Abstract
Although restoration of dystrophin expression via exon skipping in both cardiac and skeletal muscle has been successfully demonstrated in the mdx mouse, restoration of cardiac dystrophin expression in large animal models of Duchenne muscular dystrophy (DMD) has proven to be a challenge. In large animals, investigators have focused on using intravenous injection of antisense oligonucleotides (AO) to mediate exon skipping. In this study, we sought to optimize restoration of cardiac dystrophin expression in the golden retriever muscular dystrophy (GRMD) model using percutaneous transendocardial delivery of recombinant AAV6 (rAAV6) to deliver a modified U7 small nuclear RNA (snRNA) carrying antisense sequence to target the exon splicing enhancers of exons 6 and 8 and correct the disrupted reading frame. We demonstrate restoration of cardiac dystrophin expression at 13 months confirmed by reverse transcription-PCR (RT-PCR) and immunoblot as well as membrane localization by immunohistochemistry. This was accompanied by improved cardiac function as assessed by cardiac magnetic resonance imaging (MRI). Percutaneous transendocardial delivery of rAAV6 expressing a modified U7 exon skipping construct is a safe, effective method for restoration of dystrophin expression and improvement of cardiac function in the GRMD canine and may be easily translatable to human DMD patients.
Introduction
Duchenne muscular dystrophy (DMD) is a lethal, X-linked recessive disease affecting 1 in 3,500 newborn boys that results from a mutation in the dystrophin gene.1 Thousands of mutations in the 79-exon dystrophin gene have been reported with the most severe cases being associated with disruption of the reading frame and loss of functional dystrophin production.2 Boys typically present with symptoms of muscle weakness by age 5, become wheelchair-bound by early to mid-teens, and die from respiratory failure or cardiomyopathy in their late teens to early twenties. Patients with dystrophin mutations that maintain the reading frame and produce truncated but partially functional dystrophin exhibit the milder phenotype of Becker muscular dystrophy (BMD).2 Unfortunately, no effective therapy or cure exists for DMD, and novel therapeutic strategies are in need of development.
One strategy that has potential for treating and possibly curing DMD in a subset of patients is exon skipping, in which either a synthetic or ribo-oligonucleotide antisense sequence, complementary to specific cis acting signals on the primary RNA transcript interferes with splicing of a specific exon or exons. When delivered to a cell or animal, these splicing sequences interrupt splicing, and induce skipping of the exon or exons in question.3,4,5 Effective exon skipping remove the exons causing the frameshift mutation responsible for loss of functional dystrophin expression as well as additional exons as necessary to restore the reading frame. The final result will be restoration of expression of a partially truncated but functional dystrophin and amelioration of disease phenotype. In other words, exon skipping may convert the DMD phenotype to the BMD phenotype or better.
The sequences that induce exon skipping can be delivered as antisense oligonucleotides (AO) or transcribed as an antisense sequence linked to U7 or U1 small nuclear RNA (snRNA).5 Proof-of-concept studies have demonstrated successful exon skipping and restoration of dystrophin expression in cells cultured from DMD patient muscle biopsies6,7 and canine muscular dystrophy muscle biopsies8,9 using the AO approach. In addition, the AO approach has been successfully used to restore dystrophin expression in the mdx mouse model of DMD, although significant cardiac expression was not initially demonstrated.10,11 More recent studies employing AO's with a morpholino-modified backbone have reported highly efficient, systemic restoration of both skeletal muscle and cardiac dystrophin in the mdx mouse following vascular delivery.12,13,14,15 A preclinical investigation of AO-induced exon skipping in the Beagle muscular dystrophy model has demonstrated systemic skeletal muscle exon skipping and dystrophin expression with improvement in clinical symptoms 22 weeks following vascular delivery, but cardiac dystrophin production was modest, and cardiac functional evaluation was not performed.16 A second preclinical trial in healthy cynomolgus monkeys reported successful skipping of exon 50 in 25% of quadriceps muscle, but only 2% of the heart following vascular delivery, underscoring the fact that an alternative approach may be necessary for efficient cardiac exon skipping in large animals and humans.4 However, functional outcomes of exon skipping could not be determined in the normal muscles. Currently, the AO approach for single exon skipping is being evaluated in clinical trials,3,4,5 and proof-of-concept studies have already demonstrated local exon skipping and dystrophin expression following direct intramuscular injection, but efforts directed at cardiac treatment have not yet been initiated.17,18
Although the AO approach has been studied extensively and is currently being evaluated in clinical trials, it has several limitations, including the need for weekly or biweekly injections and lack of demonstration of efficient cardiac exon skipping in large animals. In addition, since more than one exon is involved in most DMD genetic lesions, multiple AO molecules would be required to restore the open reading frame. The need for repeat injections can be overcome by using viral-mediated delivery of U7 or U1 snRNA carrying the antisense sequence to mediate exon skipping. This has been successfully used with lentivirus in human DMD myoblasts19,20 and with recombinant adeno-associated virus (rAAV) in the mdx mouse.21 Significant skeletal muscle and cardiac dystrophin expression has been demonstrated in the mdx mouse as far as 74 weeks following a single vascular injection of rAAV-U7.22,23 However, large animal studies utilizing rAAV-U7 have not yet been performed, and it is possible that cardiac exon skipping following vascular delivery may be as inefficient with the rAAV-U7 approach as with the AO approach in beagles16 and cynomolgus monkeys.4 Since cardiomyopathy is a leading cause of death in DMD patients, this would be a significant therapeutic limitation.
Therefore, our goal in this study was to develop a method utililzing exon skipping to restore cardiac dystrophin expression long-term in a large animal model of DMD. We chose the golden retriever model of DMD (GRMD) because it is the largest animal of DMD available and therefore has a more severe phenotype than other smaller canine models of DMD,24 such as the Beagle16 or Cavalier King Charles Spaniel.9 It harbors a point mutation in the 3′ consensus splice acceptor site of intron 6 that causes exclusion of exon 7 and disruption of the reading frame.24 Skipping of exons 6 and 8 in this model would, in theory, restore the reading frame as well as expression of a partially truncated but functional dystrophin. We hypothesized that we could employ percutaneous transendocardial delivery of rAAV625,26 expressing a modified U7 snRNA exon skipping construct (rAAV6-U7-ESE6/8) to achieve long-term restoration of cardiac dystrophin expression in GRMD. We report successful exon skipping and restoration of cardiac dystrophin expression and improved cardiac function in GRMD canines 13 months after percutaneous transendocardial delivery of rAAV6-U7-ESE6/8.
Results
Study design
A total of nine GRMD canines were used in this study that was designed to evaluate the efficacy of rAAV6-mediated cardiac delivery of a U7 snRNA modified to skip exons 6 and 8 (U7 targeting exon splicing enhancer 6/8, or U7-ESE6/8) and restore the reading frame in the GRMD model. One dog (13 months old, 14 kg) was treated with 5 × 1011 gc/kg vector and euthanized 2 months postinjection to evaluate dystrophin expression at an early time point. The remaining five treated dogs were followed 13 months postinjection to assess long-term dystrophin expression. Of these five dogs, three were treated with 1.8 × 1012 gc/kg vector (n = 1: 5 months old, 12 kg; n = 2: 10 months old, 14–17 kg), and two were treated with 1.4 × 1013 gc/kg vector (9 months old, 15–19 kg) so that a comparison between low- and high-dose vector could be made. Three controls were euthanized at 23–25 months of age (14–20 kg).
