Abstract
The self-assembly of the predominant extracellular enamel matrix protein amelogenin plays an essential role in regulating the growth and organization of enamel mineral during early stages of dental enamel formation. The present study describes the effect of the phosphorylation of a single site on the full-length native porcine amelogenin P173 on self-assembly and on the regulation of spontaneous calcium phosphate formation in vitro. Studies were also conducted using recombinant non-phosphorylated (rP172) porcine amelogenin, along with the most abundant amelogenin cleavage product (P148) and its recombinant form (rP147). Amelogenin self-assembly was assessed using dynamic light scattering (DLS) and transmission electron microscopy (TEM). Using these approaches, we have shown that self-assembly of each amelogenin is very sensitive to pH and appears to be affected by both hydrophilic and hydrophobic interactions. Furthermore, our results suggest that the phosphorylation of the full-length porcine amelogenin P173 has a small but potentially important effect on its higher-order self-assembly into chain-like structures under physiological conditions of pH, temperature, and ionic strength. Although phosphorylation has a subtle effect on the higher-order assembly of full-length amelogenin, native phosphorylated P173 was found to stabilize amorphous calcium phosphate for extended periods of time, in sharp contrast to previous findings using non-phosphorylated rP172. The biological relevance of these findings is discussed.
Keywords: Amelogenin, Biomineralization, Calcium Phosphates, Dynamic light scattering, Enamel, Self-assembly
Introduction
Amelogenin has been shown to play an essential role in the formation of the hierarchical and intricate microstructure of dental enamel. Enamel is uniquely composed of extremely long and narrow mineral crystals packed into parallel arrays, called enamel rods, which are arranged in organized interwoven patterns. Amelogenin is the predominant component of the extracellular matrix produced by the ameloblast cell layer during enamel formation. The critical role of amelogenin in regulating enamel structure has been demonstrated through studies of the amelogenin-null mouse that exhibits a dramatic enamel phenotype with a thin layer of enamel being formed and a loss of the noted enamel rod pattern [1]. Single point mutations in the amelogenin gene have also been associated with families with amelogenesis imperfecta that display similarly malformed enamel.
In the initial secretory stage of the enamel mineral formation process, amelogenesis, long thin ribbons of mineral begin to form almost immediately as the enamel matrix is being laid down by the ameloblasts [2–4]. The initially formed mineral ribbons that occupy only 10–20% of the matrix volume are organized in parallel arrays that reflect the final organized structure of mature enamel. Although full-length amelogenin undergoes selective proteolytic processing soon after secretion, the intact full-length molecule has been shown to be exclusively located in the region of newly formed enamel mineral [5]. During the final maturation stage of amelogenesis, the organic matrix is removed almost completely by additional proteases thus allowing the mineral ribbons to grow dramatically in width and in thickness, resulting in an enamel tissue that is >95% mineral, containing only 1–2% organic matter and a small amount of water. The transient nature of the organic matrix is not common in biomineralization processes [6] and reflects an unique feature of the amelogenesis process.
A number of studies have been carried out to elucidate the role of amelogenin in enamel formation. Amelogenin and its cleavage products, which constitute 90 wt% of the extracellular enamel matrix [7,8], are characterized by a high content of proline, glutamine and histidine that comprise together about 50% of all amino acids in these molecules [9,10]. In addition, there are three discernable domains in the amelogenin molecule: a tyrosine-rich N-terminal domain of 45 amino acids, a large hydrophobic central region comprised primarily of X-Y-P repeat motives (P, proline; X and Y are mostly glutamine), and a hydrophilic C-terminus of 11 amino acids that is highly conserved among different vertebrate species [11] (Fig. 1). During enamel development, amelogenin proteolysis proceeds from the C-terminus, resulting in the loss of the hydrophilic portion of the molecule.
Figure 1.
Aligned amino acids sequences of recombinant and native porcine amelogenins used in the present study. Native proteins are phosphorylated at S-16, unlike the recombinant counterparts. Recombinant proteins also lack the N-terminal methionine. P148 and rP147 represent native and recombinant protein cleavage products, respectively, which lack 25 C-terminal amino acids, including an 11 amino acidic hydrophilic domain, as indicated.
Results from both in vitro and in vivo studies suggest that proper enamel formation is related to the capacity of amelogenin to form higher-order structures through self-assembly (for reviews, see Fincham et al., 1999; Margolis et al., 2006) [8,12]. Early in vitro studies using atomic force microscopy (AFM) and dynamic light scattering (DLS) showed that full-length amelogenin has the capacity to form spherical nanoparticles, called nanospheres, with a size range from 16 nm – 70 nm in diameter [13–16]. Based, in part, on the detection of chains of similarly sized nanometer spheres found in vivo by TEM examination of sections of forming enamel [14,17–19] it was suggested that such nanospheres further aggregate into higher-order structures that ultimately guide crystal growth and organization [20–22]. Using a combination of DLS and small angle X-ray scattering (SAXS) [23,23] data were subsequently obtained that suggest that ‘nanospheres’ of the full-length amelogenin have the ability to assemble further to form extended chain-like structures in solution. These findings are in agreement with other in vitro data that showed that amelogenin has the capacity to form chain-like structures under specified conditions [24–28]. Combined DLS and SAXS data also suggest that full-length amelogenin nanospheres consists of a dense hydrophobic core surrounded by a less-dense and hydrophilic shell comprised of the charged C-terminal domain of amelogenin, and that the charged C-terminal domain regulates the aggregation of full-length amelogenin nanoparticles [23], consistent with a recently proposed model [29].
The ability of full-length amelogenins to form higher-order assemblies may be related to their ability to guide the growth and ordered assembly of needle-like apatitic crystals. In vitro experiments have shown that amelogenins can regulate the shape of growing calcium phosphate crystals [24,30–32]. In addition, we have shown that full-length recombinant amelogenins can regulate the formation of bundles of parallel aligned apatitic crystals in vitro, similar to those seen in developing enamel, through a cooperative mechanism of simultaneous protein assembly and spontaneous crystal growth [12,24,33]. In vitro findings have also shown that full-length recombinant porcine amelogenin can transiently stabilize initially formed nanoparticles of amorphous calcium phosphate (ACP) [33], the first mineral phase found in forming enamel [34]. In the presence of the full-length porcine amelogenin, these nanoparticles were found to align to form linear needle-like particles that subsequently transformed and organized into parallel arrays of apatitic needle-like crystals. These phenomena, however, were not observed when a recombinant cleavage product that lacked 25 C-terminal amino acids (including the hydrophilic domain) was used, that instead exhibited almost no effect on calcium phosphate growth. In sharp contrast to these findings, however, when the native version of this cleavage product phosphorylated at the single serine-16 (S-16) site was used, initially formed ACP nanoparticles were stabilized for more than 24 hours and did not transform to crystalline material in the presence of the phosphorylated protein. Hence, amelogenin phosphorylation may play a critical role in the mechanism of mineralization during enamel mineral formation. The effects of phosphorylation on the self-assembly of native full-length amelogenin and its regulation of mineral formation in vitro, however, have not been elucidated.
The present study was carried out using native and recombinant porcine amelogenins to determine the effect of phosphorylation on amelogenin self-assembly in vitro, under relevant physiological conditions of temperature, pH and ionic strength. DLS and transmission electron microscopy (TEM) approaches were used, as in previous studies from our laboratory [28]. Because of its importance to self-assembly and mineralization, the role of the hydrophilic C-terminus was also investigated. In addition, the effect of phosphorylation of full-length porcine amelogenin on spontaneous calcium phosphate mineral formation in vitro was investigated. These studies were designed to provide a better understanding of the mechanisms that control the formation of the exquisite tooth enamel structure and to potentially aid the development of biomimetic approaches for mineralized tissue regeneration.
Material and Methods
Preparation of Porcine Amelogenins
Full-length recombinant and native porcine amelogenins were utilized in the present study, along with selected cleavage products. Native full-length porcine amelogenin P173 and its predominant cleavage product P148 were isolated and purified from developing tooth buds, as previously described [35,36]. Recombinant (r) analogs of these proteins, rP172 and rP147, were produced and purified as described in prior reports [37,38]. The recombinant proteins, derived from bacteria, lack a single phosphate group at serine-16 (S-16) and an N-terminal methionine that are both present in the native full-length amelogenin (P173) from pig (Fig. 1). P148 and rP147 differ from the full-length recombinant (rP172) by lacking 25 C-terminal amino acids, including the hydrophilic C-terminal domain, as indicated. Unlike rP172 and rP147, P173 and P148 have a single phosphate group at S-16 (Fig. 1).
Preparation of protein stock solutions
Protein stock solutions of 5 mg/mL were prepared by dissolving lyophilized proteins in filtered (0.2 µm), ice-cold, distilled and de-ionized water (DDW). Stock solutions had initial pH values below 4 and were stored for at least 24 hours at 4°C to ensure complete dissolution. Complete dissolution of proteins samples was confirmed using dynamic light scattering (DLS), as described below, that showed particle sizes that were between 1 nm and 2 nm, consistent with the presence of monomers or dimers [28]. Just prior to use in self-assembly and mineralization studies, protein stock solutions were centrifuged (10,900 × g, Eppendorf Centrifuge 5403) at 4°C for 20 minutes, to remove dust and particulate matter.
