Abstract
Numerous studies have suggested that an effective hepatitis C virus (HCV) vaccine must induce a strong T helper 1 (Th1) T cell response. While several therapeutic vaccine candidates have shown promise in clinical trials, response rates have been low suggesting that further optimization is important. However, such optimization has been hindered by a lack of a benchmark animal model in which to test vaccine-induced immune responses before clinical evaluation. The goal of this study was to analyze the utility of the rhesus macaque vaccination model in assessing HCV vaccine-induced T cell responses. To test this, we employed the use of a novel HCV genotype 1a/1b consensus DNA vaccine encoding both HCV nonstructural protein 3 (NS3) and nonstructural protein 4A (NS4A) proteins. Following immunization, rhesus macaques mounted HCV-specific responses strikingly similar to those reported in resolving patients, including strong NS3-specific interferon-γ (IFN-γ) responses, robust CD4+ and CD8+ T cell proliferation, and induction of polyfunctional T cells. Additionally, fine epitope mapping revealed one animal that mounted a T cell response against a known HCV NS3 human leukocyte antigen A2 (HLA-A2) epitope previously identified in humans. Taken together our findings suggest that the rhesus macaque vaccination model is a useful tool in the evaluation of immune responses induced by HCV immunogens.
Introduction
Hepatitis C virus (HCV) is a small enveloped, positive stranded RNA virus that represents a major health burden worldwide. While it is known that neutralizing antibodies against HCV can be detected within 7- to 8-week postinfection,1 they can be completely absent following resolution of infection2,3 and do not protect against reinfection.1,4,5 Instead, clearance of infection has been associated with a strong T helper 1 (Th1) T cell response directed against the more genetically conserved nonstructural proteins of the virus.6,7 More specifically, it is believed that individuals able to mount and sustain an early, multispecific CD4+ helper and CD8+ cytotoxic T cell response can clear infection.8,9,10,11
Therefore, it is becoming more likely that a T cell-based vaccine able to induce cellular immune responses targeted against the most conserved proteins of the virus may represent an effective vaccine strategy. In fact, several therapeutic vaccine candidates have shown great promise in clinical trials by transiently reducing viral loads in some chronically infected patients and improving eSVRs in others. While these vaccine candidates represent a major step forward toward an eventual HCV vaccine, the immune response rates induced were generally low suggesting that further optimization is important.
An important part of vaccine optimization is gaining a more complete understanding of the immune responses induced following vaccination. However, meaningful studies examining the immune responses induced following vaccination have been hindered by the fact that chimpanzees and humans are the only natural hosts of infection. Given both the costs and ethical concerns regarding the use of the chimpanzee model, the HCV vaccine field is in great need of a benchmark animal model in which to test, optimize, and predict vaccine induced-immune responses in human. While not a natural host for infection, the macaque has proven to be an important vaccination model for predicting successful human immune responses to multiple disease antigens because of the similarity of the macaque immune system to that of humans, as well as, the number of immune reagents available in order to dissect these responses.
Therefore, the goal of this study was to study the utility of the rhesus macaque vaccination model in assessing HCV vaccine-induced T cell responses. To accomplish this, we preformed an in-depth analysis of the specific cellular immune responses induced following immunization with a novel HCV nonstructural protein 3 (NS3)/nonstructural protein 4A (NS4A) genotype 1a/1b DNA vaccine (pConNS3/NS4A), previously described.12 Following immunization, we analyzed several features of the immune response that have been correlated with clearance of acute infection in humans including: the magnitude, the proliferative capacity, the breadth, and the polyfunctionality of HCV-specific T cell responses induced through immunization. We found that within our vaccination model, macaques were able to mount HCV-specific cellular immune responses strikingly similar to those seen in individuals who ultimately go on to clear acute infection. Analysis of both the CD4+ and CD8+ T cell response revealed these responses to be highly functional and showed that immunization was able to induce HCV-specific polyfunctional T cells, as well as, robust CD4+ and CD8+ proliferative responses. Additionally, the macaques exhibited strong anti-HCV interferon-γ (IFN-γ) T cell responses able to recognize numerous NS3 epitopes spanning the length of the construct. Perhaps the most convincing evidence for the utility of this model to predict human responses to vaccination is the fact that fine epitope mapping revealed one animal able to mount a T cell response against a known HCV NS3 human leukocyte antigen A2 (HLA-A2) epitope in humans. These results taken together suggest that the vaccination within the macaque model can produce the type of cellular immune responses characteristic of resolving patient and therefore may be an important model in which to evaluate future prophylactic and therapeutic HCV vaccines.
Results
Immunization of rhesus macaques with pConNS3/NS4A induces strong NS3-specific T cell responses
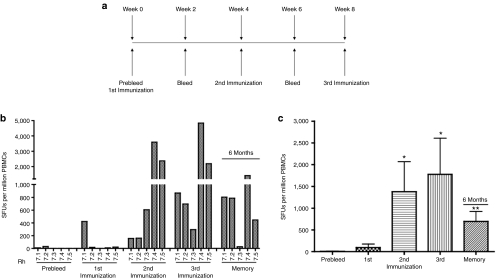
Each rhesus macaque (n = 5) received a total of three intramuscular immunization of 1 mg of pConNS3/NS4A12 followed by electroporation. The three immunizations were administered 4 weeks apart with bleeds conducted 2 weeks following each immunization (Figure 1a). Given the importance of IFN-γ production by HCV-specific T cells in the resolution of acute infection, the initial responses to each immunization with pConNS3/NS4A were monitored through the use of quantitative IFN-γ enzyme-linked immunosorbent spot (ELISpot) assays. Analysis of peripheral blood mononuclear cells (PBMCs) from each animal conducted prior to immunization revealed no preexisting immune responses to pConNS3/NS4A, as detected by IFN-γ ELISpot assay. However, following the second immunization, detectable NS3- and NS4A-specific T cell responses were seen in all five animals, ranging from 157 to 3,602 spot forming units (SFU)/106 PBMCs (Figure 1b). Following the third immunization, the majority of the animals (three of five) showed an overall increase in total NS3- and NS4A-specific T cell responses, which ranged from 295 to 4,843 SFU/106 PBMCs (Figure 1b). The total T cell response induced by pConNS3/NS4A averaged 1,381 ± 690 and 1,778 ± 830 SFU/106 PBMCs, for the second and third immunizations, respectively (Figure 1c).
