Abstract
The tyrosine kinase inhibitor imatinib is highly effective in the treatment of chronic myelogenous leukemia (CML), but primary and acquired resistance of CML cells to the drug offset its efficacy. Molecular mechanisms for resistance of CML to tyrosine kinase inhibitors are not fully understood. In the present study, we show that BCR-ABL activates the expression of the mammalian stress response gene SIRT1 in hematopoietic progenitor cells and that this involves STAT5 signaling. SIRT1 activation promotes CML cell survival and proliferation associated with deacetylation of multiple SIRT1 substrates, including FOXO1, p53, and Ku70. Imatinib-mediated inhibition of BCR-ABL kinase activity partially reduces SIRT1 expression and SIRT1 inhibition further sensitizes CML cells to imatinib-induced apoptosis. Knockout of SIRT1 suppresses BCR-ABL transformation of mouse BM cells and the development of a CML-like myeloproliferative disease, and treatment of mice with the SIRT1 inhibitor tenovin-6 deters disease progression. The combination of SIRT1 gene knockout and imatinib treatment further extends the survival of CML mice. Our results suggest that SIRT1 is a novel survival pathway activated by BCR-ABL expression in hematopoietic progenitor cells, which promotes oncogenic transformation and leukemogenesis. Our findings suggest further exploration of SIRT1 as a therapeutic target for CML treatment to overcome resistance.
Introduction
BCR-ABL activates several cell proliferation and survival pathways in hematopoietic stem/progenitor cells.1 Treatment with the BCR-ABL tyrosine kinase inhibitor imatinib mesylate results in complete cytogenetic response in most cases of chronic-phase CML, but results in poor responses in advanced phases of the disease, with frequent relapse.2 Both primary and acquired resistance contribute to recurrent disease. In chronic-phase CML, imatinib suppresses the proliferation of CML leukemic stem/progenitor cells, but does not effectively kill them,3,4 and most patients in complete cytogenetic response continue to harbor residual leukemia progenitor cells5 that may serve as a source for relapse. Despite this, BCR-ABL activity in CML stem/progenitor cells can be efficiently inhibited by tyrosine kinase inhibitors.6,7 Many other factors, including BCR-ABL mutations and gene amplification as well as BCR-ABL–independent mechanisms, may contribute to resistance, particularly for advanced phases of CML.8 Further understanding of the molecular mechanisms of the disease and resistance may help in the development of new strategies to overcome resistance and improve CML treatment.
SIRT1 is a mammalian homolog of yeast silent information regulator 2, a nicotinamide adenine dinucleotide–dependent protein deacetylase that is required for replicative lifespan extension on calorie restriction.9 SIRT1 promotes mammalian cell survival under metabolic, oxidative, and genotoxic stresses through deacetylation of multiple substrates, including p53,10,11 Ku70,12 and FOXO proteins.13–15 Overexpression of SIRT1 is detected in several primary solid tumors and hematopoietic malignancies.16–19 Inactivation of SIRT1 inhibits growth and promotes apoptosis in human cancer cells.10,11 Intriguingly, a few recent studies have shown that SIRT1 may act as a tumor suppressor in mice with germline disruptions of p53.20,21 These studies suggest complex, possibly tissue-dependent roles of SIRT1 in both tumor promotion and suppression. However, the roles of SIRT1 activation by oncogenic transformation in hematopoietic progenitor cells and CML development are unknown.
The tumor suppressor hypermethylated in cancer 1 (HIC1) is inactivated by promoter hypermethylation in CML.22 We have shown previously that the loss of HIC1 activates SIRT1 expression, which enhances cell survival under stress and DNA damage.23 We initially hypothesized that the activation of SIRT1 may play a role in the survival of CML cells for chemoresistance. We have also observed that SIRT1 expression is significantly increased in blast crisis CML cell lines KCL-22 and K562.24 In the present study, we demonstrate that SIRT1 is transcriptionally activated by BCR-ABL, providing a novel survival pathway for CML progenitor cells. SIRT1 expression is only partially reduced by imatinib treatment, and SIRT1 inhibition sensitizes CML cells to imatinib treatment. SIRT1 knockout or inhibition by a small-molecule inhibitor effectively suppresses the development of CML-like myeloproliferative disease in mice.
Methods
Cell lines, drugs, and DNA constructs
KCL-22 and K562 cells were purchased from German Collection of Cell Cultures. Imatinib mesylate was kindly provided by Novartis. Sirtinol and trichostatin A were from Sigma-Aldrich. The lentiviral shRNA vectors pSicoR PGK-puromycin (PGK-puro) and CMV–green fluorescent protein (CMV-GFP), wild-type and H363Y SIRT1–expressing retroviral vectors,11 and KRAS retroviral vector were from Addgene. Tenovin-6 was purchased from Cayman Chemical or synthesized in-house by the City of Hope Synthetic Chemistry Core.
Isolation and retroviral transduction of human CD34+ cells
The study of human samples was approved by the Institutional Review Board of City of Hope. Cord blood samples were kindly provided by StemCyte. BM and leukemia blood samples were obtained from City of Hope Tissue Bank. Mononuclear cells were isolated using Ficoll separation. CD34+ cells were isolated using a positive magnetic bead selection protocol (StemCell Technologies). Some of the purified CD34+ cells were lysed to make cell lysates and stored at −80°C until analysis; the other cells were cultured for subsequent transduction studies using retroviral vectors, as described previously.25 Cells were harvested 48 hours later and labeled with anti-CD34 allophycocyanin (APC). CD34+GFP+ cells were collected by flow cytometry. After cells were expanded in culture for 7 days, total RNA and protein lysate were prepared using standard protocols. Apoptosis and the colony-forming cell (CFC) assay were performed as described previously.5
Gene knockdown using lentiviral vectors
shRNAs were inserted into pSicoR vectors containing a puromycin or GFP expression cassette. Sequences for shRNAs are provided in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). G protein of vesicular stomatitis virus (VSV-G) pseudotyped lentiviral vectors were made using a 4-plasmid transfection system, as described previously.26 High-titer lentiviral stocks, typically 1-3 × 107 infectious units/mL, were used for transduction. Unless specified, a multiplicity of infection of approximately 5 was typically used so that nearly complete transduction was achieved. For SIRT1 knockdown in transformed CD34+ cells, SIRT1 shRNA sequences were cloned into the HIV7-SF–red fluorescent protein (HIV7-SF-RFP) lentiviral vector. MIG210 or MIGR1 (GFP) and shRNA (RFP) double-transduced cells were sorted by flow cytometry for CD34+GFP+RFP+ for the in vitro apoptosis and proliferation study. For some experiments, cells with high or low levels of GFP expression, corresponding to high or low BCR-ABL expression, respectively, were sorted separately.