Dynamic light scattering (DLS) studies
Using the described stock solutions, 2 mg/mL protein solutions were prepared with different concentrations of pH-adjusted Tris-HCl buffer (Sigma Aldrich) as a means to adjusting the ionic strength (IS) of the solution. More specifically, Tris-HCl buffers adjusted to pH 7.2 at either 25°C or 37°C were prepared to yield IS values of 163 mM, 64 mM and 15 mM (www.liv.ac.uk/buffers/buffercalc.html). An IS of 163 mM was chosen for study since it corresponds closely to the IS found for the enamel fluid phase of secretory enamel [39]. DDW, as well as all buffer stock solutions, were filtered (Whatman Syringe filter 0.2µm) before use.
DLS measurements were performed on a DynaPro MSXTC/ 12 instrument (Wyatt), equipped with a 825.2 nm gallium-arsenide diode laser (DynaPro-99-E-50) of programmable power. The instrument has a temperature-controlled sample holder (precision of 0.1°C) for a quartz cuvette of 12 µL, which was used in all experiments. Scattering data were collected at an angle of θ = 90° and processed using the software program DYNAMICS V6, version 6.3.40. A proprietary non-negative least square algorithm and cumulants were used in the calculation of mean particle size values, percent polydispersity, the baseline value, and the sum of squares. A regularization algorithm and a model for isotropic spheres was used to resolve for up to five multimodal populations and the percent polydispersity, normalized to the mean size of the peak and the hydrodynamic radius (RH).
A 12 µL aliquot of protein solution prepared on ice was transferred to the quartz cell maintained at 4°C. The quartz cell, closed with a plastic stopper, was then transferred to the DLS instrument and sealed with parafilm. Measurements started at 20 minutes after inserting the quartz cell into the temperature-controlled chamber and proceeded through a series of temperature steps, in three-degree intervals to 43°C, or until the scattering intensity went off scale, with an equilibration time of 5 minutes after each temperature increase. Four measurements, each consisting of 20 acquisitions of a 5-second duration, were performed at the same temperature in 5-minute intervals. Consequently, each sample was kept at any given temperature for slightly more than 20 minutes. We found this approach useful to monitor potential particle size changes during data collection at each temperature. This approach takes advantage of the large temperature coefficient of Tris-HCl (−0.031ΔpH/°C) to study protein assembly over a pH range between pH 8.3 and pH 7.0 by varying the temperature during the course of the experiment [28].
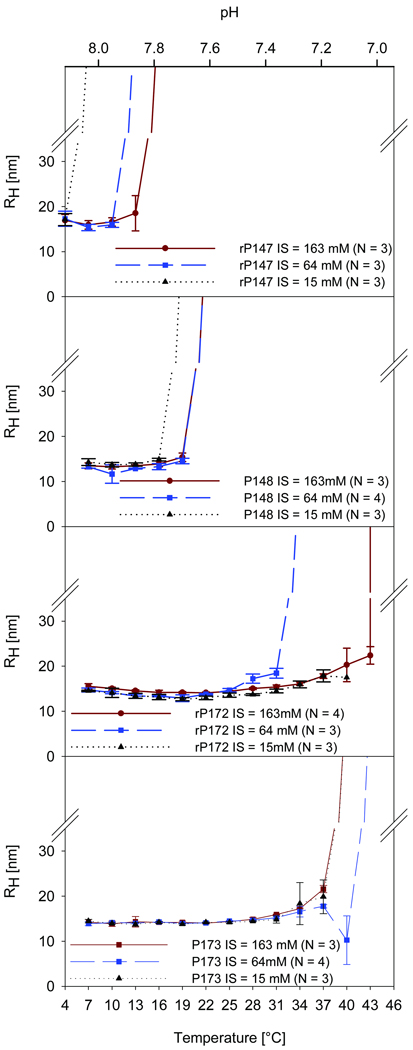
The pH at which off scale scattering occurred (Table 2) was inferred for each DLS experiment from the known relationship between temperature and pH. Reported mean values for this pH, are based on repeat experiments (N ≥ 3), taking the temperature step in the DLS experiment (and the corresponding pH value) where the scattering goes off scale in the majority of samples. It should be noted that in most experiments this occurs at the same temperature/pH in each repeat, and if any variability is observed, it is one temperature step, that is 0.08 pH units. Excellent agreement between expected and measured pH values was obtained for samples of amelogenin with concentrations of 2 mg/mL and Tris-HCl buffer concentrations at 80mM (IS = 64 mM) and 200 mM (IS = 163 mM), consistent with our previous findings using 80 mM Tris-HCl [28]. However, samples prepared with 20 mM Tris-HCl could not be studied precisely using this DLS approach, since we found that the pH of this buffer did not change in a completely predictable fashion as the temperature of the sample was increased. Measured pH values were found to be about 0.1 – 0.2 pH units below the expected pH values for this buffer concentration. Therefore, we report here the particle sizes at IS = 15mM at pH values above the point where off-scale DLS scattering was observed (Table 1) and provide an estimate of this transition pH value at IS = 15 mM for each protein (Table 2).
Table 2.
Summary of pH values where off-scale scattering begins, indicating the onset of very large assembly formation.
| Protein | rP147 | P148 | rP172 | P173 |
|---|---|---|---|---|
| IS = 15 mM | pH > 8.0 | pH > 7.7 (3) | pH < 7.2 (3) | pH 7.0 – 7.2 (3) |
| IS = 64 mM | pH 7.8 (3) | pH 7.6 (4) | pH 7.3 (3) | pH 7.0 (4) |
| IS = 163 mM | pH 7.7 (3) | pH 7.6 (3) | pH 7.0 (4) | pH 7.1 (3) |
(N) = number of experiments
Table 1.
Summary of particle sizes determined by DLS, reported as the mean hydrodynamic radius RH [nm] at pH values prior to off-scale scattering.
| Protein | rP147 | P148 | rP172 | P173 |
|---|---|---|---|---|
| IS = 15 mM | 17.1 ± 1.4 (3) | 14.1 ± 0.5 (3) | 14.5 ± 1.7 (3) | 14.6 ± 1.4 (4) |
| IS = 64 mM | 16.2 ± 1.0 (3) | 13.1 ± 1.0 (4) | 14.7 ± 1.9 (3) | 14.4 ± 1.8 (4) |
| IS = 163 mM | 17.0 ± 1.1 (3) | 13.9 ± 0.9 (3) | 16.1 ± 2.6 (4) | 15.4 ± 2.3 (3) |
(N) = number of experiments
TEM analyses of protein assemblies
Protein solutions of 2 mg/mL were prepared as described above with the desired IS regulated by Tris-HCl buffer adjusted to pH 7.2 at 37°C. Samples of P173 at pH 7.0 at 37°C were also prepared in MES buffer. Sample aliquots of 6µL were pipetted onto carbon coated Ni-grids #400 (EMS, Hartfield, PA) and incubated for 40 minutes at 37°C in a humidity chamber. Samples were then blotted on Grade 1 Whatman filter paper (Cat. No 10001-329), rinsed in filtered DDW or 20mM Tris-HCl adjusted to the desired pH 7.2 at room temperature, blotted again, and then air-dried for several minutes. For negative staining, 1% filtered phospho-tungstic acid (pH 7.2) was applied for 30 seconds. The samples were then blotted again and air-dried. Grids were prepared in duplicate. TEM analyses were carried out using a JEOL 1200 TEM microscope at 100 KV, in bright field. Images were captured using an AMT CCD camera (AMT, Danvers, MA). Image analyses (i.e., particle size measurements) were subsequently performed using ImageJ 1.64 (NIH, Bethesda, MD, http://rsb.info.nih.gov/ij/download.html).
Mineralization studies
Stock solutions of calcium (30 mM) and phosphate (3 mM) were prepared using reagent grade CaCl2.2H2O (Sigma, > 99.0 % pure) and KH2PO4 (Sigma, > 99.0 % pure). The KH2PO4 solution was adjusted to pH 7.4 at 25°C, using a small volume of KOH. With the exception of the centrifuged protein solution, all solutions were filtered (0.22 µm Isopore filters (Millipore)) prior to use. Aliquots of calcium and the pH-adjusted phosphate solution were sequentially added to protein solutions to yield final concentrations of 2.5 mM Ca, 1.5 mM P, and 2 mg/mL protein, with a final volume of 60 µL. The reaction solutions (including protein) have an initial pH value of ~ pH 7.4 upon mixing of all solution components. The samples were then placed in a thermostatic water bath adjusted to 37°C. In order to minimize evaporation, the reaction tube was tightly sealed with a cap or parafilm (America National Can, Chicago, IL). Each experiment was carried out using two identically prepared samples. In one sample, a micro-combination pH electrode (MI-410, Microelectrodes Inc., Bedford, NH) was immersed in the reaction solution to monitor changes in pH as a function of time. The other sample was used for TEM analyses, as described below.
