzymes), we focused on studying KLF16-mediated regulation of CYP1A1 in uterine cells. ChIP assays demonstrated that KLF16 bound the endogenous CYP1A1 promoter (Fig. 3b) in uterine cells, confirming this metabolic enzyme as a direct target of KLF16 in these cells (Fig. 2c). CYP1A1 is not constitutively expressed in uterine endometrium but rather is readily induced by a variety of exogenous and endogenous agents such as environmental toxins and the estrogens (16, 24). Thus, to study the effect of KLF16 on the CYP1A1 promoter, we initially induced up-regulation of this gene by treating cells with a potent inducer, TCDD. Fig. 2d shows that KLF16 repressed TCDD-induced up-regulation of CYP1A1 mRNA expression by 50% (p < 0.05). As KLF16 and TCDD regulated CYP1A1 mRNA expression by binding to distinct promoter elements, we further confirmed that endogenous KLF16 binds to the CYP1A1 BTE by ChIP, in the presence and absence of TCDD (Fig. 2e). We thereby confirmed that KLF16 has a role in direct regulation of endometrial CYP1A1 expression. To further elucidate the role of KLF16 in regulation of endometrial CYP1A1 expression, we transfected cells with siRNA against KLF16 to diminish KLF16 protein expression; transfection resulted in a 4.5-fold decrease in KLF16 protein levels (Fig. 2f, upper panel). As expected, decreased KLF16 levels resulted in a 2-fold increase in CYP1A1 mRNA expression (Fig. 2f, lower panel). Moreover, to determine whether KLF16 regulated CYP1A1 expression via its specific BTE site (−55 to −41), we performed luciferase-reporter assays using a CYP1A1-BTE-luciferase construct (Fig. 2g). KLF16 significantly repressed luciferase expression levels compared with empty vector (46%; p = 0.03), congruent with the repression of mRNA level.
FIGURE 3.
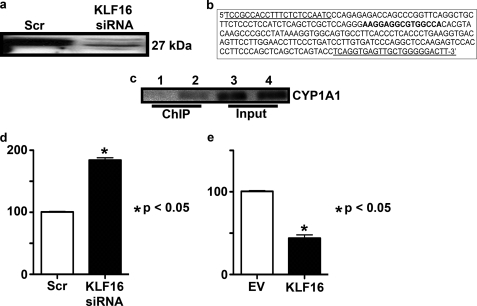
KLF16 directly repressed CYP1A1 expression in primary endometrial cells. a, primary endometrial cells were transfected with KLF16 siRNA, which resulted in >90% diminished protein levels as detected by Western blot using anti-KLF16. b, 249-bp amplicon of the KLF16 promoter showing the location of the forward and reverse primers (underlined bases) as well as the BTE (boldface type) located at base pairs −55 to −41 relative to the transcription start site. c, ChIP was performed for endogenous KLF16, which confirms that KLF16 binds the CYP1A1 promoter BTE-element in primary endometrial cells. d, real time PCR was used to determine the effect of decreased KLF16 expression on CYP1A1 mRNA expression in these cells. CYP1A1 normalized to a housekeeping gene (β2-microglobulin) was increased 2-fold (p < 0.05) in cells transfected with KLF16 siRNA compared with control, which is similar in magnitude to our findings in the uterine cell line. e, primary uterine cells were transfected with FLAG-KLF16 or corresponding empty vector along with the 6×CYP1A1-BTE luciferase construct. Results of luciferase-reporter assays normalized to protein expression revealed that KLF16 significantly repressed luciferase activity compared with empty vector control (50%; *, p = 0.01).
To evaluate the impact of KLF16 regulation on CYP1A1 at the protein level, we measured CYP1A1 enzymatic activity using a well characterized CYP reporter assay. For this purpose, we transfected uterine cells with either KLF16 or empty vector control and subsequently treated them with 1 nm TCDD. A luciferin-derived CYP1A1 substrate was added to the medium over the cells, and enzyme activity was determined by reporter activity (Fig. 2h). CYP1A1 enzymatic activity was significantly inhibited by KLF16 (60%, p < 0.001). Thus, KLF16 reduced the amount of enzymatically active CYP1A1 in endometrial cells. Results of this experiment therefore validated both PCR and reporter assays (Fig. 2, d and e, respectively).
We further confirmed KLF16 regulation of CYP1A1 in primary endometrial cells. Primary endometrial cells express KLF16, and thus we transfected the cells with KLF16 siRNA to achieve >90% diminished KLF16 protein levels (Fig. 3a). We then determined the effect of these diminished KLF16 levels using KLF16 siRNA on CYP1A1 mRNA expression in primary uterine cells using quantitative PCR and found a 2-fold increase in CYP1A1 levels with diminished KLF16 levels, confirming our findings in the uterine cell line (Fig. 3d). To determine whether endogenous KLF16 binds to the CYP1A1 promoter (Fig. 3b) BTE in primary cells, we performed ChIP with our KLF16 antibody. Fig. 3c shows that endogenous KLF16 bound the CYP1A1 promoter element in these cells. Finally, we transfected primary uterine cells with KLF16 and a 6×-BTE-luciferase reporter construct. KLF16 repressed reporter gene expression by 50% (p = 0.01) in these cells (Fig. 3e). Therefore, our results for experiments performed in primary uterine cells confirmed our observations in the uterine cell line, further supporting the role of KLF16 in CYP1A1 regulation in these cells.
CYP1A1 is a well known steroid and toxin-metabolizing enzyme in target tissues such as the uterine endometrium (25). In fact, this enzyme oxidizes several endocrine (e.g. estrogen) and environmental agents into metabolites that range in activity from inert to highly toxic, such as those that cause endometriosis (26). In combination, these results demonstrate that KLF16 regulates endogenous uterine genes involved in reproductive endocrinology via GC-rich Sp/KLF sites present in their endogenous promoters, in agreement with our biochemical experiments on DNA binding.