RNA and protein analyses
RNA was extracted with TRIzol (Invitrogen) or the PicoPure RNA isolation kit (Applied Biosystems), the first-strand DNA was synthesized with the Superscript III kit (Invitrogen), and gene expression was analyzed using the SYBR GreenER qPCR SuperMix kit (Invitrogen). Rabbit monoclonal anti–human SIRT1 (#1104-1; Epitomics), rabbit anti–mouse SIRT1 (#07-131; Upstate Biotechnology), mouse monoclonal anti–c-ABL and pY694 STAT5 (BD Pharmingen), mouse monoclonal anti-Ku70 (Neomarker), rabbit polyclonal antiacetylated p53 (Cell Signaling Technology), and rabbit polyclonal antiacetylated FOXO1 (Santa Cruz Biotechnology) were used for Western blots. To analyze Ku70 acetylation, Ku70 was pulled down from total cell lysate with conjugated anti-Ku70–agarose beads (Santa Cruz Biotechnology), followed by acetylation detection with rabbit anti-acetyl lysine Ab (Cell Signaling Technology). For the flow cytometry–based acetylation assay, cells were washed with PBS, fixed with 2% paraformaldehyde for 10 minutes, and then permeabilized with 0.1% Triton X-100. Cells were labeled with acetyl FOXO1 Ab (D-19; Santa Cruz Biotechnology) at a 1:40 dilution for 30 minutes, followed by anti–rabbit Alexa Fluor 594 (Invitrogen) for 30 minutes (1:150). Stained cells were analyzed by flow cytometry on a CyAn ADP (Dako).
Cell cycle, apoptosis, and clonogenic assays
Cell cycle, apoptosis, and clonogenic assays were performed as described previously.26 Apoptosis was analyzed with the annexin V assay kit (BD Pharmingen). For the clonogenic assay, a standard 2-layer soft agar culture was performed and colonies were scored after staining with 0.005% crystal violet.
SIRT1 mRNA stability
KCL-22 cells were seeded at 0.5 × 106/mL in 12-well plate with 2.5μM imatinib or DMSO as a mock control. Twelve hours later, actinomycin D (2 μg/mL) was added. After 0, 1, 2, 4, and 8 hours, total RNA was extracted. First-strand cDNA was reverse transcribed from 1.0 μg of total RNA using the Superscript III kit (Invitrogen). One microliter of the first-strand cDNA synthesis reaction mixture was used for PCR amplification. GAPDH was amplified with 25 cycles and SIRT1 was amplified with 30 cycles.
SIRT1 promoter assay
The 2.8-kb SIRT1 promoter-luciferase reporter and its deletion constructs were kindly provided by Toren Finkel (National Heart, Lung, and Blood Institute, NIH, Bethesda, MD).27 Because it is difficult to transfect KCL-22 cells, K562 cells were used for this assay. K562 cells were seeded at 4 × 104/well in 96-well plates and cotransfected with SIRT1 promoter-luciferase reporters and a control Renilla plasmid (Promega). LTX Lipofectamine reagent (Invitrogen) was used to transfect 0.18 μg of the reporter construct and 0.02 μg of the Renilla construct in each well per the manufacturer's instructions. Six hours after transfection, cells were treated with imatinib and harvested after another 30 hours. Luciferase activity was measured using the Dual-Luciferase Assay System (Promega) and was normalized to the Renilla reading in each lysate.
To delete STAT5 sites, the SIRT1 promoter was subcloned into MluI/XhoI sites of the pGL3 basic luciferase expression vector (Promega) for easy cloning. A standard inverse PCR protocol was applied to delete STAT5 sites with the primers provided in supplemental Table 1. Inverse PCR was performed with Pfu Ultra II fusion HS DNA polymerase (Stratagene). The PCR product was digested with DpnI (NEB) to remove the template, followed by agarose gel extraction, self-ligation, and transformation. The deletions were confirmed by DNA sequencing. The luciferase activity of the STAT5-deletion constructs was analyzed as described in the previous paragraph.
ChIP assay
The ChIP assay was performed as described previously,23 with STAT5A or 5B Ab (Cell Signaling Technology) and anti-Flag Ab (Santa Cruz Biotechnology) as controls. The primers for ChIP PCR analysis are provided in supplemental Table 1.
Animal studies
The use of animals was approved by the City of Hope Institutional Animal Care and Use Committee. For the CML tumor xenograft assay, 3 million virally transduced cells were inoculated subcutaneously into the right flank of NOD-SCID mice conditioned by 270 rad of irradiation. Tumor length, height, and width were measured weekly with a caliper. Tumor volume was estimated from the formula: V = π(L × H × W/6). Mice were euthanized when the tumor volume reached 1000 mm3.
For BM transplantation experiments, SIRT1 knockout mice28 were backcrossed to the BALB/c strain (Taconic) for at least 8 generations, and 3- to 6-month-old mice were used as donors. BALB/c mice (Taconic) at the age of 7-8 weeks were used as recipients. Retroviral transduction of BM cells and transplantation were performed as described previously with minor modifications.29 Briefly, ecotropic MIG210 and MIGR1 vectors were packaged using Phoenix-Eco cells, and viral stocks with titers higher than 2 × 106 were used for BM transduction. Donor mice were primed by IP injection with one dose of 200 mg/kg 5-fluorouracil 3 days before harvest. BM cells were harvested and cultured in prestimulation medium (DMEM, 15% heat-inactivated FBS, 5% WEHI-3B–conditioned medium, penicillin/streptomycin, 120 U/mL of recombinant murine IL-3, 500 U/mL of IL-6, and 5 U/mL of SCF [Peprotech]). After 24 hours, the cells were transduced with retroviral vectors by 2 rounds of cosedimentation transduction. Recipient mice were lethally irradiated by 2 doses of 450-rad radiation separated by 3 hours. Transduced cells were transplanted by tail vein injection.
For the in vivo drug treatment study, imatinib was prepared freshly in pure H2O and tenovin-6 was prepared in 20% cyclodextrin (wt/vol; #C0926; Sigma-Aldrich) and 10% DMSO (vol/vol) as described previously,30 and both were filtered sterile. At day 10 after transplantation, drugs were administrated continuously for 10 days. Imatinib was administrated by oral gavage with 75 mg/kg in the morning and 125 mg/kg in the afternoon. Tenovin-6 was given by IP injection at 50 mg/kg in the morning. Vehicle control animals were treated with solution containing 20% cyclodextrin and 10% DMSO.