Fourier transform-Infrared (FT-IR) spectroscopy analyses
Following each mineralization experiment, reaction mixtures were concentrated by centrifugation and the removal of 20 µl of the supernatant. A 20 µL aliquot of the concentrated sample was then placed on a KBr sheet (KBr IR Card, International Crystal Labs, Garfield, NJ) and dried overnight in a vacuum desiccator at room temperature. FT-IR spectra (4000 to 450 cm−1) of these samples were then obtained using a Perkin-Elmer, Multiscope FT-IR Microscope. Routinely, 128 scans and 4 cm−1 resolution were used.
TEM analyses of mineral products
Five (5) µL aliquots were taken for TEM analyses at specified times during the mineralization reactions. These aliquots were placed on carbon-coated Cu grids (Electron Microscopy Sciences, Hartfield, PA) for 0.5 – 1 min., blotted vertically against filter paper, quickly rinsed with distilled water, blotted again, and then air-dried. Grids were prepared in duplicate. Images were obtained in bright field and selected area electron diffraction (SAED) modes using a JEOL 1200 TEM operated at 100 KV and captured by an AMT CCD camera (AMT, Danvers, MA). The analysis of micrographs was performed using NIH ImageJ 1.64 software (NIH, Bethesda, MD).
Statistical Methods
Single factor ANOVA tests (alpha 0.01) were used to compare the results derived from TEM analyses and to test for significant differences between the sizes and dimensions of formed assemblies.
Results
Effects of phosphorylation on amelogenin self-assembly
- DLS findings
As shown in Fig. 2, for DLS experiments carried out at ionic strengths of 15 mM, 64 mM, and 163 mM, all proteins studied formed small and similarly sized nanoparticles at higher pH values, with mean hydrodynamic radii (RH) between 13 and 17 nm (Table 1), and exhibited a monomodal particle distribution with polydispersity values consistent with previously published results [40]. Differences between proteins, however, became apparent as the pH is lowered with increasing temperature where, at specific pH values, DLS scattering intensity increased sharply and dramatically beyond the detection limit of the instrument (Fig. 2) indicating further aggregation of all proteins. The pH values where off-scale scattering occurs are clearly higher in samples of rP147 and P148 compared to the full-length molecules (Fig. 2, Table 2). For example, at IS = 64 mM the cleaved proteins rP147 and P148 showed a dramatic change in scattering intensity at pH 7.8 and pH 7.6, respectively, whereas the full-length proteins rP172 and P173 remain as monomodal particle populations with RH values ~15nm until pH values reach pH 7.3 (Fig. 2). Variation in ionic strength from IS = 15 mM to the physiological value of IS = 163mM [39] did not cause significant differences in particle sizes in the pH range prior to the formation of larger assemblies in any of the studied amelogenins. Ionic strength, however, was found to have some effect on the pH at which off-scale scattering begins. This effect is more pronounced in the recombinant proteins compared to the native phosphorylated forms of amelogenin (Fig. 2). A greater IS effect was observed in samples of rP147, compared to P148, with respect to the pH at which off-scale scattering begins. A notable effect of IS is apparent in samples of rP172 (Fig. 2), where a shift of 0.3 pH units is observed for the onset of off-scale scattering from pH 7.0 at IS = 64 mM to pH 7.3 at IS = 163 mM (Fig. 2, Table 2). However, as shown in Table 2, small differences between the pH values at which DLS off-scale scattering occurs were found for the native proteins in comparison to their recombinant analogs (i.e., P148 vs. rP147; P173 vs. rP172) for all IS values.
Figure 2.
DLS results for recombinant (rP147, rP172) and native (P148, P173) porcine amelogenins at a concentration of 2mg/mL in Tris-HCl buffer, illustrating the effect of pH on the hydrodynamic radius (RH) of studied proteins at ionic strengths of IS = 15 mM, 64 mM, and 163 mM. The pH of the sample changes with temperature, as indicated in the x axes, due to the high temperature coefficient of −0.031 ΔpH/°C of Tris-HCl, as described in Materials and methods. Note that higher-order self-assembly, that is the pH/temperature where off scale scattering occurs, appears to be more affected by changes in ionic strength in recombinant amelogenins compared to the native proteins. Differences and similarities in protein behavior are discussed in the text.
- TEM findings under conditions of physiological IS = 163 mM
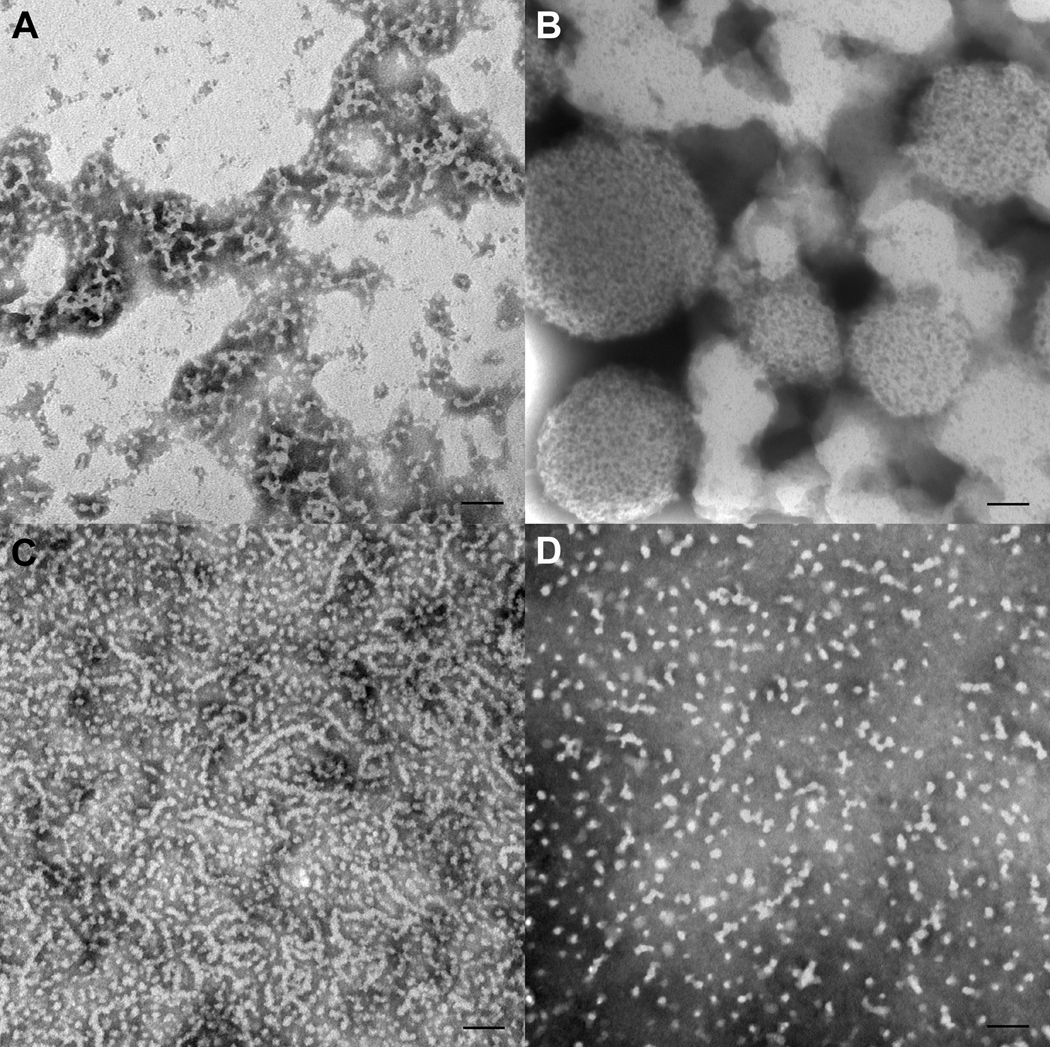
In general, TEM results obtained for samples prepared at selected IS values at pH 7.2 were consistent with observed DLS measurements of samples in solution under the same conditions. At IS = 163 mM, as shown in Fig. 3, very large aggregates were observed using TEM for rP147 and P148, the two amelogenins that exhibited off-scale scattering at pH values well above pH 7.2 (Fig. 2). The more hydrophobic molecules had a tendency to form large agglomerations. It was difficult to get an even and thin distribution of these protein samples on the TEM grids for this reason, especially for samples of P148 (Fig. 3) that formed very large sphere-like particles (200 – 500 nm in dia.), as noted in Table 3. The larger aggregates observed in experiments with P148 were comprised of smaller spheres (~5 nm in dia.) (Fig. 3). Isolated spherical nanoparticles of P148 were not observed. In contrast, in the experiments with rP147, thread-like, interconnected structures (up to ~ 100 nm long) were observed in addition to larger agglomerates (Fig. 3a). These results clearly indicated that, although both rP147 and P148 exhibited a similar tendency to form large agglomerations, the presence of single phosphate had a significant effect on the supramolecular assembly of the cleaved amelogenins studied.
Figure 3.
TEM results for recombinant and native porcine amelogenins at 2mg/mL in Tris-HCl buffer, pH 7.2 and ionic strength of IS = 163 mM. A) rP147, large aggregate and threadlike structures. B) P148 forms large agglomerates. C) rP172, elongated structures of quite variable length, some isolated spherical particles. D) P173, mostly isolated spherical particles and some small elongated structures. Scale bar = 100 nm.
Table 3.