Figure 1.
pConNS3/NS4A induces strong cellular immune responses following immunization (n = 5). (a) Immunization schedule. (b) IFN-γ ELISpot assay results for each individual animal. PBMCs were stimulated with a total of 101 HCV NS3 and NS4A 15-mer overlapping by eight amino acids and spanning the entire length of the pConNS3/NS4A protein. (c) Average responses as measured by IFN-γ ELISpot assay. Values are represented as mean + SEM, n = 5 per group. Significance was determined by Student's t-test (*P < 0.05, **P < 0.005 and ***P < 0.0005). HCV, hepatitis C virus.
The duration of the T cell response to pConNS3/NS4A was also assessed by allowing the animals to rest for 6 months following the third immunization. NS3-specific responses to pConNS3/NS4A remained detectable in four of five animals, ranging from 447 to 1,414 SFU/106 PBMCs (Figure 1c). The average response among the four macaques was 864 ± 201 SFU/106 PBMCs (Figure 1c).
pConNS3/NS4A induces robust NS3-specific CD4+ and CD8+ T cell proliferation
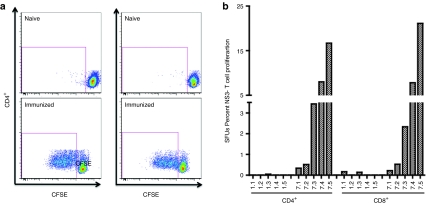
Numerous studies have found that an important correlate for both control of chronic HCV infection and clearance of acute infection is a strong and sustained anti-HCV CD4+ T cell proliferative response, especially when directed against HCV NS3.6,7,11,13 Therefore, because of this correlation, we were interested in determining the proliferative capacity of NS3-specific T cells induced in response to immunization with pConNS3/NS4A. Animals were rested for ~2 months following the third immunization at which point the proliferative response of NS3-specific T cells were assessed through CFSE dilution assays. NS3-specific proliferation was calculated as the total percentage of either CD4+ or CD8+ T cells to complete at least one round of cell division following a 5-day stimulation with NS3 peptides. Compared to the naive animals (immunized with non-HCV antigens), immunized animals mounted robust NS3-specific proliferative responses, as seen with both CD4+ and CD8+ T cells (Figure 2a). Total NS3-specific T cell proliferation averaged 5.8% ± 3% and 6.4% ± 3.9% for CD4+ and CD8+ T cells, respectively. Figure 2b shows the total percent CD4+ and CD8+ proliferation of each animal. The total percent NS3-specific proliferation did vary greatly among the five individual animals. Animals Rh7.1 and Rh7.2 showed only minimal proliferation, while animals Rh7.3, Rh7.4, and Rh7.5 showed robust NS3-specific proliferative responses (Figure 2b).
Figure 2.
Immunization with pConNS3/NS4A induces robust CD4+ and CD8+ T cell responses (n = 5). Proliferative responses were measured with CFSE dilution assay ~2 months following the last immunization. PBMCs were stimulated with HCV NS3 peptides, overlapping by eight amino acids and spanning the entire length of the NS3 protein. (a) Representative dot plots are shown for both the naive group (immunized with non-HCV antigens) and the pConNS3/NS4A immunized group. The average total percent NS3-specific T cell proliferation for each group. (b) Individual NS3-specific proliferative responses for each animal. CFSE, carboxyfluorescein diacetate, succinimidyl ester; HCV, hepatitis C virus; PBMC, peripheral blood mononuclear cell.
Immunization with pConNS3/NS4A induces polyfunctional T cell responses
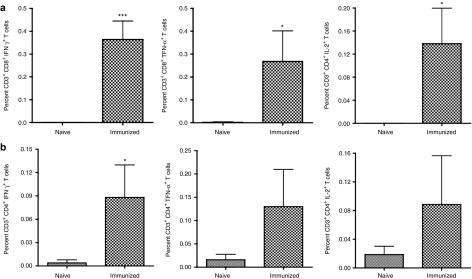
Polyfunctional T cells have been associated with the control of numerous viral infections.14,15,16 Recent studies of HCV-infected patients have suggested the importance of polyfunctional CD8+ T cells in the effective control of HCV replication and have been suggested to be an important correlate for clearance of HCV in acutely infected patients.17,18 To test for these responses, animals were rested for approximately two months following the third immunization, at which point, the phenotypic and functional characteristics of immunization induced CD4+ and CD8+ T cells were assessed through intracellular cytokine analysis by polychromatic flow cytometry. T cells were stimulated ex vivo with overlapping HCV NS3 peptides and stained intracellularly for IFN-γ, tumor necrosis factor-α (TNF-α), interleukin 2 (IL-2), and CD107A. On average, both CD8+ T cells and CD4+ T cells from immunized animals were able to produce IFN-γ, IL-2, and TNF-α in response to peptide stimulation as compared to naïve animals, (Figure 3). Following stimulation, the CD8+ T cell response for immunized animals averaged 0.36% ± 0.08%, 0.27% ± 0.13%, and 0.14% ± 0.06%, for IFN-γ, TNF-α, and IL-2 production, respectively, (Figure 3a). The CD4+ T cell response for immunized animals averaged 0.09% ± 0.04%, 0.13% ±.08%, and 0.09% ± 0.07%, for IFN-γ, TNF-α, and IL-2 production following stimulation, respectively, (Figure 3b).
Figure 3.