Vector genome detection
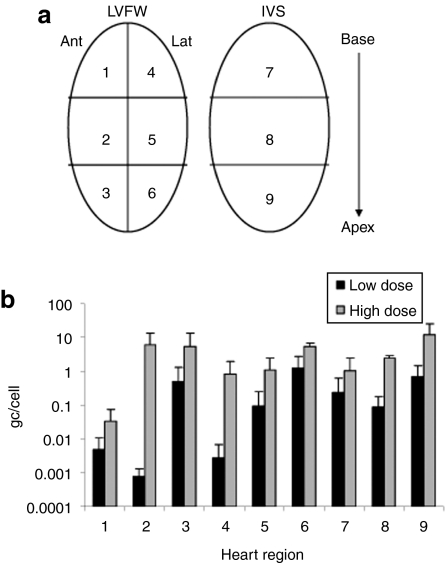
Quantitative TaqMan PCR was performed to assess the number of vector genome copies delivered to each region of the heart in GRMD canines treated with low- and high-dose vector. For analysis, the heart was divided into nine regions as depicted in Figure 1a. A dose–response relationship was observed. Escalating the vector dose by approximately eightfold increased the number of genomes delivered per cell by at least five to tenfold in all regions of the heart (Figure 1b). The lowest gene transfer efficiency was noted in region 1, the anterior base of the heart.
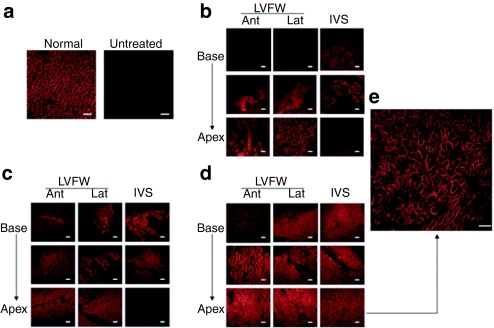
Figure 1.
Taqman analysis of vector genome distribution in the heart. (a) For analysis, biopsies were collected from the LVFW and IVS from base to apex as depicted. (b) Biodistribution of vector genomes throughout the heart in individual high- and low- dose GRMD canines. Error bars represent standard deviations. Ant, anterior; gc, genome copies; GRMD, golden retriever muscular dystrophy; IVS, interventricular septum; Lat, lateral; LVFW, left ventricular free wall.
Dystrophin mRNA analysis
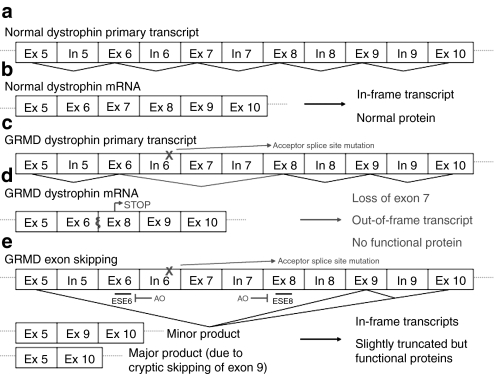
The exon skipping strategy used to restore dystrophin expression is described and depicted in Figure 2. In untreated animals, the messenger RNA (mRNA) lacks exon 7 secondary to the GRMD mutation with subsequent out-of-frame splicing of exon 6 to exon 824 (Figure 2c,d). Following successful treatment, the mRNA will lack either exons 6–8 or exons 6–9 (removal of exon 9 occurring cryptically)27 with splicing of exon 5 to either exon 9 or exon 10, both of which are in-frame (Figure 2e). An alternative product would be an mRNA lacking either exon 6 or exon 8 in addition to exon 7 and/or exon 9, all of which would be out-of-frame. Reverse transcription-PCR (RT-PCR) was used to assess exon skipping in the cardiac mRNA of GRMD dogs treated with rAAV6-U7-ESE6/8. We designed primers so that the amplified region in the untreated hearts would be 953 bp (Δ7), while the amplified region in successfully treated areas of the heart would be either 605 bp (Δ6–8) or 490 bp (Δ6–9).
Figure 2.
Description of exon skipping strategy for restoration of dystrophin expression in the GRMD canine. (a) The dystrophin gene is large, containing 79 exons. The primary RNA transcript is in normal canines is depicted schematically. For simplicity, we have focused on the region affected by the GRMD mutation. (b) Splicing of the normal primary transcript yields normal mRNA that is translated into normal dystrophin protein. (c) GRMD is caused by a point mutation in dystrophin in the acceptor splice site of intron 6. (d) Splicing of this mutant primary transcript yields an mRNA with exon 6 fused directly to exon 8. The end result is a nonsense mutation with a premature STOP in exon 8. No functional dystrophin is produced from this out-of-frame transcript. (e) Exon skipping is a strategy that can be used to restore the open reading frame, and thus dystrophin expression, in GRMD. In this study, we used rAAV6 vector to express antisense oligonucleotides (AO) to target the exon splicing enhancers (ESE) in exons 6 and 8. By inducing skipping of exons 6 and 8 in this manner, we were able to restore the open reading frame with production of a slightly truncated but functional dystrophin. It should be noted that the major product of this exon skipping strategy is an mRNA with exon 5 fused directly to exon 10 (also in-frame), as cryptic splicing of exon 9 occurs with high frequency. The resultant protein contains 75/79 of the original exons. AAV, adeno-associated virus; GRMD, golden retriever muscular dystrophy; mRNA, messenger RNA; rAAV6, recombinant AAV6.
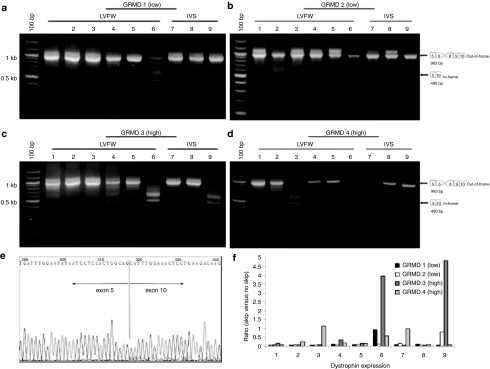
We detected evidence of successful exon skipping in all treated hearts by RT-PCR (Figure 3). When induced exon skipping was detected, the predominant band was 490 bp, representing the in-frame splicing of exon 5 to exon 10 (Figure 3a–d). This was confirmed by sequencing the splice junction (Figure 3e). It should be noted, however, that nine regions of each heart were examined (Figure 1a), and induced exon skipping was not present in all samples analyzed. Exon skipping mediated by high-dose rAAV6 was more efficient than exon skipping mediated by low-dose rAAV6 with 5–6 of 9 total samples positive per canine in the high-dose group (Figure 3c,d) versus 1–2 out of 9 total samples positive per canine in the low-dose group (Figure 3a,b). Quantitative analysis confirmed that the ratio of in-frame skipped product to out-of-frame unskipped product was higher in canines treated with high- versus low-dose rAAV6 (Figure 3f).
Figure 3.
