Background: The WRKY transcription factors recognize the W-box DNA element in target genes.
Results: We determined the NMR solution structure of the WRKY DNA-binding domain of Arabidopsis WRKY4 in complex with W-box DNA.
Conclusion: Apolar contacts by residues in the conserved WRKYGQK motif with thymine methyl groups are important in recognition of the W-box sequence.
Significance: This is the first structure of the WRKY-DNA complex.
Keywords: NMR, Plant Molecular Biology, Protein-DNA Interaction, Protein Structure, Transcription Factors
Abstract
The WRKY family transcription factors regulate plant-specific reactions that are mostly related to biotic and abiotic stresses. They share the WRKY domain, which recognizes a DNA element (TTGAC(C/T)) termed the W-box, in target genes. Here, we determined the solution structure of the C-terminal WRKY domain of Arabidopsis WRKY4 in complex with the W-box DNA by NMR. A four-stranded β-sheet enters the major groove of DNA in an atypical mode termed the β-wedge, where the sheet is nearly perpendicular to the DNA helical axis. Residues in the conserved WRKYGQK motif contact DNA bases mainly through extensive apolar contacts with thymine methyl groups. The importance of these contacts was verified by substituting the relevant T bases with U and by surface plasmon resonance analyses of DNA binding.
Introduction
The WRKY transcription factor proteins have been identified from a wide range of higher plants (1–4) and compose one of the largest families of plant-specific transcription factors (5, 6). Most WRKY proteins are involved in responses to biotic or abiotic stresses, such as pathogenic infection, injury, heat, drought, and high salinity (3, 7–12). The WRKY proteins control transcription of the target genes by binding to the promoter regions that contain a DNA element called the W-box with the core sequence TTGACY (where Y is C or T) (3, 4, 7).
An ∼60-amino acid DNA-binding domain called the WRKY domain is shared by the WRKY family, and it contains an invariant sequence, WRKYGQK, and a zinc-binding motif (5). The WRKY proteins that possess two WRKY domains are classified into group I, whereas those that possess a single WRKY domain are classified into group II or III, mainly according to the amino acid sequences of the zinc-binding motif (5). For the group I WRKY proteins, the C-terminal WRKY domain (not the N-terminal domain) is responsible for the recognition of the W-box sequence (1, 4, 7, 13).
We previously determined the solution structure of the C-terminal WRKY domain of the Arabidopsis WRKY4 protein (AtWRKY4-C)2 that comprises a four-stranded β-sheet (14). Furthermore, a crystal structure of the equivalent domain of the Arabidopsis WRKY1 protein has been reported, and it is very similar to our AtWRKY4-C structure except that it contains an additional β-strand at the N terminus of the domain (15). From the results of NMR titration analysis, we proposed a structural model for the complex between AtWRKY4-C and DNA in which the strand containing the conserved WRKYGQK sequence enters the major groove of DNA (14). Mutational analyses have indicated that this sequence is important for DNA binding (13, 15, 16). However, it was previously unknown how the WRKY domains specifically recognize the W-box sequence.
In this study, the three-dimensional structure of the complex of AtWRKY4-C with DNA containing the W-box sequence was determined by NMR spectroscopy. DNA bases were recognized mainly through apolar contacts involving the methyl groups of the T bases. The importance of the apolar contacts was verified by substituting T bases with U and by binding analyses using surface plasmon resonance (SPR).
EXPERIMENTAL PROCEDURES
Sample Preparation
The 13C/15N-labeled, 15N-labeled, and unlabeled AtWRKY4-C (Val-399–Ala-469) proteins were produced by cell-free protein synthesis with optimization for zinc-binding proteins (17, 18) and were partially purified by immobilized metal affinity chromatography using an automated system as described previously (19). The eluted protein was cleaved with tobacco etch virus protease to remove the His tag and subsequently exchanged into 20 mm Tris-HCl buffer (pH 8.0) containing 300 mm NaCl, 5 mm imidazole, 1 mm iminodiacetate, and 50 μm ZnCl2 using a HiPrep 26/10 desalting column (GE Healthcare). The protein-containing fraction was applied to a HisTrap column (GE Healthcare), and its flow-through fraction was pooled. [15N]Thy-labeled and unlabeled 16-mer double-stranded DNAs (5′-CGCCTTTGACCAGCGC-3′/5′-GCGCTGGTCAAAGGCG-3′, where the W-box core sequence is underlined) were chemically synthesized (Tsukuba Oligo Service), and [15N]thymidine phosphoramidite (CIL International, Andover, MA) was used to label the five T bases in the sequence. The concentration of the protein was determined at A280 with an extinction coefficient estimated from the amino acid sequence (20), whereas that of the double-stranded DNA was determined at A260 using an extinction coefficient that was calculated after digestion of the strands with phosphodiesterase I (Worthington). For the NMR measurements, 0.4–1.0 mm 1:1 protein-DNA complex was dissolved in 20 mm potassium phosphate buffer (pH 6.0) containing 200 mm KCl, 20 μm ZnCl2, 1 mm deuterated dithiothreitol (Isotec Inc.), 0.05 mm sodium 2,2-dimethyl-2-silapentane-5-sulfonate, and 5% D2O unless stated otherwise. For the measurement of residual dipolar couplings (RDCs), 12 mg/ml Pf1 phage (ASLA Biotech, Riga, Latvia) (21) was added. Further descriptions regarding selection of the buffer system are provided under supplemental “Materials.”