Transcriptional Regulatory Domains of KLF16 Displays Interactions with Three Sin3 Isoforms and p300
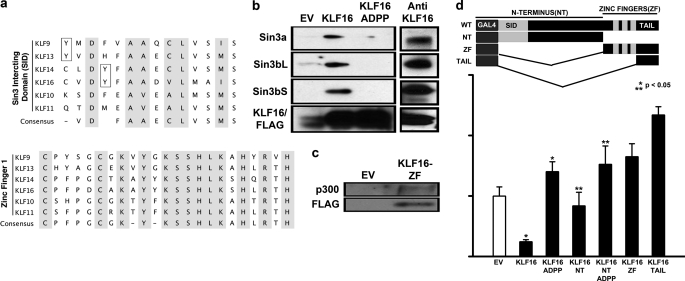
We next investigated the interaction of KLF16 with chromatin-remodeling complexes that could potentially be recruited to regulate endometrial gene promoters. KLF16 possesses a SID between residues 1 and 26 (Fig. 4a, upper panel). Mechanistically, the presence alone of this motif cannot predict which Sin3 isoform (Sin3a, Sin3bL (long form), or Sin3bS (short form)) is utilized by KLF16 for transcriptional regulation. Consequently, to determine whether KLF16 showed Sin3 isoform selectivity, we performed coimmunoprecipitation in cells cotransfected with either wild-type KLF16 or a mutant carrying a disruption in the Sin3a-binding site, A13P/D14P (KLF16ADPP) and the Sin3 isoforms. This mutant disrupts complementary folding previously described for the Sin3 PAH2-SID interaction (18). Furthermore, we additionally immunoprecipitated endogenous KLF16 from nontransfected uterine cells to determine the interaction of endogenous KLF16 with the Sin3 cofactors. Whereas both FLAG-KLF16 and endogenous KLF16 bound all three Sin3 isoforms, the ADPP mutant abolished binding (Fig. 4b). This study is the first to describe the interaction of any SID-containing KLF with the three human Sin3 isoforms. KLF16 could therefore be potentially coupled to diverse Sin3 complexes and their chromatin remodeling pathways. Differential Sin3-cofactor recruitment may ensure gene silencing under varied cellular and promoter contexts (27). As the KLF16 SID is identical to the SID in two related endometrial BTEB regulators, KLF9 and -13 (Fig. 4a, upper panel), these results have a wider mechanistic implication for delineation of the regulatory role of the BTEB-Sin3 interaction.
FIGURE 4.
KLF16 couples to different chromatin pathways involved in histone acetylation-deacetylation. a, upper panel, sequence alignment of the SID domain of BTEB-KLF9, -13, -14, and -16 reveals the discrepant position of tyrosine residues within (KLF14 and -16) and outside (KLF9 and -13) the minimal SID (shaded). Also shown are the SID domains of closely related TIEG-KLF10 and -11. Unlike KLF11, KLF16-SID does not contain a candidate phosphorylatable Ser/Thr residue. Lower panel, sequence alignment of the first zinc finger of all BTEB-KLFs. KLF16-ZF1 has high sequential homology to KLF11- and -13-ZF1 and also interacts with p300/HAT (completely conserved residues shaded). b, left panel, uterine cells transfected with either 5 μg of pCMV/FLAG vector, pCMV/FLAG-KLF16, or pCMV/FLAG-KLF16ADPP (Sin3-binding mutant) and 5 μg of either Sin3 isoforms for 48 h were subjected to immunoprecipitation using anti-FLAG-agarose beads. Immunocomplexes were analyzed by Western blot using either anti-Sin3a or anti-Sin3b. Sin3a as well as both Sin3b isoforms (long and short) were present in immunocomplexes obtained from cells transfected with FLAG-KLF16 but not from cells transfected with either the empty vector or Sin3-binding mutant FLAG/KLF16-ADPP. Right panel, immunoprecipitation of endogenous KLF16 was performed on uterine cells using an antibody to KLF16. Immunocomplexes were analyzed by Western blot using either anti-Sin3a or anti-Sin3b. Sin3a as well as both Sin3b isoforms (long and short) were detected in immunocomplexes formed by endogenous KLF16. c, uterine cells were cotransfected with 5 μg of FLAG-KLF16-ZF or FLAG-EV and 5 μg of p300. Immunoprecipitation was performed using anti-FLAG-agarose beads, and Western blot using anti-p300 showed that KLF16 bound p300/HAT via the ZF domain (d). Uterine cells were cotransfected for 48 h with either empty vector (pM), full-length, or mutation/deletion KLF16 constructs and Gal4-luciferase reporter vector. Luciferase activity normalized to lysate protein concentration showed that although full-length KLF16 and KLF16-N terminus repressed reporter expression 50% (*, **, p < 0.05, respectively), the repression was reversed when either full-length or N-terminal KLF16 constructs contained the ADPP mutation that abrogated Sin3a binding. Paradoxically, the C terminus and the C-terminal tail of KLF16 increased reporter expression.
To further characterize biochemical mechanisms underlying Sin3/KLF16-mediated silencing we focused on the best characterized Sin3 isoform Sin3a. The regulatory role of KLF16 in uterine cells was evaluated by site and deletion mutagenesis by Gal4-based reporter assays. Fig. 4d demonstrates that although the full-length KLF16 and KLF16 N terminus displayed gene silencing (50% compared with EV; p < 0.05), the silencing was reversed by the ADPP mutation in the context of either WT-KLF16 or N-terminal-KLF16. Thus, KLF16 transcriptionally repressed reporter gene expression in uterine cells via an Sin3-dependent mechanism, which was disrupted by the ADPP mutation.
Surprisingly, when isolated, both the C terminus of KLF16 as well as the C-terminal tail (comprised of the region located between the third zinc finger and the stop codon) activated gene expression, suggesting that this region may bind coactivator(s) (Fig. 4d). These data are congruent with sequence analysis, which demonstrates that KLF16 contains a p300/HAT recruitment domain that maps to the first zinc finger (Fig. 4a, lower panel). This domain has also been fully characterized in the highly related KLF13 protein (28). We show here that like KLF11 and -13, KLF16-ZF1 bound the p300 coactivator complex in uterine cells (Fig. 4c). However, when full-length KLF16 was evaluated by Gal4 assays as well as on KLF promoter sites, it behaved as a repressor (Figs. 1, e and f, 2g, and 4d). In this regard, KLF16 may work similar to KLF11 by coupling to silencing or activating chromatin-remodeling complexes under specific promoter and genomic contexts (5). Overall, these findings highlight the diverse potential capabilities of KLF16 as a repressor and activator of gene regulation in uterine cells.