RT-PCR analysis of exon skipping in GRMD canines treated with rAAV6-U7-ESE6/8. The GRMD mutation causes loss of exon 7, producing an out-of-frame transcript. Treatment with ESE6/8 will cause splicing of exon 5 to exon 9 or exon 10 (as cryptic skipping of exon 9 is a well-described phenomenon), both of which are in-frame transcripts. For analysis, biopsies were collected from the LVFW and IVS from base to apex as depicted in Figure 1a. Representative gel images are shown from GRMD canines treated with (a,b) 1.8 × 1012 gc/kg of rAAV6-U7-ESE6/8 and euthanized 13 months later (n = 3), and (c,d) 1.4 × 1013 gc/kg of rAAV6-U7-ESE6/8 and euthanized 13 months later (n = 2). Samples were considered positive for exon skipping if they contained the 490 bp band indicative of in-frame splicing of exon 5 to exon 10, as depicted in the diagram adjacent to each gel image. Samples were considered negative for exon skipping if they contained only the 953 bp band representing loss of exon 7 secondary to the GRMD mutation. Note that a greater number of samples were positive for exon skipping following treatment with high-dose rAAV6 compared to lower doses of rAAV6. (e) Sequencing of the 490 bp band confirmed splicing of exon 5 to exon 10, which produces an in-frame transcript. (f) Quantitative analysis confirmed that the ratio of in-frame skipped product to unskipped product was higher in canines treated with high-versus low-dose rAAV6. AAV, adeno-associated virus; GRMD, golden retriever muscular dystrophy; IVS, interventricular septum; LVFW, left ventricular free wall; rAAV76, recombinant AAV6; RT-PCR, reverse transcription-PCR.
Dystrophin protein analysis
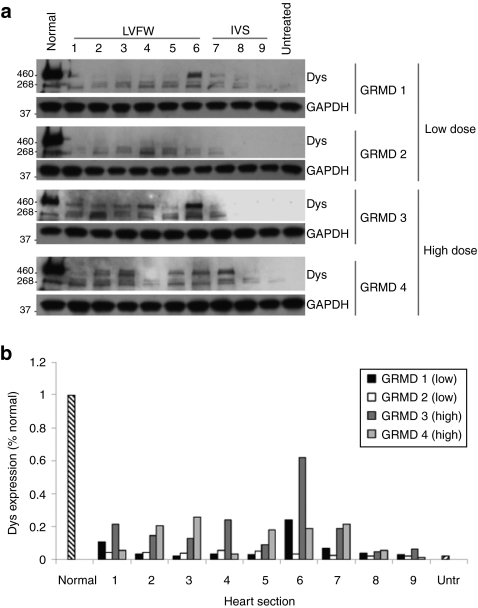
Since we had evidence of in-frame dystrophin mRNA expression from our RT-PCR analysis, we next performed immunoblotting with an antidystrophin antibody to confirm expression of dystrophin protein. The truncated dystrophin expressed in GRMD canines is slightly smaller (~409 kDa) than wild-type (427 kDa), as expected, due to loss of exons 6–9 (Figure 4a). Dystrophin protein was detected in all treated hearts (Figure 4a). However, dystrophin protein was expressed at higher levels and in a greater number of regions throughout the heart in canines treated with high-dose rAAV6 versus low-dose rAAV6 (Figure 4a). In addition, quantitative analysis revealed that dystrophin levels in treated GRMD canines reached 15–20% of normal (unaffected canine) in most regions of the heart following therapy with high-dose rAAV6 compared to only 3–5% of normal in most regions of the heart following injection with low-dose rAAV6 (Figure 4b). As a reference, dystrophin expression in untreated GRMD canines was ~2% of normal (Figure 4b).
Figure 4.
Western blot analysis of dystrophin expression in GRMD canines treated with rAAV6-U7-ESE6/8. For analysis, biopsies were collected from the LVFW and IVS from base to apex as depicted in Figure 1a. Normal, untreated unaffected canine (positive control); untreated, untreated affected GRMD canine (negative control). (a) Representative blots are shown from GRMD canines treated with either 1.8 × 1012 gc/kg of rAAV6-U7-ESE6/8 (GRMD 1, GRMD 2) or 1.4 × 1013 gc/kg of rAAV6-U7-ESE6/8 (GRMD 3, GRMD 4) and euthanized 13 months later. Note that a greater number of samples were positive for dystrophin following treatment with high-dose rAAV6 compared to low-dose rAAV6. (b) Quantitative, densitometric analysis of each western blot was performed. Dystrophin expression was higher in GRMD canines treated with high-dose rAAV6 (15–20% of normal) versus low-dose rAAV6 (3–5% of normal). AAV, adeno-associated virus; Dys, dystrophin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GRMD, golden retriever muscular dystrophy; IVS, interventricular septum; LVFW, left ventricular free wall; rAAV6, recombinant AAV6; Untr, untreated GRMD canine.
We next performed immunohistochemistry (IHC) using an antidystrophin antibody to assess tissue distribution and cellular localization of the truncated dystrophin. Dystrophin staining in a normal dog (not affected with GRMD) and in an untreated GRMD dog is shown for reference (Figure 5a). We found that dystrophin was expressed throughout the hearts of GRMD canines treated with rAAV6-U7-E6E8 (Figure 5) and that the dystrophin was localized at the cell membranes (Figure 5e). In addition, dystrophin expression was greater in canines treated with high-dose rAAV6 (Figure 5d) compared to those treated with low-dose rAAV6 (Figure 5b,c). The lowest dystrophin expression was noted in the anterior base of the heart.
Figure 5.
Immunohistochemical (IHC) staining for dystrophin in GRMD canines treated with rAAV6-U7-ESE6/8. For analysis, sections were prepared from the LVFW and IVS from base to apex as depicted in Figure 1a. (a) Images from normal (untreated, unaffected) (positive control) and untreated (untreated, affected GRMD) (negative control) are shown for reference. Representative images are shown from GRMD canines treated with (b) 5 × 1011 gc/kg of rAAV6-U7-ESE6/8 and euthanized 2 months later (n = 1), (c) 1.8 × 1012 gc/kg of rAAV6-U7-ESE6/8 and euthanized 13 months later (n = 3), and (d) 1.4 × 1013 gc/kg of rAAV6-U7-ESE6/8 and euthanized 13 months later (n = 2). (e) The inset demonstrates membrane localization of dystrophin in an image taken at high power (×40 objective). Bar, 200 µm. Note that a greater area of myocardium is positive for dystrophin expression following treatment with high-dose rAAV6 compared to the lower doses of rAAV6. AAV, adeno-associated virus; Ant, anterior; GRMD, golden retriever muscular dystrophy; IVS, interventricular septum; Lat, lateral; LVFW, left ventricular free wall; rAAV6, recombinant AAV6.
Fibrosis
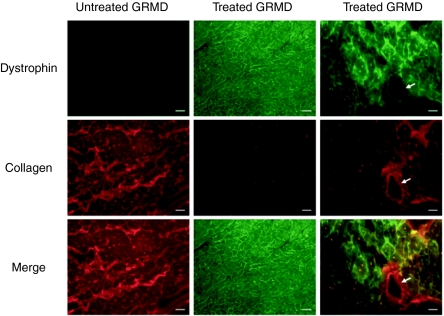
We assessed cardiac fibrosis by performing IHC for collagen. We also co-stained for dystrophin so that a relationship between dystrophin expression and fibrosis could be established. In untreated GRMD canines, dystrophin expression was absent, and collagen content was high (Figure 6). In treated GRMD canines, dystrophin expression was restored, and collagen content was reduced (Figure 6). Merging of collagen and dystrophin stained images revealed that collagen content was reduced specifically in regions where dystrophin expression was restored (Figure 6). The presence of collagen in a blood vessel wall confirmed specificity of collagen staining (Figure 6).
Figure 6.
Analysis of fibrosis in GRMD canines treated with rAAV6-U7-ESE6/8. Myocardium was co-stained for dystrophin and collagen I. Representative slides are shown from GRMD canines treated with 1.4 × 1013 gc/kg of rAAV6-U7-ESE6/8 and euthanized 13 months later. An untreated GRMD is shown as control. Note that collagen content is high when dystrophin staining is low or absent. The white arrow depicts collagen staining in a blood vessel wall. Bar, 200 µm. AAV, adeno-associated virus; GRMD, golden retriever muscular dystrophy; rAAV6, recombinant AAV6.