NMR Analyses and Structure Determination
NMR spectra were recorded on a Bruker DMX-750 (750.13 MHz for 1H and 76.02 MHz for 15N) or DMX-500 (500.13 MHz for 1H, 125.76 MHz for 13C, and 50.68 MHz for 15N) spectrometer at 298 or 303 K. The protein backbone and side chain resonance assignments were partly obtained from the previous DNA titration experiments (14) and completed here by a series of triple-resonance experiments (22). The resonances of DNA were assigned using the typical base-sugar connectivities for DNA strands appearing in NOESY spectra at a mixing time of 100 ms (supplemental Fig. S1) (23). Chemical shifts are referenced to internal sodium 2,2-dimethyl-2-silapentane-5-sulfonate directly (1H) or indirectly (13C and 15N) as recommended in the Biological Magnetic Resonance Data Bank. The completeness of the assignments in the region used for the structure determination was 93.1% for non-labile and backbone amide protons of the protein or 97.4% for non-labile protons of DNA excluding H4′, H5′, and H5" atoms. A heteronuclear single-quantum correlation (HSQC) spectrum of a sample dissolved in 99.96% D2O (Isotec Inc.) after lyophilization was recorded at 283 K to identify the hydrogen bonds in the protein.
For evaluation of 1H-15N RDCs, the in-phase/anti-phase (IPAP) HSQC (24) spectra were recorded, where the IPAP subspectra were obtained in an interleaved manner. The two subspectra were processed by addition or subtraction to yield the other two subspectra containing upfield or downfield components of the cross-peaks. The RDCs for the protein backbone amides, arginine side chain Nϵ-Hϵ groups, and DNA T base imides were obtained in separately measured spectra, where the parameters were optimized so as to maximize the intensities of the respective peaks. The structure was calculated using the CNS program (25) as described under supplemental “Materials.”
Structure Analyses
Intermolecular contacts were analyzed by in-house Fortran programs and Insight II (Accelrys, San Diego, CA). A hydrogen bond was defined by a hydrogen donor and acceptor distance that was <3.5 Å and a donor-hydrogen-acceptor angle that was >110°. An electrostatic attraction was defined by a distance between a side chain nitrogen of Arg/Lys and a phosphate oxygen that was <5.0 Å. An apolar contact representing both hydrophobic and van der Waals forces was defined by a C-C distance that was <5.0 Å.
SPR
Experiments were performed at 298 K using a Biacore X apparatus (GE Healthcare) as described previously (14). Potassium phosphate (20 mm; pH 6.0) containing 100 mm KCl, 20 μm ZnCl2, and 0.005% Tween 20 was used as the running buffer unless stated otherwise. In total, 590, 623, 606, 622, 580, and 600 resonance units of double-stranded DNAs (5′-bio-CGCCTTTGACCAGCGC-3′/5′-GCGCTGGTCAAAGGCG-3′ (where the W-box consensus sequence is underlined, and bio indicates biotinylation at the 5′-end), 5′-bio-CGCCdUTTGACCAGCGC-3′/5′-GCGCTGGTCAAAGGCG-3′, 5′-bio-CGCCTdUTGACCAGCGC-3′/5′-GCGCTGGTCAAAGGCG-3′, 5′-bio-CGCCTTdUGACCAGCGC-3′/5′-GCGCTGGTCAAAGGCG-3′, 5′-bio-CGCCTTTGACCAGCGC-3′/5′-GCGCdUGGTCAAAGGCG-3′, and 5′-bio-CGCCTTTGACCAGCGC-3′/5′-GCGCTGGdUCAAAGGCG-3′, respectively) were immobilized on Sensor Chip SA surfaces (GE Healthcare) in one (flow cell 2) of the two flow cells, and the other was treated as the control. Solutions containing AtWRKY4-C at concentrations of 10 nm to 1 μm were injected into the flow cells at 20 μl/min for 5 min. The equilibrium binding constants were obtained by fitting the equilibrium response values at different protein concentrations to the simple 1:1 binding model using BIAevaluation 3.0 software (GE Healthcare). For data that did not reach a plateau level even at the highest protein concentration, i.e. for DNAs with a substitution at T6 or T9′, the maximum response values were fixed to those expected when the protein molecules were bound to all of the immobilized DNA molecules at a 1:1 stoichiometry.
RESULTS AND DISCUSSION
Structure Determination
To determine the structure, we used a WRKY domain and a 16-mer double-stranded DNA with the same sequences as those used for previous protein structure determination and DNA titration analysis, for which a 1:1 binding stoichiometry was revealed by SPR (14). Assignments of the protein backbone resonances in the complex have been essentially completed by the previous DNA titration analysis and three-dimensional NMR analyses. Therefore, in this study, resonance assignments of the protein side chains and DNA chains were performed. In the spectra of the complex, intermolecular NOEs were observed, and these directly defined the geometry of the molecular interface (supplemental Fig. S1).
