Abstract
Haptoglobin is an acute phase inflammatory marker. Its main function is to bind hemoglobin released from erythrocytes to aid its elimination, and thereby haptoglobin prevents the generation of reactive oxygen species in the blood. Haptoglobin levels have been repeatedly associated with a variety of inflammation-linked infectious and non-infectious diseases, including malaria, tuberculosis, human immunodeficiency virus, hepatitis C, diabetes, carotid atherosclerosis, and acute myocardial infarction. However, a comprehensive genetic assessment of the inter-individual variability of circulating haptoglobin levels has not been conducted so far.
We used a genome-wide association study initially conducted in 631 French children followed by a replication in three additional European sample sets and we identified a common single nucleotide polymorphism (SNP), rs2000999 located in the Haptoglobin gene (HP) as a strong genetic predictor of circulating Haptoglobin levels (Poverall = 8.1×10−59), explaining 45.4% of its genetic variability (11.8% of Hp global variance). The functional relevance of rs2000999 was further demonstrated by its specific association with HP mRNA levels (β = 0.23±0.08, P = 0.007). Finally, SNP rs2000999 was associated with decreased total and low-density lipoprotein cholesterol in 8,789 European children (Ptotal cholesterol = 0.002 and PLDL = 0.0008).
Given the central position of haptoglobin in many inflammation-related metabolic pathways, the relevance of rs2000999 genotyping when evaluating haptoglobin concentration should be further investigated in order to improve its diagnostic/therapeutic and/or prevention impact.
Introduction
Human haptoglobin (Hp) is an acute phase inflammatory glycoprotein essentially synthesized by the liver and up-regulated by cytokines [1]. Hp is polymorphic with two co-dominant alleles, Hp1 and Hp2 encoded by the Haptoglobin (HP) gene located in chromosome 16 and resulting in three common isoforms: Hp1-1, Hp2-2 and Hp2-1 (called HP ‘common polymorphism’) [2]. In normal physiological conditions, Hp protein concentration in blood ranges between 0.3 and 2.0 g/L in adults [3] but significant fall in its level during the first decade of life [4]. The main property of Hp is to scavenge circulating hemoglobin (Hb) released by hemolysis or normal red blood cells turnover [5]. The resulting circulating Hp-Hb complexes are eliminated by Kupffer's cell(s) in the liver, preventing the generation of reactive oxygen species [2], [6]. Therefore, Hp plays an important role in preventing renal damage and iron loss that can occur following an intravascular hemolysis. Hp is also able to bind apolipoprotein (Apo) A-I [7] to protect the Apo A-I effector domain of lecithin-cholesterol acyltransferase against oxidative stress, and Hp consequently modulates the high-density lipoprotein (HDL) function [8]. Furthermore, Hp can bind Apo E and the resulting complexes influence cholesterol esterification [9]. These functional characteristics confer to Hp a major role in the reverse transport of cholesterol between peripheral cells and the liver for degradation.
Hp levels and HP rs72294371 ‘common polymorphism’ (Data S1) have been consistently associated with inflammatory-linked infectious [10], [11] and non-communicable diseases [11], [12]. Malaria caused by Plasmodium falciparum, which is associated with extensive intravascular hemolysis, decreases Hp to undetectable levels as the Hb-scavenging system is saturated [13].13 In malaria-endemic areas, hypohaptoglobinemia has been proposed as an indirect biochemical indicator of malaria [14]. HP ‘common polymorphism’ should be considered at diagnosis of tuberculosis. Eisaev and colleagues [15] described an increased recurrence of pulmonary tuberculosis with worse prognosis in Hp2-2 Caucasians. Furthermore, this HP ‘common polymorphism’ contributes to mortality and viral load in Human immunodeficiency virus (HIV) infection [16]. Hp2-2 HIV carriers have a more pronounced viral replication rate and a worse prognosis compared to Hp1-1 or Hp2-1 HIV carriers [16], [17]. Hepatitis C infection has also been associated with low serum Hp concentrations [18] and an overrepresentation of the Hp1-1 phenotype has been associated with high risk for chronic hepatitis C [19], [20]. The HP ‘common polymorphism’ has also an effect on various other infectious diseases [21], [22], [23], [24].
In Type 2 diabetes patients, the Hp2-2 phenotype has been suggested to confer greater risk of cardiovascular events [12], [25], [26] and of carotid atherosclerosis [27]. Moreover, high Hp level is a risk factor for acute myocardial infarction, stroke and heart failure [28], [29]. Therefore, the routine measurement of Hp level has been suggested to be incorporated in daily medical practice to evaluate cardiovascular risk [29].
Despite these findings, the basis of Hp level inter-individual variability is still unknown. To identify genetic variants modulating physiological levels of Hp, we analyzed genome-wide association study (GWAS) data generated in European children for whom no age-related disease may influence Hp concentrations. We also assessed the association between the identified variants and cardiovascular risk factors (total, HDL and low density lipoprotein-cholesterol, Apolipoproteins A1 and B).
Results
Table 1 shows the phenotypic characteristics of the studied populations. Hp levels in children were low, in accordance with its reference distribution [4].
Table 1. Phenotypic characteristics of the studied populations.
| Population | Subsample | Sample size (% female) | Age (years) [95% CI] | BMI (kg/m2) [95% CI] | Haptoglobin (g/L) [95% CI] | LDL-cholesterol (mmol/L) [95% CI] | Total cholesterol (mmol/L) [95% CI] |
| SFS | Discovery cohort: Phase 1 vs Hp | 631 (50.9%) | 11.93 [11.76–12.11] | 17.66 [17.49–17.84] | 0.65 [0.62–0.68] | 2.99 [2.94–3.05] | 4.79 [4.74–4.85] |
| Families: Phase 2 vs Hp and lipids | 2,957 (49.2%) | 29.84 [29.38–30.30] | 22.66 [22.52–22.81] | 0.95 [0.94–0.97] | 3.43 [3.40–3.46] | 5.30 [5.26–5.34] | |
| Obese children | Replication cohort: Phase 2 vs Hp and lipids | 1,015 (52.3%) | 11.07 [10.86–11.27] | 28.24 [27.84–28.64] | 1.18 [1.15–1.22] | 2.74 [2.70–2.79] | 4.47 [4.42–4.52] |
| GENDAI | Replication cohort: Phase 2 vs Hp and lipids | 419 (53.1%) | 11.16 [11.10–11.23] | 19.76 [19.46–20.07] | 0.81 [0.76–0.85] | 3.12 [3.06–3.17] | 4.79 [4.73–4.86] |
| NFBC1986 | Replication cohort: Phase 2 vs lipids | 5,310 (50.8%) | 16 | 21.27 [21.17–21.37] | NA | 2.54 [2.52–2.56] | 4.27 [4.24–4.29] |
| VERONA cohort | Replication cohort: Phase 2 vs lipids | 401 (43.4%) | 10.90 [10.75–11.04] | 18.01 [17.78–18.23] | NA | 2.41 [2.40–2.47] | 4.21 [4.15–4.28] |
CI: confidence interval; BMI: body mass index; LDL: low-density lipoprotein.