Immunization with pConNS3/NS4A induces NS3-specific T cells able to produce numerous cytokines. PBMCs were stimulated with HCV NS3 peptides, overlapping by eight amino acids and spanning the entire length of the NS3 protein. PBMCs were stained intracellularly for IFN-γ, IL-2, and TNF-α ~2 months following the last immunization. (a) Average NS3-specific CD8+ T cell responses for each group. (b) Average NS3-specific CD4+ T cell responses for each group. Values are represented as mean + SEM, n = 5 per group. Significance was determined by Student's t-test (*P < 0.05, **P < 0.005, and ***P < 0.0005). HCV, hepatitis C virus; PBMC, peripheral blood mononuclear cell.
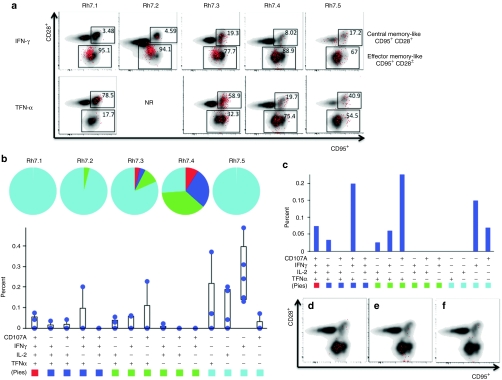
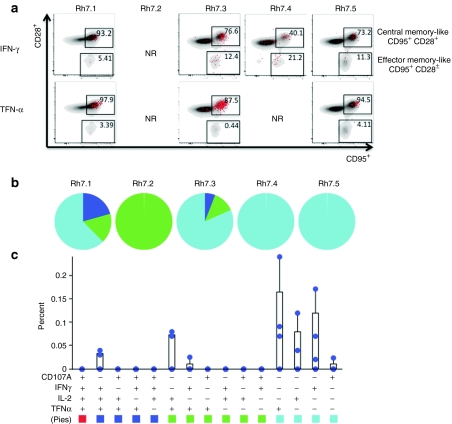
Next, we performed a detailed analysis of both NS3-specific CD4+ T cell responses and NS3-specific CD8+ T cell responses. For all five animals, the predominant cytokine produced by CD8+ T cells in response to peptide stimulation was IFN-γ. TNF-α was also commonly expressed and was identified in four out of five animals. While an important correlation to progression of HCV infection to the chronic state is loss of effector function suggesting the importance of sustained effector function in viral control, an effective vaccine must also have the ability to induce potent memory responses in order to be protective long-term. Therefore, both CD8+ IFN-γ+ and CD8+ TNF-α+ responses were further characterized by their expression of both CD28 and CD95. CD8+ IFN-γ+ T cells were mostly CD28− CD95+, which is suggestive of a more effector memory-like phenotype, while the majority of CD8+ TNF-α+ T cells were more evenly distributed between CD28− CD95+ and CD28+ CD95+ populations, suggesting that these cells display both an effector memory-like and a central memory-like phenotype, (Figure 4a).
Figure 4.
Analysis of CD8+ NS3-specific T cell responses. PBMCs were stimulated with HCV NS3 peptides, overlapping by eight amino acids and spanning the entire length of the NS3 protein. (a) Analysis of CD8+ IFN-γ+ and CD8+ TNF-α+ T cell responses (red) based on CD28 and CD95 staining. (b) PBMCs were stained intracellularly for IFN-γ, TNF-α, IL-2, and CD107A and were analyzed for polyfunctionality. Pie charts represent the proportion of NS3-specific CD8+ T cells for each animal that have one function (light blue), two functions (green), three functions (dark blue), and four functions (red). The graph is the average polyfunctional NS3-specific CD8+ T cell response among all five animals. Values are represented as mean + SEM, n = 5 per group. (c) Graph of the polyfunctional NS3-specific CD8+ T cell response of Rh7.4. (d–f) The three largest NS3-specific polyfunctional CD8+ T cell responses (red) of Rh7.4 were plotted based on CD28 and CD95 expression. (d) TNF-α+ CD107A+, (e) TNF-α+ CD107A+ IFN-γ+, (f) TNF-α+ CD107A+ IL-2+ IFN-γ+. HCV, hepatitis C virus; NR, no response; PBMC, peripheral blood mononuclear cell.
Polyfunctionality was analyzed based on four different functions, IFN-γ, TNF-α, IL-2, and CD107A. In Figure 4b, each pie chart represents the total NS3-specific CD8+ T cell response for each animal as graphed by function. Light blue, green, dark blue and red represent one, two, three and four functions, respectively. Three out of five immunized animals were able to mount CD8+ T cells with two or more functions, while two animals were able to mount a vaccine specific CD8+ T cells able to perform all four functions. The graph in Figure 4b represents the average CD8+ T cell responses for all five animals. While some animals were able to mount a polyfunctional CD8+ T cell response, the average CD8+ T cell response among the five animals was predominantly a monofunctional IFN-γ or TNF-α response. Animal Rh7.4, however, was able to mount a much more diverse T cell response as compared with the other four animals; CD8+ T cells able to produce numerous combinations of functions in response to immunization were detected. Figure 4c shows the individual NS3-specific CD8+ T cell responses of Rh7.4. The three most dominant multi-functional responses were further characterized based on the markers CD28 and CD95, (Figure 4d,e,f). CD8+ T cells able to express both TNF-α and CD107A (Figure 4d) and CD8+ T cells able to express TNF-α, CD107A and IFN-γ (Figure 4e) were predominately CD28− CD95+, suggesting that these two types of responses are mainly effector memory-like. On the other hand, CD8+ T cells able to produce all four functions, TNF-α, CD107A, IL-2, and IFN-γ, fell into both CD28− CD95+ and CD28+ CD95+ populations, suggesting that this type of response is both effector and central memory-like, (Figure 4f).