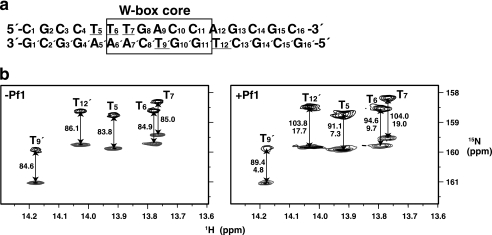
In addition to the distance- and dihedral angle-based restraints, we employed RDCs (26, 27) to restrain the relative angles between the bond vectors and the overall molecular alignment axes. For this purpose, 15N-labeled T bases were introduced into the DNA, and IPAP-HSQC spectra (24) were measured for the complex with uniformly 15N-labeled protein with or without the partial alignment induced by filamentous phage (Fig. 1) (21). The complex structure was calculated in a sequential manner as described under supplemental “Materials.” The structure thus obtained satisfied the experimental restraints, possessed idealistic stereochemical properties, and showed a good convergence (Fig. 2a, Table 1, and supplemental Fig. S2). The z axis of the alignment tensor generated by the phage was found to be approximately parallel to the DNA helical axis, whereas the x axis, which is the longer of the two rhombic axes, was nearly parallel to the vector from the center of the DNA to that of the protein (Fig. 2a). Therefore, the RDCs appeared to restrict the individual 1H-15N vectors relative to the axes that were essentially defined by the DNA helical structure and the protein-DNA interaction.
FIGURE 1.
RDCs for 15N-labeled thymine bases of the W-box DNA. a, sequence of the double-stranded DNA used in the present NMR analysis. The W-box core sequence is boxed. The underlined T bases were 15N-labeled for the RDC analysis. b, IPAP-HSQC spectra of the complex between AtWRKY4-C and the W-box DNA containing the 15N-labeled T bases with (right panel) or without (left panel) 12 mg/ml Pf1 phage. The upfield and downfield components are shown in black and gray, respectively, and the coupling values are indicated in Hz. The RDCs are shown below the coupling values in the right panel.
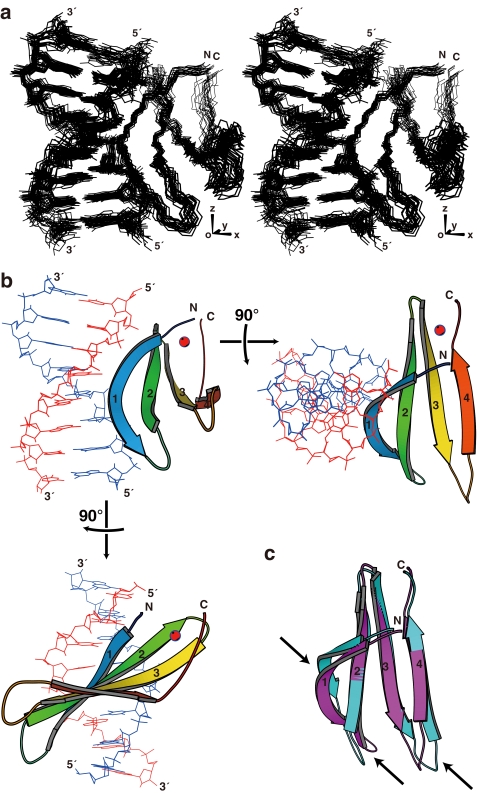
FIGURE 2.
Structure of the complex of AtWRKY4-C and the W-box DNA. a, ensemble of the 20 selected structures in stereo view, which were fitted using the atoms shown, i.e. backbone non-hydrogen atoms in the protein and all non-hydrogen atoms in DNA. b, ribbon diagram of the representative structure with the lowest energy. c, comparison of the AtWRKY4-C structures in the complex with DNA (magenta) and the free protein (cyan; Protein Data Bank code 1WJ2). In a, the axes regarding the alignment tensor are indicated. In b, the four β-strands are numbered from the N terminus, and the zinc ion is shown as a red sphere. In c, the arrows indicate regions with a relatively large structure perturbation caused by DNA binding. The region of the protein presented is Tyr-412–His-465, and that of the DNA presented is C4–C11/G4′–G11′. The figures were produced by MolScript (40).
TABLE 1.
Structural statistics
| Structural constraintsa | |
| Protein, zinc-bound | |
| Sequential NOEs | 260 |
| Medium-range NOEs (2 ≤ |i − j| ≤ 4) | 81 |
| Long-range NOEs (|i − j| > 4) | 313 |
| Hydrogen bonds | 38 |
| Torsion (φ) angles by TALOS+b | 24 |
| Torsion (ψ) angles by TALOS+b | 24 |
| RDC | 58 |
| Theoreticalc | 14 |
| DNA | |
| Intranucleoside NOEsd | 93 |
| Sequential NOEs | 122 |
| Interstrand NOEs | 32 |
| RDC | 5 |
| Theoreticale | 194 |
| Protein·DNA | |
| NOEs | 69 |
| Total | 1327 |
| Characteristics of 20 selected structures | |
| r.m.s.d.f from constraints | |
| Distances (Å) | 0.013 ± 0.001 |
| Torsion angles | 0.13 ± 0.03º |
| van der Waals energy (kcal/mol)g | 50.3 ± 4.1 |
| r.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.0018 ± 0.0001 |
| Bond angles | 0.407 ± 0.013º |
| Improper angles | 0.29 ± 0.05º |
| Average r.m.s.d. to mean structure (Å)h | |
| Non-hydrogen atoms in complex | 1.08 ± 0.12 |
| Non-hydrogen atoms in protein | 0.95 ± 0.08 |
| Backbone N, Cα, and C atoms in protein | 0.46 ± 0.10 |
| Non-hydrogen atoms in DNA | 0.77 ± 0.21 |
| Ramachandran plot for proteini | |
| Most favored region (%) | 84.6 |
| Additionally allowed region (%) | 14.6 |
| Generously allowed region (%) | 0.8 |
| Disallowed region (%) | 0.1 |
a Constraints used in the final calculation (see supplemental “Materials”).