Patterns of family correlations for serum Hp concentrations were assessed following both unadjusted values (Table 2). Model 1, which was not adjusted, did not show any family correlation. Model 2, which took into account age and body mass index (BMI) as covariates showed significant correlations for all the various pairs of relatives. Model 3, which hypothesized no effect of gender on family correlations, showed significant father-mother, father-son and son-son correlations (Table 2).
Table 2. Estimates of familial correlations ± standard error for serum haptoglobin concentration (656 familles/2680 individuals).
| Model 1 | Model 2 | Model 3 | Model 4 | |
| Adjustment for | None | Age & BMI | Age & BMI | Age, BMI & rs2000999 allelic frequency |
| Father - Mother (FM) | 0.126±0.038 | 0.111±0.038** | 0.111±0.038** | 0.156±0.038*** |
| Father - Son (FS) | 0.277±0.037 | 0.264±0.037*** | 0.230±0.021*** | 0.206±0.022*** |
| Father - Daughter (FD) | 0.277±0.036 | 0.265±0.037*** | [0.230] | [0.206] |
| Mother - Son (MS) | 0.227±0.036 | 0.204±0.037*** | [0.230] | [0.206] |
| Mother - Daughter (MD) | 0.184±0.039 | 0.188±0.040*** | [0.230] | [0.206] |
| Son - Son (SS) | 0.213±0.060 | 0.186±0.064** | 0.274±0.036*** | 0.239±0.036*** |
| Son - Daughter (SD) | 0.273±0.047 | 0.271±0.047*** | [0.274] | [0.239] |
| Daughter - Daughter (DD) | 0.355±0.067 | 0.386±0.064*** | [0.274] | [0.239] |
| Loge L2 (Logarithm of likelihood function) | 1458.83 | 1338.20 | 1342.20 | 1215.13 |
| Alternate model | Model 1 | Model 2 | Model 3 | |
| χ2 (df) | 241.26(8) | 8.00(5) | 254.14(4) | |
| P | *** | NS | *** |
* P≤0.05, ** P≤0.01, *** P≤0.001: compared to zero.
Model 1 estimated all eight correlations without adjustment.
Model 2 estimated all eight correlations with adjustment for age and BMI,
Model 3 estimated three correlations with adjustment for age and BMI with no gender effect on parent or offspring correlations: FS = MS = MD = FD and SS = SD = DD.
Model 4 estimated three correlations with adjustment for age, BMI and rs2000999 allelic frequency with no gender effect on parent or offspring correlations: FS = MS = MD = FD and SS = SD = DD.
Values in brackets were constrained to be equal to a preceding value according to the hypotheses of the model.
BMI: body mass index.
We then assessed the components of variance attributable to additive genetic effects, shared household effects and residual environmental factors (including assay variability) in 656 nuclear bi-parental families (2,680 individuals) from the STANISLAS Family Study (SFS) cohort (Table 3). Model 2, which included the three components after adjustment for age and BMI gave a better description of the variance decomposition than model 1 which was not adjusted. Hp genetic variance represented 26% (P<0.001) of the total variance. Shared (i.e. within families) and random environmental variances were 11.6 and 62.4% respectively (Table 3).
Table 3. Variance components of serum haptoglobin concentrations (656 families/2680 individuals, rs2000999 & rs4788597 as allelic frequency).
| Model 1 | Model 2 | Model 3 | |
| Adjustment for | None | Age & BMI | Age, BMI & rs2000999 allelic frequency |
| Polygenic variance: σ2 G | 0.0478±0.0120***, 24.9% | 0.0460±0.0109***, 26.0% | 0.0226±0.0101*, 14.2% |
| Household variance: σ2 H | 0.0250±0.0065***, 13.0% | 0.0206±0.0059***, 11.6% | 0.0233±0.0053***, 14.7% |
| Residual variance: σ2 E | 0.1194±0.0078***, 62.1% | 0.1103±0.0072***, 62.4% | 0.1130±0.0069***, 71.1% |
| Loge L2 (Logarithm of likelihood function) | −968.90 | −1074.58 | −1200.04 |
| Alternate model | - | Model 1 | Model 2 |
| χ2 (df) | - | 211.36 (8) | 250.92 (4) |
| P | - | *** | *** |
P≤0.05,
P≤0.01,
*** P≤0.001.
BMI: body mass index.
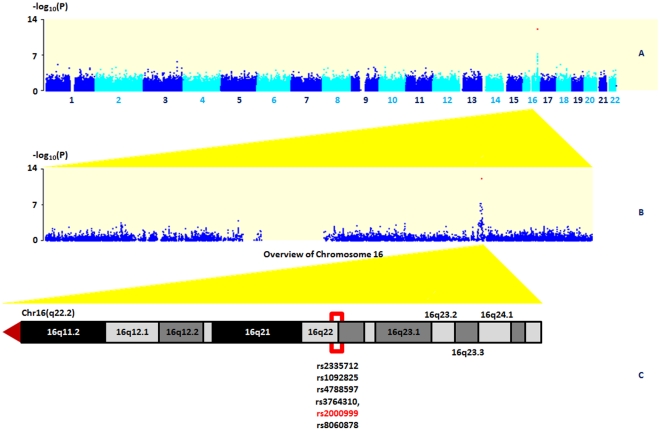
Our GWAS based on 631 unrelated children of the SFS cohort showed strongest association signal for Hp levels in a 218-kb linkage disequilibrium (LD) block on chromosome 16 that includes the HP gene (Figure 1). Using the square-root transformed Hp measurement adjusted for gender, age and z-BMI under the additive model, we identified in this region two significant association signals 90-kb apart: rs2000999 (with A as allele effect: β = −0.123, standard error [SE] = 0.017, P = 6.32×10−13; Table 4) and rs10492825 (with C as effect allele: β = −0.0876, SE = 0.016, P = 5.50×10−08; Table 4). Both SNPs rs2000999 and rs10492825 display moderate LD (r2 = 0.48, HapMap CEU release #27). In order to assess the redundancy between these two signals, we ran conditional regression analyses for both SNPs adjusted for each other and found that rs2000999 alone drove the association observed at the HP locus (rs2000999: Prs10492825 adjusted = 1.95×10−7, rs10492825: Prs2000999 adjusted = 0.91, Data S2, Table S1).