Molecular Mechanisms Underlying the Interaction of KLF16 with Sin3 Reveal That Silencing by This Transcription Factor Is Regulated Rather than Constitutive
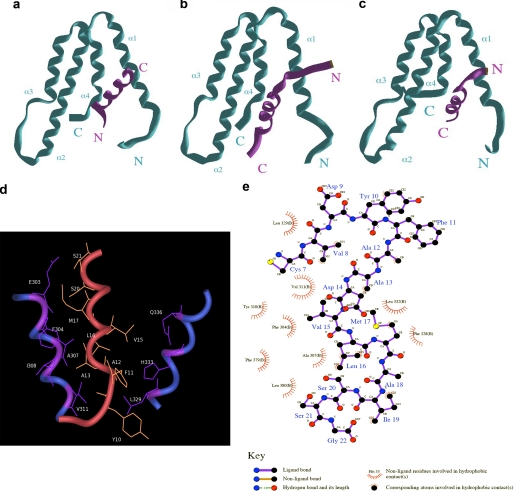
The availability of previously described structures for two different types of SIDs has advanced our understanding on how Sin3-HDAC complexes are recruited by well known transcriptional repressor proteins, such as MAD1 and HBP1 (19, 29). These structures have shown that although both of these SIDs bind to a hydrophobic pocket formed by the 4 helix-bundle fold formed by the PAH2 domain of Sin3, they adopt a reverse N to C terminus orientation. Notably, alignment of the KLF16, MAD1, and HBP1 SIDs, as shown in Fig. 5a, further clarifies the need of performing these careful studies because all three peptides, previously defined experimentally as α-helices by NMR, CD, and/or x-ray crystallography, can be aligned in different manners (18, 19, 29). These alignments were carefully evaluated and scored (maximal similarity score) in a pairwise fashion using an edit distance algorithm focused on the α-helical nature of the peptides in question so as to reward absolute conservation of residues as well as conservation of residue character. Residues were classified as hydrophobic (Gly, Ala, Val, Leu, Ile, Met, Phe, Trp, and Pro), hydrophilic (Ser, Thr, Cys, Tyr, Asn, and Gln), polar positive/hydrophilic (His, Lys, and Arg), or polar negative/hydrophobic (Asp and Glu). The algorithm was constructed as follows: for sequence X (x1, x2 … xi) and sequence Y (y1, y2 … yi), opening gap penalty δ = −10, gap extension penalty φ = −5, residue match αij = +100 (absolute) or +50 (hydrophobic class match) or +25 (hydrophilic class match), and absolute mismatch ζj = 0. Therefore, the maximal similarity score (MSS) between sequences X and Y = Σαj + Σζj + Σδ + Σφ. Interestingly, to achieve a maximal alignment score when all three SID peptides are oriented N to C terminus, the MAD1 SID must be moved to the left within the matrix, introducing gap penalties for two of three pairwise comparisons (MSSMAD1/HBP1 = 28.5, MSS HBP1/KLF16 = 36.8, and MSSMAD1/KLF16 = 21.1). However, when the MAD1 peptide is aligned in a reverse orientation to the other peptides (Fig. 5b), the scoring of each pairwise alignment improves significantly (MSSMAD1/HBP1 = 32.4, MSS HBP1/KLF16 = 36.8, and MSSMAD1/KLF16 = 41.2). Moreover, the overall similarity-identity between the MAD1 and KLF16 SID was obtained by this reversed alignment, raising the possibility that these peptides might adopt different orientations within the PAH2 domain, as shown previously for HBP1 (29). Because SIDs form amphipathic α-helices, we subsequently built these peptides using helical wheels to define the orientation of their hydrophobic surfaces. Fig. 5 (c–f) shows that, congruent with the SID alignments, these wheels support the idea that the KLF16 SID adopts an orientation in solution, which is similar to HBP1 but different from MAD1. More importantly, there data show that the KLF16 SID may dock into the PAH2 four helices bundle in an N to C terminus orientation with its hydrophobic face upward within the Sin3 hydrophobic pocket. These features are important not only for understanding the mechanisms underlying transcriptional silencing by KLF16 but also provide useful information for the future design of small molecules that can therapeutically modulate this function. The KLF16 SID has been previously demonstrated to adopt an α-helix configuration in solution (18). These data, along with the valuable information gained from SID alignments, led us to develop homology-based structural models based upon previously solved structures for the MAD1 SID-Sin3 PAH2 and the HBP1 SID-Sin3 PAH2 (19, 29). The three-dimensional structure of the KLF16 SID-Sin3 PAH2 complex derived from molecular modeling using the PDB code 1S5R structure as template is depicted in Fig. 6. The Sin3a-KLF16 complex (Fig. 6c) consists of the PAH2 domain that adopts a left-handed, up-and-down, four-helix bundle structure with residues in all four helices, as well as in the turn regions, defining a compact structural domain with an extensive hydrophobic core and an amphipathic α-helix for the KLF16 SID. Interestingly, the KLF16 SID interacts with Sin3a in similar manner to HBP1, which adopts a reversed orientation relative to the Sin3a-MAD1 complex (Fig. 6, a–c). A 2.0-ns MD simulation reveals that the KLF16 SID prefers to interact with Sin3a with its α-helix buried between the hydrophobic pocket formed by the four-helix bundle structure formed by the Sin3a-PAH2 domain, as shown in Fig. 6, c and d. However, experiments performed by docking the KLF16 SID in the reverse orientation, as performed for the Sin3a-MAD1 complex by our laboratory, was disrupted before completion of the 2-ns MD simulation (data not shown) (18). Thus, the model presented here reflects the most plausible structural configuration of the KLF16 SID-Sin3A PAH2 complex. The most critical interactions between the KLF16 SID and Sin3A-PAH2 domain shown in Fig. 6e are hydrophobic in nature and include the following: Sin3 Leu-329 and SID Tyr-10; Sin3 His-333 and SID Tyr-10, SID Phe-11 and SID Ala-12; Sin3 Gln-336 and SID Val-15; Sin3 Glu-303 and SID Ser-20, SID Ser-21; Sin3 Phe-304 and SID Met-17; Sin3 Ala-307 and SID Leu-16; Sin3 Ile-308 and SID Ala-13; Sin3 Val-311 and SID Leu-16. Mutational interference with key SID residues supports the validity of this model. Pro substitutions at Ala-13 and Asp-14 disrupt the SID function (as shown in Figs. 6e and 4d) likely because of the ability of this residue to destabilize helix formation. In addition, careful analysis of the KLF16 SID-Sin3 PAH2 model predict that modification of Tyr-10, such as phosphorylation, may alter the stability of this complex, as this residue contributes hydrophobic bonds to the binding. Similarly, because, as demonstrated by NMR (19, 29), the formation of the SID-PAH2 complex follows an induced fitting mechanism, modifications of this residue may interfere with complex formation. This analysis, along with the fact that KLF16 contains a Pro-rich binding site for the Src homology 3 domain Src-type kinases (amino acids 58–63; Fig. 7b) in close proximity to its SID, predicts that rather than functioning constitutively, the KLF16 SID may be regulated by Tyr modifications. If true, KLF16 would be the first SID-containing protein whose mechanism is amenable to this type of modification. Therefore, molecular modeling combined with molecular dynamic simulations shed insights into potential mechanisms responsible for the functional coupling of KLF16 to the Sin3-HDAC complex at atomic resolution and provide predictions of potential mechanistic importance, as experimentally tested below.
FIGURE 6.
Predictive structural model for the Sin3a-PHA2-KLF16 SID complex. a, MAD1-SID-PAH2 complex. The MAD1 SID α-helix adopts an N to C orientation within the hydrophobic pocket form by the four helices bundle of PAH2. b, HBP1-SID-PAH2 complex. The HBP1 SID α-helix adopts a reverse C to N orientation within the hydrophobic pocket formed by the four helices bundle of PAH2. c, KLF16 SID-PHA2 complex. The KLF16 SID α-helix adopts an HBP1-like N to C orientation within the hydrophobic pocket formed by the four helices bundle of PAH2. Similar experiments performed by docking of the KLF16 SID in a reverse MAD1-like orientation were not stable after MD simulation. d, close up view displaying the primary hydrophobic contacts that contribute to the KLF16 SID-PAH2 complex. e, simplified view of the most important bonds responsible for the formation and function of the KLF16 SID. This model predicts that modifications of Tyr-10, which contributes to the formation and/or stability of the KLF16 SID-PAH2 complex, may impact on the regulation of the silencing activity of this KLF protein.