b Ref. 39.
c Distance, angle, and planarity restraints applied for zinc ions and coordinating atoms (see supplemental “Materials”).
d NOEs between base and sugar protons.
e Distance, dihedral angle, and planarity restraints for DNA base pairs and backbone (see supplemental “Materials”).
f r.m.s.d., root mean square deviation.
g Value for the repel function in the CNS package (25).
h Values calculated with Tyr-412–His-465 of the protein and/or C4–C11/G4′–G11′ of DNA.
i Values calculated with Tyr-412–His-465.
Structure of the Complex
The structure of the protein moiety of the complex consists of a four-stranded β-sheet (β1, Trp-414–Val-422; β2, Tyr-427–Thr-436; β3, Cys-439–Arg-447; and β4, Val-455–Glu-460), which is similar to that of the protein not bound to the DNA (14), with a backbone root mean square deviation of 1.9 Å (Fig. 2, b and c). The DNA is in the B-form with a slight bent toward the protein. The β-sheet plane is almost perpendicular to the DNA helical axis but is slightly tilted to fit the rim of the sheet into the major groove. The rim strand, i.e. the β1-strand that contains the invariant WRKYGQK sequence, composes the major molecular interface. The revealed binding mode, which is largely consistent with the previous model that was based on the DNA titration analysis (14), is called a β-wedge in this study, as described below.
It has been suggested that Gly-418 of the WRKYGQK sequence was irregularly inserted into the typical antiparallel β-sheet, and this induced a kink in this strand (14). The present structure revealed that the kink created the convex curvature of the β-sheet rim and thereby enabled the close contact of this strand with DNA bases. In addition, the formation of the complex significantly altered the relative position of this strand to the others (Fig. 2c). This appeared to influence the structure of the loop connecting the β1- and β2-strands and that connecting the β3- and β4-strands as well and thereby slightly altered the length of the strands.
Contacts in the Molecular Interface
Eight bases in 7 consecutive bp are contacted by Arg-415, Lys-416, Tyr-417, Gly-418, Gln-419, and Lys-420 of the β1-strand or all of the residues in the invariant WRKYGQK sequence, except Trp-414, through apolar and hydrogen-bonding interactions (Fig. 3a). All of the contacts, including those with the sugar phosphate backbone, cover the range of 8 bp (Fig. 3a). The details are described below.
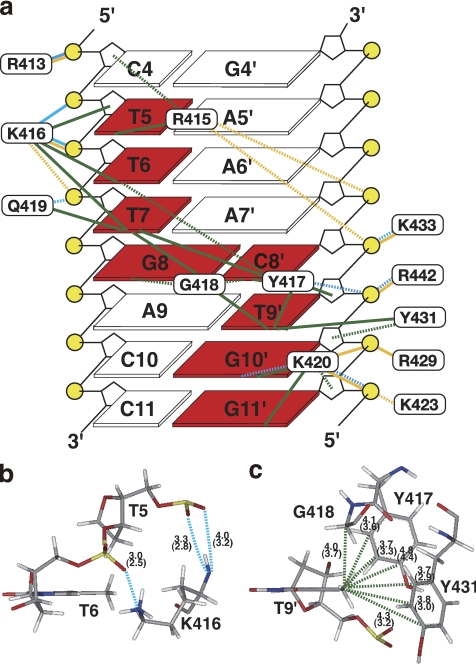
FIGURE 3.
Intermolecular contacts. a, summary of microscopic forces identified in the interface between AtWRKY4-C and the W-box DNA (bases in squares are numbered as in Fig. 1a), which were drawn looking from the major groove side of DNA. DNA bases, the sugar backbone, and phosphates are shown by squares, black lines, and yellow circles, respectively. Bases contacted by the protein are highlighted in red. Cyan, yellow, and green lines represent hydrogen bonds, contacts by electrostatic attraction, and apolar contacts, respectively, as defined under “Experimental Procedures.” Solid and dashed lines indicate that these contacts were identified in ≥75% and ≥40% of the structures, respectively. b, hydrogen bonds formed by backbone and side chain nitrogen atoms of Lys-416 are indicated by dashed cyan lines. The average and minimal (in parentheses) distances between donor nitrogen and acceptor oxygen atoms in the selected structures are shown. c, apolar contacts with the methyl group of T9′ (dashed green lines), with the average and minimal (in parentheses) C-C distances indicated. The figures in b and c were produced using the Insight II molecular display program.
The side chain carbon atoms of Arg-415 form extensive apolar contacts with the T5 base carbons, mainly of the methyl groups, and the backbone sugar carbons of C4. At the same time, it is possible for the guanidyl group of Arg-415 to form electrostatic interactions with the A7′ and C8′ phosphates.