Mouse hematopoietic stem/progenitor cell isolation and analysis
The following Abs were used for cell analysis and sorting: CD150-APC (clone TC15-12F12.2; BioLegend), CD41-PE-Cy7 (clone eBioMWReg30; eBioscience), CD48-PE-Cy7 (clone HM48-1; eBioscience), cKit-APC (clone 2B8; BD Pharmingen), Sca1-PE-Cy7 (clone D7; BD Pharmingen), and BD Pharmingen PE-labeled lineage Abs (Gr-1, clone RB6-8C5; Mac-1, clone M 1/70; B220, clone RA3-6B2; Ter119, clone TER-199; and CD3e, clone 145-2C11). After depleting lineage-positive cells using EasySep (StemCell Technologies), cells were labeled with PE-lineage cocktail and other cell markers. Mouse hematopoietic stem cells in the BABL/c strain were analyzed by flow cytometry using the Lin−CD150+CD41−CD48− combination, as described previously.31 The cell-cycle status for hematopoietic stem cells was further analyzed using Hoechst 33342 and pyronin Y staining, as described previously.32 The BM progenitor cell colony formation assay was performed using M3434 methylcellulose medium (StemCell Technologies).
Statistical analysis
For animal studies, Kaplan-Meier survival analysis was performed and statistical significance was calculated using the log-rank test. For other data analysis, the t test was performed. Two-tailed analysis was used in all cases and P < .05 was considered statistically significant.
Results
SIRT1 is activated by BCR-ABL transformation
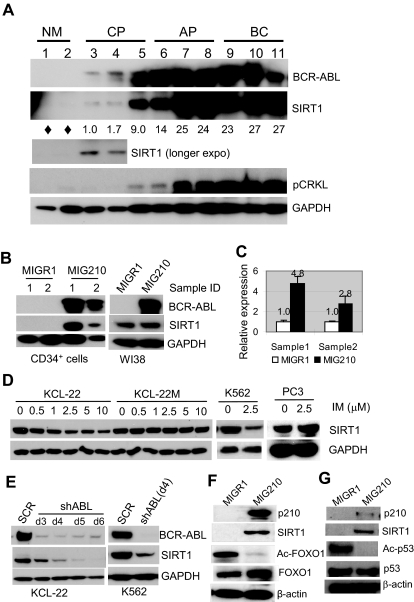
Given that SIRT1 is repressed by HIC1 and is overexpressed in CML cell lines,23,24 in the present study, SIRT1 expression was examined in primary human CML cells. Normal and CML CD34+ progenitor cells were isolated. SIRT1 expression was barely detectable in normal CD34+ progenitor cells by Western blot, but was increased in chronic CML progenitor cells and further increased toward the advanced phases of CML, and this was was correlated with increasing BCR-ABL expression and tyrosine kinase activity as measured by CRKL phosphorylation (Figure 1A). This finding prompted the determination of the change of SIRT1 expression during BCR-ABL transformation. Normal CD34+ cells were transduced with the BCR-ABL retroviral vector MIG210.29 The SIRT1 protein was up-regulated by BCR-ABL transduction along with SIRT1 mRNA level (Figure 1B-C). No significant change of HIC1 expression was found after BCR-ABL expression in CD34+ cells (not shown), suggesting the SIRT1 activation occurs early during transformation without altering HIC1 expression. Intriguingly, BCR-ABL did not increase SIRT1 expression in normal human fibroblasts WI38 (Figure 1B) and several other cell lines in which SIRT1 was already expressed (supplemental Figure 1), indicating that BCR-ABL activation of SIRT1 in human hematopoietic progenitor cells may be a selective outcome of its transformation of these cells.
Figure 1.
BCR-ABL expression activates SIRT1 in human hematopoietic progenitor cells. (A) SIRT1 protein in CD34+ normal and CML cells. Chronic phase (CP) and advanced-phase CML (AP, accelerated phase; BC, blast crisis) were from unrelated patients with active disease. Normal CD34+ cells (NM) were from the peripheral blood of healthy adult donors after BM mobilization by GM-CSF treatment. Three CP patients had not received prior treatment. AP and BC patients received IFN, hydroxyurea, or combination treatment and samples were obtained after at least 2 weeks off chemotherapy, with the exception that BC patient 9 had not received prior treatment. Densitometry of SIRT1 expression was quantified after normalizing to GAPDH. Diamonds indicate no significant reading above background. (B-C) SIRT1 protein (B) and mRNA (C) levels after BCR-ABL transduction (MIG210) in normal CD34+ cells from 2 independent healthy donors compared with transduction of WI38 cells. Empty vector MIGR1 was used as a control. (D) Change of SIRT1 protein levels on imatinib (IM) treatment for 48 hours in the CML cell lines KCL-22, K562, and KCL-22M, and in prostate cancer PC3 cells. (E) SIRT1 expression on BCR-ABL knockdown by shABL lentiviral vector at 3-6 days after initial transduction. SCR indicates scrambled shRNA. (F) Representative Western blot analysis of FOXO1 acetylation change in CD34+ cells after MIG210 transduction. (G) Representative Western blot analysis of p53 acetylation. CD34+ cells were treated with 10μM nutlin-3 and 0.5μM trichostatin A for 6 hours, followed by p210 transduction.
When the CML cell lines KCL-22 and K562 cells were treated with imatinib, SIRT1 expression decreased in a drug concentration-dependent manner, but was not depleted even with 10μM imatinib (Figure 1D), which almost completely suppressed tyrosine kinase activity in these cells (supplemental Figure 2). In contrast, the treatment did not change SIRT1 expression in KCL-22M cells that express the imatinib-resistant T315I mutant BCR-ABL26 or in a prostate cancer cell line. Similarly, when BCR-ABL was knocked down, SIRT1 protein levels decreased (Figure 1E). These data further support that BCR-ABL activates SIRT1 expression in CML cells.
FOXO1, a SIRT1 target protein, was deacetylated in CD34+ cells on BCR-ABL expression (Figure 1F). Another known SIRT1 substrate p53, is typically expressed at low levels and difficult to detect in normal CD34+ cells (not shown). CD34+ cells were treated with the MDM-2 inhibitor nutlin-3 to stabilize p53 and its deacetylation was blocked by the class I/II histone deacetylase inhibitor trichostatin A, which does not inhibit SIRT1. Under these settings, we observed that activation of SIRT1 by BCR-ABL efficiently deacetylated p53 in nutlin-3– and trichostatin A–treated cells (Figure 1G). Therefore, SIRT1 activation results in deacetylation of its cellular targets in progenitor cells.
SIRT1 activation by BCR-ABL involves STAT5 signaling
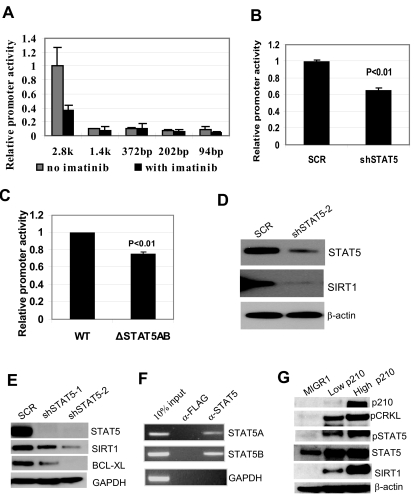
SIRT1 expression is subjected to multiple levels of regulation, including protein phosphorylation,33 mRNA stability,34 and transcription.23,27,35,36 BCR-ABL did not affect SIRT1 protein stability per se or the half-life of SIRT1 mRNA (supplemental Figure 3). Therefore, BCR-ABL regulation of SIRT1 transcription was examined. By transfecting K562 cells with a 2.8-kb SIRT1 promoter luciferase reporter and its deletion constructs,27 a major SIRT1 promoter activation region was identified between −1484 and −2850 bp, and this activation was repressed by imatinib treatment in K562 cells (Figure 2A). This regulatory region is beyond the sequence for previously identified transcriptional factor binding sites.23,27,35,36
Figure 2.