Figure 1. Manhattan plot of the GWAS of the discovery cohort comprising 631 children.
A, A Manhattan plot showing the −log10(P values) of SNPs from the association analysis of the 631 SFS children from stage 1. B, An overview of the −log10(P values) of Chromosome 16. C, The genomic region of the 618 LD block displayed in UCSC Genome Browser.
Table 4. Discovery and replication of rs2000999 association data for Hp levels.
| Haptoglobin (g/L) | ||||||||||
| Population | Stage | N | MAF | GG | GA | AA | β | Standard error | raw P | P adjusted with rs10492825 |
| SFS - unrelated children | Discovery | 631 | 0.21 | 0.736±0.406 | 0.532±0.321 | 0.361±0.180 | −0.123 | 0.017 | 6.32×10−13 | 1.95×10−07 |
| SFS - families | Replication | 2,957 | 0.20 | 1.056±0.467 | 0.819±0.445 | 0.549±0.330 | −0.138 | 0.007 | 1.17×10−75 | |
| Obese children | Replication | 1,015 | 0.22 | 1.261±0.492 | 1.064±0.455 | 1.002±0.534 | −0.088 | 0.013 | 1.20×10−11 | |
| GENDAI | Replication | 419 | 0.25 | 0.811±0.450 | 0.813±0.558 | 0.731±0.319 | −0.021 | 0.021 | 0.306 | |
| Replication sample | Replication | 4,391 | 0.22 | 3.49×10−41 | ||||||
| Overall meta-analysis | Replication | 5,022 | 8.09×10−59 |
N: sample size; MAF: Minor Allele Frequency; β:beta coefficient for the effect allele A.
We confirmed the association between SNP rs2000999 and circulating Hp levels in three additional independent European cohorts: the GENDAI study of Greek children, a subset of obese children from the East of France (Ntotal = 1,434), and a familial subset (Ntotal = 2,957) of the SFS cohort (Preplication = 3.49×10−41, Poverall = 8.09×10−59; Table 4).
Accounting for the rs2000999 allelic frequency, the pattern of familial correlation (Table 2, model 4) decreased from 0.230 to 0.206 and from 0.274 to 0.239 for sibling and child-parent respectively, whereas the adequacy of the model was significantly improved. Additional adjustment for rs2000999 for the components of variance attributable to additive genetic effects, shared household effects and residual environmental factors (Table 3, model 3) significantly improved the likelihood function and the proportion of phenotypic variability accounted for by genetic effects decreased (26.0% to 14.2%, in comparison to model 2). Moreover, the component attributable to household factors increased (11.6% to 14.7%). We thus determined that rs2000999 is the major genetic determinant of Hp levels accounting for 11.8% of Hp global variance and 45.4% of the genetic variance of this trait.
In order to assess the degree of independence of rs2000999 from the HP rs72294371 ‘common polymorphism’ (Data S1), we genotyped the latter in the GWAS first stage children (SFS cohort) by using a PCR-based method and a gel reading. Only a subset of 341 out of 631 samples was successfully genotyped after independent readings by two readers. In this sample set, we found no evidence for LD between HP ‘common polymorphism’ and rs2000999 (r2 = 0.135) nor with the other SNPs that were genotyped by the Illumina array within the 218 kb LD block that includes both HP ‘common polymorphism’ and rs2000999 (0.001<r2<0.137; N = 31 SNPs). Furthermore, the HP ‘common polymorphism’ (minor allele frequency [MAF] = 0.46) and rs2000999 (MAF = 0.20) were both highly associated with Hp levels, as expected (P = 4×10−7 and P = 1×10−7, respectively). When both variants were included in the same regression model, we found that they significantly and independently contributed to the increased Hp levels (PHP rs72294371 ‘common polymorphism’ = 0.001 and Prs2000999 = 5×10−5) indicating that the association with rs2000999 would be novel and not redundant with the HP ‘common polymorphism’. However, it is noteworthy that despite strong efforts, we did not succeed by far in genotyping all samples. We used two other technologies: a pre-designed TaqMan copy number assay (Applied Biosystems) and a PCR-based method with another design than previously used. Unfortunately, we did not find a good concordance (<70%) between the three methods. We conclude that given the state of art, we cannot definitively conclude that the present signal of association is not related to the HP ‘common polymorphism’ genotype.
In order to validate our main results, we secondary assessed the effect of SNP rs2000999 on HP gene expression in subcutaneous adipose tissue sample from 194 non-obese subjects ascertained from the Swedish SibPair cohort (Data S2). We found a significant contribution of rs2000999 to HP expression (β = 0.23±0.08; P = 0.007; PBayesian = 0.006).
We finally assessed by additive model the effect of SNP rs2000999 on total, HDL and low-density lipoprotein (LDL) cholesterol, Apolipoproteins A1 and B in five independent European pediatric cohorts totaling 8,789 children. Total cholesterol was ln-transformed and we normalized the LDL cholesterol by computing the square root. All measurements were adjusted for gender, age (excepting the NFBC1986) and z-score BMI. Our data showed that rs2000999, with A as allele effect, was associated with total cholesterol (β = −0.011, SE = 0.003, P = 0.002; Table 5) and LDL-cholesterol (β = −0.017, SE = 0.004, P = 0.0008; Table 5). The association with HDL-cholesterol and Apolipoproteins A1 and B are displayed in Table S2.