The side chain and backbone carbon atoms of Lys-416 form apolar contacts with the T6 and T7 methyl groups and T5 sugar atoms. At the same time, the backbone amide and side chain amino groups of Lys-416 form hydrogen bonds with the phosphates of T5 and T6, respectively (Fig. 3b). The hydrogen bonding by the backbone amide is consistent with the previous observation that binding to the DNA induced a very large downfield shift (∼1.8 ppm) of the amide proton resonance (14). In addition, the hydrogen bond between the Lys-416 amino and T6 phosphate groups is strengthened by electrostatic attraction. This amino group also forms an electrostatic interaction with the T7 phosphate.
The aromatic rings of Tyr-417 and Tyr-431 and the backbone of Gly-418 surround the T9′ methyl group and form extensive apolar contacts (Fig. 3, a and c); this is consistent with the observed intermolecular NOEs and the upfield-shifted resonances of the T9′ base protons (supplemental Fig. S1). Moreover, the aromatic carbons of Tyr-417 contact the T6 base, T7 base (mainly the methyl groups), C8′ base, and T9′ sugar carbons by apolar interactions, and the hydroxyl group of the same residue simultaneously contacts the T9′ phosphate by hydrogen-bonding interactions. Gly-418 also forms extensive apolar contacts with the T7, G8, and G8′ base carbons, which is enabled by the deep entrance of this residue into the DNA groove.
The side chain amide nitrogen and side chain/backbone carbons of Gln-419 form a hydrogen bond with the T7 phosphate and apolar contacts with the T7 base, respectively. Lys-420 contacts the G10′ base, G11′ base, and G11′ sugar carbons by apolar interactions and simultaneously forms hydrogen bonds with the N7/O6 atoms of G10′ and/or the phosphate oxygens of G11′. In addition, it contacts the phosphates of G10′ and/or G11′ by electrostatic attraction.
In addition to the above residues, Arg-413, Lys-423, Arg-429, Lys-433, and Arg-442 form hydrogen bonds and/or electrostatic contacts with phosphate groups (Fig. 3a). Arg-429 and Lys-433 are located on the same strand, i.e. β2-strand, but protrude to the opposite sides of the β-sheet to each other. Arg-442 is located on the β3-strand, considerably distant from the major contacting strand, i.e. β1-strand. These contacts appear to be enabled after the close fitting of the β-sheet to the DNA groove and therefore to contribute significantly to the fixing of the binding geometry.
Verification of Importance of Apolar Contacts with T Bases
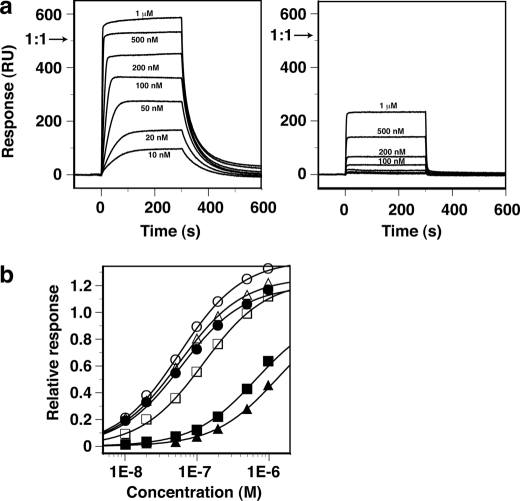
As described above, the recognition of the W-box sequence by AtWRKY4-C was achieved mainly by extensive apolar contacts with the methyl groups of the T bases (Fig. 3). We verified the importance of these contacts by substitution of the T bases with U (namely, elimination of the methyl groups) and evaluation of binding affinities by SPR (Fig. 4 and Table 2). The equilibrium SPR response value appeared to reach the maximum of that expected when the proteins bound to all of the immobilized DNAs at a 1:1 stoichiometry (Fig. 4a, left panel, arrow) or slightly more for the 16-mer W-box DNA without substitution. A binding constant of 1.9 × 107 m−1 was obtained by fitting the data to the simple 1:1 binding model (Fig. 4b and Table 2). In contrast, binding to the DNA by substitution of T9′ with U appeared too weak to reach a plateau level within the protein concentration range of the present experiment (Fig. 4a, right panel), for which an ∼25-fold weaker binding constant was obtained by data fitting (Fig. 4b and Table 2). These results verified the importance of the extensive apolar contacts involving the T9′ methyl group (Fig. 3c). NMR revealed that even in the relevant weak complex, the framework of protein-DNA interaction was probably conserved (supplemental Fig. S1).
FIGURE 4.
Binding of AtWRKY4-C to W-box DNAs observed by SPR. a, SPR difference sensorgrams (with the responses in the control flow cell subtracted) for binding of AtWRKY4-C to a double-stranded 16-mer W-box DNA (see Fig. 1a) without (left panel) and with (right panel) substitution of a T9′ base with U, where the protein concentrations were 10 nm to 1 μm as indicated. The protein solutions were injected over a period of 0–300 s. The arrows indicate the maximum response value expected when the protein molecules bind to all of the immobilized DNA molecules at a 1:1 stoichiometry. b, equilibrium response values relative to the above expected maximum values as a function of protein concentration. The DNAs that were used were the 16-mer without substitution (●) and with substitution of T with U at T5 (○), T6 (■), T7 (□), T9′ (▴), and T12′ (△). Fitting curves to the simple 1:1 binding model are shown.
TABLE 2.