BCR-ABL activation of SIRT1 involves STAT5 signaling. (A) Luciferase reporter constructs of SIRT1 promoter (2852-94 bp) were transfected into K562 cells, followed by mock or imatinib treatment for 30 hours. Relative promoter activity was calculated by normalizing luciferase activity to the Renilla control. (B) Effect of STAT5 knockdown on SIRT1 promoter luciferase activity in K562 cells. (C) Effect of deleting both STAT5-binding sites on SIRT1 promoter luciferase activity in K562 cells. (D-E) Effect of STAT5 knockdown on endogenous SIRT1 expression in K562 (D) and KCL-22 cells (E). (F) ChIP assay of STAT5A and 5B on SIRT1 promoter. Anti-Flag Ab was used as a ChIP control. (G) BCR-ABL dose effect on SIRT1 expression. High- and low-GFP–expressing CD34+ cells were sorted (supplemental Figure 5) for analysis. BCR-ABL levels were validated by CRKL phosphorylation, and blots were reprobed for SIRT1 and STAT5 expression and modifications.
Searching for transcriptional factors binding at this region that are related to BCR-ABL functions, 2 putative binding sites for STAT5A and 5B were found at −1838 and −2235 (supplemental Figure 4). STAT5 is a key signaling molecule that is phosphorylated and activated by BCR-ABL, and is essential for BCR-ABL transformation.37 We found that STAT5 knockdown significantly reduced SIRT1 promoter activity (Figure 2B) and, similarly, deletion of both STAT5-binding sites reduced SIRT1 promoter activity (Figure 2C). STAT5 knockdown reduced SIRT1 expression in both K562 and KCL-22 cells, which was accompanied by reduction of another STAT5 target gene, BCL-XL38 (Figure 2D-E). The ChIP assay showed that both STAT5A and 5B were associated with the promoter (Figure 2F), further supporting STAT5 regulation of SIRT1. Consistently, BCR-ABL dose-dependent activation of SIRT1 in CD34+ cells was accompanied by increased STAT5 activity and the activation of other STAT5 target genes, including RAD51, PIM1, and BCL-XL38,39 (Figure 2G and supplemental Figure 5). These results suggest that BCR-ABL activates SIRT1 transcription in CML cells, and that this involves the STAT5-signaling pathway.
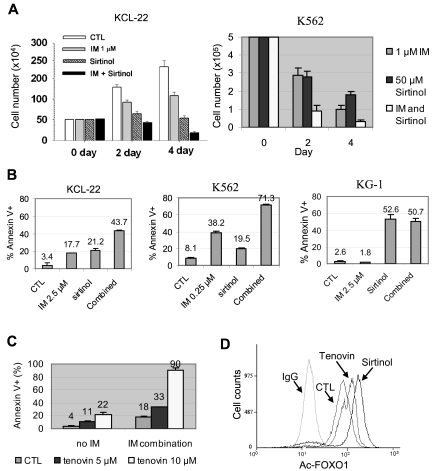
SIRT1 inhibition induces apoptosis and suppresses growth and transformation of CML cells
To determine the consequence of SIRT1 activation, KCL-22 and K562 cells were treated with sirtinol, a small-molecule inhibitor of SIRT1, with or without imatinib. We showed previously that whereas K562 cells are sensitive to 1μM imatinib, killing KCL-22 cells requires higher concentrations of imatinib and these cells can grow in 1μM imatinib at a reduced rate.26 Sirtinol alone inhibited the growth of both cell lines, and the combination of sirtinol with 1μM imatinib enhanced their inhibitory effects (Figure 3A). Sirtinol treatment induced apoptosis in both K562 and KCL-22 cells, and further sensitized these cells to imatinib-induced apoptosis; however, such a sensitizing effect was not seen in the BCR-ABL–negative leukemia cell line KG-1 (Figure 3B). Sirtinol and imatinib both affected the cell cycle of CML cells, reducing the S/G2/M populations and increasing the sub-G1 population in KCL-22 cells, whereas the same treatment rapidly increased the sub-G1 and apoptotic fractions in K562 cells (supplemental Figure 6). Similar to sirtinol, another sirtuin inhibitor, tenovin-6, also sensitized KCL-22 cells to imatinib (Figure 3C). In agreement with these effects, sirtinol and tenovin-6 treatment increased FOXO1 acetylation in CML cells (Figure 3D).
Figure 3.
Effects of pharmacologic inhibition of SIRT1 on CML cell survival. (A) KCL-22 and K562 cells were treated with 1μM imatinib mesylate (IM) with or without 50μM sirtinol. Surviving cells were counted at 2 and 4 days after the treatment. (B) Apoptosis was analyzed 2 days after drug treatment. KCL-22 cells and BCR-ABL–negative KG-1 cells were treated with 2.5μM IM, whereas K562 cells were treated with 0.25μM IM in the presence or absence of 50μM sirtinol. DMSO was used as control (CTL). (C) KCL-22 cells were treated with DMSO (CTL) or 5 or 10μM tenovin-6 in the absence or presence of 2.5μM IM. Apoptosis was analyzed 3 days after drug treatment. (D) Flow cytometry–based analysis of FOXO1 acetylation in KCL-22 cells after mock, 2.5μM tenovin-6, or 50μM sirtinol treatment. Cells were labeled with anti-acetyl FOXO1 Ab or normal IgG as a control.