Like the CD8+ T cell response, the predominant cytokine responses for CD4+ T cells were also IFN-γ and TNF-α. Four out of five animals mounted an NS3-specific CD4+ IFN-γ+ response, while three out of five mounted a CD4+ TNF-α+ response. Analysis of cell surface expression of both CD28 and CD95 revealed that the majority of CD4+ IFN-γ T cells were CD28+ CD95+, suggesting the majority of these T cells displayed a more central memory-like phenotype, (Figure 5a). However, both animal Rh7.3, Rh7.4, and Rh7.5 showed a smaller, but a distinct population of CD28− CD95+ CD4+ IFN-γ+ T cells, suggesting that these animals were also able to mount a CD4+ IFN-γ+ effector memory-like response, as well. Similarly, the majority of CD4+ TNF-α T cells displayed central memory-like phenotype, falling mostly into the CD95+ CD28− population, (Figure 5a).
Figure 5.
Analysis of CD4+ NS3-specific T cell responses. PBMCs were stimulated with HCV NS3 peptides, overlapping by eight amino acids and spanning the entire length of the NS3 protein. (a) Analysis of CD4+ IFN-γ+ and CD4+ TNF-α+ T cell responses (red) based on CD28 and CD95 staining. (b,c) PBMCs were stained intracellularly for IFN-γ, TNF-α, and IL-2 and were analyzed for polyfunctionality. (b) Pie charts representing the proportion of NS3-specific CD4+ T cells for each animal that have one function (light blue), two functions (green), three functions (dark blue), and four functions (red). (c) Average polyfunctional response among all five animals (NR, no response). Values are represented as mean + SEM, n = 5 per group. HCV, hepatitis C virus; PBMC, peripheral blood mononuclear cell.
Finally, the polyfunctionality of the NS3-specific CD4+ T cell response was analyzed. Figure 5b represents the total CD4+ T cell response of each animal as graphed by function. Three out of five animals had CD4+ T cells able to perform two or more functions, while two animals had CD4+ T cells responses able to perform three functions; IFN-γ, IL-2, TNF-α. Figure 5c represents the average NS3-specific CD4+ T cell response. As seen with the CD8+ T cell responses, the average CD4+ T cell response is predominantly monofunctional, with the highest average responses being TNF-α or IFN-γ.
Identification of novel HCV NS3 rhesus macaque epitopes
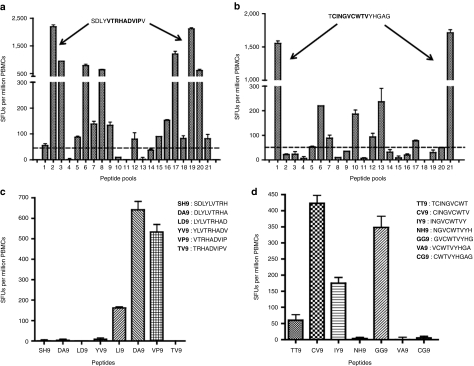
It is believed that a successful immune responses against HCV includes a strong CD8+ T cell response able to recognize and target multiple epitopes within the viral genome, while persistent infection is often marked by low frequencies of HCV-specific CD8+ T cells able to recognize only a small number of epitopes.8,10,19,20,21,22 We were therefore interested in the breadth of the NS3-specific T cell response induced in the animals following immunization. Therefore, the T cell response of the two highest responders, Rh7.4 and Rh7.5, were mapped using a matrix epitope mapping technique and IFN-γ ELISpot assays (described in the methods section), (Figure 6a,b). Supplementary Table S1 lists all the possible reactive 15-mer peptides identified for each animal. As seen in Supplementary Table S1, Rh7.4 and Rh7.5 both mounted broad T cell responses in response to immunization and were able to recognize numerous peptides spanning the length of the HCV NS3 protein.
Figure 6.
Epitope mapping of Rh7.4 and Rh7.5 with IFN-γ ELISpot assays. PBMCs were stimulated with 21 separate pools of HCV NS3 and NS4A peptides. Each pool contained ~10 of the 101 HCV NS3 and NS4A 15-mer overlapping peptides spanning the length of pConNS3/NS4A. Each individual peptide was represented in two pools of the 21 pools. (a,b) Matrix epitope mapping of animals Rh7.4 and Rh7.5. (a) Responses of Rh7.4. (b) Responses of Rh7.5. (c,d) The dominant 15-mer epitopes were fine mapped with overlapping 9-mers. (c) Responses for Rh7.4. (d) Responses for Rh7.5. Values are represented as mean + SEM, n = 3 per group. HCV, hepatitis C virus; PBMC, peripheral blood mononuclear cell.
The dominant epitopes of our two highest responders were further characterized with fine epitope mapping, (Figure 6c,d). The dominant 15-mer peptides for animals Rh7.4 and Rh7.5 were peptide 18 and peptide 10, respectively. In order to identify the dominant epitopes within peptide 18 and peptide 10, seven 9-mer peptides spanning the length of each dominant peptide were generated and responses to each 9-mer were measured by IFN-γ ELISpot assays. Rh7.4 responded to three of the 9-mer peptides: LI9 (LVTRHADVI), VP9 (VTRHADVIP), and TV9 (TRHADVIPV, (Figure 6c). Animal Rh7.5 also responded to three 9-mers: CV9 (CINGVCWTV), IY9 (INGVCWTVY), and GG9 (GVCWTVYHG), (Figure 6d). Rh7.5 generated the highest response against the epitope CV9, which interestingly, is a known human HLA-A2 epitope for NS3.