Echocardiography
Echocardiography (echo) was performed on GRMD canines pre- and post-treatment and was also performed on age-matched, untreated GRMD canines to assess cardiac function and geometry. Data are summarized in Supplementary Table S1. The cardiac function and geometry of the animals in this study varied widely and did not correlate well with age or treatment. For example, the youngest dog in this study, DM74 (15 months old at post-treatment echo), had the most dilated ventricle (left ventricular inner diameter in diastole (LVIDd): 4.4 cm) and most depressed contractility (fractional shortening: 15%) despite treatment. In contrast, some of the older dogs in the study, DM109 and DM110 (23–24 months old at post-treatment echo) had LV chamber diameter (LVIDd: 3.1 cm) and contractility (fractional shortening: 30–32%) values in the low normal range without treatment. As a result of this variability, no significant differences in cardiac function and geometry were noted in the treated animals compared to control.
Cardiac MRI
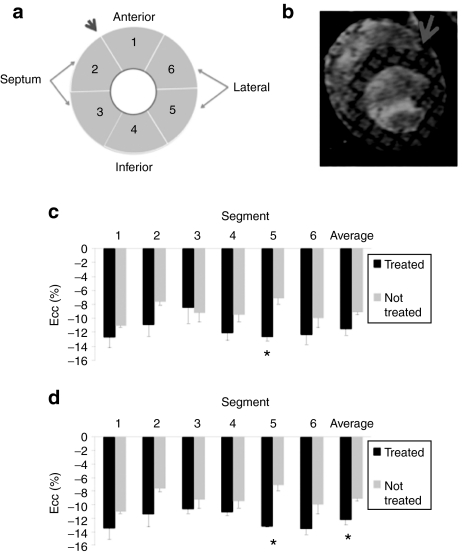
Average peak circumferential strain was greater (P < 0.01) in the treated (−12.7 ± 0.5%) versus untreated (−7.2 ± 0.8%) GRMD dogs in the lateral LV free wall region of the heart (Figure 7c). Other regions showed greater variability among the treated dogs, with some dogs demonstrating greater strain than untreated and others showing similar values as the untreated dogs. Overall, there was general agreement between dystrophin expression and the circumferential strain measures. For example, expression was lowest in GRMD 78 of the treated dogs and this was also the dog that showed the lowest average peak circumferential myocardial strain of the treated dogs. Furthermore, when GRMD 78 was omitted from the comparison between groups due to the low dystrophin expression levels in the heart, the average peak circumferential strain was greater (P = 0.027) in treated (−12.2 ± 0.8%) versus untreated (−9.1 ± 0.4%) GRMD dogs (Figure 7d).
Figure 7.
Cardiac MRI assessment of function. (a) Left ventricular myocardium in the short-axis view was divided into six equal segments with the first segment placed in the anterior region. (b) The slice analyzed was located in the mid-papillary region and (c) the average peak circumferential strain (Ecc) was measured in the treated and untreated GRMD canines. (d) Note that both segment 5 and average Ecc are significant if the animal with the lowest dystrophin expression is removed from analysis. *P < 0.05. Error bars represent standard deviations. GRMD, golden retriever muscular dystrophy; MRI, magnetic resonance imaging.
Discussion
This is the first report demonstrating long-term restoration of cardiac dystrophin expression in a large animal model of DMD. rAAV6-U7-ESE6/8 was utilized to skip exons 6 and 8 and restore the dystrophin reading frame in GRMD canines. We confirmed the presence of exon skipping using RT-PCR and sequencing and confirmed expression and membrane localization of dystrophin using western blot and IHC in GRMD canines 13 months following percutaneous transendocardial delivery of the exon skipping vector.
Although efficient and long-term cardiac exon skipping has been previously described in the mdx mouse,22,23 there are very few reports of cardiac exon skipping in large animal models of DMD. For example, weekly to biweekly intravenous injections of AO's were used to induce exon skipping in the Beagle canine X-linked muscular dystrophy (CXMD) model.16 Although efficient skeletal muscle exon skipping was demonstrated, cardiac exon skipping and dystrophin expression was present at only minimal levels. Similar results were reported for exon skipping in the cynomolgus monkey, with exon skipping product detected in 25% of quadriceps but only 2% of the heart following weekly intravenous injection of AO's.4 Since cardiomyopathy is a leading cause of death in DMD patients, correction of the skeletal muscle defect without the cardiac defect would not be a viable therapeutic option and would limit the application of exon skipping as a novel treatment for DMD.
The results of these large animal studies suggested to us that a specific delivery strategy may be needed to target the heart. We have previously demonstrated that percutaneous transendocardial delivery of rAAV6 can be used to achieve efficient cardiac transgene expression in the canine.25,26 We therefore developed a strategy in this investigation to utilize our minimally invasive cardiac gene transfer protocol to deliver a rAAV6-U7-snRNA exon skipping construct to restore dystrophin expression in the GRMD heart. Our novel approach overcomes the two main limitations of AO-mediated exon skipping in large animals: lack of significant cardiac exon skipping and the need for at least weekly injections to maintain continued exon skipping. Indeed, we demonstrated cardiac exon skipping and dystrophin expression 13 months following the initial treatment. Such a strategy could be used to complement the already established intravascular delivery method for AO-mediated exon skipping that targets skeletal muscle efficiently.4,16 Alternatively, an investigator may choose to utilize one of the vascular delivery methods currently being employed in large animals to target skeletal muscle with delivery of a rAAV-U7 exon skipping construct so that repeat injections of AO's would not be necessary.28,29 Applied translationally, this strategy would allow a DMD patient to undergo percutaneous transendocardial rAAV delivery to target the heart and vascular rAAV delivery to target the skeletal muscle once in the same treatment session, which would offer a definite advantage over strategies requiring multiple treatment sessions.
The other leading gene therapy-based approach for dystrophin replacement employs rAAV to deliver mini- or micro-dystrophin constructs.30,31 This strategy is currently being evaluated in large animal models29,30 and human trials.3 In this approach, ~14 kb dystrophin complementary DNA (cDNA) is truncated to under 5 kb so that it will fit within the packaging capacity of the rAAV vector.30 The goal is to convert the severe phenotype of DMD to the much milder BMD phenotype by inducing expression of a truncated but partially functional dystrophin.31 Although this strategy can be universally applied to DMD patients, we believe there are circumstances where exon skipping may provide an advantage over gene replacement. For example, the exon skipping product in this investigation, Δ6–9, lacks only ~0.5 kb of the 14 kb dystrophin cDNA, and it is estimated that a large percentage of DMD patients would benefit from the skipping of a similar number of exons, most of which are located in the hot spot region of exons 43–55.3 This skipped product represents >95% of the wild-type dystrophin cDNA compared to mini- and micro-dystrophins, which represent only ~40% of the cDNA.30 Since restoration of dystrophin expression via exon skipping leads to production of a protein much closer in size to wild-type dystrophin than the mini- or micro-dystrophin approach and since the severity of phenotype correlates to some extent with the size of in-frame deletions,2 one may expect DMD patients treated with exon skipping to display a milder BMD phenotype than those treated with mini- or micro-dystrophin.