Table 5. Association of rs2000999 with lipid traits.
| Total cholesterol | LDL-cholesterol | |||||||
| N | MAF | β | Standard error | p-value | β | Standard error | p-value | |
| SFS children | 1,644 | 0.202 | −0.003 | 0.008 | 0.73 | 0.0004 | 0.010 | 0.967 |
| Obese Children | 1,015 | 0.216 | −0.008 | 0.011 | 0.439 | −0.009 | 0.013 | 0.507 |
| GENDAI | 419 | 0.253 | −0.023 | 0.012 | 0.055 | −0.031 | 0.013 | 0.022 |
| NFBC1986 | 5,310 | 0.186 | −0.013 | 0.004 | 0.002 | −0.022 | 0.006 | 8.96E-05 |
| Verona Cohort | 401 | 0.200 | −0.011 | 0.014 | 0.446 | −0.011 | 0.016 | 0.515 |
| Overall meta-analysis | 8,789 | −0.011 | 0.003 | 0.002 | −0.017 | 0.004 | 0.0008 |
N: sample size; MAF: Minor Allele Frequency; β:beta coefficient for the effect allele A.
Discussion
We first determined in 656 nuclear families that 26% of the Hp plasma level variance was under genetic control. Then, using a GWAS in 631 children from the same population and replicating in three independent populations, we identified rs2000999 as the major genetic determinant of Hp levels. This genetic variant alone explained 45.4% of the genetic variance of this trait (11.8% of Hp global variance). SNP rs2000999 is located in the intronic region of HP gene, in a region previously believed to be the HPR gene (encoding the haptoglobin-related protein) which shares more than 90% nucleotide sequence homology with HP [30]. It is 17 kb apart a duplication of 59 α chain amino acid residues resulting to an intragenic duplication of 1.7 kb and which is known as the HP ‘common polymorphism’ [31].
Our study shows that SNP rs2000999 also modulated expression levels of the Hp mRNA in human adipose tissue suggesting that this SNP (or a SNP in very strong LD with this one) is indeed functional. It is noteworthy that SNP rs2000999 has been previously reported to associate with total cholesterol in 4,200 adults from the EUROSPAN consortium [32] and with both total and LDL-cholesterol in 100,000 adults of European and non-European ancestry [33]. Interestingly, we confirmed the effect of this SNP on these lipid traits in European children.
Increased plasma levels of several inflammatory markers correlate with higher incidence and prognosis of various cardiovascular diseases [34], [35], [36], [37]. Hp level measurement has been recently shown to improve the predictive information for major cardiovascular events [29]. As rs2000999 is also associated with lipid levels, this marker links inflammation and cardiovascular risk. It is noteworthy that the impact of rs2000999 association on lipids occurs early in life and is consistent with previous findings that the precursors of cardiovascular diseases originate in childhood [38], [39].
Interestingly, the effect of rs2000999 on Hp levels is more important in our discovery cohort which includes healthy children having low Hp concentration (0.65 g/L±0.39). As shown in the analyses for other diseases [40], the statistical power of GWAS can be increased in healthy homogeneous controls.
In addition, the effect of aging and of the environment is minimized in children. Then, by using healthy pediatric populations, we were able to assess more accurately the effect of the SNP rs2000999 on Hp levels.
We tried to assess the degree of independence between rs2000999 and the HP ‘common polymorphism’. Three different methods were evaluated to genotype the HP ‘common polymorphism’ in our whole GWAS sample set. Unfortunately, we found no concordance between the three methods, which underlie a major difficulty to carry out an accurate genotyping of this polymorphism. Even if this difficulty was not clearly discussed and not published to our knowledge, it is admitted in the scientific field and it should also be present in the clinical diagnosis setting. In contrast, SNP rs2000999 can be accurately and easily genotyped.
Our findings should be further replicated in non-European adults, especially in those affected by infectious diseases. More generally, rs2000999 should be assessed in cohorts of patients affected by the large variety diseases associated with Hp levels. It is not a trivial task, as Hp is a trait that has been infrequently measured in cohorts used for genetic studies. Given the major effect of rs2000999 on Hp gene expression and on Hp levels, Mendelian randomization approach would be of interest to test the causative effect of this SNP on infectious and non-communicable phenotypes in order to assess its clinical relevance.
Materials and Methods
Ethics Statement
All the populations involved in the present study were recruited in accordance with the latest version of the Declaration of Helsinki for Ethical Principles for Medical Research Involving Human Subjects. All participants and their parents gave a written informed consent. Genetic studies protocols were approved by the local ethics committees for the protection of subjects for biomedical research: the Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale (CCPPRB).
Study populations
The STANISLAS Family Study (SFS)
The SFS is a 10-year longitudinal survey involving 1,006 volunteer families of European ancestry whose members were free of chronic disease (cardiovascular or cancer) with recruitment taking place from 1993–95 [41]. The SFS samples and data are part of the Biological Resources Centre (BRC) “Interactions Gène-Environnement en Physiopathologie CardioVasculaire” (IGE-PCV) in Nancy, France. Genome-wide genotyping was performed on a subset of 631 unrelated children (mean age 11.93 years [11.76–12.11]) constituting the discovery cohort [42] after screening for latent population substructure (Data S2). The 2,957 remaining individuals after quality control were analysed in the replication studies (mean age 29.84 [29.38–30.30]). Hp levels, BMI and the cardiovascular risk traits including total, high density lipoprotein (HDL) and low density lipoprotein (LDL)-cholesterol (calculated by the Friedewald formula [43]), Apolipoprotein A1 and B were available for all participants.
Obese Children
We studied obese children (defined as BMI>97th percentile for age and sex according to a French cohort [44]) ascertained from 449 nuclear families with at least one obese offspring, recruited in the Paediatric Endocrine Unit of Jeanne de Flandres Hospital of Lille, France or through a national media campaign. We analyzed 1,015 children (mean age 11.07 years [10.86–11.27]) for whom Hp, BMI, total, HDL and LDL-cholesterol, Apolipoprotein A1 and B measurements were available.
The GeNe and Diet Attica Investigation (GENDAI)
The GENDAI pediatric cohort was recruited from children living in the Attica region of Greece [45]. From November 2005 to June 2006, 1,138 peri-adolescent children were recruited from randomly selected elementary schools of Attica. We analyzed 419 children (mean age 11.16 years [11.10–11.23]) for whom Hp, BMI, total, HDL and LDL-cholesterol, Apolipoprotein A1 and B measurements were available.