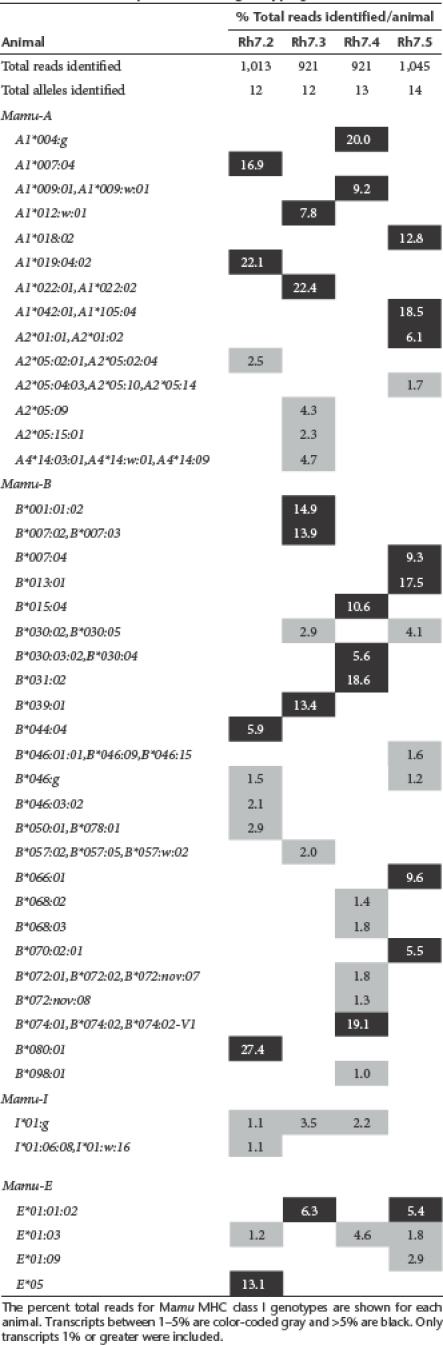
To help indentify whether animal Rh7.5 possessed an unique MHC that may explain the restriction of this epitope, high resolution MHC class I genotypes were determined for four study animals by cDNA/PCR pyrosequencing; Rh7.1 was excluded due to limited sample availability. This analysis revealed diverse profiles of MHC class I transcripts (Table 1) that were essentially distinct for each animal and consistent with previous observations for Chinese-origin rhesus macaques.23,24 Interestingly, the repertoire of class I transcripts identified in Rh7.5 included the Mamu-A1*018:02 allele (accession no. AM295899) which was only identified in Rh7.5 and is 91% identical at the nucleotide level to HLA-A*0210 in humans (accession no. Z23071). Despite considerable differences between the predicted gene products encoded by these macaque and human class I alleles (only 68 % amino acid identity over the region sequenced which contains most of the peptide-binding domain encoded by exons 2 and 3), Mamu-A1*018:02 may be a plausible candidate for restriction of the CD8+ T cell response mounted by Rh7.5 against the HCV NS3 CV9 epitope (Figure 6d) since this same epitope was described previously for a human HLA-A2 allele.25,26 The class I genotyping results presented in Table 1 can be used to help identify the restriction of the various immunodominant HCV NS3 responses described here, however rigorous characterization of these restricting alleles is beyond the scope of the current study.
Table 1. Rhesus macaque MHC class I genotyping.
Discussion
Perhaps one of the greatest impediments in the development of HCV vaccines has been the lack of easily accessible, cost effective and predictive models in which to test HCV vaccines. Regardless of this, several promising therapeutic vaccines have been tested in the clinics and have shown great promise. However, while these vaccines have been shown to reduce viral load in some individuals, the response rates have generally been low and viral load reductions have been transient suggesting that further optimization is merited before mass clinical adoption.27,28,29,30,31 A critical part of this optimization depends on fully understanding the immune responses induced following vaccination, which would be greatly aided with the use of a benchmark animal model in which to predict and optimize T cell responses to HCV antigens before clinical studies in humans. While not a natural host of infection, the rhesus macaques, due to its immunological similarities with humans, has been an important model for understanding vaccine-induced responses in numerous disease models.
Therefore, in this study our goal was to determine the utility of the macaque model in assessing HCV vaccine-induced T cells responses. In order to accomplish this, we used a novel NS3/NS4A genotype 1a/1b consensus DNA vaccine (pConNS3/NS4A).12 We chose the DNA vaccine platform due to the fact that antigen is expressed and processed intracellularly, which allows for better T cell presentation. We focused our study primarily on the T cell responses induced following immunization with NS3 because of several studies showing the importance of NS3-specific T cell responses in the clearance of acute infection.6,7 We were particularly interested in how T cell responses induced within the rhesus macaque vaccination model compared to those previously reported in patients with resolving infection and therefore following immunization, the magnitude, breadth, proliferative capacity and the polyfunctionality of the HCV-specific T cell response was monitored. The results of immune analysis revealed that the macaques were able to mount HCV-specific T cell responses extremely similar to those seen in acutely infected resolving patients.
Results from the ELISpot assays revealed that all five animals were able to mount strong HCV-specific IFN-γ T cell responses following immunization. Production of IFN-γ has been shown to have antiviral effects against HCV in cell culture32 and has also been associated with large reductions in viral load during acute infection in humans.13 Likewise, analysis of both HCV-specific CD4+ and CD8+ proliferative responses revealed that immunization was able to generate very robust proliferative T cell responses in three out of five animals. Numerous studies have shown that the loss of CD4+ proliferative responses is predictive of progress to chronic infection, while the continued presence of strong CD4+ proliferative responses, especially those directed against NS3, has been shown to be an important correlate for viral control and clearance.6,7,13 Additionally, analysis of the cytokine profile revealed that both HCV-specific CD4+ and CD8+ T cells were able to produce IFN-γ, TNF-α and IL-2 in response to NS3 peptide stimulation. Since resolution of infection can occur in the absence of elevated serum transaminase levels, cytokine production, such as IFN-γ and TNF-α, from HCV-specific T cells haven been suggested to play an important role in control and clearance of acute infection. While the percentage of cytokine producing NS3-specific T cells detected by ICS may at first seem low compared to the total amount of NS3-specific T cell proliferation detected in the CFSE proliferation assay, given the vast differences in how these two assays are performed, as well as the differences in the NS3-specific T cell populations they are measuring, these two assays cannot necessarily be compared to one another.33
When HCV-specific T cells responses were further characterized based on CD28 and CD95 expression it was determined that immunization induced both effector-memory like and central-memory like CD4+ and CD8+ T cells able to produce either IFN-γ or TNF-α. The results of the memory phenotyping were further validated by IFN-γ ELISpot assays performed six months following immunization, which showed that immunization had indeed produced a large pool of memory HCV-specific T cells, which has been shown to be important for protecting convalescent animals from chronic infection following reexposure to the virus.