Summary of amelogenin features observed by TEM at pH 7.2, as a function of ionic strength (IS)
| Protein | pH 7.2 IS 15 mM | pH 7.2 IS 15 mM | pH 7.2 IS 64 mM | pH 7.2 IS 64 mM | pH 7.2 IS 163 mM | pH 7.2 IS 163 mM |
|---|---|---|---|---|---|---|
| Feature | Particle Size (nm) |
Feature | Particle Size (nm) |
Feature | Particle Size (nm) |
|
| rP147 | Few isolated spherical particles. | D: 8.2 ± 2.2 (112) | Interconnected spherical particles of varying size | D: 27.4 ± 11.2 (150) | Isolated spherical particles. | D: 8.2 ± 2.0 (150) |
| Irregularly shaped particles of variable size and some elongated, connected structures | D: 45.7 ± 15.9 (150) | Small thread-like structures | L: up to 116.3 | |||
| W: 8.0 ± 1.7 (149) | W: 7.8 ± 1.6 (149) | |||||
| Large aggregates | D: 158.5 ± 100.3 | |||||
| P148 | Large agglomerates | D: 110.1 ± 54.5 (144) | Irregularly shaped large particles | D: 27.5 ± 19.9 (150) | Very large agglomerates | D: 353.4 ± 115.2 (26) |
| Large agglomerates made up of apparently elongated subunits | W: 7.9 ± 1.8 (150) | Large spherical agglomerates, often too thick for imaging | D: from 6.1 to 129.9 | Small spherical particles within large agglomerates | D: 4.8 ± 1.5 (150) | |
| rP172 | Isolated spherical particles | D: 10.2 ± 2.5 (143) | Small spherical aggregates | D: 14.4 ± 4.3 (150) | Isolated spherical particles. | D: 10.0 ± 2.1 (75) |
| Elongated chain-like structures | L: up to 269.9 (47) | Chain-like structures, highly variable in length | L: up to 281.2 (35) | |||
| W: 10.9 ± 1.9 (150) | W: 10.2 ± 2.1 (150) | |||||
| P173 | Isolated spherical particles | D: 17.7 ± 4.4 (150) | Isolated spherical particles | D: 10.5 ± 2.0 (150) | Isolated spherical particles. | D: 10.9 ± 2.5 (150) |
| Elongated chain-like structures | L: up to 333.6 (45) | Elongated chain-like structures | L: up to 266.6 (91) | Some small elongated particles. | W: 10.0 ± 2.2 (150) | |
| W: 16.6 ± 3.5 (140) | W: 11.4 ± 2.6 (150) | L/W: 3.2 ± 1.2 (150) | ||||
| L/W: 5.1 ± 2.3 (138) | L/W: 5.6 ± 2.7 (91) | |||||
D: diameter of spherical particles or large sphere-like agglomerates in nm; W: width of elongated structures in nm; L: length of elongated structure in nm; L/W: mean ratio L to W – unit less; (N): number of measurements. Clearly predominant features and their size are shown in bold.
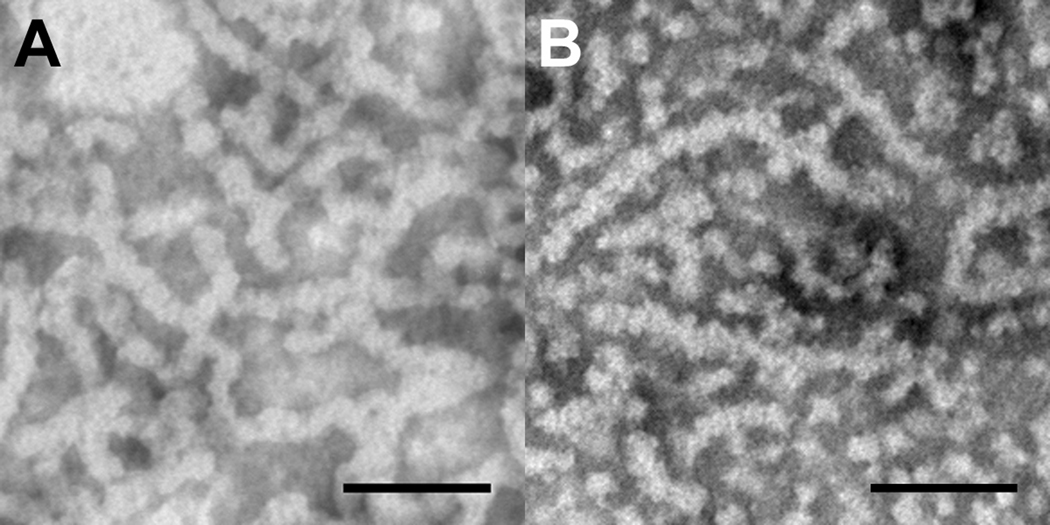
In contrast to the cleaved molecules, the full-length amelogenins that exhibited off-scale scattering below pH 7.2 at IS=163 mM, did not form large aggregates at this pH value. More specifically, P173 appeared at pH 7.2 as either isolated spheres or as very small, elongated aggregates of these spheres (~2–4 particles long), as shown in Fig. 3 and described in Table 3. The full-length amelogenin rP172, however, showed the presence of some very long elongated structures (up to ~ 300 nm long) at pH 7.2, IS=163 mM, along with isolated spherical particles and smaller elongated aggregates (Fig. 3). The elongated rP172 structures were comprised of smaller spherical particles ~ 10 nm in diameter (Table 3). However, as shown in Fig. 4, when P173 was prepared at pH 7.0 (IS=163 mM), instead of pH 7.2, very long chains (up to ~270 nm) of spherical nanoparticles (9.9 ± 2.1 nm in dia.) similar to those seen with rP172 appeared to predominate, although isolated spheres of similar size (9.4 ± 2.6 nm in dia.) were also seen. For comparative purposes, rP172 at pH 7.2 is also shown in Fig. 4 at higher magnification. These TEM data suggest that both full-length molecules form similar elongated chain-like structures although at slightly different pH values.
Figure 4.
TEM micrographs of A) P173 at pH = 7.0 in MES buffer, ionic strength IS = 163mM and B) rP172 at pH = 7.2 in Tris-HCl buffer, ionic strength IS = 163mM. Scale bar = 100nm. Note that both full-length native and recombinant amelogenin form elongated structures made up of smaller apparently spherical subunits.
- TEM findings under conditions of lower IS of 15 and 64 mM
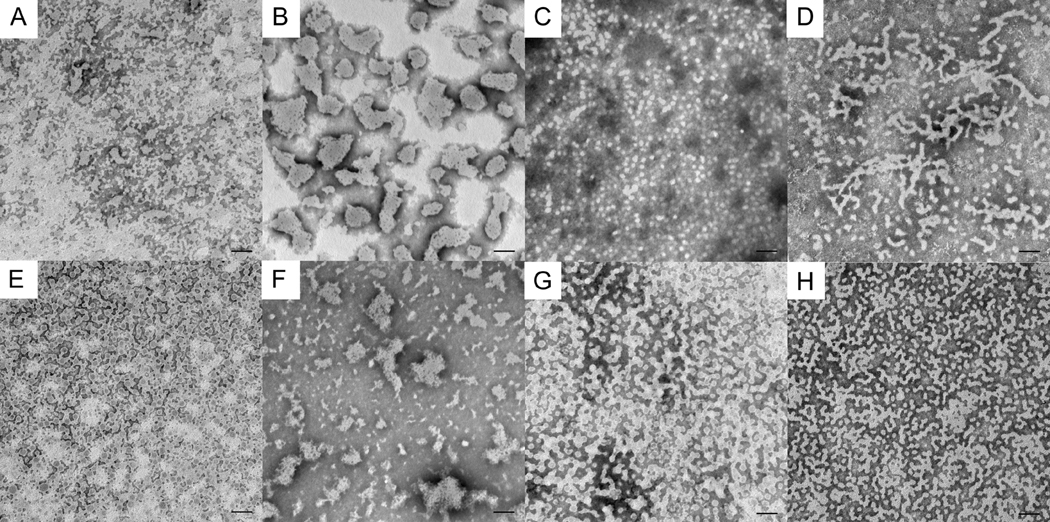
As shown in Fig. 5, at an IS of 64 mM, elongated assemblies of rP172 were also observed at pH 7.2. These structures again extended up to 300 nm in length (Table 3), as was seen for rP172 at higher IS (163 mM), although on average (data not shown) chain-lengths were shorter at IS = 64 mM (Fig. 3). At a lower IS of 15 mM and pH 7.2, rP172 appeared to be present primarily as isolated spheres (Fig. 5). On the other hand, full-length native amelogenin P173 under IS = 64 mM conditions was found to form some elongated structures at pH 7.2, unlike that observed at the higher IS of 163 mM. At yet lower IS of 15 mM, P173 was again found to form elongated structures, unlike what was observed for rP172. As seen at higher IS at pH 7.2 (Fig. 3), P148 tended to form large unstructured aggregates at lower IS of 15 and 64 mM at pH 7.2, that appeared to be made up of elongated sub-units structures (Fig. 5). At both 15 and 64 mM IS at pH 7.2, rP147 exhibited interconnected spherical particles. Like pH, variations in IS had a notable effect on the structural organization of the full-length amelogenins. A summary of TEM data obtained at pH 7.2 as a function of IS values studied is provided in Table 3.