The Northern Finland 1986 Birth Cohort (NFBC1986)
The NFBC1986 is a prospective birth cohort including all Finnish mothers of European ancestry with children whose expected date of birth fell between July 1, 1985 and June 30, 1986 in the two northernmost provinces in Finland [46]. Clinical examination at 15–16 years follow-up was conducted between August 2001 and June 2002. All cohort members living in Finland with known address (n = 9,215) were invited, and 6,798 participated (74%). We analyzed 5,310 adolescents successfully genotyped in the NFBC1986 cohort for whom BMI, total, HDL and LDL-cholesterol, Apolipoprotein A1 and B measurements were available.
The Verona cohort
The Verona cohort consists of Italian children recruited from the general population of Verona, Italy, whose families were randomly chosen from the registry office database of the town, and contacted by post. We analysed 401 children (mean age 10.90 years [10.75–11.04]) successfully genotyped for whom at least BMI, total, HDL and LDL-cholesterol, Apolipoprotein A1 and B measurements were available.
The SibPair cohort
The SibPair cohort comprises 154 nuclear families (732 subjects) from Sweden, each containing an obesity-discordant sib pair (at least 10 kg/m2 difference in BMI). Gene expression and genetic variation were analysed in 194 non-obese subjects from the SibPair cohort.
Genotyping
Genomewide genotypes were generated for the 631 unrelated SFS children using the Illumina Human CNV370-Duo array [42]. Briefly,750 ng of genomic DNA was processed using Illumina's protocol for the BeadStation genotyping platform (Illumina), followed by GenCall software analysis(Illumina) to automatically cluster, call genotypes, and assign confidence scores using the GenTrain clustering algorithm (Illumina). We discarded a total of 2,552 SNPs due to the following reasons: extreme Hardy-Weinberg disequilibrium (P<0.001), low genotyping call rates (<95%) or low minor-allele frequencies (<1%). We retained 318,237 SNPs for analysis. Genomic control λGC was 1.01.
We used the Applied Biosystems SNPlex™ technology to replicate the association of genome-wide significant genetic variants in the SFS replication set, obese children and GENDAI, NFBC1986 and Verona cohorts.
SNPlex is based on the Oligonucleotide Ligation Assay (OLA) combined with multiplex PCR target amplification and was carried out as per the manufacturer's instructions (http://www.appliedbiosystems.com). Allelic discrimination was performed by capillary electrophoresis analysis using an Applied Biosystems 3730xl DNA Analyzer and GeneMapper 3.7 software. Genotyping call rate was above 95% in all populations studied and genetic variants were in HW equilibrium (p>0.001).
We used a PCR-based method [47] to genotype for the HP ‘common polymorphism’ in the 631 children of the discovery cohort (SFS cohort) in order to determine any linkage disequilibrium with regard to genome-wide significant variants identified in the analysis. Only genotypes that were concordant following a double blind genotyping call by two independent readers were retained for statistical analyses (N = 341). Two additional genotyping methods for HP ‘common polymorphism’ were used in order to validate the above-method: a custom TaqMan copy number assay (Applied Biosystems) following the manufacturer's recommendations and another PCR-based method using the following oligonucleotide primers : 5′-CTCTCCTTTCTCCCTTCCTGTC-3′ and 5′-TTTATCCACTGCTTCTCATTGT-3′. We didn't obtain correspondence between the banding patterns and the Hp genotypes.
Haptoglobin measurement
Blood samples were collected between 8:00 and 9:00 am or 11:30 and 12:30 pm by venipeuncture after overnight fasting. Hp protein levels were measured in blood plasma samples by high sensitivity immunophotometry analyses using the BN™II Siemens analyzer (Siemens, Marburg, Germany) and Siemens reagents and following the manufacturer's instructions.
Lipids measurements
Total cholesterol, HDL-cholesterol and apolipoproteins A1 and B were assayed using enzymatic methods (AU640 [Olympus, Watford, UK]) and LDL-cholesterol was calculated using the Friedewald formula [43].
Statistical analyses
Heritability estimate of Hp levels in the SFS
Intra-familial correlations were estimated by using maximum likelihood techniques [48] with and without adjustment for covariates. This statistical approach allowed adjustment for covariates within models, simultaneously and separately for fathers, mothers, sons and daughters. The significance of various familial correlations, or sex and generation differences in correlations, was tested using the log-likelihood ratio test. Correlations were computed under two sets of hypotheses: gender effects on correlations for parents and children and no gender effect for all correlations.
Variance component analysis was applied in order to assess the relative contributions of genetic, common household factors and individual specific environment in familial aggregation of serum haptoglobin concentrations. The variable used to estimate variance component was adjusted for age and BMI, separately for fathers, mothers, sons and daughters. The analysis was conducted by using a multivariate normal model for pedigree analysis as described by Lange and colleagues [49], [50]. with the software FISHER, which also performed tests of goodness-of-fit of the underlying multinormal distribution. The general model assumed that the studied trait was the result of the sum of three independent random components: a polygenic component (G) representing additive genetic factors, household factors common to individuals within a family (H) and unmeasured environmental factors particular to an individual (including measurement error) (E). These three components were assumed to be normally distributed with mean equal to 0 and variance equal to σ2G, σ2H and σ2E, respectively.
The hypothesis of no polygenic component or no household effect was checked by comparing a model including σ2G, σ2H and σ2E with a model including only σ2H and σ2E or σ2G and σ2E, respectively. In addition, possible effects of covariates (age and BMI) and genome-wide significant variants' allelic frequency on these variance components were tested.
Comparison of nested models was based on the likelihood ratio criteria. Eventually, the best parsimonious model was selected. The percentage contributions of the three components, additive genetic factors (heritability), household factors and residual environmental, to residual phenotypic variance (after adjustment for covariates) were determined.
Genome-wide association and replication analyses
We carried out genome-wide association and replication analyses on Hp levels using linear mixed regression models under the additive genetic model with one degree of freedom, adjusting for age, gender and BMI and using PLINK [51]. The summary statistics were combined in the meta-analyses (Data S2), using the inverse normal method with equal weight for each population. In this method, P values of each study are transformed into their inverse normal z score and the weighted sum, over all studies, is compared to a normal N (0, 1), provided the sum of squared weights equals 1. The estimates of variants effects on Hp and their standard errors for each separate analysis were combined in the meta-analysis using the weighted inverse normal method, and the overall effect and its confidence interval were estimated using the inverse variance method implemented in the ‘meta.summaries’ function of the R RMETA package (http://cran.r-project.org/web/packages/rmeta/index.html). No major heterogeneity in effects was observed (P<0.02). The same mixed model and the same software were used to analyse the association of genetic variants with lipid traits.