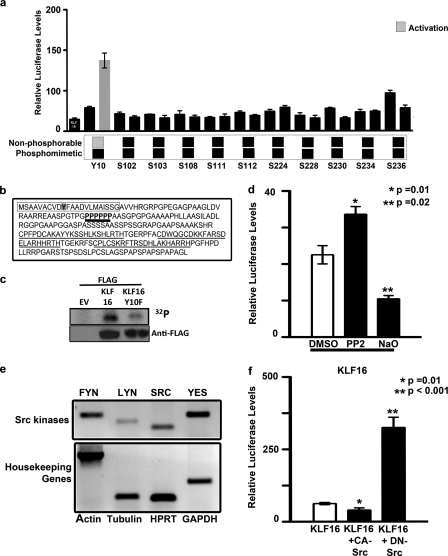
FIGURE 7.
Gene silencing activity KLF16 is regulated by tyrosine modifications. a, uterine cells were cotransfected with constructs from a library of in vitro mutagenized KLF16-Gal4 proteins where individual serine, threonine, and tyrosine residues were replaced with either alanine (phenylalanine for tyrosine) or aspartic acid as indicated. The Gal4-luciferase reporter system was used to evaluate the transcriptional role of each mutant. Whereas all of the serine/threonine mutants repressed reporter expression analogous to WT-KLF16, the KLF16Y10F mutant (where tyrosine 10 in the WT protein was replaced by nonphosphorylatable phenylalanine) reversed repression. b, N terminus of KLF16 contains a proline-rich domain (boldface type, black underline). Also indicated are the Sin3-interacting domain (box) containing tyrosine 10 (shaded), and the three zinc fingers (gray underline). The proline-rich domain is located close to the KLF16-SID. c, to detect in vivo phosphorylation of KLF16, Ishikawa cells were transfected with EV, FLAG-KLF16, or FLAG-KLF16Y10F and subsequently labeled with [32P]orthophosphate. Phosphorylation of KLF16 was detected by immunoprecipitation using anti-FLAG followed by exposure to autoradiography film. d, uterine cells transfected with 10 μg of FLAG-KLF16 were treated with either Src tyrosine kinase-specific inhibitor PP2 or tyrosine phosphorylase-specific inhibitor sodium orthovanadate. PP2 reversed reporter repression, compared with vehicle-treated control (*, p = 0.01). In contrast, sodium orthovanadate augmented repression (**, p = 0.02). e, RT-PCR was used to evaluate the expression of Src family members in uterine cells. Four out of eight were expressed: FYN, LYN, SRC, and YES kinases. Human β-actin, β-tubulin, HPRT, and GAPDH were used as internal controls. f, luciferase reporter assays were performed in uterine cells transfected with either 5 μg of FLAG-KLF16 or corresponding EV and 5 μg of either a constitutively active or dominant negative (kinase-inactive) SRC kinase construct along with 2.5 μg of 6×-BTE-luciferase reporter construct. Luciferase levels normalized to protein concentration and parent vector showed that whereas luciferase reporter was repressed by KLF16 and constitutively active SRC (*, p = 0.01), repression was reversed when KLF16 was cotransfected with kinase inactive DN-SRC (**, p < 0.001) indicating the role of Src kinase in post-translational regulation of KLF16.
To better understand whether SID-KLF16-mediated repression was regulated or constitutive, we performed in vitro mutagenesis of all serine, threonine, and tyrosine residues that were predicted (Scansite, Netphos, and ELM databases) to be targets of kinase signaling. These residues were mutagenized to either nonphosphorylatable alanine (phenylalanine for tyrosine) or phosphomimetic aspartic acid. Transcriptional activity of all resultant mutants was tested using the Gal4-luciferase system (Fig. 7a). Mutation of Tyr-10 to nonphosphorylatable phenylalanine reversed transcriptional repression compared with either WT-KLF16 or phosphomimetic KLF16Y10D in uterine cells (Fig. 7a). This result therefore demonstrated that mutations that mimic changes induced by tyrosine phosphatase/kinase pathways interfere with KLF16-SID function. To determine whether KLF16 is phosphorylated in vivo, we transfected Ishikawa cells with EV, FLAG-KLF16, or FLAG-KLF16Y10F and subsequently utilized [32P]orthophosphate for cell labeling. Immunoprecipitation of FLAG-tagged KLF16 demonstrated phosphorylation of this protein, as shown by 32P labeling. Although this method detects phosphorylation of serine, threonine, and tyrosine residues, diminished intensity of phosphorylation with the KLF16Y10F mutant suggested that this site indeed is a phosphorylated site in vivo (Fig. 7c). Subsequently, to investigate the involvement of tyrosine phosphatase/kinase pathways, we treated uterine cells with a pharmacological Src kinase-specific inhibitor PP2 (4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine), tyrosine phosphorylase-specific inhibitor (sodium orthovanadate), and genetic inhibitors of Src kinases (Fig. 7d). PP2 significantly reversed silencing activity compared with vehicle control (p = 0.01). Congruently, sodium orthovanadate enhanced gene silencing compared with WT-KLF16 (47% of vehicle control; p = 0.02). To complement these pharmacological investigations with molecular approaches, we evaluated the expression profile of members of the Src family in uterine cells. Uterine cells expressed FYN, LYN, SRC, and YES kinases (Fig. 7e), suggesting that any of these kinases may act as a candidate to modulate important cell processes in uterine endometrium. We focused subsequent genetic studies on the most extensively characterized member of this family, SRC kinase itself, because its role in the endometrium is well documented (30–34). To determine the effects of SRC on KLF16 activity, uterine cells were cotransfected with KLF16, a 6×BTE-luciferase reporter, and either a constitutively active or dominant negative SRC construct. Whereas constitutive SRC facilitated KLF16-mediated silencing, the dominant form of SRC significantly activated luciferase (64% of control versus 300% of control; p = 0.01 and p < 0.001, respectively; Fig. 7f). In conclusion, as predicted from our molecular modeling experiments, both pharmacological and genetic experiments reported here support a role for SRC kinase in modulating the gene silencing function of KLF16 in uterine cells.