Binding to W-box DNAs with and without T-to-U base substitutions
| Position of substitution | KAa | Decrease |
|---|---|---|
| m−1 | -fold | |
| No substitution | (1.9 ± 0.4) ×107 | |
| T5 to U | (1.7 ± 0.1) ×107 | 1.1 |
| T6 to U | (1.8 ± 0.3) ×106 | 11 |
| T7 to U | (8.4 ± 0.3) ×106 | 2.3 |
| T9′ to U | (7.5 ± 2.5) ×105 | 25 |
| T12′ to U | (1.7 ± 0.1) ×107 | 1.1 |
a S.D. values in four repeated experiments were used as error levels.
In addition, the substitution of T6 or T7 significantly decreased the affinity by 11- or 2.3-fold (Table 2). T6 and T7, as well as T9′, are the conserved T bases in the W-box core sequence (TTGACY) and are each contacted by two amino acids or more of AtWRKY4-C (Fig. 3a). The other T bases in the DNA used, i.e. T5 and T12′, which are outside of the W-box sequence, showed no sensitivity to the substitution (Fig. 4b and Table 2). T12′ is not contacted by the domain, whereas T5 is contacted by only a single residue in AtWRKY4-C (Fig. 3a).
Implications for Preference for DNA Bases
At the position upstream of the W-box core sequence (TTGACY), i.e. the base equivalent to T5 of the present DNA, a G base is most preferred (appreciably better than A or T), as shown by a systematic study on the promoter sequences of the target genes (16). A simple model based on the present complex structure showed that Arg-415 could form a hydrogen-bonding contact with the G base at this position (supplemental Fig. S3). This contact is essentially in the manner typical of the recognition of the G base by Arg (28) and may explain the preference for DNA bases.
This preference is significant for AtWRKY6 and AtWRKY11, which belong to groups IIb and IId, respectively, but not for AtWRKY26, AtWRKY38, and AtWRKY43, which belong to groups I, III, and IIc, respectively, as revealed by experiments using base substitutions (16). It should be noted that, for AtWRKY26 and AtWRKY43, Arg is conserved at a position that is equivalent to Arg-413 of AtWRKY4, but the equivalent residue is Gln or Ser for AtWRKY6 and AtWRKY11. Therefore, we suggest the possibility that the hydrogen-bonding contact between Arg-413 and a phosphate (Fig. 3a), which is reinforced by electrostatic attractions, ensures the stability of the complex without the contact between Arg-415 and the G base. In contrast, Gln or Ser at this position is not capable of gaining electrostatic attraction but is capable of forming hydrogen bonds with phosphate, as simply modeled based on the present complex structure (data not shown). A basic residue, i.e. Arg or Lys, is conserved at this position in all group I and IIc WRKY proteins, and Gln and Ser are conserved in group IIb and IId proteins, respectively (5). Therefore, the above explanation is valid for all proteins belonging to these WRKY groups. However, AtWRKY38 possesses Leu at this position, and it may not form a hydrogen bond or gain electrostatic attractions. This protein, as well as other group III WRKY proteins, possesses basic residues in several different sites compared with the proteins of other WRKY groups and therefore may possess a slightly different binding framework, which consequently may not require the Arg-G contact. As discussed above, the differences in amino acids (particularly those capable of contacting DNA) among the WRKY groups may lead to a preference for bases that flank the TTGACY core motif and thereby enable selective and concerted control of many target genes of the WRKY family of transcription factors.
Implications for Requirement of Amino Acids
The involvement of the WRKYGQK sequence in DNA binding was investigated by mutational experiments (13, 15, 16). Among the residues, Trp, Tyr, and two Lys residues were clearly indicated as indispensable in DNA binding (13, 15). The Trp residue, i.e. Trp-414 of AtWRKY4 (residue numbers in AtWRKY4 are used for the equivalent residues in this section), forms the structural core of the domain (14), so the mutation to Ala (13) may disrupt the correct structure that is necessary for DNA binding. Lys-416, Tyr-417, and Lys-420 directly contact DNA bases and/or the sugar phosphate backbone (Fig. 3), and mutations of Lys-416 to Ala, Tyr-417 to Ala or Arg, and Lys-420 to Ala abolished DNA-binding activity (13, 15). It should be noted that mutations of Tyr-417 to Phe (15, 16) and Lys-416 to Arg (15) only partially impaired the activity, which is consistent with the present complex structure in that the Tyr-to-Phe mutation should maintain the apolar contacts between the aromatic ring and the T9′ methyl group and in that the Lys-to-Arg mutation maintains hydrogen-bonding/electrostatic interactions with the phosphate. Furthermore, the importance of Gly-418 was clear, as large decreases were observed in affinity because of a mutation to Ala or Phe (13, 15). Probably, the addition of side chain disables the deep entrance of Gly-418 into the DNA groove.
In contrast, mutations of Arg-415 or Gln-419 did not affect complex formation (13, 15), although contacts involving these residues were observed in the present complex structure (Fig. 3a). These results indicate that the contacts by the above residues are not indispensable for forming the complex, at least under the conditions of the relevant electrophoretic experiments. In addition, in this study, substitution of the T5 base with U did not impair the affinity (Fig. 4 and Table 2), indicating that the apolar contacts between Arg-415 and the T5 methyl group are not indispensable. However, we hypothesize that, under a cellular condition that would be considerably different from the experimental ones, contacts, including the presumable contact between Arg-415 and the G base (supplemental Fig. S3), may contribute to ensuring the binding and the preference for DNA bases.
