Abstract
ArgR is the regulator of arginine biosynthesis genes in Streptomyces species. Transcriptomic comparison by microarrays has been made between Streptomyces coelicolor M145 and its mutant S. coelicolor ΔargR under control, unsupplemented conditions, and in the presence of arginine. Expression of 459 genes was different in transcriptomic assays, but only 27 genes were affected by arginine supplementation. Arginine and pyrimidine biosynthesis genes were derepressed by the lack of ArgR, while no strong effect on expression resulted on arginine supplementation. Several nitrogen metabolism genes expression as glnK, glnA and glnII, were downregulated in S. coelicolor ΔargR. In addition, downregulation of genes for the yellow type I polyketide CPK antibiotic and for the antibiotic regulatory genes afsS and scbR was observed. The transcriptomic data were validated by either reverse transcription-PCR, expression of the gene-promoter coupled to the luciferase gene, proteomic or by electrophoresis mobility shift assay (EMSA) using pure Strep-tagged ArgR. Two ARG-boxes in the arginine operon genes suggest that these genes are more tightly controlled. Other genes, including genes encoding regulatory proteins, possess a DNA sequence formed by a single ARG-box which responds to ArgR, as validated by EMSA.
Introduction
Arginine metabolism is feedback repressed by arginine in different Gram-positive and Gram-negative bacteria. This effect is mediated by ArgR, a hexameric protein that represses arginine biosynthesis genes, using L-arginine as co-repressor, in Escherichia coli [1] and Pseudomonas [2]. A similar effect is exerted by the homologous regulatory protein AhrC in Bacillus subtilis [3] or Lactococcus [4] and by ArgR in Corynebacterium [5].
Characterization of argR, encoding the ArgR repressor [6] and the use of AhrC protein permitted to understand the arginine biosynthesis cluster regulation in Streptomyces clavuligerus and to locate ARG-boxes, for ArgR binding, upstream of several arginine biosynthesis genes [7]. This provided the basis to study arginine regulation in other Streptomyces species. This group of soil-dwelling bacteria produces many secondary metabolites that use arginine, or arginine-related molecules, as precursors. Streptomycin, mitomycin, streptothricin or clavulanic acid are metabolites that contain moieties of guanidine, carbamoyl groups, ornithine or arginine, all of which are compounds related to the arginine biosynthesis or catabolism [8]–[11]. The C5 moiety of clavulanic acid, produced by S. clavuligerus, derives directly from an arginine molecule [11]; in addition, undecylprodigiosin [12] and other metabolites contain proline, which as arginine derives from glutamate, and might share common regulatory mechanisms with arginine.
DNA microarrays and proteomic studies are useful tools to understand metabolic pathway regulation in microorganisms. In Pseudomonas, which has four arginine catabolic pathways: arginine deiminase (ADI), arginine succinyl transferase (AST), arginine decarboxylase (ADC) and arginine dehydrogenase (ADH), the transcriptomic studies identified 38 genes related to the ADH pathway that are induced by arginine in the absence of ArgR, as well as 27 arginine-induced genes of the AST pathway [2], [13]. Using transcriptomic studies Caldara et al. [14] have also found that arginine uptake systems encoded by the art and the hisJQMP genes are repressed by arginine in E. coli.
Transcriptomic studies have been very useful to understand the molecular mechanisms of global control by high hierarchy regulators, for example, A-factor regulation in Streptomyces griseus [15] and phosphate and nitrogen regulation in S. coelicolor [16]–[17]. These studies allowed us to understand networks interconnecting phosphate and nitrogen metabolism in Streptomyces [18], carbon regulation [19] and the connection between the global regulator AfsR, a protein controlling antibiotic production in S. coelicolor, and the phosphate regulation exerted by PhoP, the response regulator of the two components of the PhoRP system that controls the pho regulon [20].
Therefore, it was convenient to analyze gene expression under control and arginine-supplemented conditions, to obtain better knowledge about arginine transport and catabolism in this model microorganism, and to determine whether ArgR is a regulatory protein involved only in arginine control, or if it has wider regulatory functions.