Polyfunctional analysis of immunization induced HCV-specific CD4+ and CD8+ T cells revealed that all animals with the exception of one, were able to mount either a polyfunctional HCV-specific CD4+ or CD8+ T cell response, suggesting that not only did immunization induce a potent T cell response, but a highly functional response as well. Recent studies of HCV-infected patients have suggested the importance of polyfunctional CD8+ T cells in the effective control of HCV replication and have been shown to be an important correlate for clearance of HCV in acutely infected patients.17,18
Finally, the overall HCV-specific T cell response was further characterized by epitope mapping to determine the breadth of the immune response. Matrix epitope mapping of the two highest responders revealed that these animals were not only able to induce very strong T cell responses to immunization, but that these responses were directed against a large number of different NS3 epitopes spanning the entire length of the protein. Numerous studies of human infection have shown that an effective cellular immune response against HCV includes T cell responses able to recognize and target multiple MHC class I-restricted epitopes, while persistent infection is often marked by a T cell responses corresponding to a small number of epitopes.8,10
Perhaps the most convincing finding for the utility of the rhesus macaque in predicting HCV-specific T cell responses in humans is the fact that animal Rh7.5 was able to recognize a HLA-A2 epitope identified in humans. This is the first study to report a conserved HCV epitope between humans and rhesus macaques. Besides highlighting the striking similarities between the macaque and human immune response, the dominant epitope identified in animal Rh7.5 may serve as a useful tool in future rhesus macaque studies by allowing for the development of CINGVCWTV tetramers which would make it possible to more easily follow vaccine-induced HCV-specific T cells and to study the immune response in more depth. According to unpublished data from the Wisconsin National Primate Research Center at the University of Wisconsin–Madison, expression of members of the Mamu-A1*018 allele lineage is relatively common in Chinese-origin rhesus macaque cohorts with 38 of 292 chromosomes positive (13%). In contrast, only 1.3% (6/470 chromosomes examined to date) of the predominantly Indian-origin rhesus macaques express Mamu-A*018 alleles. Provided that fine characterization of the restricting alleles confirms that Mamu-A1*018:02 is in fact responsible for the restriction of the immunodominant CINGVCWTV epitope seen in animal Rh7.5, based on the relative abundance of this allele within the rhesus macaque population it would be possible to easily identify and enrich for animals carrying this allele in future HCV immunization studies.
Taken together, the results of this study show that immunization in the nonhuman primate, rhesus macaque vaccination model is able to induce HCV-specific T cell responses strikingly similar to those observed in resolving patients which further validates the use of this vaccination model to help predict potential immune responses in humans following vaccination. Given the similarities between the rhesus macaque and human immune system, results from this study also suggest that DNA immunization with the nonstructural proteins of HCV may induce the type of protective T cell responses needed to protect individuals from the progression of HCV infection to the chronic state and merits further study in the context of future HCV T cell-based vaccines.
Materials and Methods
pConNS3/NS4A. The generation of pConNS3/NS4A, along with its expression in vitro and immunogenicity in mice, has been previously described.12
Animals. The 10 Chinese rhesus macaques used in the study were housed at BIOQUAL (Rockville, MD). These animals were used and maintained in accordance with the Guide for the Care and Use of Laboratory Animals.
Immunization. The pConNS3/NS4A plasmid was prepared by VGXI (The Woodlands, TX). The plasmid was subsequently diluted in sterile water formulated with 1% (wt/wt) poly--glutamate sodium salt (MW = 10.5 kDa) (Sigma-Aldrich, St. Louis, MO). Each macaque was intramuscularly injected with 1 mg of pConNS3/NS4A diluted in a total volume of 0.75 ml. The site of injection was electroporated using the CELLECTRA adaptive constant current electroporation device (three pulses of 0.5 Amp constant current, 1 second apart and 52 ms in length). Each animal received a total of three immunizations, 4 weeks apart.
Blood collection and PBMC isolation. Animals were bled once prior to immunization to test for preexisting NS3-specific T cell responses and were subsequently bled 2 weeks following each of the three immunizations. Prior to each bleed, animals were anesthetized with a mixture of ketamine (10 mg/kg) and acepromazine (0.1 mg/kg), and the blood was collected in EDTA tubes. PBMCs were isolated using standard Ficoll-Hypaque density gradient centrifugation resuspended in complete media (RPMI 1640 with 2 mmol/l -glutamine supplemented with 10% heat-inactivated FBS, 1× antibiotic/antimycotic, and 55 µmol/l β-mercaptoethanol).
IFN-γ ELISpot assays. IFN-γ ELISpot assays were performed as previously described34 using detection and capture antibodies from MabTech, Nacka Strand, Sweden. PBMCs were stimulated with a total of 101 HCV NS3 and NS4A 15-mer peptides (Invitrogen), overlapping by eight amino acids and spanning the entire length of the pConNS3/NS4A protein. These peptides were pooled into five different stimulation pools at a concentration of 2 µg/ml/peptide. The PBMCs were plated at a concentration of 200,000 cells per well. The average number of SFU was adjusted to 1 × 106 splenocytes for graphing purposes.
Matrix epitope mapping: The T cell response to pConNS3/NS4A was mapped using a matrix epitope mapping technique. The 101 HCV NS3 and NS4A 15-mer overlapping peptides spanning the length of pConNS3/NS4A were pooled into 21 separate pools. Each individual peptide was represented in two pools of the 21 pools. These pools were then used to stimulation PBMCs in an IFN-γ ELISpot assay as described above.