Cytoplasm-to-Nucleus Shuttling and Euchromatic Compartmentalization Constitute an Additional Key Mechanism for Regulating KLF16 Function
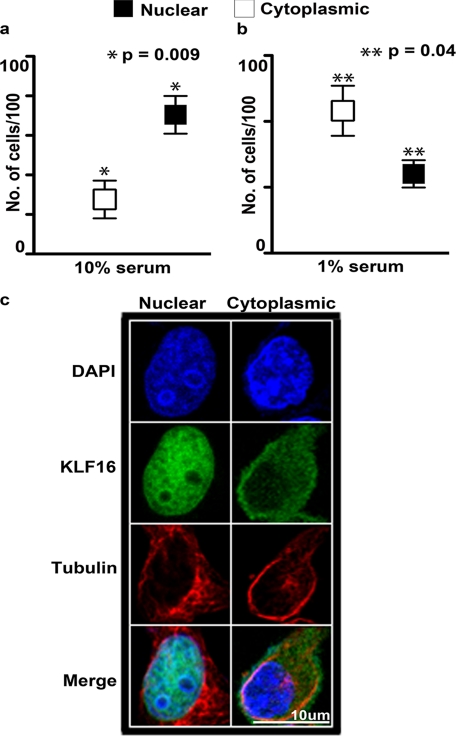
Numerous proteins also respond to signaling pathways via nucleocytoplasmic shuttling. However, information on regulation of KLF proteins by cytoplasm-to-nuclear shuttling as well as their localization within a specific chromatin compartment has remained elusive. Using immunofluorescence, we observed that KLF16 localized to both cytoplasm and nuclei in uterine cells in a serum-responsive manner, used here as a model for activation of multiple signaling cascades (Fig. 8, a–c). In the absence of serum, which keeps signaling quiescently, KLF16 localized to nuclei in 22% of cells (43% cytoplasmic and 35% both; p = 0.04) (Fig. 8b). In contrast, under serum-replete (10%) conditions, nuclear KLF16 significantly increased to 72% (18% cytoplasmic and 10% both; p = 0.009) (Fig. 8a). As cytoplasmic KLF16 cannot regulate gene expression, inhibition of shuttling in quiescent cells suggests that serum growth factors or cytokines regulate nuclear access of KLF16. Therefore, these results strongly support nuclear localization as a key regulatory mechanism of KLF16 in gene expression.
FIGURE 8.
Membrane-to-nucleus signaling regulates the accessibility of KLF16 in the nucleus. Localization of KLF16 in endometrial cells was determined by immunofluorescence using anti-KLF16 in uterine cells treated with media supplemented with either 10% (a) or 0% (b) fetal bovine serum. KLF16 demonstrated preferential nuclear localization compared with cytoplasmic localization under normal serum conditions (a, 70.4% versus 27.5%, respectively; *, p = 0.009). In contrast, KLF16 demonstrated preferential cytoplasmic localization under serum-free conditions (b, 24% versus 43.2%, respectively; **, p = 0.04). c, representative cells demonstrating immunolocalization of KLF16 in uterine cells. Cells stained with anti-KLF16 and nuclear (DAPI) as well as cytoplasmic (anti-α-tubulin) markers. Blue, DAPI; green, KLF16; red, α-tubulin.
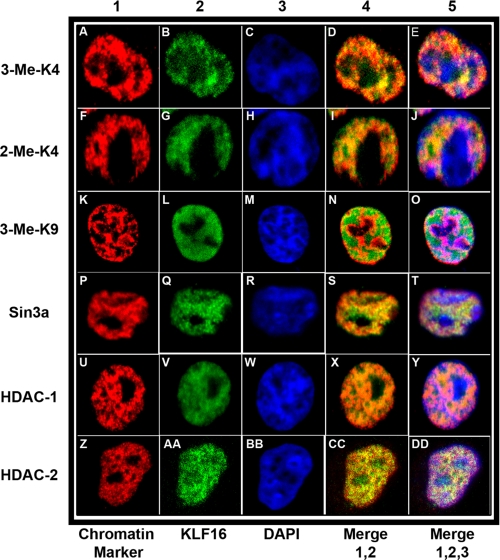
Subsequent to nuclear translocation, KLF16 predominantly localized to the euchromatic compartment. Euchromatin is enriched in cofactors used by KLF16 to regulate gene expression. Selective localization of KLF16 to euchromatin was supported by colocalization data demonstrating endogenous KLF16 alongside specific markers for nuclear domains (Fig. 9). These markers included trimethyl histone H3 K4 (Fig. 9, A–E) and dimethyl histone H3 K4 (Fig. 9, F–J) for euchromatin, as well as trimethyl histone H3 K9 for heterochromatin (Fig. 9, K–O). Additionally, we also evaluated colocalization of endogenous KLF16 with its chromatin regulators Sin3a (Fig. 9, P–T), HDAC1 (Fig. 9, U–Y), and HDAC2 (Fig. 9, Z–DD). Colocalization of KLF16 with Sin3a and HDAC1–2 within euchromatin is consistent with our biochemical data from Fig. 3d. Fig. 9 also demonstrates that KLF16 was largely excluded from heterochromatin. These data describe for the first time nuclear translocation as an important regulatory mechanism. Additionally, we show that KLF16 is a euchromatic protein that colocalized with short term gene silencing complexes, Sin3-histone-deacetylases. Collectively, these results demonstrate that nuclear translocation in response to signaling as well as euchromatic targeting is a defining functional feature of KLF16, revealing a second important mechanism for regulation of this KLF protein.
FIGURE 9.
KLF16 compartmentalization to euchromatin with chromatin cofactors in cultured endometrial cells. For coimmunolocalization of KLF16 in uterine cells, representative images are shown. All cells were stained with DAPI (blue, column 3, panels C, H, M, R, W, and BB) to visualize the DAPI-light euchromatic and DAPI-intense heterochromatic regions. All cells were also stained with anti-KLF16 (green, column 2, panels B, G, L, Q, V, and AA). For chromatin colocalization, cells were additionally stained with specific monoclonal antibodies (red, column 1, panels A, F, K, P, U, and Z) to either euchromatic markers trimethyl H3K4 or dimethyl H3K4 (panels A–E and F–J, respectively) or a heterochromatic marker trimethyl H3K9 (panels K–O). Overlay of corresponding KLF16 and individual chromatin markers (column 4, panels D, I, N, S, X, and CC) and of KLF16, individual chromatin markers and DAPI (column 5, panels E, J, O, T, Y, and DD), respectively, is shown in the figure, in columns 4 and 5 are labeled merge 1,2 and merge 1, 2, 3, respectively. KLF16 preferentially colocalized with euchromatin markers (A–J) compared with the heterochromatic marker (K–O). To evaluate colocalization with the Sin3a corepressor complex, cells were stained with monoclonal antibodies to Sin3a, HDAC1, and HDAC2 (panels P–T, U–Y, and Z–DD, respectively). KLF16 extensively colocalized with Sin3a, HDAC1, and HDAC2 (P–DD).