Figure 5.
Comparison between native and recombinant pig amelogenins at 2mg/mL in Tris-HCl buffer, pH = 7.2 and ionic strengths of 15 mM (A – D) and 64 mM (E – H). A) and E) rP147; B) and F) P148; C) and G) rP172; D) and H) P173. Scale bar = 100 nm. Note that rP172 prevails as isolated spherical particles at low ionic strength of 15 mM but forms elongated structures at higher ionic strength at the same pH. The change in ionic strength appears to have the opposite effect in P173, with more and longer elongated features at low ionic strength and more isolated spherical particles at higher ionic strength (see also Fig. 2). Additional details are provided in the text and in Table 3.
Effect of full-length native amelogenin on the spontaneous formation of calcium phosphate
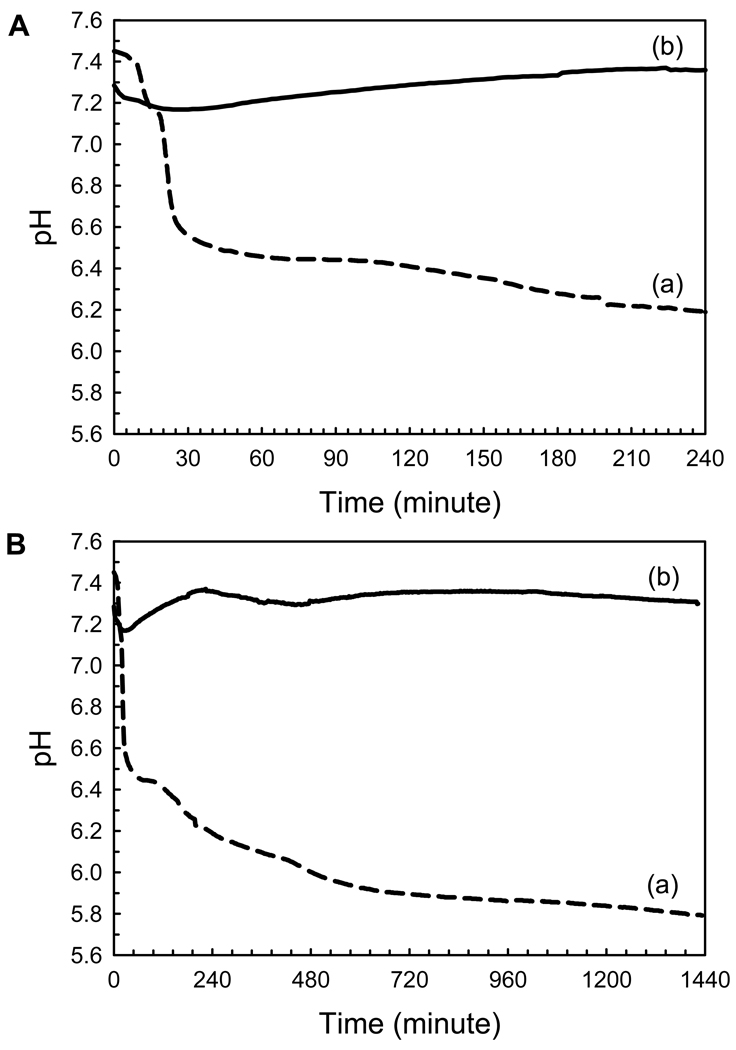
The effect of P173 on the rate of spontaneous calcium phosphate precipitation was monitored via changes in pH. In the absence of protein (control), a decrease in pH was observed to take place in three distinct steps, corresponding to an initial induction period of ~ 15 min associated with the formation of amorphous calcium phosphate (ACP), as shown below, followed by rapid and slower apatitic crystal growth phases (Fig. 6). In sharp contrast to these findings, however, in the presence of P173, relatively little pH change was observed for 1 day (Fig. 6) and even longer (i.e., up to 3 days (data not shown)). At the beginning of the reaction, the pH decreased slightly (ΔpH= ~ 0.2) during the first hour and then slowly increased to almost the initial pH value that remained relatively constant for up to 24 h, as shown. Marked changes in pH as seen in the control were not observed. These results show that P173 has a profound effect on mineralization, as clarified by TEM and FT-IR, as described below. Similar results were obtained for multiple repeats (at least n=3) using 5 different preparations of P173.
Figure 6.
Changes in pH as a function of time observed during mineralization experiments, in the absence (a, control) and presence of P173 (b), monitored over a 24-h period (B). Changes in pH are presented in an expanded scale (A) for better clarity. Similar results were obtained for multiple repeats (at least n=3) using 5 different preparations of P173. The significance of observed differences is discussed in the text.
- TEM and SAED analyses of mineral phases
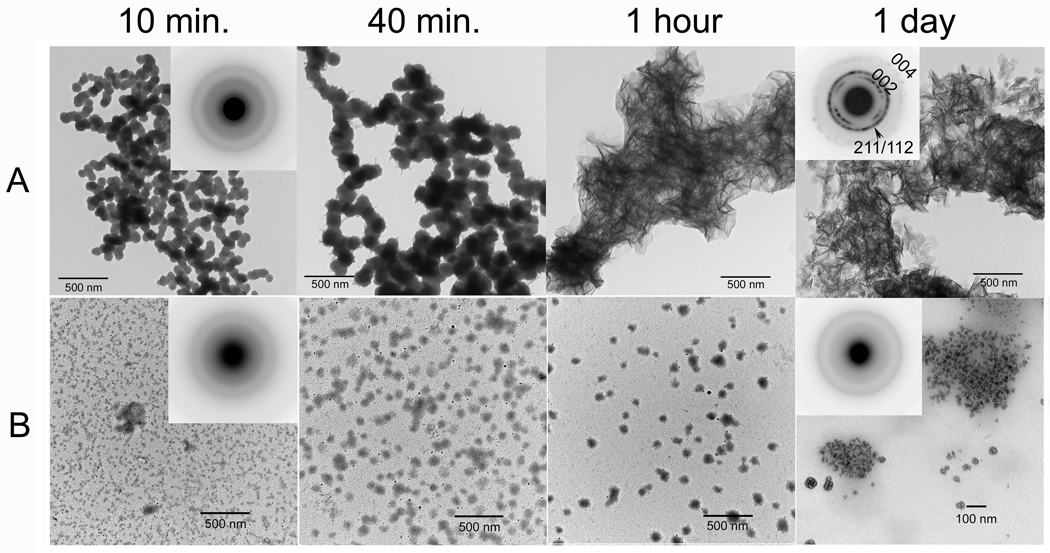
In the absence of amelogenin (control), mono-dispersed spherical particles (d = 75.9 ± 9.8 nm, n = 60) of ACP were observed by TEM within the first 15 min, as confirmed by SAED (Fig. 7A, 15 min, inset). These nanoparticles then transformed to randomly arranged thin plate-like crystals (l = 122.7 ± 25.1 nm, n = 71) after 1 d (Fig. 7A, 1d). SAED analyses (Fig. 7A, 1d, inset) were consistent with the formation of hydroxyapatite (HA) crystals. In marked contrast to the control, mono-dispersed particles and/or networks of ACP were observed by TEM in samples containing P173 at time points from 15 min (d = 9.3 ± 2.0 nm, n= 90) to 1 d (d = 12.4 ± 3.5 nm, n= 103) (Fig. 7B). These particles were much smaller in diameter than ACP particles initially formed in the control. SAED patterns of 1 d samples were characteristic of ACP, consistent with the lack of pH change observed during the mineralization experiments in the presence of P173.
Figure 7.
TEM micrographs of calcium phosphate mineral products formed in the absence (A, control) and presence (B) of P173 examined at selected times (10 min, 40 min, 1 hour, and 1 day), as described in Materials and methods. As shown (A and B) at 10 min, amorphous calcium phosphate (ACP) was initially formed in the control and in the presence of P173, based on the observed (insets) selected area electron diffraction (SAED) patterns. Subsequent changes in mineral particle shape and organization with time are described in the text. As described, in the control ACP particles form networks that appear to undergo transformation within 1 hour. After 1 day, as shown, randomly arranged plate-like apatitic crystals were found in the control (A, inset – showing circular distribution of apatitic reflections). In contrast, ACP was observed in the presence of P173 even after 1 day (B, inset).