In addition to the WRKYGQK residues, Arg-429 and Lys-433 are important in the binding (15), which is consistent with the present structure (Fig. 3a). In particular, it was demonstrated that Arg and Lys were interchangeable at position 433 (15), and both could form hydrogen bonds and electrostatic attractions simultaneously, as observed in the present structure.
DNA-binding Domains Containing β-Sheets as the Molecular Interface
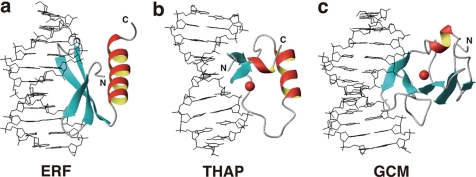
The majority of the DNA-binding domains utilize α-helices to contact DNA, whereas a relative minority of them utilize β-sheets. Many of the latter, such as MetJ-Arc repressors (29, 30), the AtERF1 ERF domain (Fig. 5a) (31), Tn916 integrase (32), and the THAP zinc finger (Fig. 5b) (33, 34), have two- or three-stranded β-sheets that fit into the major groove of DNA. For these, the β-sheet plane is approximately parallel to the DNA helical axis around the molecular interface. The contacting amino acid side chains are located on one side of the β-sheet plane that is supported by α-helices on the other side.
FIGURE 5.
DNA-binding domains contacting DNA by utilizing β-sheets. a, ERF domain of Arabidopsis AtERF1 (Protein Data Bank code 1GCC). b, THAP zinc finger domain of human THAP1 (code 2KO0). c, larger subdomain of the GCM domain of Drosophila GCM (code 1ODH). Red spheres in b and c indicate zinc ions. The DNA-binding mode in c, as well as that of the WRKY domain, is termed the β-wedge (see “Results and Discussion”). The figures were produced by MOLMOL (41).
In contrast, the WRKY domain presented here and the GCM domain (Fig. 5c) (35) utilize four- or five-stranded β-sheets, which are too large to fit into the DNA groove in the manner described above. Instead, β-sheets enter the groove with planes that are approximately perpendicular to the DNA helical axis. The proteins utilize the rim of the β-sheet, so the side chains that are located on both sides of the plane are involved in the contacts. An α-helix-supporting β-sheet was not identified for the WRKY domain (14, 15), whereas a short α-helix appeared to support the relevant β-sheet of the GCM domain, although it was located apart from the molecular interface. We refer to this atypical binding mode as a β-wedge because the β-sheet appears to sharply cut into the DNA groove.
Together with the similarity in the arrangement of the zinc-binding residues, it has been proposed that the WRKY and GCM domains are evolutionarily related, although the latter is shared only by animals (14, 36, 37). It should be pointed out that the β1-strand (the rim strand) of the WRKY domain is kinked in the middle by the insertion of Gly-418, whereas that of the GCM domain is short enough to be included in the major groove without a kink.
For the NAC domain, which composes another major family of plant-specific transcription factors, the β-sheet structure possesses a basic rim strand with a kink that was induced by the insertion of a Gly residue (38), which is similar to the WRKY domain. Therefore, NAC may adopt the β-wedge mode of DNA binding and may be evolutionarily related to the WRKY and GCM transcription factors.
Supplementary Material
Acknowledgments
We thank T. Harada, T. Nagira, M. Ikari, and Y. Tomo (RIKEN) for technical support in sample preparation and F. Delaglio (National Institutes of Health) for providing TALOS+ software.
This work was supported in part by the RIKEN Structural Genomics/Proteomics Initiative (RSGI) and the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science, and Technology.

This article contains supplemental “Materials” and Figs. S1–S5.
The atomic coordinates and structure factors (code 2LEX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The 1H, 13C, and 15N chemical shifts are available in the Biological Magnetic Resonance Data Bank under BMRB accession number 17732.
- AtWRKY4-C
- Arabidopsis thaliana WRKY4 protein C-terminal WRKY domain
- SPR
- surface plasmon resonance
- RDC
- residual dipolar coupling
- HSQC
- heteronuclear single-quantum correlation
- IPAP
- in-phase/anti-phase.