In summary, our studies characterized the DNA binding selectivity, transcriptional regulatory properties, and additionally discovered two novel mechanisms for regulating KLF16-mediated gene silencing. Functional studies in endometrial cells demonstrate that this transcription factor utilizes these new mechanisms for the regulation of genes involved in reproductive endocrinology. Because the structural domains of KLF16 are highly related to those from KLF9, KLF13, KLF14, KLF10, and KLF11, the mechanisms derived by our biochemical studies are highly likely applicable to also understanding these other highly related KLF family members.
DISCUSSION
This study provides insight into the mechanisms used by KLF16 to function as a novel transcription factor involved in reproductive endocrinology. Our biochemical studies demonstrate the following for KLF16: 1) recognized three distinct KLF binding elements; 2) regulated the expression of key endometrial genes involved in metabolism and endocrine function; 3) possessed the ability to either repress or activate transcription; 4) functioned by coupling to two antagonistic chromatin-mediated pathways, the Sin3a-HDAC and HAT systems; 5) complexed with the Sin3a-HDAC system by binding to the PAH2 domain, primarily by hydrophobic interactions in a manner that differ from the MAD1-SID but highly resemble the HBP1-SID; 6) interacted with all three Sin3 isoforms (Sin3a, Sin3bL, and Sin3bS); 7) its SID is the first domain of its type whose transcriptional activity is regulated by tyrosine modification; 8) underwent cytoplasmic-to-nuclear shuttling to enhance gene expression; and 9) colocalized, after translocation, with its chromatin cofactors in euchromatin to ensure proper execution of its transcriptional function. Thus, these results significantly advance our understanding of the mechanisms deployed by KLF16 to regulate gene expression that is important for maintaining metabolic and endocrine functions in uterine cells. Several of the novel findings and conceptual interpretations from this study will aid in understanding the function of other highly related KLF proteins (KLF9, KLF10, KLF11, KLF13, and KLF14), making the data more widely applicable than previously anticipated.
Once in the proximity of DNA, KLF16 recognized at least three cis-regulatory elements. ROB screening revealed that KLF16 selects a unique GC-rich sequence, related to yet distinct from the SP1 GC box (14). KLF16 not only preferred this highly GC-rich sequence (Fig. 1, b and c) but additionally also bound the BTE found in the promoter of its endometrial target gene CYP1A1. Surprisingly, KLF16 displayed minimal binding and reporter activity via the CACCC box, which is believed to be the preferred sequence for most other KLF proteins (3). These results illustrate how highly related zinc finger DNA binding domains as those found in different Sp/KLF members select distinct sequence cores, embedded within the overall framework of GC-rich genomic regions. Furthermore, different KLF transcription factors may be further classified on the basis of their binding specificity to GC-rich elements versus the CA box. Thus, these data will aid ongoing efforts in other laboratories aimed at deciphering a binding code for KLF proteins by applying molecular biophysics and structural biology techniques.
An important aspect of this study is the validation of our biochemical data using an endogenous KLF16 gene target, the estrogen-metabolizing enzyme CYP1A1, an optimal model for studying the relevance of this transcription factor to endometrial cell biology. CYP1A1 catalyzes an NADPH-mediated oxidation of its substrates to corresponding hydroxyl derivatives, which are often functionally active, facilitating a spectrum of biological effects of estrogens (25, 35–37). The endometrium is a key target tissue for estrogen, which enables cyclic endometrial regeneration. Prolonged estrogen stimulation, however, causes endometrial hyperproliferation and neoplasia (38, 39). Impaired estrogen signaling in the endometrium results in atrophy, leading to infertility (40, 41). Although endometrial cells respond to estrogen levels, they do not synthesize this hormone endogenously, relying instead on the dual processes of uptake and metabolism to maintain estrogen bioavailability. CYP1A1 also metabolizes environmental agents such as dioxin, a toxin that has been linked to endometriosis, a debilitating disease affecting 10% of females in reproductive age (26, 42–44). Direct alterations in CYP1A1 expression also associate with endometriosis (45). Indeed, using EMSA, ChIP, and reporter assays, we demonstrated that KLF16 binding to the CYP1A1-BTE repressed this gene with concomitant reduction in mRNA, reporter, and enzymatic activity (Figs. 2 and 3). In summary, the ability of KLF16 to repress endometrial CYP1A1 expression and lower its enzymatic activity, as demonstrated here, suggests a role for this KLF protein in uterine physiology and potentially pathobiology.
Gal4-based transcriptional regulatory assays (Fig. 4d) demonstrate that KLF16 contains intrinsic repressor and activator functions, which involve the coupling of KLF16 to either HDAC or HAT systems, respectively (Fig. 4, b and c). Such a dual mechanism would endow KLF16 with the ability to mediate forward as well as reverse chromatin acetylation. The N-terminal domain contains a SID motif that recruited all human Sin3 proteins, which in turn complex with HDACs to mediate gene silencing. The C-terminal activation domain, in contrast, recruited histone acetylases, in particular p300. These activities are located at two opposite ends of the protein, separated by regions amenable to extensive post-translational modifications, which may influence the function of both domains independently. Thus, these results shed light into mechanisms by which KLF16 couples to chromatin remodeling, a type of knowledge that is scanty for many KLF proteins.
Interestingly, KLF16-mediated repression was regulated rather than constitutive, as reflected by the ability of the Src tyrosine kinase to modulate this function. Initial evidence for the existence of this phenomenon was gathered through molecular modeling with molecular dynamic simulations and extensive protein-wide site-directed mutagenesis of predicted phosphorylation sites within KLF16. Thus, it is important to discuss the building, complex formation, and stability of the three-dimensional structural model for the Sin3 PAH2-KLF16 SID complex. Both careful sequence alignment and comparative helical wheel analysis of the KLF16 SID amphipathic α-helix suggest its similarity with the HBP1 SID but different from the MAD1 SID (Fig. 5). Noteworthy, molecular modeling of the KLF16 SID-Sin3 PAH2 domain complex based on homology to the Sin3a-PAH2-MAD1 SID structure was unstable when refined by molecular dynamic simulation. This approach however led to successful generation of the first three-dimensional model for the KLF16 SID-PAH2 complex, which remained stably bound under similar simulations, a result that supports good stereochemistry and a proven predictive value for this model (Fig. 6). Analysis of this model, for instance, suggests that Tyr-10 modification, which is amenable to undergo phosphorylation by SRC-type kinases, may compromise the stability of the complex. Indeed, mutational analysis showed that a KLF16 Y10F mutation, which mimics constitutive dephosphorylation of this residue, significantly reversed repression (Fig. 7a). Conversely, a phosphomimetic mutation of this residue (Y10D) did not do so, suggesting involvement of tyrosine phosphorylation/dephosphorylation in the regulation of the silencing activity of KLF16. We further confirmed that the Tyr-10 residue of KLF16 is phosphorylated in uterine cells (Fig. 7c). Additional experiments using both pharmacological and dominant negative inhibitors of Src further supported a role for this tyrosine kinase in modulating the activity of KLF16 (Fig. 7, d and f). Notably, similar predictions and experimental approaches had previously led us to define that the SID of the highly related protein, KLF11, is regulated instead by Ser/Thr phosphorylation via AKT/ERK1 (4). Thus, together the results of both studies reveal that, although similar SIDs are shared by members of two subfamilies of highly related KLF proteins, namely the TIEG (KLF10 and KLF11) and the BTEB groups (KLF9, KLF13, KLF14, and KLF16), their activity is differentially regulated by distinct kinase systems (Ser/Thr versus Tyr kinases). These results should further aid elegant ongoing efforts in the field of structural biology of SID-containing proteins and their interaction with the PAH2 domain of different Sin3 isoforms (19, 29).