- FT-IR analyses of mineral phases formed
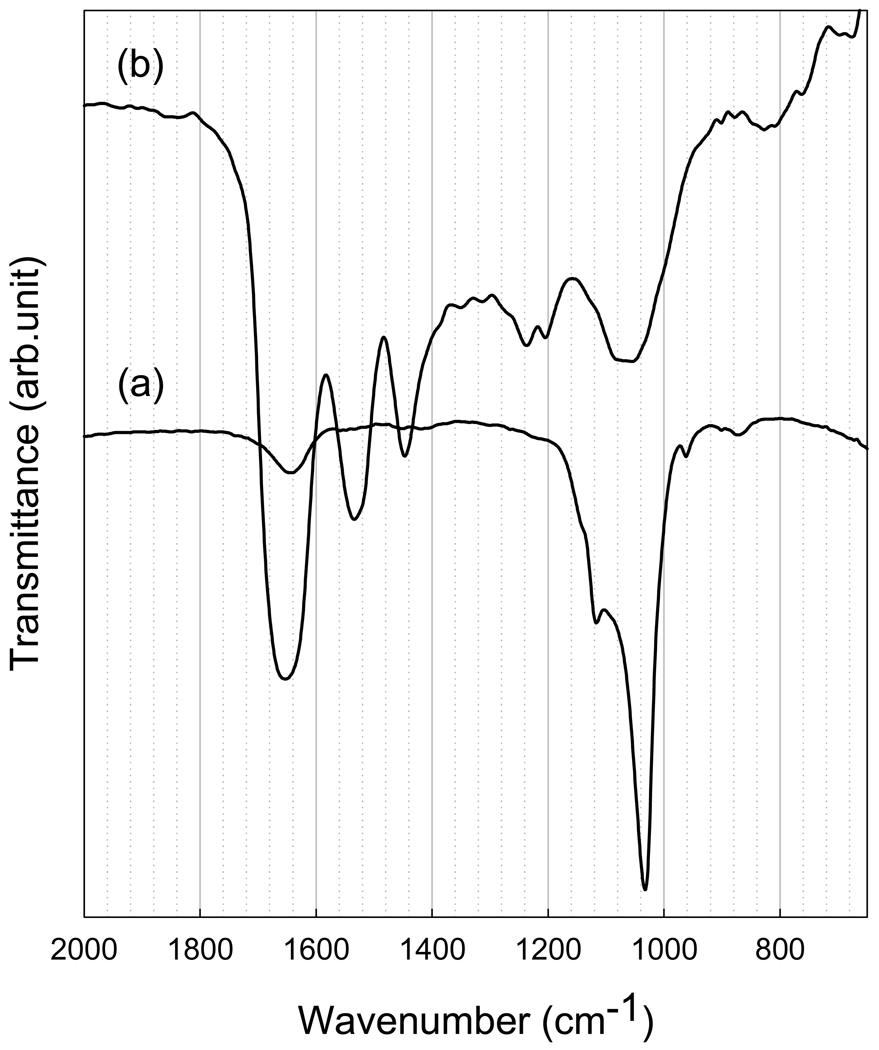
In the absence of protein (Fig. 8(a)), the FT-IR spectra of 1-d samples were found to correspond to a poorly crystalline apatitic phase, with a major absorbance peak at about 1033 cm−1 and a shoulder at 1117 cm−1 arising from asymmetrical ν3 stretching vibrations of PO4 3− [41]. The fact that these bands are not well resolved to reveal additional splitting, reflect the small size of the mineral particles formed and/or suggest that the resulting minerals have low crystallinity. In contrast, in samples prepared in the presence of P173 (Fig. 8(b)), a broad, featureless band was observed at 950 to 1150 cm−1 in the ν3 phosphate region, characteristic of ACP [41]. In samples containing P173, Amide I and Amide II bands in the 1600–1700cm−1 and 1500–1600 cm−1, respectively, were observed, indicating the presence of proteins in the reaction products. The band between 1400 and 1500 cm−1 represents protein side-chain vibrations. A band at 1650 cm−1, seen clearly in the control, is most likely the hydroxyl group from water or HA. FT-IR analyses of the mineralization reaction products are consistent with SAED results obtained for 1-d control samples and samples prepared in the presence of P173 (Fig. 7B), and provide further evidence that P173 is a potent stabilizer of ACP.
Figure 8.
FT-IR spectra of calcium phosphate minerals (after 1 day) produced in the absence (a, control) and presence (b) of P173. FT-IR data for the control are consistent with a poorly crystalline apatitic phase (a), while the spectrum obtained for samples produced in the presence of P173 is characteristic of ACP (b). As discussed in the text, Amide I and Amide II bands can also be seen in the latter spectrum. FT-IR results are consistent with noted SAED findings.
Discussion
The present results show for the first time that the full-length native (phosphorylated) amelogenin P173 has the capacity in vitro (e.g., Fig. 4) under physiological conditions of ionic strength (IS=163 mM) and temperature (37°C) to form higher order chain-like structures that are similar to those observed for its recombinant (non-phosphorylated) analog rP172. However, although amelogenin phosphorylation does not significantly alter the ability of full-length amelogenin to form higher order chain-like structures, our results show that phosphorylation at a single site (S-16) has a small but potentially important effect on the pH at which the onset of higher-order assembly of full-length amelogenin takes place. Based on both DLS (Fig. 2, Table 2) and TEM (Figs 3 and 4) findings, the formation of higher-order amelogenin assemblies from similarly sized spherical nanoparticles (Table 1) is very sensitive to pH. As shown in vitro at pH 7.2 under physiological conditions (Fig. 3c,d), recombinant and native full-length amelogenins have markedly different self-assembly structures. P173 appears primarily as isolated spherical particles, while rP172 predominantly forms very long chain-like structures. At pH 7.0, however, P173 similarly forms elongated chain-like assemblies that are nearly identical in appearance (Fig. 4) and size (Table 3) to rP172 structures formed at pH 7.2 under the same physiological conditions of IS and temperature in vitro. These findings suggest that both the structure and possible biological function of full-length amelogenin are regulated by tightly controlled pH values found in developing rat (pH 7.23) and pig (pH 7.26) enamel during the secretory stage of amelogenesis [4,39,42,43]. In contrast to the full-length amelogenins, the predominant native (phosphorylated) amelogenin cleavage product found during the secretory stage of amelogenesis [35,44], P148, and its recombinant analog rP147, exhibited tendencies to form large aggregates, under the same experimental physiological conditions (Table 3), although rP147 also tended to form smaller interconnected structures, unlike P148. The formation of larger aggregates of these more hydrophobic amelogenins that lack the hydrophilic C-terminus domain takes place at substantially higher pH values of pH 7.6 – 7.7 (Table 3, Fig. 2). These results are consistent with our previously reported findings at IS = 64 mM [28] and again demonstrate that the hydrophilic C-terminal domain of both native and recombinant full-length amelogenin plays a critical role in the formation of higher order chain-like assemblies, as found in other studies using recombinant amelogenins [23,26,27].
Despite the fact that we have found in the present study that P173 and rP172 behave similarly with respect to their ability to form chain-like structures in vitro (e.g., Fig. 4), albeit at somewhat different pH values, the single phosphate group was found to have a dramatic effect on calcium phosphate formation. P173 was found to stabilize ACP for long periods of time and prevent its transformation to HA, unlike that observed using rP172 under the same in vitro conditions [33]. In contrast to P173, rP172 was found to stabilize ACP for relatively short periods of time and subsequently guide ACP transformation and the organization of forming HA crystals. These findings have important biological implications, as they are supported by earlier electron microscopy studies that suggested that ACP is the first mineral phase formed in secretory enamel [45,46]. Consistent with these latter reports, a recent study using multiple characterization techniques has conclusively demonstrated that ACP is the first enamel mineral formed during the secretory stage of amelogenesis that subsequently transforms into apatitic crystals [34]. Importantly, ACP mineral particles were found to be organized in parallel arrays in newly deposited enamel that were similar in morphology and organization to regions of older enamel that were comprised of mineral crystals. These observations indicate that mineral morphology and organization in developing enamel are established by the organic matrix prior to its crystallization, consistent with our in vitro findings using rP172 [33]. We hypothesize that the capacity of rP172 to organize the arrangement of growing crystals in vitro is related to its ability to form higher order chain-like structures. Furthermore, in our recent study on in vitro collagen fibril mineralization in the presence of amelogenin, the first mineral phase formed was ACP that was again transiently stabilized by amelogenin [47]. Strong evidence has also been obtained to show that transient mineral phases, including ACP, are involved in bone formation [48–50].
Based on the results of the present study showing that the full-length native amelogenin P173 has the capacity to form higher-order chain-like structures similar to those formed by recombinant porcine full-length molecule [13,23,24,28,32] that can guide the formation of organized arrays of apatitic crystals via ACP nanoparticle precursors [33], it is likely that the native full-length protein plays a similar role in vivo. This notion is further supported by the fact that full length amelogenin and organized arrays of ACP particles are both present only in the youngest layers of enamel in a close proximity to ameloblasts [34] and the decrease in the full length amelogenin in the matrix due to its proteolytic cleavage [51,52] coincides with ACP phase transformation in the older layers of enamel [34]. Along these lines, it has been proposed that the linear assemblies of spherical structures found in developing enamel contain enamel matrix proteins and amorphous mineral particles that subsequently fuse and transform into long apatitic crystals upon the proteolytic processing of enamel matrix components [53,54]. Although such a mechanism is worthy of consideration, no direct evidence has yet been provided in support of this proposal. The fact that P173 was found in the present study to be an effective stabilizer of ACP in vitro is consistent with the noted in vivo findings [34] that suggest that ACP persists in developing enamel for substantial periods of time prior to transformation to apatitic crystals. Studies are currently underway in our laboratory to explore relevant factors that can affect the transformation of ACP to HA in the presence of the full-length native amelogenin P173. Despite the mechanism involved, however, the present results clearly show that phosphorylation is an important functional element of the amelogenin structure. Previously we found that the predominant native porcine degradation product P148 that lacks 25 C-terminal amino acids and the hydrophilic C-terminus similarly stabilizes ACP formation and prevents its transformation to HA in vitro [33]. However, unlike the non-phosphorylated analog of rP172, rP147 had almost no influence on calcium phosphate formation in vitro. In agreement with this latter observation, our present findings show that P148 and rP147 do not tend to form higher-order chain-like structures like those found for P173 and rP172. Hence, organized mineral formation in developing enamel is likely to be guided primarily by full-length amelogenin through its ability to form organized assemblies. At the same time, phosphorylated P148 may play an important role through its capacity to stabilize ACP and inhibit of HA formation [33]. As the major protein component found in the secretory stage of developing pig enamel [35,36], this capacity would serve to prevent unwanted crystal formation throughout the majority of the extracellular matrix during the secretory stage of amelogenesis in which nascent enamel mineral particles occupy only 10 – 20% of its total volume [55,56]. Based on present and prior findings, we propose that P173 and P148 play distinctly different roles in the process of amelogenesis that reflect, in part, difference in their capacities for self-assembly.