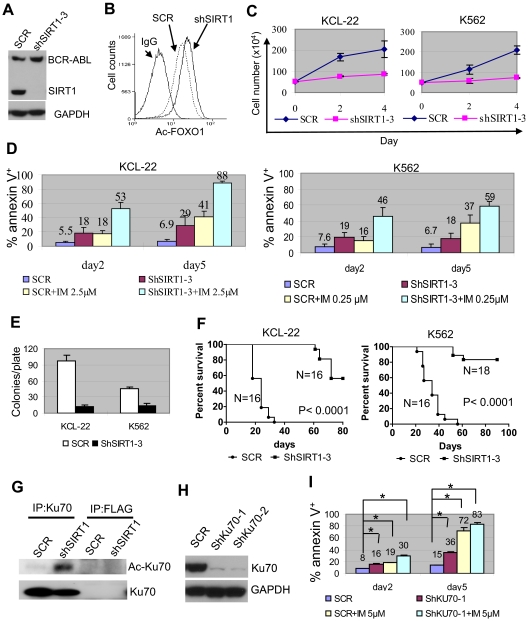
SIRT1 knockdown increased FOXO1 acetylation without affecting BCR-ABL expression (Figure 4A-B), suppressed the proliferation of KCL-22 and K562 cells, and enhanced imatinib-induced apoptosis (Figure 4C-D). Substantial SIRT1 knockdown was required to achieve such effects (supplemental Figure 7), which explained why SIRT1 inhibition could sensitize CML cells to imatinib even though the drug partially reduced the SIRT1 level (Figure 1D). Intriguingly, overexpression of SIRT1 failed to protect CML cells from the apoptosis induced by imatinib or by BCR-ABL knockdown, although it protected CML cells from oxidative stress (supplemental Figure 8). The reason for this difference is unclear, but may be that BCR-ABL activates multiple survival pathways1 or that SIRT1 expression in these cells reaches a plateau level needed for survival. SIRT1 knockdown inhibited soft agar colony formation of KCL-22 and K562 cells significantly (Figure 4E) and the growth of xenografted CML cells in immunodeficient mice (Figure 4F).
Figure 4.
Effects of SIRT1 knockdown on CML cell survival. (A) Knockdown of SIRT1 in KCL-22 cells using lentiviral vector shSIRT1-3. (B) Effect of SIRT1 knockdown on FOXO1 acetylation in KCL-22 3 days after transduction. (C) Three days after scrambled shRNA (mock) or shSIRT1-3 transduction using a multiplicity of infection of 5, cells were plated directly (without sorting) in triplicate in 24-well plates and viable cells were counted on the days indicated. (D) Three days after mock or shSIRT1-3 infection, 5 × 105 cells were cultured with or without 2.5μM imatinib (IM) for KCL-22 or 0.25μM IM for K562 cells for another 2 or 5 days for apoptosis analysis. The percentage of annexin V+ cells was plotted. Error bars are standard deviations. (E) Soft agar colony formation assay. Three days after mock or shSIRT1-3 transduction, 500 cells per plate were seeded on standard 2-layer soft agar. The colonies were counted after 21 days. (F) Survival curves of mice receiving xenografted CML cells. After overnight infection with shRNA vectors, 3 million cells each were inoculated into NOD-SCID mice. Mice were euthanized when the tumor volume reached 1000 mm3. (G) Effect of SIRT1 knockdown on Ku70 acetylation. FLAG Ab was used as control. (H-I) The effect of Ku70 knockdown (H) on apoptosis of KCL-22 cells (I). Three days after scrambled shRNA (SCR) or shKU70 transduction, 5μM imatinib was added for another 2 or 5 days, and apoptosis was analyzed. Two sets of shRNAs showed similar results and data from one set are shown. *P < .05.
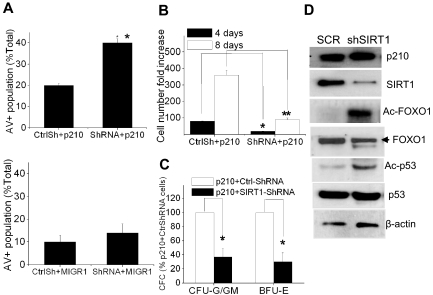
To determine the effect of SIRT1 inhibition on BCR-ABL transformation in primary CD34+ cells, the cells were coinfected with MIG210 and SIRT1 shRNA. SIRT1 knockdown increased apoptosis of BCR-ABL–transduced CD34+ cells but had a minimal effect on normal CD34+ cells (Figure 5A). SIRT1 knockdown suppressed proliferation and CFC formation of the transduced CD34+ cells (Figure 5B-C) and induced FOXO1 and p53 acetylation (Figure 5D).
Figure 5.
SIRT1 inhibition suppresses growth and induces apoptosis of BCR-ABL–transduced CD34+ cells. (A) Apoptosis of MIG210- or MIGR1-transduced normal CD34+ cells with mock (CtrlSh) or SIRT1 (shRNA) knockdown. The double-transduced CD34+ cells were sorted and then analyzed for annexin V+ cells after 48 hours of culture in low concentrations of growth factors similar to those present in long-term BM culture stroma-conditioned medium. (B-C) Analysis of total cell numbers (B) and CFC formation (C) of MIG210-transduced normal CD34+ cells with mock or SIRT1 knockdown. *P < .05; **P < .01. (D) Increased acetylation of FOXO1 and p53 after SIRT1 knockdown in MIG210-transduced normal CD34+ cells.
Deacetylation of p53 plays an important role in SIRT1-mediated cell survival.11,12 However, p53 is deleted in the blast crisis CML cell lines KCL-22 and K562, so the effect of SIRT1 must be through additional downstream factors in these cells. Ku70 is another effector that is deacetylated and activated by SIRT1 for enhancing cell survival.12 In the present study, Ku70 was acetylated after SIRT1 knockdown in KCL-22 cells (Figure 4G). Ku70 knockdown induced apoptosis and sensitized CML cells to imatinib treatment (Figure 4H-I). These results suggest that SIRT1 knockdown suppresses the growth and transformation of human CML cells, and that this may involve multiple downstream effectors.
SIRT1 knockout suppresses development of a CML-like disease in a mouse model
To further understand the role of SIRT1 in CML, the impact of SIRT1 knockout on CML disease development was examined in a mouse BM transduction/transplantation model.29 Because the disease penetrance for this model is approximately 100% in the BALB/c strain, SIRT1 knockout mice28 were backcrossed to the BALB/c strain for at least 8 generations. Some of the SIRT1−/− mice survived through adulthood with relatively normal development, although smaller body stature and closed eye lids occurred in some, as described previously.28 Consistent with a previous study,40 no significant differences were found between SIRT1−/− and SIRT1+/+ mice in hematopoietic stem cell frequency, cell cycle, colony formation, and blood lineage differentiation (supplemental Figure 9), or in engraftment and hematologic profiles in transplantation recipients (supplemental Figure 10).