Results
Comparison of S. coelicolor strains grown in the presence and absence of arginine
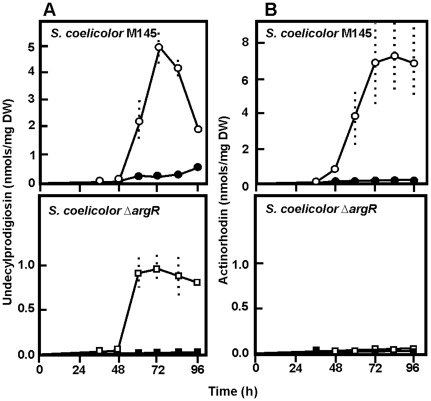
Experiments were performed to define the optimal conditions to achieve similar growth kinetics in MG medium, as well as good reproducibility, for the parental strain S. coelicolor M145 and the argR (SCO1576)-deleted mutant S. coelicolor ΔargR. A high arginine concentration is required in Streptomyces to produce a clear effect on enzymes of the arginine biosynthesis pathway in MG medium [6], [21], therefore, the MG cultures were supplemented with 25 mM arginine. The pattern of arginine utilization was similar for both strains; approximately 15% of the arginine was consumed at 32 h of growth and no arginine was left at 96 h. Antibiotic onset in liquid MG medium using S. coelicolor M145 occurred at about 40 h for actinorhodin and 50 h for undecylprodigiosin. Under non-supplemented conditions, S. coelicolor ΔargR produced only about 20% undecylprodigiosin in relation to the parental strain and lacked completely actinorhodin formation. Arginine supplementation strongly impaired the production of the pigments in both strains (Fig. 1).
Figure 1. Growth and antibiotic production by S. coelicolor M145 and S. coelicolor ΔargR.
(A) Undecylprodigiosin production. (B) Actinorhodin production. Antibiotic production in MG medium by S. coelicolor M145 (white circles) and S. coelicolor ΔargR (white squares) and in MG supplemented with 25 mM arginine (black circles and squares). Vertical bars show standard deviation of three replicates.
Transcription profiles of S. coelicolor M145 and the ΔargR mutant in response to arginine
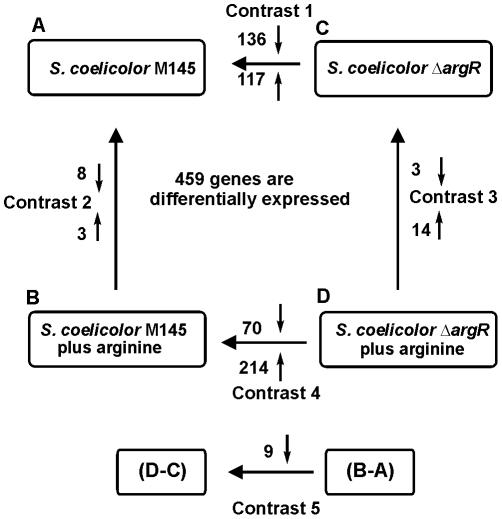
The statistical analysis of four biological replicates for each experimental condition indicated in Materials and Methods identified 459 genes with significant differential transcription (Dataset S1) in at least one out of the five contrasts shown in Fig. 2. Only 27 genes (6.1%) were differentially expressed in the arginine-supplemented conditions, with respect to the control, indicating a weak transcriptional response to the presence of arginine (Fig. 2, contrasts 2 and 3). Most of the 459 differentially expressed genes corresponded to the comparisons between the wild-type strain and the ΔargR mutant in control (contrast 1) or arginine-supplemented (contrast 4) cultures. About 50% of the genes differentially expressed encode membrane proteins, secreted proteins or proteins with unknown functions.
Figure 2. Scheme showing the number of genes differentially expressed in the five contrasts.
S. coelicolor M145 and S. coelicolor ΔargR were grown in absence (A and C) or in presence of 25 mM arginine (B and D). The arrow orientation refers to the comparison between two conditions. Differential transcription values were obtained by subtracting the Mg values of the first condition from those of the second condition (corresponding to the arrowheads). The small vertical arrows indicate the number of genes up-expressed (up-oriented arrows) or down-expressed (down-oriented arrows) in each of the contrasts. Contrast 5 is the interaction contrast that reflects the differential response to the arginine supplementation of the two strains.