Gene-expression investigation
To investigate the effect of genome-wide significant variants on gene expression (Data S2), we used data from 194 non-obese individuals from the SibPair cohort [52]. Gene expression data for HP was measured in subcutaneous adipose tissue [53] from 347 siblings using the Affymetrix Human U133 Plus 2.0 platform (208470_s_at and 208471_at, respectively). DNA was isolated from peripheral blood and genotypes were generated using Illumina 610-Quad arrays.
We used a linear mixed model (Pinheiro and Bates, 2000) to assess association of SNPs with gene expression. Log-transformed expression level was regressed on the random-effect term, which accommodates the family pedigree structure, and on the fixed-effect terms i.e. sex, age, BMI level and the SNP of interest (recoded as 0 = AA; 1 = AG; 2 = GG according to an additive model). Analysis was carried out using the R function lmer() (package lme4) with p-values obtained from the t-statistic.
Significance of the fixed effects was further investigated in the Bayesian set-up using the R function mcmcsamp() (package lme4) that generates Monte Carlo Markov Chain samples from the posterior distribution of the parameters of a linear mixed model. The prior on the fixed effects parameters is taken to be locally uniform while the prior on the variance-covariance matrices of the random effects is taken to be the locally non-informative prior. Based on 100,000 samples drawn from the posterior distribution, we calculated the smallest p such that the (1−p) credible interval does not contain the value 0. This parameter was finally used to assess the p-value obtained from the t-statistic: if smaller than p, it was considered anticonservative and its value discarded.
Supporting Information
HP rs72294371 ‘common polymorphism’ flanking sequence. Source: 1000Genome. (http://browser.1000genomes.org/Homo_sapiens/Variation/Summary?r=16:72090747-72091766v=rs72294371vdb=variationvf=13544249).
(DOC)
Supplementary Methods. 1. Screening of latent population substructure. 2. Conditional analysis. 3. Meta-analysis. 4. Gene-expression analysis.
(DOC)
Conditional regression within the recombination hotspot around rs2000999. UNADJ: unadjusted p-values, SNP: single nucleotide polymorphism, add: additive model.
(XLS)
Association of rs2000999 with HDL-cholesterol and Apolipoproteins A1 and B. N: sample size; MAF: Minor Allele Frequency; β: beta coefficient for the effect allele A.
(DOC)
Acknowledgments
The authors would like to thank the study participants and their families for their time and effort to help create the present study. They would like to thank also all the field investigators for the recruitment and examinations of the populations involved in this study.
We thank Sidonie Vivequin for the double reading of all gels that were generated for the genotyping of the HP ‘common polymorphism’, Jean-Claude Chèvre and Franck De Greave for bioinformatics support, Marianne Deweirder and Michèle Pfister for DNA preparation.
Footnotes
Competing Interests: The GENDAI cohort was funded by a research grant from Coca-Cola Hellas. The commercial funders gave general support for the recruitment or realization of some measurements in the populations and have no relevant declarations relating to employment, consultancy, patents, products in development or marketed products, etc. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The STANISLAS Family Study was supported by the “Caisse Nationale d'Assurance Maladies des Travailleurs Salariés” (CNAM), the “Institut National de la Santé et de la Recherche Médicale” (INSERM), the “Région Lorraine”, the “Communauté Urbaine du Grand Nancy”, the Henri Poincaré University of Nancy 1, and the “BioIntelligence” project; the Verona and French Obese children cohorts were supported by funding from “Contrat Plan Etat Région” entitled “Genetic dissection of polygenic obesity using GWA approach” and the “Fond Européen pour le Développement Régional”; the NFBC1986 has received financial support from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu, Finland, the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643), the National Heart, Lung, and Blood Institute grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), National Institutes of Health/National Institute of Mental Health (5R01MH63706:02), ENGAGE project and grant agreement HEALTH-F4-2007-201413, and the Medical Research Council (G0500539, G0600705, PrevMetSyn/SALVE). The DNA extractions, sample quality controls, biobank up-keeping and aliquotting was performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki; The GENDAI cohort was funded by a research grant from Coca-Cola Hellas; The Sib Pair cohort was supported by the Swedish Research Council (K2010-55X-11285-13), the Swedish Foundation for Strategic Research to Sahlgrenska Centre for Cardiovascular and Metabolic Research and the Swedish federal government under the LUA/ALF agreement. The haptoglobin measurements of replication populations were supported by SIEMENS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 2.Bowman BH, Kurosky A. Haptoglobin: the evolutionary product of duplication, unequal crossing over, and point mutation. Adv Hum Genet. 1982;12:453-184. doi: 10.1007/978-1-4615-8315-8_3. [DOI] [PubMed] [Google Scholar]
- 3.Dati F, Schumann G, Thomas L, Aguzzi F, Baudner S, et al. Consensus of a group of professional societies and diagnostic companies on guidelines for interim reference ranges for 14 proteins in serum based on the standardization against the IFCC/BCR/CAP Reference Material (CRM 470). International Federation of Clinical Chemistry. Community Bureau of Reference of the Commission of the European Communities. College of American Pathologists. Eur J Clin Chem Clin Biochem. 1996;34:517–520. [PubMed] [Google Scholar]