Flow cytometry
Flow cytometry reagents: The following directly conjugated antibodies were used: anti-rhesus CD3− allophycocyanin cyanine dye 7 (APC-Cy7) [clone SP34-2], anti-rhesus CD4− peridinin chlorophyll protein 5.5 (PerCP5.5) [clone L-200], antihuman CD8− allophycocyanin (APC) [clone SK1], antihuman CD107A− fluorescein isothiocyanate (FITC) [clone H4A3], antihuman CD95− phycoerythryin cyanine dye 5 (PE-Cy5) [clone DX2], antihuman TNF-α- phycoerythryin cyanine dye 7 (PE-Cy7) [clone Mab11], antihuman CD16-Pacific Blue [clone 3G8], antihuman IFN-γ-Alexa Fluor 700 [clone B27], antihuman IL-2− phycoerythryin (PE) [clone MQ1-17H12], and antihuman CD14-Pacific Blue [clone M5E2] (all from BD Biosciences, San Jose, CA); antihuman CD28-Texas Red [clone CD28.2] (Beckman Coulter, Brea, CA) and antihuman CD20-Pacific Blue [clone 2H7] (eBioscience, San Diego, CA). Aqua Live/Dead fixable dead cell Stain Kit (Molecular Probes, Eugene, OR) was used according to manufacturer's protocol to identify live cells.
Samples were collected on a LSRII flow cytometer (BD Biosciences, Franklin Lakes, NJ). BD CompBeads (BD Biosciences, San Jose, CA) and single fluorochromes were used for compensation. Data were analyzed using FlowJo software, version 8.7.1 for Mac (Tree Star, Ashland, OR). In order to obtain polyfunctional data, Boolean gates were created in FlowJo. Data were exported to PESTLE, version 1.6.2, for background subtraction and further analyzed with SPICE, version 4.3; both programs were obtained from M. Roederer (National Institutes of Health, Bethesda, MD).
T cell proliferation: PBMCs were pelleted and resuspended in 1 ml CFSE (Molecular Probes, Eugene, OR) diluted 1:2000 in phosphate-buffered saline. PBMCs were incubated at 37 °C for 10 minutes, quenced with 10 ml complete media, and pelleted. Cells were then resuspended in complete media at a concentration of 106 PBMCs/100 µl and plated in round bottom 96 well plates. Cells were then stimulated with 100 µl complete media containing either 2 µg/ml NS3/NS4A peptides, 2.5 µg/ml concavlin A (positive control) or 0.1% dimethyl sulfoxide (negative control). Cell cultures were incubated for 5 consecutive days and then stained for flow cytometry analysis. PBMCs were first stained with a live/dead marker according to manufacturer's protocol, washed with phosphate-buffered saline, and then stained with the following extracellular antibodies, CD3, CD4, and CD8 for 30 minutes at 4 °C. PBMCs washed twice with phosphate-buffered saline and fixed with 1% paraformaldehyde. Fifty thousand CD3+ events were collected per sample.
Intracellular cytokine staining: PBMCs were resuspended in complete media at a concentration of 1 × 106 cells/100 µl and plated in a round bottom 96-well plate along with anti-CD107a. Cells were stimulated for 5 hours at 37 °C with 100 µl of either 2 µg/ml HCV NS3/NS4A peptides, 1 µg/ml Staphylococcus enterotoxin B (positive control; Sigma-Aldrich) or 0.1% dimethyl sulfoxide (negative control) diluted in complete media supplemented with GolgiStop and GolgiPlug (BD Biosciences, San Jose, CA). Following incubation, cells were washed three times with phosphate-buffered saline and stained for viability and surface markers with anti-CD3, CD4, CD8, CD95, CD28, CD14, CD16, and CD20 for 30 minutes at 4 °C. Cells were permeabilized and washed using BD Cytofix/Cytoperm Solution Kit (BD Bioscience) and then stained intracellularly with anti-IFN-γ, IL-2, and TNF-α for 45 minutes at 4 °C. Following staining, cells were washed and then fixed with 1% paraformaldehyde and stored at 4 °C until analysis. The percent of specific function was calculated as the percent function of the peptide-stimulated group minus the percent function of the 0.1% dimethyl sulfoxide-stimulated group for each animal. The threshold for positive responses was set at 0.02%.
MHC class I genotyping. RNA was isolated from frozen PBMC samples, and 568-bp primary cDNA/PCR amplicons were generated spanning the highly polymorphic peptide-binding domain of class I molecules, as previously described.35 The primary amplicons, which can be distinguished by bar codes incorporated in the primers were normalized, pooled, and subjected to emulsion PCR. These emPCR products were pyrosequenced in a Roche/454 Titanium run. After amplicon image processing and base calling, BLAT was used to align the resulting sequences to previously described class I alleles.
SUPPLEMENTARY MATERIAL Table S1 Dominant and subdominant peptides.
Acknowledgments
This work was supported by the Cancer Research Institute training grant “Predoctoral Emphasis Pathway in Tumor Immunology” (KLK). This manuscript was supported in part by funding from the NIH to DBW and from Inovio Pharmaceuticals. Grant Support: Cure Grant—PA Department of Health—7 August 2007. The authors note possible commercial conflicts associated with this work, which may include Wyeth, Inovio, BMS, Virxsys, Ichor, Merck, Althea, and Aldeveron.
Supplementary Material
Dominant and subdominant peptides.