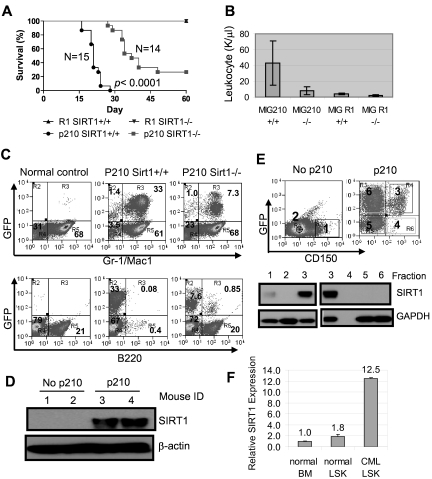
BM cells from 3-month-old SIRT1−/− and SIRT1+/+ mice were transduced with BCR-ABL (MIG210) or control vector (MIG R1), both bearing GFP as a marker, and 1 × 105 to 2.5 × 105 transduced cells each were transplanted into lethally irradiated BALB/c recipients. The transduction and engraftment efficiency for both wild-type and knockout cells were similar (supplemental Figure 10A). Mice receiving BCR-ABL–transformed SIRT1+/+ BM cells developed CML-like disease in 3-4 weeks consisting of signature characteristics of the disease29: markedly elevated WBCs predominated by granulocytes, splenomegaly, and multiple organ involvement (Figure 6A-B and supplemental Figure 11). Mice receiving transformed SIRT1−/− cells showed significantly delayed disease development, with markedly lower peripheral blood WBC counts (P < .0001; Figure 6B). In BM, dramatic expansion of Gr-1/Mac-1 lineage cells occurred in mice receiving transformed SIRT1+/+ cells and altered BM cell composition, whereas SIRT1 knockout inhibited the myeloid expansion (Figure 6C). As a consequence, mice receiving transformed knockout cells survived much longer than mice receiving transformed SIRT1+/+ cells (Figure 6A and supplemental Figure 11A). No difference was observed with empty vector transduction (Figure 6A).
Figure 6.
SIRT1 knockout suppresses the development of CML-like disease in a mouse model. (A-B) Survival curves (A) for mice receiving 2.5 × 105 mononuclear cells transduced by BCR-ABL MIG210 vector (p210) or empty vector (R1) and total leukocyte counts (B) for mice at 18 days after transplantation. In R1-transduced mouse controls, there were 8 mice receiving SIRT1+/+ donor cells and 6 mice receiving SIRT1−/− cells. (C) BM cell lineage analysis in normal BALB/c (control) and mice receiving MIG210 transformed SIRT1+/+ or SIRT1−/− cells. (D) SIRT1 protein expression in total BM mononuclear cells from normal BALB/c and mice receiving MIG210-transformed SIRT1+/+ cells (n = 2 in each group). (E) BM progenitor cells from normal BALB/c and mice receiving MIG210-transformed SIRT1+/+ cells were enriched by EasySep to remove lineage cells and then sorted for GFP and CD150 expression. Bold numbers indicated the sorted fractions that were used for analysis of SIRT1 protein expression. GFP+CD150+ (fraction 3) cells were also loaded with fractions 1 and 2 from BALB/c mice for comparison. (F) SIRT1 mRNA levels in normal BALB/c BM and LSK cells compared with LSK cells purified from CML mice.
SIRT1 expression was poor in mouse BM cells but was dramatically up-regulated in those receiving BCR-ABL–transformed cells (Figure 6D). Accordingly, SIRT1 knockout only moderately increased FOXO1 acetylation in BM progenitor cells (supplemental Figure 12). Interestingly, SIRT1 activation by BCR-ABL occurred selectively in lineage-depleted and transformed GFP+CD150+ cells (fraction 3 in Figure 6E), but not in GFP+CD150− cells (fraction 6 in Figure 6E). Although most lineage cells were depleted by EasySep, both the GFP+CD150+ and the GFP+CD150− compartments may still have had some cells expressing low lineage markers (Linlo), and therefore they were mixed populations containing progenitor and committed precursor cells. It is unclear why SIRT1 was not activated in the GFP+CD150− compartment. GFP+CD150−Linlo/− cells could be derived from BCR-ABL transduction of CD150−Linlo/− cells directly or by differentiation of GFP+CD150+Linlo/− cells. It is possible that CD150−Linlo/− cells are nonpermissive for SIRT1 activation by BCR-ABL or that SIRT1 expression is lost after differentiation.
CML progenitor cells in C57BL/6 mice are enriched in the same cell pool as normal progenitor cells identified by Lin−Sca1+cKit+ (LSK); however, the identity of CML progenitor cells in the BALB/c strain is not well characterized. LSK cells in BALB/c mice contain 25% BM-repopulating progenitors.41 To examine SIRT1 expression in these progenitor cells, LSK cells from CML mice were sorted (supplemental Figure 13) and SIRT1 RNA was also found to be significantly elevated in LSK cells (Figure 6F). Therefore, our data indicate that SIRT1 is activated by BCR-ABL in BALB/c mouse progenitor cells within CD150+ and LSK pools, which is consistent with its activation in human CD34+ cells. However, the increased SIRT1 response to oncogenic transformation is not universal. As shown in supplemental Figure 14, the SIRT1 level was also increased in mouse BM cells on KRAS expression, but not in KRAS-transformed NIH-3T3 cells, suggesting a certain specificity of the SIRT1 response in hematopoietic cells.
Combination of SIRT1 inhibition with imatinib for treatment of mouse CML-like disease
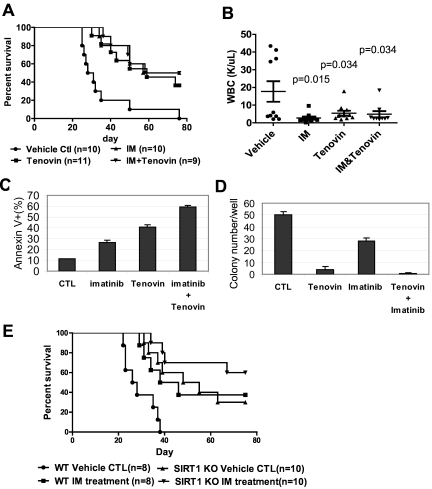
The present study also investigated whether treatment of CML mice with a SIRT1 inhibitor would deter the disease. Tenovin-6, administrated at 50 mg/kg/d, as described previously,30 significantly reduced WBC counts and extended the survival of CML mice (P = .0002, Figure 7A-B). No obvious hematologic toxicity or body weight loss was detected with tenovin-6 alone or in combination with imatinib (supplemental Figure 15). However, there was no increased survival advantage conferred by combining tenovin-6 with imatinib compared with individual drug treatment. To determine whether this result is caused by species difference, BM cells from CML mice were isolated and analyzed for their sensitivity to both drugs in vitro. Tenovin-6 did sensitize mouse cells to imatinib-induced apoptosis and further suppressed CFC formation (Figure 7C-D). Therefore, it is possible that metabolism of these drugs may cause unfavorable interaction in vivo or that the doses of 2 drugs may not be optimal for in vivo additive effects in this model. Alternatively, specificity of tenovin-6 may play a role because it also inhibits SIRT2.30
Figure 7.