As we have previously discussed [28], changes in solution pH can induce protein assembly through the alteration of charges on amino acid side-chains that result in a net reduction in electrostatic repulsion between similarly charged proteins. Our present results are consistent with other findings that support this general concept [57] and previous work from our laboratory [28]. These results show that the onset of aggregation of amelogenin nanoparticles to form larger protein assemblies takes place, in general, at pH values near the respective protein isoelectric points, consistent with other recent reports [58,59]. Hence, these collective findings suggest that hydrophobic interactions between amelogenins play an important role in amelogenin self-assembly. A similar conclusion was drawn from studies on the adsorption of recombinant full-length mouse amelogenin (rM179) onto positively charged polyelectrolyte films [60]. Given the fact that amelogenin primarily contains hydrophobic residues, it is reasonable that hydrophobic interactions represent an important factor with respect to the regulation of protein-protein interactions. Accordingly, it is also reasonable that, in most cases, large variations in ionic strength (i.e., 15 – 163 mM) had a relatively small impact on 1) the size of amelogenin nanoparticles observed prior to off-scale DLS scattering, i.e. prior to further protein aggregation at relatively lower pH values (Table 1) and 2) the pH value at which amelogenins formed these larger aggregates (Table 2). In summary, our results indicate that hydrophobic interactions play an essential role in amelogenin self-assembly.
Similarly, like pH, IS had a notable effect on the structures of the “larger aggregates” of the full-length amelogenins that formed in the vicinity of pH values (i.e., pH 7.2) where the onset of higher-order amelogenin assembly was observed for the full-length molecules (Fig. 2). As reported here (Figs. 3 and 5; Table 3), the tendency for rP172 to form chain-like structures at pH 7.2 increased with a decrease in IS, while the opposite was observed for P173. Although the reason for the difference in the observed effect of IS on the behavior of P173 and rP172 is not apparent, this difference is likely related to the presence of the single phosphate group in the native protein. Thus, as demonstrated here by the observed effect of pH, IS, and the presence of specific charged protein residues (i.e., the hydrophilic C-terminal domain and the single phosphate group on S-16) on the formation of chain-like amelogenin structures, ionic interactions also clearly play an important role in amelogenin assembly. A recent collaborative study from our laboratory [40] using DLS and small-angle X-ray scattering has found that full-length recombinant amelogenins (rP172 and rM179) in solution are actually best described as non-spherical oblate nanostructures and not as spherical nanoparticles. These anisotropic structures may reflect asymmetries in surface properties (e.g., surface charge) that help to explain why full-length amelogenin nanoparticles form anisotropic elongated chain-like structures.
Conclusion
In summary, the present study shows that the phosphorylation of the single site on full-length porcine amelogenin P173 has a small but potentially important effect on its higher-order self-assembly into chain-like structures under physiological conditions of pH, temperature and ionic strength. This importance is related to the fact that higher-order amelogenin self-assembly is very sensitive to pH. Our findings also suggest that amelogenin self-assembly is affected by both hydrophilic and hydrophobic interactions. Despite the fact that the presence of the single phosphorylation site on native full-length porcine amelogenin P173 has a subtle effect on self-assembly, it was found to have a profound effect on calcium phosphate formation in vitro.
Acknowledgements
This work was supported by grant DE-016376 (HCM) from the National Institute of Dental and Craniofacial Research. S-YK was also partially supported by grant T32 DE-007327.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 2.Nylen MU, Omnell KA, Eanes ED. Crystal Growth in Rat Enamel. J. Cell Biol. 1963;18:109–123. doi: 10.1083/jcb.18.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arsenault AL, Robinson BW. The dentino-enamel junction: a structural and microanalytical study of early mineralization. Calcif. Tiss. Int. 1989;45:111–121. doi: 10.1007/BF02561410. [DOI] [PubMed] [Google Scholar]
- 4.Smith CE. Cellular and chemical events during enamel maturation. Crit. Rev. Oral Biol. Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 5.Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K, Kobayashi S. Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13–17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochem. 1991;96:129–138. doi: 10.1007/BF00315983. [DOI] [PubMed] [Google Scholar]
- 6.Weiner S. Organization of extracellularly mineralized tissues: a comparative study of biological crystal growth. CRC Crit. Rev. Biochem. 1986;20:365–408. doi: 10.3109/10409238609081998. [DOI] [PubMed] [Google Scholar]
- 7.Termine JD, Belcourt AB, Miyamoto MS, Conn KM. Properties of dissociatively extracted fetal tooth matrix proteins. II. Separation and purification of fetal bovine dentin phosphoprotein. J. Biol. Chem. 1980;255:9769–9772. [PubMed] [Google Scholar]
- 8.Fincham AG, Moradian-Oldak J, Simmer JP. The structural biology of the developing dental enamel matrix. J. Struct. Biol. 1999;126:270–299. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 9.Toyosawa S, O'Huigin C, Figueroa F, Tichy H, Klein J. Identification and characterization of amelogenin genes in monotremes, reptiles, and amphibians. Proc.Natl.Acad.Sci. U.S.A. 1998;95:13056–13061. doi: 10.1073/pnas.95.22.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Clarkson BH, Sun K, Mansfield JF. Self-assembly of synthetic hydroxyapatite nanorods into an enamel prism-like structure. J. Coll. Interf. Sci. 2005;288:97–103. doi: 10.1016/j.jcis.2005.02.064. [DOI] [PubMed] [Google Scholar]
- 11.Sire JY, Delgado S, Fromentin D, Girondot M. Amelogenin: lessons from evolution. Arch. Oral Biol. 2005;50:205–212. doi: 10.1016/j.archoralbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 13.Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J. Struct. Biol. 1994;112:103–109. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 14.Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P, Jr, Slavkin HC. Evidence for amelogenin "nanospheres" as functional components of secretory-stage enamel matrix. J. Struct. Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 15.Moradian-Oldak J, Leung W, Fincham AG. Temperature and pH-dependent supramolecular self-assembly of amelogenin molecules: a dynamic light-scattering analysis. J. Struct. Biol. 1998;122:320–327. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- 16.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J. Struct. Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 17.Bai P, Warshawsky H. Morphological-Studies on the Distribution of Enamel Matrix Proteins Using Routine Electron-Microscopy and Freeze-Fracture Replicas in the Rat Incisor. Anat. Rec. 1985;212:1–16. doi: 10.1002/ar.1092120102. [DOI] [PubMed] [Google Scholar]
- 18.Robinson C, Fuchs P, Weatherell JA. The Appearance of Developing Rat Incisor Enamel Using A Freeze Fracturing Technique. J. Crys. Growth. 1981;53:160–165. [Google Scholar]
- 19.Smales FC. Structural subunit in prisms of immature rat enamel. Nature. 1975;258:772–774. doi: 10.1038/258772a0. [DOI] [PubMed] [Google Scholar]
- 20.Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light scattering. Biopol. 1994;34:1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 21.Moradian-Oldak J, Simmer JP, Lau EC, Diekwisch T, Slavkin HC, Fincham AG. A review of the aggregation properties of a recombinant amelogenin. Connect. Tiss. Res. 1995;32:125–130. doi: 10.3109/03008209509013714. [DOI] [PubMed] [Google Scholar]
- 22.Fincham AG, Luo W, Moradian-Oldak J, Paine ML, Snead ML, Zeichner-David M, Teaford MF, Smith MM, Ferguson MWJ. Development, Function and Evolution of Teeth. Cambridge, UK: Cambridge University Press; 2000. Enamel biomineralization: the assembly and disassembly of the protein extracellular organic matrix; pp. 37–61. [Google Scholar]