- 4.Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O, Ledue TB, et al. Reference distributions for the positive acute phase serum proteins, alpha1-acid glycoprotein (orosomucoid), alpha1-antitrypsin, and haptoglobin: a practical, simple, and clinically relevant approach in a large cohort. J Clin Lab Anal. 2000;14:284–292. doi: 10.1002/1098-2825(20001212)14:6<284::AID-JCLA7>3.0.CO;2-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quaye IK. Haptoglobin, inflammation and disease. Trans R Soc Trop Med Hyg. 2008;102:735–742. doi: 10.1016/j.trstmh.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki T, Yanagisawa Y, Nagai T. Analysis of the affinity of each haptoglobin polymer for hemoglobin by two-dimensional affinity electrophoresis. Clin Chim Acta. 1997;258:137–144. doi: 10.1016/s0009-8981(96)06468-6. [DOI] [PubMed] [Google Scholar]
- 7.Braeckman L, De Bacquer D, Delanghe J, Claeys L, De Backer G. Associations between haptoglobin polymorphism, lipids, lipoproteins and inflammatory variables. Atherosclerosis. 1999;143:383–388. doi: 10.1016/s0021-9150(98)00330-x. [DOI] [PubMed] [Google Scholar]
- 8.Balestrieri M, Cigliano L, Simone ML, Dale B, Abrescia P. Haptoglobin inhibits lecithin-cholesterol acyltransferase in human ovarian follicular fluid. Mol Reprod Dev. 2001;59:186–191. doi: 10.1002/mrd.1021. [DOI] [PubMed] [Google Scholar]
- 9.Salvatore A, Cigliano L, Carlucci A, Bucci EM, Abrescia P. Haptoglobin binds apolipoprotein E and influences cholesterol esterification in the cerebrospinal fluid. J Neurochem. 2009;110:255–263. doi: 10.1111/j.1471-4159.2009.06121.x. [DOI] [PubMed] [Google Scholar]
- 10.Kasvosve I, Speeckaert MM, Speeckaert R, Masukume G, Delanghe JR. Haptoglobin polymorphism and infection. Adv Clin Chem. 2010;50:23–46. doi: 10.1016/s0065-2423(10)50002-7. [DOI] [PubMed] [Google Scholar]
- 11.Langlois MR, Delanghe JR. Biological and clinical significance of haptoglobin polymorphism in humans. Clin Chem. 1996;42:1589–1600. [PubMed] [Google Scholar]
- 12.Asleh R, Marsh S, Shilkrut M, Binah O, Guetta J, et al. Genetically determined heterogeneity in hemoglobin scavenging and susceptibility to diabetic cardiovascular disease. Circ Res. 2003;92:1193–1200. doi: 10.1161/01.RES.0000076889.23082.F1. [DOI] [PubMed] [Google Scholar]
- 13.Rother RP, Bell L, Hillmen P, Gladwin MT. The clinical sequelae of intravascular hemolysis and extracellular plasma hemoglobin: a novel mechanism of human disease. Jama. 2005;293:1653–1662. doi: 10.1001/jama.293.13.1653. [DOI] [PubMed] [Google Scholar]
- 14.Mohapatra MK, Mohanty S, Mohanty BK, Sahu GN. Hypohaptoglobinaemia as a biochemical and epidemiological marker of falciparum malaria. J Assoc Physicians India. 1999;47:874–877. [PubMed] [Google Scholar]
- 15.Eisaev BA. [Results of the treatment of patients with recurrence of pulmonary tuberculosis with different types of haptoglobin]. Probl Tuberk. 1995:20–22. [PubMed] [Google Scholar]
- 16.Delanghe JR, Langlois MR, Boelaert JR, Van Acker J, Van Wanzeele F, et al. Haptoglobin polymorphism, iron metabolism and mortality in HIV infection. Aids. 1998;12:1027–1032. [PubMed] [Google Scholar]
- 17.Friis H, Gomo E, Nyazema N, Ndhlovu P, Krarup H, et al. Iron, haptoglobin phenotype, and HIV-1 viral load: a cross-sectional study among pregnant Zimbabwean women. J Acquir Immune Defic Syndr. 2003;33:74–81. doi: 10.1097/00126334-200305010-00011. [DOI] [PubMed] [Google Scholar]
- 18.Bacq Y, Schillio Y, Brechot JF, De Muret A, Dubois F, et al. [Decrease of haptoglobin serum level in patients with chronic viral hepatitis C]. Gastroenterol Clin Biol. 1993;17:364–369. [PubMed] [Google Scholar]
- 19.Louagie HK, Brouwer JT, Delanghe JR, De Buyzere ML, Leroux-Roels GG. Haptoglobin polymorphism and chronic hepatitis C. J Hepatol. 1996;25:10–14. doi: 10.1016/s0168-8278(96)80321-7. [DOI] [PubMed] [Google Scholar]
- 20.Van Vlierberghe H, Delanghe JR, De Bie S, Praet M, De Paepe A, et al. Association between Cys282Tyr missense mutation and haptoglobin phenotype polymorphism in patients with chronic hepatitis C. Eur J Gastroenterol Hepatol. 2001;13:1077–1081. doi: 10.1097/00042737-200109000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Delanghe J, Langlois M, Ouyang J, Claeys G, De Buyzere M, et al. Effect of haptoglobin phenotypes on growth of Streptococcus pyogenes. Clin Chem Lab Med. 1998;36:691–696. doi: 10.1515/CCLM.1998.122. [DOI] [PubMed] [Google Scholar]
- 22.Rohde KH, Dyer DW. Analysis of haptoglobin and hemoglobin-haptoglobin interactions with the Neisseria meningitidis TonB-dependent receptor HpuAB by flow cytometry. Infect Immun. 2004;72:2494–2506. doi: 10.1128/IAI.72.5.2494-2506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calderoni DR, Andrade Tdos S, Grotto HZ. Haptoglobin phenotype appears to affect the pathogenesis of American trypanosomiasis. Ann Trop Med Parasitol. 2006;100:213–221. doi: 10.1179/136485906X86356. [DOI] [PubMed] [Google Scholar]
- 24.Speeckaert R, Speeckaert MM, Padalko E, Claeys LR, Delanghe JR. The haptoglobin phenotype is associated with the Epstein-Barr virus antibody titer. Clin Chem Lab Med. 2009;47:826–828. doi: 10.1515/CCLM.2009.193. [DOI] [PubMed] [Google Scholar]
- 25.Asleh R, Guetta J, Kalet-Litman S, Miller-Lotan R, Levy AP. Haptoglobin genotype- and diabetes-dependent differences in iron-mediated oxidative stress in vitro and in vivo. Circ Res. 2005;96:435–441. doi: 10.1161/01.RES.0000156653.05853.b9. [DOI] [PubMed] [Google Scholar]
- 26.Levy AP, Roguin A, Hochberg I, Herer P, Marsh S, et al. Haptoglobin phenotype and vascular complications in patients with diabetes. N Engl J Med. 2000;343:969–970. doi: 10.1056/NEJM200009283431313. [DOI] [PubMed] [Google Scholar]
- 27.Ryndel M, Behre CJ, Brohall G, Prahl U, Schmidt C, et al. The haptoglobin 2-2 genotype is associated with carotid atherosclerosis in 64-year old women with established diabetes. Clin Chim Acta. 2010;411:500–504. doi: 10.1016/j.cca.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Holme I, Aastveit AH, Hammar N, Jungner I, Walldius G. Haptoglobin and risk of myocardial infarction, stroke, and congestive heart failure in 342,125 men and women in the Apolipoprotein MOrtality RISk study (AMORIS). Ann Med. 2009;41:522–32. doi: 10.1080/07853890903089453. [DOI] [PubMed] [Google Scholar]