To engage in chromatin remodeling, KLF16 must reside in the cell nucleus and bind DNA. We found that KLF16 shuttled from the cytoplasm to the nucleus in a serum-dependent manner suggesting that growth factors or cytokines in serum may regulate the accessibility of KLF16 to chromatin (Fig. 8). Like KLF11, KLF16 was regulated by cell signaling rather than operating in a constitutive manner (46). Once inside the nucleus, KLF16 along with its cofactors Sin3a, HDACs, and p300 colocalized to euchromatin (Fig. 9) (28). Euchromatic localization facilitates the use of both chromatin-remodeling systems by KLF16 in a nuclear region where gene expression is regulated transiently. These results are also compatible with a model whereby signaling-induced events are important in the regulation of KLF16 function. Interestingly, functional regulation by nuclear localization, essential for other transcription factors (NFκB), has not been examined for any KLF protein. These studies therefore underscore the importance of cell compartmentalization as a mechanism that could regulate the function of members of this important transcription factor family.
In conclusion, this study significantly expands our understanding of new molecular mechanisms underlying the biochemical function of KLF16 in particular, as well as provides information applicable to other members of this protein family. These biochemical mechanisms have been validated and studied in a cellular context where the regulation of gene expression is important for uterine cell biology, making this information highly relevant to the field of biochemistry and reproductive endocrinology.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK52913 (to R. U.). This work was also supported by the Mayo Foundation.

This article contains supplemental Table 1 and Fig. 1.
- KLF
- Krüppel-like factor
- MD
- molecular dynamics
- SID
- Sin3-interacting domain
- EV
- empty vector
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- ROB
- random oligonucleotide binding
- TCDD
- 2,3,7,8 tetrachlorodibenzo-p-dioxin
- MSS
- maximal similarity score
- BTE
- basic transcription element.
REFERENCES
- 1. Philipsen S., Suske G. (1999) A tale of three fingers. The family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27, 2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lomberk G., Urrutia R. (2005) The family feud. Turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 392, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bieker J. J. (2001) Krüppel-like factors. Three fingers in many pies. J. Biol. Chem. 276, 34355–34358 [DOI] [PubMed] [Google Scholar]
- 4. Buttar N. S., DeMars C. J., Lomberk G., Rizvi S., Bonilla-Velez J., Achra S., Rashtak S., Wang K. K., Fernandez-Zapico M. E., Urrutia R. (2010) Distinct role of Krüppel-like factor 11 in the regulation of prostaglandin E2 biosynthesis. J. Biol. Chem. 285, 11433–11444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernandez-Zapico M. E., van Velkinburgh J. C., Gutiérrez-Aguilar R., Neve B., Froguel P., Urrutia R., Stein R. (2009) MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet beta cells. J. Biol. Chem. 284, 36482–36490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao S., Fernandez-Zapico M. E., Jin D., Puri V., Cook T. A., Lerman L. O., Zhu X. Y., Urrutia R., Shah V. (2005) KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding protein-induced transcriptional activation of caveolin-1 in response to cholesterol signaling. J. Biol. Chem. 280, 1901–1910 [DOI] [PubMed] [Google Scholar]
- 7. Gutiérrez-Aguilar R., Froguel P., Hamid Y. H., Benmezroua Y., Jørgensen T., Borch-Johnsen K., Hansen T., Pedersen O., Neve B. (2008) Genetic analysis of Krüppel-like zinc finger 11 variants in 5864 Danish individuals. Potential effect on insulin resistance and modified signal transducer and activator of transcription-3 binding by promoter variant -1659G>C. J. Clin. Endocrinol. Metab. 93, 3128–3135 [DOI] [PubMed] [Google Scholar]
- 8. Zhang D., Zhang X. L., Michel F. J., Blum J. L., Simmen F. A., Simmen R. C. (2002) Direct interaction of the Krüppel-like family (KLF) member, BTEB1, and PR mediates progesterone-responsive gene expression in endometrial epithelial cells. Endocrinology 143, 62–73 [DOI] [PubMed] [Google Scholar]
- 9. Small K. S., Hedman A. K., Grundberg E., Nica A. C., Thorleifsson G., Kong A., Thorsteindottir U., Shin S. Y., Richards H. B., Soranzo N., Ahmadi K. R., Lindgren C. M., Stefansson K., Dermitzakis E. T., Deloukas P., Spector T. D., McCarthy M. I. (2011) Identification of an imprinted master transregulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 43, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simmen R. C., Eason R. R., McQuown J. R., Linz A. L., Kang T. J., Chatman L., Jr., Till S. R., Fujii-Kuriyama Y., Simmen F. A., Oh S. P. (2004) Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Krüppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J. Biol. Chem. 279, 29286–29294 [DOI] [PubMed] [Google Scholar]
- 11. Gebelein B., Urrutia R. (2001) Sequence-specific transcriptional repression by KS1, a multiple zinc finger-Krüppel-associated box protein. Mol. Cell. Biol. 21, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lomberk G., Bensi D., Fernandez-Zapico M. E., Urrutia R. (2006) Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8, 407–415 [DOI] [PubMed] [Google Scholar]
- 13. Cook T., Gebelein B., Belal M., Mesa K., Urrutia R. (1999) Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J. Biol. Chem. 274, 29500–29504 [DOI] [PubMed] [Google Scholar]
- 14. Gebelein B., Fernandez-Zapico M., Imoto M., Urrutia R. (1998) KRAB-independent suppression of neoplastic cell growth by the novel zinc finger transcription factor KS1. J. Clin. Invest. 102, 1911–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaczynski J., Zhang J. S., Ellenrieder V., Conley A., Duenes T., Kester H., van Der Burg B., Urrutia R. (2001) The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J. Biol. Chem. 276, 36749–36756 [DOI] [PubMed] [Google Scholar]
- 16. Fujii-Kuriyama Y., Imataka H., Sogawa K., Yasumoto K., Kikuchi Y. (1992) Regulation of CYP1A1 expression. FASEB J. 6, 706–710 [DOI] [PubMed] [Google Scholar]