In this study, we evaluated exon skipping mediated by rAAV vector using a dose escalation approach. The first dog enrolled in the study was treated with rAAV6 at a dose of 5 × 1011 gc/kg as this dose of rAAV6-mediated efficient expression of the green fluorescent protein (GFP) reporter in the canine heart in our previous report.25 Surprisingly, this dose of rAAV6-U7-ESE6/8 did not mediate significant exon skipping or restoration of dystrophin expression. Since that dose was based on data from GFP expression driven by the chicken β-actin promoter with CMV enhancer (CB), we hypothesized that the poor efficiency of exon skipping observed at the same dose in this study could be due to either (i) reduced activity of the U7 promoter versus the CB promoter or (ii) increased complexity involved in achieving successful exon skipping and restoration of dystrophin expression versus relative ease of inducing GFP expression via traditional transcription and translation (iii) or a combination of the two. We are unaware of any studies addressing these issues, but we reasoned that increased vector dose would lead to increased exon skipping if our hypothesis were true. Therefore, we increased the dose of rAAV6 ~3.5-fold (1.8 × 1012 gc/kg), based on vector availability, but only observed a modest increase in exon skipping and dystrophin expression.
At this point, we were still concerned that we had not yet reached the threshold vector dose to saturate the rAAV6 cellular receptors and increase the degree of exon skipping. In order to increase the dose of rAAV significantly, we would have to pool multiple vector preps per animal based on the yield from the triple transfection method of rAAV production, which would impose a financial burden. Therefore, in order to dose escalate, we utilized high-dose rAAV6 produced using baculovirus expression vectors in insect cell culture.32,33,34 The baculovirus expression vectors system is a novel alternative to rAAV vector production using traditional triple transfection in 293 cells, the method by which the rAAV vector used to treat the initial canines in this study was produced.35 The baculovirus expression vectors system is capable of producing vector yields 1–2 logs greater per prep than the traditional triple transfection method of rAAV production. Therefore, we were able to evaluate rAAV6 at a dose of 1.4 × 1013 gc/kg, which is approximately eightfold higher than the previous dose of rAAV6 used in this study. We found that high-dose rAAV6 was superior to low-dose rAAV6 in terms of exon skipping evaluated by RT-PCR and dystrophin expression evaluated by both western blot and IHC. In addition, Taqman analysis confirmed that escalating the vector dose by approximately eightfold increased the number of genomes delivered per cell by at least five to tenfold in all regions of the heart. Future investigation will be needed to determine if further dose escalation will result in increased vector genome delivery and exon skipping since it is unclear if our high dose was sufficient to saturate cellular rAAV6 receptors and/or intracellular exon skipping machinery.
It should also be noted that the lowest gene transfer, both in terms of vector genome content and restoration of dystrophin expression, was observed in the anterior base of the heart. This was a result of the technical difficulty associated with accessing this region of the heart following advancement of the injection catheter into the LV retrograde via the aorta. Future effort will be directed towards identifying a strategy to increase gene transfer to this region.
We analyzed cardiac function at 13 months by both echocardiography and magnetic resonance imaging (MRI). We did not detect a significant improvement in cardiac function in treated GRMD canines compared to untreated controls via echocardiography. It is probable that the inherent variability in GRMD cardiomyopathy makes it difficult to detect significant changes in cardiac function via echocardiography. For example, two recent studies were unable to detect evidence of LV dilation and dysfunction by standard echocardiography in a total of 21 GRMD canines aged 6 months to 2 years, which spans the age–range of the dogs in our study.36,37 A historical study reported LV dysfunction in 1/7 GRMD canines by 1 year of age,38 while another report describes hyperechoic LV lesions on echocardiography in 12/13 GRMD canines.39 Finally, a preliminary study with n = 1 at 6 months and 1 year reported depressed fractional shortening compared to n = 1 control at 6 months and 1 year.40 Indeed, as demonstrated in Supplementary Table S1, cardiac function and geometry, did not correlate with treatment or age in the dogs in our study.
However, using cardiac MRI, we were able to detect increased LV circumferential strain in the lateral free wall segment indicating improved regional cardiac function. In addition, if the dog with lowest dystrophin expression was removed, the average peak circumferential strain was greater in treated versus untreated GRMD dogs. Cardiac MRI is more sensitive than echocardiography in analyzing cardiac function, especially in DMD. Both Hor41 and Ashford42 demonstrated that MRI strain analysis can be used to detect progressive occult cardiovascular dysfunction in DMD patients with normal global function, and previous studies have shown a reduction of circumferential cardiac strain with disease progression in mdx mice43 and children with DMD.44 Furthermore, tagged imaging with circumferential strain analysis has been shown to be a sensitive measure to monitor improvement following transendocardial gene transfer of vascular endothelial growth factor (VEGF) in a canine model of ischemic cardiomyopathy.45 Similarly, the rAAV-mediated exon skipping strategy implemented in this study seems to be effective in ameliorating the degree of cardiac dysfunction as determined by MRI measurements of circumferential strain.
Although we report promising preclinical data describing the use of rAAV6-mediated exon skipping to restore cardiac dystrophin expression in GRMD, there are several considerations which must be discussed and/or addressed before clinical translation to DMD patients. Safety is a critical issue, and although the procedure was well tolerated in canines for the 13-month duration of this study, it is unclear if any long-term toxicity will result from either the procedure itself or from chronic expression of antisense RNA. For example, cardiac fibrosis resulting from the multiple injections required for global cardiac gene transfer could lead to arrhythmogenesis later in life, and long-term, constitutive expression of antisense RNA could lead to disruption of off- target, endogenous gene expression. In addition, any complication associated with cardiac catheterization, including periprocedural bleeding and infection, could occur each time the protocol is utilized. It should also be noted that while the canines used in the study may closely approximate the size of newly diagnosed DMD patients, further scaling up of vector doses may be necessary due to interspecies variation in gene transfer efficiency and may also be necessary to achieve improvement in cardiac function.
In summary, this is the first report demonstrating long-term restoration of cardiac dystrophin expression in a large animal model of DMD. We employed percutaneous transendocardial delivery of rAAV6-U7-ESE6/8 to induce exon skipping and demonstrated evidence of successful exon skipping and dystrophin expression 13 months following treatment. We also demonstrated reduced fibrosis and improved cardiac function via cardiac MRI in treated animals. Further investigation will be directed towards treating GRMD canines at an earlier age and/or increasing vector dose so that increased exon skipping and additional improvement in cardiac function may be appreciated.
Materials and Methods
Vector design and production. Each vector was designed to express modified U7 snRNA (U7SmOPT)21 carrying antisense sequence to target the canine exon splicing enhancers of exons 6 and 8 (ESE6/8) under control of the U7 promoter. The optimal antisense sequences were determined previously by in vitro screening in GRMD myotubes with subsequent confirmation of function in GRMD skeletal muscle (Published abstract: Vulin, A et al. (2006). Neuromuscul Disord 16: 722–723. doi:10.1016/j.nmd.2006.05.249). rAAV6 vector used for the low-dose treatment group was produced using the previously described pseudotyping protocol by the Vector Core of the Children's Hospital of Philadelphia.35 Briefly, rAAV genomes containing rAAV2 inverted terminal repeats were packaged by triple transfection of 293 cells with a cis-plasmid containing the U7-ESE6/8 exon skipping construct, an adenovirus helper plasmid, and a chimeric trans-helper plasmid containing the rAAV2 rep gene fused to the capsid gene of rAAV6. rAAV6 vector used for the high-dose treatment group was produced using the BEV system in insect cell culture.32,33,34
U7 Promoter: 5′CACATACGCGTTTCCTAGGAAACCAGAGAA GATCAAAGCCCCTCTCACACACCGGGGAGCGGGGAAGAGAA CTGTTTTGCTTTCATTGTAGACCAGTGAAATTGAGGGG TTTTCCGACCGAAGTCAGAAAACCTG 3′
U7SmOPT: 5′ CTCCAAAAATT 3′
ESE 6: 5′ CTGGGTCCGACAATCAACTCGTAATTATCCACAGG TTAATGT 3′
ESE 8: 5′ TGTATATCACATCACTCTTCCAAGTTTTGCCTCAAC AAGT 3′
Animal use and vector delivery protocol. All animals were handled in compliance with National Institute of Health and institutional guidelines that were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Nine affected GRMD canines were used in this study. One dog (13 months old, 14 kg) was treated with 5 × 1011 gc/kg vector and euthanized 2 months postinjection to evaluate dystrophin expression at an early time point. The remaining five treated dogs were followed 13 months postinjection to assess long-term dystrophin expression. Of these five dogs, three were treated with 1.8 × 1012 gc/kg vector (n = 1: 5 months old, 12 kg; n = 2: 10 months old, 14–17 kg), and two were treated with 1.4 × 1013 gc/kg vector (9 months old, 15–19 kg) so that a comparison between low- and high-dose vector could be made. Three untreated controls were euthanized at 23–25 months of age (14–20 kg).