REFERENCES
- 1. Ishiguro S., Nakamura K. (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571 [DOI] [PubMed] [Google Scholar]
- 2. Rushton P. J., Macdonald H., Huttly A. K., Lazarus C. M., Hooley R. (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 29, 691–702 [DOI] [PubMed] [Google Scholar]
- 3. Rushton P. J., Torres J. T., Parniske M., Wernert P., Hahlbrock K., Somssich I. E. (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- 4. de Pater S., Greco V., Pham K., Memelink J., Kijne J. (1996) Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 24, 4624–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206 [DOI] [PubMed] [Google Scholar]
- 6. Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O. J., Samaha R. R., Creelman R., Pilgrim M., Broun P., Zhang J. Z., Ghandehari D., Sherman B. K., Yu G. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110 [DOI] [PubMed] [Google Scholar]
- 7. Eulgem T., Rushton P. J., Schmelzer E., Hahlbrock K., Somssich I. E. (1999) Early nuclear events in plant defense signaling: rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C., Chen Z. (2000) Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 42, 387–396 [DOI] [PubMed] [Google Scholar]
- 9. Hara K., Yagi M., Kusano T., Sano H. (2000) Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol. Gen. Genet. 263, 30–37 [DOI] [PubMed] [Google Scholar]
- 10. Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., Kamiya A., Nakajima M., Enju A., Sakurai T., Satou M., Akiyama K., Taji T., Yamaguchi-Shinozaki K., Carninci P., Kawai J., Hayashizaki Y., Shinozaki K. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold, and high salinity stresses using a full-length cDNA microarray. Plant J. 31, 279–292 [DOI] [PubMed] [Google Scholar]
- 11. Rizhsky L., Liang H., Mittler R. (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 130, 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pnueli L., Hallak-Herr E., Rozenberg M., Cohen M., Goloubinoff P., Kaplan A., Mittler R. (2002) Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 31, 319–330 [DOI] [PubMed] [Google Scholar]
- 13. Maeo K., Hayashi S., Kojima-Suzuki H., Morikami A., Nakamura K. (2001) Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 65, 2428–2436 [DOI] [PubMed] [Google Scholar]
- 14. Yamasaki K., Kigawa T., Inoue M., Tateno M., Yamasaki T., Yabuki T., Aoki M., Seki E., Matsuda T., Tomo Y., Hayami N., Terada T., Shirouzu M., Tanaka A., Seki M., Shinozaki K., Yokoyama S. (2005) Solution structure of an Arabidopsis WRKY DNA-binding domain. Plant Cell 17, 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duan M. R., Nan J., Liang Y. H., Mao P., Lu L., Li L., Wei C., Lai L., Li Y., Su X. D. (2007) DNA-binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 35, 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ciolkowski I., Wanke D., Birkenbihl R. P., Somssich I. E. (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY domain function. Plant Mol. Biol. 68, 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kigawa T., Yabuki T., Yoshida Y., Tsutsui M., Ito Y., Shibata T., Yokoyama S. (1999) Cell-free production and stable isotope labeling of milligram quantities of proteins. FEBS Lett. 442, 15–19 [DOI] [PubMed] [Google Scholar]
- 18. Matsuda T., Kigawa T., Koshiba S., Inoue M., Aoki M., Yamasaki K., Seki M., Shinozaki K., Yokoyama S. (2006) Cell-free synthesis of zinc-binding proteins. J. Struct. Funct. Genomics 7, 93–100 [DOI] [PubMed] [Google Scholar]
- 19. Aoki M., Matsuda T., Tomo Y., Miyata Y., Inoue M., Kigawa T. (2009) Automated system for high throughput protein production using the dialysis cell-free method. Protein Expr. Purif. 68, 128–136 [DOI] [PubMed] [Google Scholar]
- 20. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen M. R., Mueller L., Pardi A. (1998) Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 5, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 22. Bax A. (1994) Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 4, 738–744 [Google Scholar]
- 23. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York [Google Scholar]
- 24. Ottiger M., Delaglio F., Bax A. (1998) Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 25. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26. Tolman J. R., Flanagan J. M., Kennedy M. A., Prestegard J. H. (1995) Nuclear magnetic dipole interactions in field-oriented proteins: information for structure determination in solution. Proc. Natl. Acad. Sci. U.S.A. 92, 9279–9283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tjandra N., Grzesiek S., Bax A. (1996) Magnetic field dependence of nitrogen-proton J splittings in N-15-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J. Am. Chem. Soc. 118, 6264–6272 [Google Scholar]
- 28. Pabo C. O., Sauer R. T. (1992) Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61, 1053–1095 [DOI] [PubMed] [Google Scholar]
- 29. Somers W. S., Phillips S. E. (1992) Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature 359, 387–393 [DOI] [PubMed] [Google Scholar]
- 30. Raumann B. E., Rould M. A., Pabo C. O., Sauer R. T. (1994) DNA recognition by β-sheets in the Arc repressor-operator crystal structure. Nature 367, 754–757 [DOI] [PubMed] [Google Scholar]
- 31. Allen M. D., Yamasaki K., Ohme-Takagi M., Tateno M., Suzuki M. (1998) A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box-binding domain in complex with DNA. EMBO J. 17, 5484–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wojciak J. M., Connolly K. M., Clubb R. T. (1999) NMR structure of the Tn916 integrase-DNA complex. Nat. Struct. Biol. 6, 366–373 [DOI] [PubMed] [Google Scholar]
- 33. Campagne S., Saurel O., Gervais V., Milon A. (2010) Structural determinants of specific DNA recognition by the THAP zinc finger. Nucleic Acids Res. 38, 3466–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sabogal A., Lyubimov A. Y., Corn J. E., Berger J. M., Rio D. C. (2010) THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat. Struct. Mol. Biol. 17, 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cohen S. X., Moulin M., Hashemolhosseini S., Kilian K., Wegner M., Müller C. W. (2003) Structure of the GCM domain-DNA complex: a DNA-binding domain with a novel fold and mode of target site recognition. EMBO J. 22, 1835–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Babu M. M., Iyer L. M., Balaji S., Aravind L. (2006) The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 34, 6505–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamasaki K., Kigawa T., Inoue M., Watanabe S., Tateno M., Seki M., Shinozaki K., Yokoyama S. (2008) Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol. Biochem. 46, 394–401 [DOI] [PubMed] [Google Scholar]
- 38. Ernst H. A., Olsen A. N., Skriver K., Larsen S., Lo Leggio L. (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kraulis P. J. (1991) MolScript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 41. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.