Combination of SIRT1 inhibition with imatinib for treatment of mouse CML. (A-B) Survival curves (A) and total blood leukocyte counts (B) for CML mice treated with drugs. Ten days after receiving 1 × 105 BM cells transduced by MIG210, mice were treated by vehicle, imatinib (200 mg/kg/d), tenovin-6 (50 mg/kg/d), or combination for 10 days. Total blood leukocyte counts were analyzed at day 20 after transplantation. (C) BM cells from CML mice were cultured with 2.5μM imatinib, 1μM tenovin-6, or combination for 3 days, and apoptosis was analyzed for GFP+ cell fraction. DMSO was used as reagent control (CTL). (D) After treating GFP-sorted BM cells from CML mice with imatinib, tenovin-6, or combination for 24 hours, cells were washed and plated in methylcellulose medium without drugs for CFC assay. (E) Effect of combination of SIRT1 knockout and imatinib on CML mouse survival. Wild-type or SIRT1−/− donor BM cells were transduced with MIG210 and 2 × 105 cells were transplanted for each recipient. Imatinib or vehicle was given as described in panel A.
Finally, it was examined whether SIRT1 knockout may improve imatinib treatment. BCR-ABL–transduced wild-type or SIRT1 knockout BM cells were transplanted, and the recipient mice were treated with vehicle or imatinib. Compared with control mice receiving wild-type cells and vehicle treatment, imatinib extended the survival of CML mice (P = .011) in a manner similar to SIRT1 knockout, whereas SIRT1 knockout plus imatinib treatment provided a further survival advantage for CML mice (P < .0001; Figure 7E). These results indicate that specific SIRT1 inhibition enhances imatinib treatment. These results suggest that activation of SIRT1 is required for efficient BCR-ABL transformation of hematopoietic progenitor cells and that inhibition of SIRT1 suppresses CML disease development.
Discussion
Overexpression of SIRT1 is detected in many cancers, but it is unknown whether SIRT1 activation is required for any tumorigenesis or if it is merely a consequence of tumorigenesis. Our study provides strong evidence that in hematopoietic progenitor cells, SIRT1 activation by BCR-ABL is essential for efficient transformation and CML development. STAT5 appears to be a key BCR-ABL intermediate for SIRT1 transcriptional activation. STAT5 is involved in a wide array of cytokine signaling for stem/progenitor cells, epithelial cells, and the immune system.42 It is also activated in certain forms of acute myelogenous leukemia (AML) and acute lymphoid leukemia37 and may serve as a common signaling pathway for SIRT1 activation in human CD34+ progenitor cells in response to oncogenic and environmental stress. In agreement with this notion, SIRT1 is overexpressed in certain forms of AML,17 which is likely an outcome of transformation by AML oncogenes.
The identification of BCR-ABL activation of the SIRT1 pathway increases our understanding of CML and expands the previously known BCR-ABL–regulated molecular network that controls CML cell survival and proliferation.1 Intriguingly, inhibition of BCR-ABL kinase activity only partially reduces SIRT1 expression even with up to 10μM imatinib, which can reduce the tyrosine kinase activity almost completely. Consistently, knockdown of STAT5 or deletion of both STAT5 binding sites only partially reduces SIRT1 promoter activity, and STAT5 knockdown also only partially reduces the expression of endogenous SIRT1. In addition, BCR-ABL knockdown appears to reduce SIRT1 expression more completely than imatinib treatment. These findings suggest the likely existence of additional pathways for SIRT1 activation that are not fully dependent on BCR-ABL kinase activity. It is known that BCR-ABL can promote certain cellular activities independently of tyrosine kinase function, for example, cellular localization and adhesion regulation.43 As a stress response gene, SIRT1 may still be activated in response to those kinase-independent cellular changes. Given that a high degree of SIRT1 knockdown is required for induction of apoptosis, it is possible that the SIRT1-mediated survival pathway remains active even though BCR-ABL kinase activity is suppressed. Therefore, inhibition of SIRT1 can sensitize CML cells to imatinib treatment. Our finding parallels the fact that human CML progenitor cells are resistant to apoptosis even though BCR-ABL kinase activity is effectively blocked.6,7 Consistently, in a separate study, we have found that inhibition of SIRT1 sensitizes human CML progenitor cells to imatinib treatment, inducing apoptosis and suppressing CFC formation (data not shown).
SIRT1 knockout clearly inhibits myeloid expansion of BCR-ABL–transformed cells. However, it is a bit surprising that SIRT1 knockout has little impact on normal hematopoietic stem/progenitor cell functions. This finding could be because of low SIRT1 expression in progenitor cells in association with low cell-cycling status, particularly under nonstress conditions. Accordingly, SIRT1 knockout only moderately increases acetylation of its target FOXO1 in mouse BM progenitor cells, which is in agreement with the previous observation that little change in p53 acetylation occurs in mouse thymocytes after SIRT1 knockout.28 Alternatively, SIRT1−/− mice might have gained a survival advantage independently of SIRT1, given that only a portion of these mice survived. However, such gain of function cannot override BCR-ABL oncogenic stress response, because we show that loss of SIRT1 suppresses CML development.
The results of the present study improve our understanding of the roles of SIRT1 in tumorigenesis and suggest a rationale for targeting SIRT1 to overcome CML resistance to BCR-ABL inhibitors. However, caution is needed in using such an approach in light of recent studies showing potential tumor suppressor functions of SIRT1. First, in their effort to mimic effects of calorie restriction, Oberdoerffer et al showed that SIRT1 overexpression or resveratrol treatment reduces thymic lymphoma in the p53+/− background.20 Intriguingly, however, the incidence of adenocarcinoma, leukemia, and sarcoma increases in SIRT1-overexpressing or resveratrol-treated mice.20 In contrast, calorie restriction delays tumor formation but does not change tumor spectrum and incidence in p53+/− mice.44 Second, Wang et al demonstrated that heterozygous loss of SIRT1 drastically accelerates tumorigenesis in the p53+/− background.21 Surprisingly, in that study, the control p53+/− mice developed few tumors over 20 months of observation, which is in sharp contrast to numerous studies showing that p53+/− mice are highly susceptible to tumorigenesis within a similar time frame.44–47 Wang et al also suggested that SIRT1 is a haploinsufficient tumor suppressor.21 However, no notable tumor formation has been reported in 2 aging studies using SIRT1−/− mice over 18-24 months48,49 and SIRT1 homozygous knockout does not increase intestinal polyp load in Apcmin/+ mice.50 Given the results of the present study, it is likely that tissue types may influence SIRT1 functions in cancer. Nevertheless, our study reveals SIRT1 activation as a novel mechanism for CML cell proliferation and survival, and therefore targeting SIRT1 may be valuable in CML treatment to overcome resistance.
Supplementary Material
Acknowledgments
The authors thank Frederick Alt for SIRT1 knockout mice, Warren Pear for the mouse BM transduction/transplantation protocol, Toren Finkel for the SIRT1 promoter constructs, Bing Wang for technical assistance, and Kenichi Yakushijin and Fumiko Yakushijin at the City of Hope Biopolymer and Synthetic Chemistry Core for synthesis of tenovin-6 and sirtinol for in vivo study.