The vector delivery procedure was performed under general anesthesia, and dogs were placed in left lateral recumbency. Heart rate, respiratory rate, systolic blood pressure, electrocardiogram, and oxygen saturation were monitored throughout the anesthetic period. A right carotid arterotomy was performed, and a 7 French introducer was placed in the vessel, followed by insertion of the injection catheter. This catheter was a steerable injection catheter with an adjustable length core needle (MyoCath; Bioheart, Sunrise, FL), which has previously been employed to deliver rAAV vector in our laboratory.25,26 The catheter was flushed with heparinized blood before vector infusion to prevent inactivation of the virus.46 Next, under fluoroscopic guidance, the catheter was advanced into the LV cavity, and by steering the needle tip and adjusting the needle length, ~60 transendocardial injections of 250 µl each were performed to target the LV free wall and interventricular septum from base to apex and from endocardium to epicardium with rAAV vector. Contrast media was added to the vector solution so that injection sites could be visualized. This allowed us to differentiate between injected and uninjected regions of the heart and helped to ensure that the vector solution was distributed globally throughout the myocardium.
For each procedure vector was mixed with 2 cc of sterile contrast solution (Omnipaque; Nycomed Imaging, Oslo, Norway) and diluted with sterile saline to produce 15 cc for injection. Lidocaine (2 mg/kg as a bolus followed by a constant rate infusion at 50 µg/kg/min) was initiated if ventricular tachycardia developed during the procedure. After recovery, dogs were treated with carprofen for 2 days and amoxicillin–clauvolonic acid for 5 days.
TaqMan PCR. To determine the number of vector genome copies delivered per cell, samples were snap-frozen in liquid nitrogen at the time of euthanasia. Following DNA extraction, genome copy (gc) titres were quantified by TaqMan PCR (Applied Biosystems, Foster City, CA) using primers and probes designed against the exon skipping construct. The amplicon was 64 base pairs. 1 µg of template DNA was used per reaction, and several controls were performed to confirm the specificity and accuracy of the PCR. A spike control was performed in which the test sample was spiked with exogenous test assay target to rule out PCR inhibition. The assay was also performed on samples from the negative control, saline-injected animal to determine background signal of the assay, which was negligible (<1 gc per 5,000 cells, or >1 log lower than the minimum gc detected in experimental samples).
Forward primer: CCTGCTCCAAAAATTTGTATATCAC
Reverse primer: CTTCCGCAAACTTGTTGAG
Probe: 6FAM-TCA CTC TTC CAA GTT TTG-MGBNFQ
RT-PCR and DNA sequencing. At the time of euthanasia, cardiac biopsies were collected as depicted in Figure 1 and snap-frozen in liquid nitrogen. Tissue was crushed with a mortar and pestle on dry ice, and total RNA was isolated using the TRIzol extraction method (Invitrogen, Carlsbad, CA). One hundred fifty nanograms of RNA from each sample was subjected to single-strand reverse transcription and 5µl of the generated cDNA was used as the template in the subsequent nested PCR reactions to detect evidence of exon skipping (Gene Amp RNA PCR Core Kit, Roche Applied Biosystems, Branchburg, NJ). The dystrophin primers used for the nested PCRs were designed to amplify exons 1–10. DNA sequencing to confirm splicing of exon 5 to exon 10 was performed by the DNA sequencing core of the Children's Hospital of Philadelphia.
Reaction 1
Exon 1: 5′ CAT CAG AGA AAA ACG AAT AGG 3′
Exon 10: 5′ AAT CTC TCC TTG GGC TTG CAG 3′
Reaction 2
Exon 1: 5′ GTG GGA AGA AGT AGA GGA CTG 3′
Exon 10: 5′ CTC AGC TGA AAG AAG CCA CGA 3′
Immunoblot. Cardiac biopsies were obtained for western blotting as depicted in Figure 1 and snap-frozen in liquid nitrogen. Specimens were pulverized, homogenized in 10 volumes of triple-detergent lysis buffer (50 mmol/l Tris, pH 8.0, 0.1% SDS, 1.0% Triton X-100, 0.5% DOC, 5 mmol/l EDTA, 50 m,ol/l DTT, 0.4 tablet/10 ml Complete Protease Inhibitor Cocktail Tablets (Roche, Indianapolis, IN)), and centrifuged at 13,000 rpm for 5 minutes. Protein concentration of the supernatant was then determined using the BioRad Protein Assay (Bio-Rad Laboratories, Hercules, CA), and a 50 µg aliquot of each sample was denatured using an equal volume of 2× sample loading buffer (130 mmol/l Tris, pH = 8.0, 20% glycerol, 4.6% SDS, 2% DTT, 0.02% bromophenol blue) at 100 °C (5 minutes) then fractionated electrophoretically on a 2% SDS-polyacrylamide gel (Lonza, Rockland, ME). Proteins were then electroblotted onto a polyvinylidene fluoride membrane (Immobilon-P; Millipore, Bedford, MA) using the iBlot transfer apparatus (Invitrogen) after a 30 minute incubation in NuPAGE transfer buffer with 10% methanol (Invitrogen). The membrane was subsequently blocked by incubating in Tris-buffered saline containing 5% nonfat dry milk and 0.05% Tween 20. Immunoblotting was performed to detect dystrophin using a rabbit polyclonal antibody raised against a C-terminal epitope (1:2,000; Abcam, Cambridge, MA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:4,000; Santa Cruz Biotechnology, Santa Cruz, CA) was used as a loading control. Secondary antibody (anti-rabbit; GE Healthcare, Buckinghamshire, UK) was used at a dilution of 1:2,000. Detection was performed using the Super Signal West Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL).
IHC. At the time of euthanasia, tissues were harvested for evaluation of dystrophin expression. The heart was divided into sections as depicted in Figure 1; each section was frozen in OCT embedding compound (Tissue Tek, Torrance, CA), and 10 µm cryosections were prepared. Sections were blocked in 5% bovine serum albumin (BSA)/phosphate-buffered saline (PBS) and subsequently incubated overnight at 4 °C in 5% BSA/PBS containing a rabbit polyclonal antibody raised against a C-terminal epitope of dystrophin (1:2,000; Abcam) or collagen I (1:100; Abcam). Following washes in PBS, sections were incubated in 5% BSA/PBS containing Alexa Fluor IgG secondary antibody (1:2,000; Invitrogen) for 1 hour in the dark at room temperature. Slides were then mounted with Vectashield DAPI media (Vector Laboratories, Burlingame, CA) and examined for fluorescence using a Leitz DMRBE fluorescent microscope (Leica, Bannockburn, IL) equipped with a Micro Max digital camera (Princeton Instruments, Trenton, NJ) interfaced with Image Pro Plus software (Media Cybernetics, Bethesda, MD). Exposure times were equal for all samples. The threshold for detection was set above background levels measured in the untreated canine specimens.