- 17. Fernandez-Zapico M. E., Mladek A., Ellenrieder V., Folch-Puy E., Miller L., Urrutia R. (2003) An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. EMBO J. 22, 4748–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J. S., Moncrieffe M. C., Kaczynski J., Ellenrieder V., Prendergast F. G., Urrutia R. (2001) A conserved α-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol. 21, 5041–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brubaker K., Cowley S. M., Huang K., Loo L., Yochum G. S., Ayer D. E., Eisenman R. N., Radhakrishnan I. (2000) Solution structure of the interacting domains of the Mad-Sin3 complex. Implications for recruitment of a chromatin-modifying complex. Cell 103, 655–665 [DOI] [PubMed] [Google Scholar]
- 20. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 21. Pang Y. P., Kumar G. A., Zhang J. S., Urrutia R. (2003) Differential binding of Sin3 interacting repressor domains to the PAH2 domain of Sin3A. FEBS Lett. 548, 108–112 [DOI] [PubMed] [Google Scholar]
- 22. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR. Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 23. Narayan V. A., Kriwacki R. W., Caradonna J. P. (1997) Structures of zinc finger domains from transcription factor Sp1. Insights into sequence-specific protein-DNA recognition. J. Biol. Chem. 272, 7801–7809 [DOI] [PubMed] [Google Scholar]
- 24. Whitlock J. P., Jr. (1999) Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 39, 103–125 [DOI] [PubMed] [Google Scholar]
- 25. Fotsis T., Zhang Y., Pepper M. S., Adlercreutz H., Montesano R., Nawroth P. P., Schweigerer L. (1994) The endogenous oestrogen metabolite 2-methoxyoestradiol inhibits angiogenesis and suppresses tumour growth. Nature 368, 237–239 [DOI] [PubMed] [Google Scholar]
- 26. Johnson K. L., Cummings A. M., Birnbaum L. S. (1997) Promotion of endometriosis in mice by polychlorinated dibenzo-p-dioxins, dibenzofurans, and biphenyls. Environ. Health Perspect. 105, 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vermeulen M., Walter W., Le Guezennec X., Kim J., Edayathumangalam R. S., Lasonder E., Luger K., Roeder R. G., Logie C., Berger S. L., Stunnenberg H. G. (2006) A feed-forward repression mechanism anchors the Sin3/histone deacetylase and N-CoR/SMRT corepressors on chromatin. Mol. Cell. Biol. 26, 5226–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song C. Z., Keller K., Murata K., Asano H., Stamatoyannopoulos G. (2002) Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J. Biol. Chem. 277, 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swanson K. A., Knoepfler P. S., Huang K., Kang R. S., Cowley S. M., Laherty C. D., Eisenman R. N., Radhakrishnan I. (2004) HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 11, 738–746 [DOI] [PubMed] [Google Scholar]
- 30. Maruyama T., Yoshimura Y., Yodoi J., Sabe H. (1999) Activation of c-Src kinase is associated with in vitro decidualization of human endometrial stromal cells. Endocrinology 140, 2632–2636 [DOI] [PubMed] [Google Scholar]
- 31. Maruyama T., Yoshimura Y., Sabe H. (1999) Tyrosine phosphorylation and subcellular localization of focal adhesion proteins during in vitro decidualization of human endometrial stromal cells. Endocrinology 140, 5982–5990 [DOI] [PubMed] [Google Scholar]
- 32. Shiozawa T., Shih H. C., Miyamoto T., Feng Y. Z., Uchikawa J., Itoh K., Konishi I. (2003) Cyclic changes in the expression of steroid receptor coactivators and corepressors in the normal human endometrium. J. Clin. Endocrinol. Metab. 88, 871–878 [DOI] [PubMed] [Google Scholar]
- 33. Shah Y. M., Rowan B. G. (2005) The Src kinase pathway promotes tamoxifen agonist action in Ishikawa endometrial cells through phosphorylation-dependent stabilization of estrogen receptor (α) promoter interaction and elevated steroid receptor coactivator 1 activity. Mol. Endocrinol. 19, 732–748 [DOI] [PubMed] [Google Scholar]
- 34. Shimizu A., Maruyama T., Tamaki K., Uchida H., Asada H., Yoshimura Y. (2005) Impairment of decidualization in SRC-deficient mice. Biol. Reprod. 73, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 35. Honkakoski P., Negishi M. (2000) Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem. J. 347, 321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee A. J., Cai M. X., Thomas P. E., Conney A. H., Zhu B. T. (2003) Characterization of the oxidative metabolites of 17β-estradiol and estrone formed by 15 selectively expressed human cytochrome P450 isoforms. Endocrinology 144, 3382–3398 [DOI] [PubMed] [Google Scholar]
- 37. Tsuchiya Y., Nakajima M., Yokoi T. (2005) Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 227, 115–124 [DOI] [PubMed] [Google Scholar]
- 38. Cavalieri E., Frenkel K., Liehr J. G., Rogan E., Roy D. (2000) Estrogens as endogenous genotoxic agents—DNA adducts and mutations. J. Natl. Cancer Inst. Monographs 27, 75–93 [DOI] [PubMed] [Google Scholar]
- 39. Yager J. D., Liehr J. G. (1996) Molecular mechanisms of estrogen carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 36, 203–232 [DOI] [PubMed] [Google Scholar]
- 40. Hewitt S. C., Harrell J. C., Korach K. S. (2005) Lessons in estrogen biology from knockout and transgenic animals. Annu. Rev. Physiol. 67, 285–308 [DOI] [PubMed] [Google Scholar]
- 41. Korach K. S. (1994) Insights from the study of animals lacking functional estrogen receptor. Science 266, 1524–1527 [DOI] [PubMed] [Google Scholar]
- 42. Cummings A. M., Metcalf J. L. (1995) Induction of endometriosis in mice. A new model sensitive to estrogen. Reprod. Toxicol. 9, 233–238 [DOI] [PubMed] [Google Scholar]
- 43. Mayani A., Barel S., Soback S., Almagor M. (1997) Dioxin concentrations in women with endometriosis. Hum. Reprod. 12, 373–375 [DOI] [PubMed] [Google Scholar]
- 44. Cummings A. M., Metcalf J. L. (1995) Effects of estrogen, progesterone, and methoxychlor on surgically induced endometriosis in rats. Fundam. Appl. Toxicol. 27, 287–290 [DOI] [PubMed] [Google Scholar]
- 45. Hadfield R. M., Manek S., Weeks D. E., Mardon H. J., Barlow D. H., Kennedy S. H. (2001) Linkage and association studies of the relationship between endometriosis and genes encoding the detoxification enzymes GSTM1, GSTT1, and CYP1A1. Mol. Hum. Reprod. 7, 1073–1078 [DOI] [PubMed] [Google Scholar]
- 46. Cook T., Gebelein B., Mesa K., Mladek A., Urrutia R. (1998) Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-β-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem. 273, 25929–25936 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.