REFERENCES
- Bassett SE, Guerra B, Brasky K, Miskovsky E, Houghton M, Klimpel GR.et al. (2001Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection Hepatology 331479–1487. [DOI] [PubMed] [Google Scholar]
- Pawlotsky JM. Diagnostic tests for hepatitis C. J Hepatol. 1999;31 Suppl 1:71–79. doi: 10.1016/s0168-8278(99)80378-x. [DOI] [PubMed] [Google Scholar]
- Farci P, Alter HJ, Govindarajan S, Wong DC, Engle R, Lesniewski RR.et al. (1992Lack of protective immunity against reinfection with hepatitis C virus Science 258135–140. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Chavez D, Bigger C, Brasky KM, Wang XH.et al. (2004Cross-genotype immunity to hepatitis C virus J Virol 781575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., and, Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature. 2005;436:961–966. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C.et al. (1996Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response J Clin Invest 98706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepolder HM, Zachoval R, Hoffmann RM, Wierenga EA, Santantonio T, Jung MC.et al. (1995Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection Lancet 3461006–1007. [DOI] [PubMed] [Google Scholar]
- Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY.et al. (1999Analysis of a successful immune response against hepatitis C virus Immunity 10439–449. [DOI] [PubMed] [Google Scholar]
- Post JJ, Pan Y, Freeman AJ, Harvey CE, White PA, Palladinetti P, Hepatitis C Incidence and Transmission in Prisons Study (HITS) Group et al. Clearance of hepatitis C viremia associated with cellular immunity in the absence of seroconversion in the hepatitis C incidence and transmission in prisons study cohort. J Infect Dis. 2004;189:1846–1855. doi: 10.1086/383279. [DOI] [PubMed] [Google Scholar]
- Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P.et al. (2000Analysis of successful immune responses in persons infected with hepatitis C virus J Exp Med 1911499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach JT, Diepolder HM, Jung MC, Gruener NH, Schraut WW, Zachoval R.et al. (1999Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C Gastroenterology 117933–941. [DOI] [PubMed] [Google Scholar]
- Lang KA, Yan J, Draghia-Akli R, Khan A., and, Weiner DB. Strong HCV NS3- and NS4A-specific cellular immune responses induced in mice and Rhesus macaques by a novel HCV genotype 1a/1b consensus DNA vaccine. Vaccine. 2008;26:6225–6231. doi: 10.1016/j.vaccine.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimme R, Oldach D, Chang KM, Steiger C, Ray SC., and, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari A, Bart PA, Stöhr W, Tapia G, Garcia M, Medjitna-Rais E.et al. (2008An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses J Exp Med 20563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari A, Cellerai C, Enders FB, Köstler J, Codarri L, Tapia G.et al. (2007Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype Proc Natl Acad Sci USA 10416233–16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA., and, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- Badr G, Bédard N, Abdel-Hakeem MS, Trautmann L, Willems B, Villeneuve JP.et al. (2008Early interferon therapy for hepatitis C virus infection rescues polyfunctional, long-lived CD8+ memory T cells J Virol 8210017–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffreda D, Comte D, Cavassini M, Giostra E, Bühler L, Perruchoud M.et al. (2008Polyfunctional HCV-specific T-cell responses are associated with effective control of HCV replication Eur J Immunol 382665–2677. [DOI] [PubMed] [Google Scholar]
- Shoukry NH, Grakoui A, Houghton M, Chien DY, Ghrayeb J, Reimann KA.et al. (2003Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection J Exp Med 1971645–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B, Chang KM, McHutchison JG, Kokka R, Houghton M., and, Chisari FV. Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J Clin Invest. 1996;98:1432–1440. doi: 10.1172/JCI118931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J.et al. (2003HCV persistence and immune evasion in the absence of memory T cell help Science 302659–662. [DOI] [PubMed] [Google Scholar]
- Lauer GM, Barnes E, Lucas M, Timm J, Ouchi K, Kim AY.et al. (2004High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection Gastroenterology 127924–936. [DOI] [PubMed] [Google Scholar]
- Otting N, Heijmans CM, van der Wiel M, de Groot NG, Doxiadis GG., and, Bontrop RE. A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics. 2008;60:507–514. doi: 10.1007/s00251-008-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, de Vos-Rouweler AJ, Heijmans CM, de Groot NG, Doxiadis GG., and, Bontrop RE. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics. 2007;59:367–375. doi: 10.1007/s00251-007-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny A, McHutchison JG, Pasquinelli C, Brown ME, Brothers MA, Grabscheid B.et al. (1995Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif J Clin Invest 95521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel MJ, Dudley D, Afdhal N, Grakoui A, Rice CM, Choo QL.et al. (1995HLA class I-restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterization of patterns of cytokine release J Clin Invest 962311–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habersetzer F, Baumert TF., and, Stoll-Keller F. GI-5005, a yeast vector vaccine expressing an NS3-core fusion protein for chronic HCV infection. Curr Opin Mol Ther. 2009;11:456–462. [PubMed] [Google Scholar]
- Schlaphoff V, Klade CS, Jilma B, Jelovcan SB, Cornberg M, Tauber E, IC41 Study Group et al. Functional and phenotypic characterization of peptide-vaccine-induced HCV-specific CD8+ T cells in healthy individuals and chronic hepatitis C patients. Vaccine. 2007;25:6793–6806. doi: 10.1016/j.vaccine.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Firbas C, Jilma B, Tauber E, Buerger V, Jelovcan S, Lingnau K.et al. (2006Immunogenicity and safety of a novel therapeutic hepatitis C virus (HCV) peptide vaccine: a randomized, placebo controlled trial for dose optimization in 128 healthy subjects Vaccine 244343–4353. [DOI] [PubMed] [Google Scholar]
- Klade CS, Wedemeyer H, Berg T, Hinrichsen H, Cholewinska G, Zeuzem S.et al. (2008Therapeutic vaccination of chronic hepatitis C nonresponder patients with the peptide vaccine IC41 Gastroenterology 1341385–1395. [DOI] [PubMed] [Google Scholar]
- Inovio biomedical reveals safety results of Tripep's ChronVac-C delivered using Inovio's electroporation delivery systems 2008 ). < http://www.tradingmarkets.com/.site/news/ BREAKING%20NEWS/1209773/?hcode=relatednews >. Accessed 26 April 2006.
- Frese M, Schwärzle V, Barth K, Krieger N, Lohmann V, Mihm S.et al. (2002Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs Hepatology 35694–703. [DOI] [PubMed] [Google Scholar]
- Gattinoni L, Klebanoff CA, Palmer DC, Wrzesinski C, Kerstann K, Yu Z.et al. (2005Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells J Clin Invest 1151616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JD, Robinson TM, Kutzler MA, Parkinson R, Calarota SA, Sidhu MK.et al. (2005SIV DNA vaccine co-administered with IL-12 expression plasmid enhances CD8 SIV cellular immune responses in cynomolgus macaques J Med Primatol 34262–270. [DOI] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O'Leary CE, Lank SM, Tuscher JJ.et al. (2009Major histocompatibility complex genotyping with massively parallel pyrosequencing Nat Med 151322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dominant and subdominant peptides.