Echocardiography. Echocardiography was performed on the treated dogs before gene delivery and again before euthanasia and on three untreated, affected dogs using a Philips Sonos 7500 (Philips, Wilmington, MA) and a 2–4 MHz transducer. The same sonographer performed all studies, which included 2D, M-mode, spectral doppler, and tissue doppler of the lateral mitral valve annulus.
Cardiac MRI. Anesthesia was induced in dogs with an intravenous infusion of propofol (1.0–2.0 mg/min/kg) and fentanyl (0.005 mg/kg/min) via the cephalic vein, and maintained using reduced concentrations (propofol: 0.2 mg/kg/min; fentanyl: 0.7 mcg/kg/min). MR data were acquired in apnea by turning off the ventilator, along with infusion of a bolus of cisatracurium (0.1 mg/kg). Respiration, electrocardiogram, O2 saturation, and blood pressure were monitored during the MR scanning protocol.
For image acquisition, dogs were placed in the dorsal position in the bore of the magnet of a GE 1.5 Telsa Signa MR system (GE Healthcare, Milwaukee, WI). A torso array receive-only coil was positioned over the thoracic region. Cardiac imaging was performed with retrospective electrocardiogram gating. Cardiac-gated tagged images were acquired using a fast spoiled gradient recalled sequence (field of view 24 × 24 cm; acquisition matrix 256 × 128; repetition time 9.2 ms; echo time 5.5 ms; flip angle 20°). Grid tag spacing was 7 mm. Images were acquired in the short-axis trans-ventricular view (4–5 slices, 8 mm slice thickness, no slice gap). Positioning of the short-axis images was achieved using sagittal localizers and long-axis four-chamber images of the heart.47 Triggering utilized a single cardiac phase cycle with minimum trigger delay and 16 frames per cardiac cycle.
Analysis of the tagged images was performed using harmonic phase analysis (HARP; Diagnosoft, Palo Alto, CA) for peak circumferential myocardial strain.44,48 The short-axis slice in the mid-papillary region of the left ventricle was chosen for analysis. In a middle-frame image, epicardial and endocardial traces were drawn and then automatically propagated to create a mesh dividing the myocardium into six regions in the short-axis view of all 16 frames acquired in the cardiac cycle. For consistency, the first segment was placed starting at the anterior region of the ventricular septum. The average peak circumferential myocardial strain from each segment was calculated for the mid-wall using Eulerian strain algorithms.49
SUPPLEMENTARY MATERIAL Table S1. Echocardiography data from GRMD canines pre- and post-treatment with AAV6-U7-ESE6/8.
Acknowledgments
This work was supported by a grant from the NHLBI (P01-HL059407) (to H.L.S. and J.M.W.), from the Parent Project Muscular Dystrophy (to H.L.S.), by a Wellstone Muscular Dystrophy Cooperative Center Grant (U54-AR052646) (to H.L.S.), by NIH (T32-HL-007748) (to L.T.B.) and (RR02512) (to Mark Haskins), and by the International Collaborative Effort (ICE) for DMD (to H.L.S., R.M.K., and L.G.). Support was also provided by the Division of Intramural Research of the National Heart, Lung, and Blood Institute (NIH) (R.M.K.). We thank the Vector Core of the Children's Hospital of Philadelphia and Katherine High, MD for vector production. We thank Mark Haskins, VMD, PhD (University of Pennsylvania) for support of the canine colony. J.M.W. is an inventor on patents that have been licensed to various biopharmaceutical companies. Portions of the technology described in this report are covered by United States and European patents assigned to the Secretary of the Department of Health and Human Services. A fraction of the licensing fees and royalty payments made to the NIH are distributed to the inventors in accordance with US Government and NIH policy.
Supplementary Material
Echocardiography data from GRMD canines pre- and post-treatment with AAV6-U7-ESE6/8.
REFERENCES
- Emery AE. The muscular dystrophies. Lancet. 2002;359:687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ., and, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- Miyagoe-Suzuki Y., and, Takeda S. Gene therapy for muscle disease. Exp Cell Res. 2010;316:3087–3092. doi: 10.1016/j.yexcr.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Moulton HM., and, Moulton JD. Morpholinos and their peptide conjugates: therapeutic promise and challenge for Duchenne muscular dystrophy. Biochim Biophys Acta. 2010;1798:2296–2303. doi: 10.1016/j.bbamem.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Partridge T. The potential of exon skipping for treatment for Duchenne muscular dystrophy. J Child Neurol. 2010;25:1165–1170. doi: 10.1177/0883073810371130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, den Dunnen JT.et al. (2001Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells Hum Mol Genet 101547–1554. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F.et al. (2003Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients Hum Mol Genet 12907–914. [DOI] [PubMed] [Google Scholar]
- McClorey G, Moulton HM, Iversen PL, Fletcher S., and, Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, Burke MM, Nagel N, Holder A.et al. (2010A duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient cavalier king charles spaniels is amenable to exon 51 skipping PLoS ONE 5e8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA.et al. (2003Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse Nat Med 91009–1014. [DOI] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J.et al. (2005Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles Proc Natl Acad Sci USA 102198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Li Y, Morcos PA, Doran TJ, Lu P., and, Lu QL. Octa-guanidine morpholino restores dystrophin expression in cardiac and skeletal muscles and ameliorates pathology in dystrophic mdx mice. Mol Ther. 2009;17:864–871. doi: 10.1038/mt.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Lu P, Benrashid E, Malik S, Ashar J, Doran TJ.et al. (2010Dose-dependent restoration of dystrophin expression in cardiac muscle of dystrophic mice by systemically delivered morpholino Gene Ther 17132–140. [DOI] [PubMed] [Google Scholar]
- Jearawiriyapaisarn N, Moulton HM, Sazani P, Kole R., and, Willis MS. Long-term improvement in mdx cardiomyopathy after therapy with peptide-conjugated morpholino oligomers. Cardiovasc Res. 2010;85:444–453. doi: 10.1093/cvr/cvp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jearawiriyapaisarn N, Moulton HM, Buckley B, Roberts J, Sazani P, Fucharoen S.et al. (2008Sustained dystrophin expression induced by peptide-conjugated morpholino oligomers in the muscles of mdx mice Mol Ther 161624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S.et al. (2009Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs Ann Neurol 65667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C.et al. (2009Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study Lancet Neurol 8918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M.et al. (2007Local dystrophin restoration with antisense oligonucleotide PRO051 N Engl J Med 3572677–2686. [DOI] [PubMed] [Google Scholar]