- 29.Holme I, Aastveit AH, Hammar N, Jungner I, Walldius G. Inflammatory markers, lipoprotein components and risk of major cardiovascular events in 65,005 men and women in the Apolipoprotein MOrtality RISk study (AMORIS). Atherosclerosis. 2010;213:299–305. doi: 10.1016/j.atherosclerosis.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 30.Maeda N. Nucleotide sequence of the haptoglobin and haptoglobin-related gene pair. The haptoglobin-related gene contains a retrovirus-like element. J Biol Chem. 1985;260:6698–6709. [PubMed] [Google Scholar]
- 31.Koda Y, Soejima M, Yoshioka N, Kimura H. The haptoglobin-gene deletion responsible for anhaptoglobinemia. Am J Hum Genet. 1998;62:245–252. doi: 10.1086/301701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igl W, Johansson A, Wilson JF, Wild SH, Polasek O, et al. Modeling of environmental effects in genome-wide association studies identifies SLC2A2 and HP as novel loci influencing serum cholesterol levels. PLoS Genet. 2010;6:e1000798. doi: 10.1371/journal.pgen.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zethelius B, Berglund L, Sundstrom J, Ingelsson E, Basu S, et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med. 2008;358:2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 35.Melander O, Newton-Cheh C, Almgren P, Hedblad B, Berglund G, et al. Novel and conventional biomarkers for prediction of incident cardiovascular events in the community. JAMA. 2009;302:49–57. doi: 10.1001/jama.2009.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner SJ, Ketch TR, Gandhi SK, Sane DC. Routine hematologic clinical tests as prognostic markers in patients with acute coronary syndromes. Am Heart J. 2008;155:806–816. doi: 10.1016/j.ahj.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 37.Marcovina SM, Crea F, Davignon J, Kaski JC, Koenig W, et al. Biochemical and bioimaging markers for risk assessment and diagnosis in major cardiovascular diseases: a road to integration of complementary diagnostic tools. J Intern Med. 2007;261:214–234. doi: 10.1111/j.1365-2796.2006.01734.x. [DOI] [PubMed] [Google Scholar]
- 38.Raitakari OT, Juonala M, Kahonen M, Taittonen L, Laitinen T, et al. Cardiovascular risk factors in childhood and carotid artery intima-media thickness in adulthood: the Cardiovascular Risk in Young Finns Study. Jama. 2003;290:2277–2283. doi: 10.1001/jama.290.17.2277. [DOI] [PubMed] [Google Scholar]
- 39.Mahoney LT, Burns TL, Stanford W, Thompson BH, Witt JD, et al. Coronary risk factors measured in childhood and young adult life are associated with coronary artery calcification in young adults: the Muscatine Study. J Am Coll Cardiol. 1996;27:277–284. doi: 10.1016/0735-1097(95)00461-0. [DOI] [PubMed] [Google Scholar]
- 40.Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 41.Visvikis-Siest S, Siest G. The STANISLAS Cohort: a 10-year follow-up of supposed healthy families. Gene-environment interactions, reference values and evaluation of biomarkers in prevention of cardiovascular diseases. Clin Chem Lab Med. 2008;46:733–747. doi: 10.1515/CCLM.2008.178. [DOI] [PubMed] [Google Scholar]
- 42.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 43.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 44.Rolland-Cachera MF, Cole TJ, Sempe M, Tichet J, Rossignol C, et al. Body Mass Index variations: centiles from birth to 87 years. Eur J Clin Nutr. 1991;45:13–21. [PubMed] [Google Scholar]
- 45.Papoutsakis C, Vidra NV, Hatzopoulou I, Tzirkalli M, Farmaki AE, et al. The Gene-Diet Attica investigation on childhood obesity (GENDAI): overview of the study design. Clin Chem Lab Med. 2007;45:309–15. doi: 10.1515/CCLM.2007.080. [DOI] [PubMed] [Google Scholar]
- 46.Jarvelin MR, Elliott P, Kleinschmidt I, Martuzzi M, Grundy C, et al. Ecological and individual predictors of birthweight in a northern Finland birth cohort 1986. Paediatr Perinat Epidemiol. 1997;11:298–312. doi: 10.1111/j.1365-3016.1997.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 47.Koch W, Latz W, Eichinger M, Roguin A, Levy AP, et al. Genotyping of the common haptoglobin Hp 1/2 polymorphism based on PCR. Clin Chem. 2002;48:1377–1382. [PubMed] [Google Scholar]
- 48.Donner A, Koval JJ. A multivariate analysis of family data. Am J Epidemiol. 1981;114:149–154. doi: 10.1093/oxfordjournals.aje.a113162. [DOI] [PubMed] [Google Scholar]
- 49.Lange K, Weeks D, Boehnke M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 50.Lange K, Westlake J, Spence MA. Extensions to pedigree analysis. III. Variance components by the scoring method. Ann Hum Genet. 1976;39:485–491. doi: 10.1111/j.1469-1809.1976.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 51.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlsson LM, Jacobson P, Walley A, Froguel P, Sjostrom L, et al. ALK7 expression is specific for adipose tissue, reduced in obesity and correlates to factors implicated in metabolic disease. Biochem Biophys Res Commun. 2009;382:309–314. doi: 10.1016/j.bbrc.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trayhurn P, Wood IS. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br J Nutr. 2004;92:347–355. doi: 10.1079/bjn20041213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HP rs72294371 ‘common polymorphism’ flanking sequence. Source: 1000Genome. (http://browser.1000genomes.org/Homo_sapiens/Variation/Summary?r=16:72090747-72091766v=rs72294371vdb=variationvf=13544249).
(DOC)
Supplementary Methods. 1. Screening of latent population substructure. 2. Conditional analysis. 3. Meta-analysis. 4. Gene-expression analysis.
(DOC)
Conditional regression within the recombination hotspot around rs2000999. UNADJ: unadjusted p-values, SNP: single nucleotide polymorphism, add: additive model.
(XLS)
Association of rs2000999 with HDL-cholesterol and Apolipoproteins A1 and B. N: sample size; MAF: Minor Allele Frequency; β: beta coefficient for the effect allele A.
(DOC)