This study was supported by a career development award from the STOPCANCER (to W.Y.C.) Foundation; by grant W81XWH-06-1-0268 from the US Department of Defense (to W.Y.C.); by the National Institutes of Health (grants R01 CA143421 to W.Y.C. and R01 CA95684 to R.B.); and by a translational research grant from the V-Foundation (to W.Y.C. and R.B.). The core facilities used in this study were supported by NCl grant P30 CA033572.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.Y., Z.W., L.L., H.Z., and H.M. designed and carried out the experiments; D.H. synthesized the SIRT1 inhibitors; and R.B. and W.Y.C. conceived and designed the studies and wrote the manuscript.
Conflict-of-interest disclosure: This manuscript is a part of a patent application filed by City of Hope.
The current affiliation for H.Y. is Department of Stem Cell and Regenerative Medicine, Beijing Institute of Transfusion Medicine, Beijing, China.
Correspondence: WenYong Chen, PhD, Department of Cancer Biology, Beckman Research Institute/City of Hope, 1500 E Duarte Rd, Duarte, CA 91010; e-mail: wechen@coh.org; or Ravi Bhatia, MD, Division of Hematopoietic Stem Cell and Leukemia Research, City of Hope, 1500 E Duarte Rd, Duarte, CA 91010; e-mail: rbhatia@coh.org.
References
- 1.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 2.Deininger MW, Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol Rev. 2003;55(3):401–423. doi: 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- 3.Graham SM, Jorgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 4.Holtz MS, Slovak ML, Zhang F, Sawyers CL, Forman SJ, Bhatia R. Imatinib mesylate (STI571) inhibits growth of primitive malignant progenitors in chronic myelogenous leukemia through reversal of abnormally increased proliferation. Blood. 2002;99(10):3792–3800. doi: 10.1182/blood.v99.10.3792. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia R, Holtz M, Niu N, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101(12):4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- 6.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121(1):396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konig H, Holyoake TL, Bhatia R. Effective and selective inhibition of chronic myeloid leukemia primitive hematopoietic progenitors by the dual Src/Abl kinase inhibitor SKI-606. Blood. 2008;111(4):2329–2338. doi: 10.1182/blood-2007-05-092056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8(11):1018–1029. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 9.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 10.Luo J, Nikolaev AY, Imai S, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 11.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 12.Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 13.Motta MC, Divecha N, Lemieux M, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 14.Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 15.Daitoku H, Hatta M, Matsuzaki H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101(27):10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffman DM, Grizzle WE, Bamman MM, et al. SIRT1 Ii significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67(14):6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 17.Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19(10):1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 18.Jang KY, Hwang SH, Kwon KS, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am J Surg Pathol. 2008;32(10):1523–1531. doi: 10.1097/PAS.0b013e31816b6478. [DOI] [PubMed] [Google Scholar]
- 19.Nosho K, Shima K, Irahara N, et al. SIRT1 histone deacetylase expression is associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2009;22(7):922–932. doi: 10.1038/modpathol.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberdoerffer P, Michan S, McVay M, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang RH, Sengupta K, Li C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issa JP, Zehnbauer BA, Kaufmann SH, Biel MA, Baylin SB. HIC1 hypermethylation is a late event in hematopoietic neoplasms. Cancer Res. 1997;57(9):1678–1681. [PubMed] [Google Scholar]
- 23.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30(8):907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 25.Ramaraj P, Singh H, Niu N, et al. Effect of mutational inactivation of tyrosine kinase activity on BCR/ABL-induced abnormalities in cell growth and adhesion in human hematopoietic progenitors. Cancer Res. 2004;64(15):5322–5331. doi: 10.1158/0008-5472.CAN-03-3656. [DOI] [PubMed] [Google Scholar]
- 26.Yuan H, Wang Z, Gao C, et al. BCR-ABL gene expression is required for its mutations in a novel KCL-22 cell culture model for acquired resistance of chronic myelogenous leukemia. J Biol Chem. 2010;285(7):5085–5096. doi: 10.1074/jbc.M109.039206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306(5704):2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 28.Cheng HL, Mostoslavsky R, Saito S, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100(19):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92(10):3780–3792. [PubMed] [Google Scholar]
- 30.Lain S, Hollick JJ, Campbell J, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13(5):454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 32.Shen H, Boyer M, Cheng T. Flow cytometry-based cell cycle measurement of mouse hematopoietic stem and progenitor cells. In: Bunting K, editor. Hematopoietic stem cell protocols. Vol 430. Totowa, NS: Springer; 2008. pp. 77–86. [DOI] [PubMed] [Google Scholar]
- 33.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7(19):3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 34.Abdelmohsen K, Pullmann R, Jr, Lal A, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185(2):203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, Chen L, Hou X, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8(9):1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 37.Van Etten RA. Aberrant cytokine signaling in leukemia. Oncogene. 2007;26(47):6738–6749. doi: 10.1038/sj.onc.1210758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gesbert F, Griffin JD. Bcr/Abl activates transcription of the Bcl-X gene through STAT5. Blood. 2000;96(6):2269–2276. [PubMed] [Google Scholar]
- 39.Slupianek A, Schmutte C, Tombline G, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8(4):795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 40.Narala SR, Allsopp RC, Wells TB, et al. SIRT1 acts as a nutrient-sensitive growth suppressor and its loss is associated with increased AMPK and telomerase activity. Mol Biol Cell. 2008;19(3):1210–1219. doi: 10.1091/mbc.E07-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spangrude GJ, Brooks DM. Mouse strain variability in the expression of the hematopoietic stem cell antigen Ly-6A/E by bone marrow cells. Blood. 1993;82(11):3327–3332. [PubMed] [Google Scholar]
- 42.Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22(6):711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wertheim JA, Forsythe K, Druker BJ, Hammer D, Boettiger D, Pear WS. BCR-ABL-induced adhesion defects are tyrosine kinase-independent. Blood. 2002;99(11):4122–4130. doi: 10.1182/blood.v99.11.4122. [DOI] [PubMed] [Google Scholar]
- 44.Berrigan D, Perkins SN, Haines DC, Hursting SD. Adult-onset calorie restriction and fasting delay spontaneous tumorigenesis in p53-deficient mice. Carcinogenesis. 2002;23(5):817–822. doi: 10.1093/carcin/23.5.817. [DOI] [PubMed] [Google Scholar]
- 45.Chen WY, Cooper TK, Zahnow CA, et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 2004;6(4):387–398. doi: 10.1016/j.ccr.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 46.Venkatachalam S, Shi YP, Jones SN, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17(16):4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harvey M, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5(3):225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 48.Boily G, Seifert EL, Bevilacqua L, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3(3):e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8(1):38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boily G, He XH, Pearce B, Jardine K, McBurney MW. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009;28(32):2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.