- Incitti T, De Angelis FG, Cazzella V, Sthandier O, Pinnarò C, Legnini I.et al. (2010Exon skipping and duchenne muscular dystrophy therapy: selection of the most active U1 snRNA antisense able to induce dystrophin exon 51 skipping Mol Ther 181675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A, Babbs A, van Ommen GJ, Garcia L., and, Davies KE. Enhanced exon-skipping induced by U7 snRNA carrying a splicing silencer sequence: Promising tool for DMD therapy. Mol Ther. 2009;17:1234–1240. doi: 10.1038/mt.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L.et al. (2004Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping Science 3061796–1799. [DOI] [PubMed] [Google Scholar]
- Denti MA, Incitti T, Sthandier O, Nicoletti C, De Angelis FG, Rizzuto E.et al. (2008Long-term benefit of adeno-associated virus/antisense-mediated exon skipping in dystrophic mice Hum Gene Ther 19601–608. [DOI] [PubMed] [Google Scholar]
- Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C.et al. (2006Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model Proc Natl Acad Sci USA 1033758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp NJ, Kornegay JN, Van Camp SD, Herbstreith MH, Secore SL, Kettle S.et al. (1992An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy Genomics 13115–121. [DOI] [PubMed] [Google Scholar]
- Bish LT, Sleeper MM, Brainard B, Cole S, Russell N, Withnall E.et al. (2008Percutaneous transendocardial delivery of self-complementary adeno-associated virus 6 achieves global cardiac gene transfer in canines Mol Ther 161953–1959. [DOI] [PubMed] [Google Scholar]
- Bish LT, Sleeper MM., and, Sweeney HL. Percutaneous transendocardial delivery of self-complementary adeno-associated virus 6 in the canine. Methods Mol Biol. 2011;709:369–378. doi: 10.1007/978-1-61737-982-6_24. [DOI] [PubMed] [Google Scholar]
- Reiss J., and, Rininsland F. An explanation for the constitutive exon 9 cassette splicing of the DMD gene. Hum Mol Genet. 1994;3:295–298. doi: 10.1093/hmg/3.2.295. [DOI] [PubMed] [Google Scholar]
- Kornegay JN, Li J, Bogan JR, Bogan DJ, Chen C, Zheng H.et al. (2010Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs Mol Ther 181501–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino-Klapac LR, Montgomery CL, Bremer WG, Shontz KM, Malik V, Davis N.et al. (2010Persistent expression of FLAG-tagged micro dystrophin in nonhuman primates following intramuscular and vascular delivery Mol Ther 18109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, Phelps SF.et al. (2002Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy Nat Med 8253–261. [DOI] [PubMed] [Google Scholar]
- Wang Z, Chamberlain JS, Tapscott SJ., and, Storb R. Gene therapy in large animal models of muscular dystrophy. ILAR J. 2009;50:187–198. doi: 10.1093/ilar.50.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete A., and, Kotin RM. Large-scale production of recombinant adeno-associated viral vectors. Methods Mol Biol. 2008;433:79–96. doi: 10.1007/978-1-59745-237-3_5. [DOI] [PubMed] [Google Scholar]
- Smith RH, Levy JR., and, Kotin RM. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther. 2009;17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag T, Cecchini S., and, Kotin RM. Producing recombinant adeno-associated virus in foster cells: overcoming production limitations using a baculovirus-insect cell expression strategy. Hum Gene Ther. 2009;20:807–817. doi: 10.1089/hum.2009.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog RW, Yang EY, Couto LB, Hagstrom JN, Elwell D, Fields PA.et al. (1999Long-term correction of canine hemophilia B by gene transfer of blood coagulation factor IX mediated by adeno-associated viral vector Nat Med 556–63. [DOI] [PubMed] [Google Scholar]
- Chetboul V, Carlos C, Blot S, Thibaud JL, Escriou C, Tissier R.et al. (2004Tissue Doppler assessment of diastolic and systolic alterations of radial and longitudinal left ventricular motions in Golden Retrievers during the preclinical phase of cardiomyopathy associated with muscular dystrophy Am J Vet Res 651335–1341. [DOI] [PubMed] [Google Scholar]
- Chetboul V, Escriou C, Tessier D, Richard V, Pouchelon JL, Thibault H.et al. (2004Tissue Doppler imaging detects early asymptomatic myocardial abnormalities in a dog model of Duchenne's cardiomyopathy Eur Heart J 251934–1939. [DOI] [PubMed] [Google Scholar]
- Valentine BA, Cooper BJ, de Lahunta A, O'Quinn R., and, Blue JT. Canine X-linked muscular dystrophy. An animal model of Duchenne muscular dystrophy: clinical studies. J Neurol Sci. 1988;88:69–81. doi: 10.1016/0022-510x(88)90206-7. [DOI] [PubMed] [Google Scholar]
- Moise NS, Valentine BA, Brown CA, Erb HN, Beck KA, Cooper BJ.et al. (1991Duchenne's cardiomyopathy in a canine model: electrocardiographic and echocardiographic studies J Am Coll Cardiol 17812–820. [DOI] [PubMed] [Google Scholar]
- Devaux JY, Cabane L, Esler M, Flaouters H., and, Duboc D. Non-invasive evaluation of the cardiac function in golden retriever dogs by radionuclide angiography. Neuromuscul Disord. 1993;3:429–432. doi: 10.1016/0960-8966(93)90090-7. [DOI] [PubMed] [Google Scholar]
- Hor KN, Wansapura J, Markham LW, Mazur W, Cripe LH, Fleck R.et al. (2009Circumferential strain analysis identifies strata of cardiomyopathy in Duchenne muscular dystrophy: a cardiac magnetic resonance tagging study J Am Coll Cardiol 531204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford MW, Jr, Liu W, Lin SJ, Abraszewski P, Caruthers SD, Connolly AM.et al. (2005Occult cardiac contractile dysfunction in dystrophin-deficient children revealed by cardiac magnetic resonance strain imaging Circulation 1122462–2467. [DOI] [PubMed] [Google Scholar]
- Li W, Liu W, Zhong J., and, Yu X. Early manifestation of alteration in cardiac function in dystrophin deficient mdx mouse using 3D CMR tagging. J Cardiovasc Magn Reson. 2009;11:40. doi: 10.1186/1532-429X-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor KN, Gottliebson WM, Carson C, Wash E, Cnota J, Fleck R.et al. (2010Comparison of magnetic resonance feature tracking for strain calculation with harmonic phase imaging analysis JACC Cardiovasc Imaging 3144–151. [DOI] [PubMed] [Google Scholar]
- Dicks D, Saloner D, Martin A, Carlsson M., and, Saeed M. Percutaneous transendocardial VEGF gene therapy: MRI guided delivery and characterization of 3D myocardial strain. Int J Cardiol. 2010;143:255–263. doi: 10.1016/j.ijcard.2009.02.030. [DOI] [PubMed] [Google Scholar]
- Marshall DJ, Palasis M, Lepore JJ., and, Leiden JM. Biocompatibility of cardiovascular gene delivery catheters with adenovirus vectors: an important determinant of the efficiency of cardiovascular gene transfer. Mol Ther. 2000;1 5 Pt 1:423–429. doi: 10.1006/mthe.2000.0059. [DOI] [PubMed] [Google Scholar]
- Mai W, Weisse C., and, Sleeper MM. Cardiac magnetic resonance imaging in normal dogs and two dogs with heart base tumor. Vet Radiol Ultrasound. 2010;51:428–435. doi: 10.1111/j.1740-8261.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- Osman NF, Kerwin WS, McVeigh ER., and, Prince JL. Cardiac motion tracking using CINE harmonic phase (HARP) magnetic resonance imaging. Magn Reson Med. 1999;42:1048–1060. doi: 10.1002/(sici)1522-2594(199912)42:6<1048::aid-mrm9>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman NF, McVeigh ER., and, Prince JL. Imaging heart motion using harmonic phase MRI. IEEE Trans Med Imaging. 2000;19:186–202. doi: 10.1109/42.845177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Echocardiography data from GRMD canines pre- and post-treatment with AAV6-U7-ESE6/8.