- 23.Aichmayer B, Margolis HC, Sigel R, Yamakoshi Y, Simmer JP, Fratzl P. The onset of amelogenin nanosphere aggregation studied by small-angle X-ray scattering and dynamic light scattering. J. Struct. Biol. 2005;151:239–249. doi: 10.1016/j.jsb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J. Struct. Biol. 2005;149:182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Habelitz S, Kullar A, Marshall SJ, DenBesten PK, Balooch M, Marshall GW, Li W. Amelogenin-guided crystal growth on fluoroapatite glass-ceramics. J. Dent. Res. 2004;83:698–702. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- 26.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 27.Moradian-Oldak J, Du C, Falini G. On the formation of amelogenin microribbons. Eur. J. Oral Sci. 2006;114 Suppl 1:289–296. doi: 10.1111/j.1600-0722.2006.00285.x. discussion 327-289, 382. [DOI] [PubMed] [Google Scholar]
- 28.Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. J. Struct. Biol. 2007;160:57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukae M, Yamamoto R, Karakida T, Shimoda S, Tanabe T. Micelle structure of amelogenin in porcine secretory enamel. J. Dent. Res. 2007;86:758–763. doi: 10.1177/154405910708600814. [DOI] [PubMed] [Google Scholar]
- 30.Iijima M, Moriwaki Y, Takagi T, Moradian-Oldak J. Effects of bovine amelogenins on the crystal morphology of octacalcium phosphate in a model system of tooth enamel formation. J. Crys. Growth. 2001;222:615–626. [Google Scholar]
- 31.Iijima M, Moriwaki Y, Wen HB, Fincham AG, Moradian-Oldak J. Elongated growth of octacalcium phosphate crystals in recombinant amelogenin gels under controlled ionic flow. J. Dent. Res. 2002;81:69–73. doi: 10.1177/002203450208100115. [DOI] [PubMed] [Google Scholar]
- 32.Iijima M, Du C, Abbott C, Doi Y, Moradian-Oldak J. Control of apatite crystal growth by the co-operative effect of a recombinant porcine amelogenin and fluoride. Eur. J. Oral Sci. 2006;114 Suppl. 1:304–307. doi: 10.1111/j.1600-0722.2006.00324.x. [DOI] [PubMed] [Google Scholar]
- 33.Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Litman A, Margolis HC. Role of 20 kDa amelogenin (P148) phosphorylation on calcium phosphate formation in vitro. J. Biol. Chem. 2009;284:18972–18977. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beniash E, Metzler RA, Lam RS, Gilbert PU. Transient amorphous calcium phosphate in forming enamel. J. Struct. Biol. 2009;166:133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamakoshi Y, Tanabe T, Fukae M, Shimizu M. Porcine amelogenins. Calcif. Tiss. Int. 1994;54:69–75. doi: 10.1007/BF00316293. [DOI] [PubMed] [Google Scholar]
- 36.Yamakoshi Y, Hu JC-C, Ryu OH, Tanabe T, Oida S, Fukae M, Simmer JP. A Comprehensive Strategy for Purifying Pig Enamel Protein. In: Kobayashi I, Ozawa H, editors. Proceedings of the 8th International Symposium on Biomineralization. Kanagawa: Tokai Univ. Press; 2003. pp. 326–332. [Google Scholar]
- 37.Simmer JP, Lau EC, Hu CC, Aoba T, Lacey M, Nelson D, Zeichner-David M, Snead ML, Slavkin HC, Fincham AG. Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif. Tiss. Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- 38.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, Simmer JP. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J. Dent. Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 39.Aoba T, Moreno EC. The enamel fluid in the early secretory stage of porcine amelogenesis: chemical composition and saturation with respect to enamel mineral. Calcif. Tiss. Int. 1987;41:86–94. doi: 10.1007/BF02555250. [DOI] [PubMed] [Google Scholar]
- 40.Aichmayer B, Wiedemann-Bidlack FB, Gilow C, Simmer JP, Yamakoshi Y, Emmerling F, Margolis HC, Fratzl P. Amelogenin nanoparticles in suspension: Deviations from spherical shape and pH-dependent aggregation. Biomacromol. 2010;11:369–376. doi: 10.1021/bm900983b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gadaleta S, Paschalis E, Betts F, Mendelsohn R, Boskey A. Fourier transform infrared spectroscopy of the solution-mediated conversion of amorphous calcium phosphate to hydroxyapatite: New correlations between X-ray diffraction and infrared data. Calcif. Tiss. Int. 1996;58:9–16. doi: 10.1007/BF02509540. [DOI] [PubMed] [Google Scholar]
- 42.Smith CE, Issid M, Margolis HC, Moreno EC. Developmental changes in the pH of enamel fluid and its effects on matrix-resident proteinases. Adv. Dent. Res. 1996;10:159–169. doi: 10.1177/08959374960100020701. [DOI] [PubMed] [Google Scholar]
- 43.Smith CE, Chong DL, Bartlett JD, Margolis HC. Mineral acquisition rates in developing enamel on maxillary and mandibular incisors of rats and mice: Implications to extracellular acid loading as apatite crystals mature. J. Bone Min. Res. 2005;20:240–249. doi: 10.1359/JBMR.041002. [DOI] [PubMed] [Google Scholar]
- 44.Tanabe T, Fukae M, Uchida T, Shimizu M. The localization and characterization of proteinases for the initial cleavage of porcine amelogenin. Calcif. Tiss. Int. 1992;51:213–217. doi: 10.1007/BF00334549. [DOI] [PubMed] [Google Scholar]
- 45.Diekwisch TG. Subunit compartments of secretory stage enamel matrix. Connect. Tiss. Res. 1998;38:101–111. doi: 10.3109/03008209809017026. [DOI] [PubMed] [Google Scholar]
- 46.Bodier-Houllé P, Steuer P, Meyer JM, Bigeard L, Cuisinier FJG. High-resolution electron-microscopic study of the relationship between human enamel and dentin crystals at the dentinoenamel junction. Cell Tiss. Res. 2000;301:389–395. doi: 10.1007/s004410000241. [DOI] [PubMed] [Google Scholar]
- 47.Deshpande AS, Fang PA, Simmer JP, Margolis HC, Beniash E. Amelogenin-collagen interactions regulate calcium phosphate mineralization in vitro. J. Biol. Chem. 2010;285:19277–19287. doi: 10.1074/jbc.M109.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crane NJ, Popescu V, Morris MD, Steenhuis P, Ignelzi JMA. Raman spectroscopic evidence for octacalcium phosphate and other transient mineral species deposited during intramembranous mineralization. Bone. 2006;39:434–442. doi: 10.1016/j.bone.2006.02.059. [DOI] [PubMed] [Google Scholar]
- 49.Mahamid J, Sharir A, Addadi L, Weiner S. Amorphous calcium phosphate is a major component of the forming fin bones of zebrafish: Indications for an amorphous precursor phase. Proc. Natl. Acad. Sci. USA. 2008;105:12748–12753. doi: 10.1073/pnas.0803354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahamid J, Aichmayer B, Shimoni E, Ziblat R, Li C, Siegel S, Paris O, Fratzl P, Weiner S, Addadi L. Mapping amorphous calcium phosphate transformation into crystalline mineral from the cell to the bone in zebrafish fin rays. Proc. Natl. Acad. Sci. USA. 2010;107:6316–6321. doi: 10.1073/pnas.0914218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida T, Tanabe T, Fukae M, Shimizu M. Immunocytochemical and immunochemical detection of a 32 kDa nonamelogenin and related proteins in porcine tooth germs. Arch. Histol. Cytol. 1991;54:527–538. doi: 10.1679/aohc.54.527. [DOI] [PubMed] [Google Scholar]
- 52.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol. Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson C, Shore RC, Wood SR, Brookes SJ, Smith DA, Wright JT, Connell S, Kirkham J. Subunit structures in hydroxyapatite crystal development in enamel: implications for amelogenesis imperfecta. Connect. Tiss. Res. 2003;44 Suppl 1:65–71. [PubMed] [Google Scholar]
- 54.Robinson C. Self-oriented assembly of nano-apatite particles: a subunit mechanism for building biological mineral crystals. J. Dent. Res. 2007;86:677–679. doi: 10.1177/154405910708600801. [DOI] [PubMed] [Google Scholar]
- 55.Robinson C, Kirkham J, Hallsworth AS. Volume distribution and concentration of protein, mineral and water in developing bovine enamel. Arch. Oral. Biol. 1988;33:159–162. doi: 10.1016/0003-9969(88)90040-4. [DOI] [PubMed] [Google Scholar]
- 56.Fukae M. Amelogenesis - Three dimensional structure of amelogenin micelles and their degradation by a cascade system. The Proceedings of International Symposium of Oral Science in Tsurumi University School of Dental Medicine. Tsurumi. 2002:73–78. [Google Scholar]
- 57.Yu SY, Yao P, Jiang M, Zhang GZ. Nanogels prepared by self-assembly of oppositely charged globular proteins. Biopolymers. 2006;83:148–158. doi: 10.1002/bip.20539. [DOI] [PubMed] [Google Scholar]
- 58.He X, Li W, Habelitz S. The cooperative self-assembly of 25 and 23 kDa amelogenins. J. Struct. Biol. 2008;164:314–321. doi: 10.1016/j.jsb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uskokovic V, Castiglione Z, Cubas P, Zhu L, Li W, Habelitz S. Zeta-potential and Particle Size Analysis of Human Amelogenins. J. Dent. Res. 2010;89:149–153. doi: 10.1177/0022034509354455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gergely C, Szalontai B, Moradian-Oldak J, Cuisinier FJG. Polyelectrolyte-Mediated Adsorption of Amelogenin Monomers and Nanospheres Forming Mono- or Multilayers. Biomacromol. 2007;8:2228–2236. doi: 10.1021/bm070088+. [DOI] [PMC free article] [PubMed] [Google Scholar]