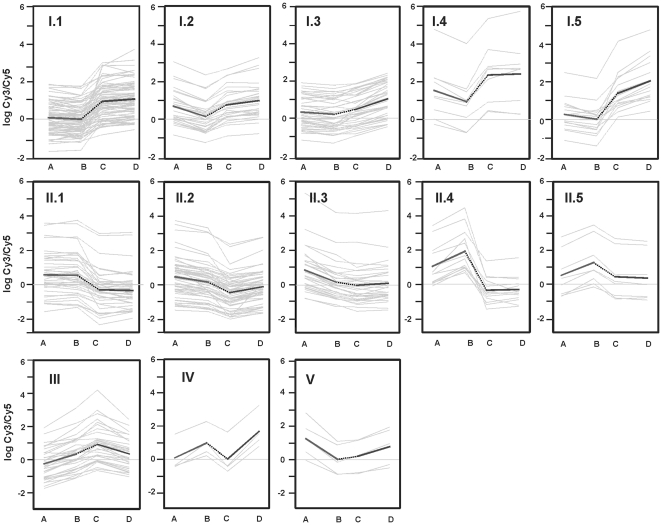
The transcriptional profiles analysis permitted to establish a classification of the genes into five types. The patterns of genes differentially transcribed in the two strains in unsupplemented or supplemented conditions fell mainly into types I and II (Fig. 3) with different modulations of the expression (subtypes 1–5). Forty-three genes showed the pattern of expression indicated in types III to V (Fig. 3). Here we will analyze general genes included in the five different types, while genes with specific functions will be analyzed in subsequent sections.
Figure 3. Transcription profiles.
Genes differentially expressed in MG cultures of S. coelicolor M145 (A); S. coelicolor M145 supplemented with arginine (B); S. coelicolor ΔargR (C) and S. coelicolor ΔargR supplemented with arginine (D). The genes have been grouped in types and subtypes according with their expression profile. Each grey line corresponds to the change in transcription of a given gene. The thicker line is the transcription mean profile for all the genes in the proposed group.
Type I genes
For type I genes, ArgR appeared to act as a repressor protein. Arginine itself did not affect expression of 76 genes (subtype I.1) and produced a small negative effect in the presence of ArgR or a stimulatory effect in the absence of ArgR in the other 69 genes (subtypes I.2 and I.3, respectively), which was stronger in subtypes I.4 and I.5. The arginine effect, if detected, was negative in the parental strain, but positive in the mutant strain (subtypes I.2–I.5). Thus, arginine appeared to function as a co-repressor for the genes classified into types I.2, I.4 and I.5. Surprisingly, subtypes I.1–I.3 comprise 14 genes for regulatory proteins including regulators of the TetR, DeoR, AraC, GntR or LipR families and sigma, anti-sigma or anti-sigma antagonist factors (Table S1). Other genes of type I encode proteases, peptidases or aminotransferases, adenosine deaminases and glutamyl-tRNA-charging amidotransferases, or are related to amino acid metabolism, such as those for serine hydroxymethyltransferase (SCO4837), NAD-dependent glutamate dehydrogenase (SCO2999), glutamate binding protein (SCO5776), and the B12-dependent and the B12-independent methionine synthases MetH and MetE (SCO1657 and SCO0985, respectively). Two genes involved in sporulation regulation, and five genes for aerial mycelium formation are also included in this group (Table S1).
Type II genes
For type II genes, ArgR behaved as an activator. Addition of arginine (contrasts 2 and 3) did not modify these genes expression (42 genes in type II.1) or produced a slightly positive or negative differential expression in the presence or absence of ArgR, respectively (56 and 32 genes in subtypes II.2 and II.3). Genes encoding regulators of AsnC, AraC, PadR, AbaA, TetR or DeoR type or members of two component systems are present in these subtypes. Type II includes genes for proteases, peptidases and protease inhibitors (Table S1). Some secondary metabolism genes, such as those encoding the putative actinorhodin transporter (SCO5083) and the butyrolactone-binding protein ScbR (SCO6265), belong to this type.
Subtypes II.4 and II.5 include 20 genes that were upregulated by arginine supplementation in the parental strain (contrast 1), but not in the argR mutant. The most interesting genes in these subtypes are the sigma-like afsS gene (SCO4425), a gene that encodes a tetracenomycin-like transporter (SCO2373) and a member of the two component system for nitrate sensing (SCO1160).
Type III genes
This type comprises 34 genes whose transcription was stimulated by arginine in the strain containing ArgR but downregulated in the absence of ArgR (i.e. the opposite behaviour to that of types I.4 and I.5). Genes of relevance in this type are transcriptional regulators (SCO2184 and SCO3769), a member of a two-component system (SCO6421), and the ATP-dependent polyphosphate kinase (SCO1781).
Type IV genes
The expression of the four genes of this type was stimulated by arginine, independently on the presence of ArgR. Two of these genes encode, respectively, an amidinotransferase (SCO1222) and the γ-aminobutyric acid aminotransferase (gabT, SCO5676).
Type V genes
The five genes in this type showed patterns similar to those of types I.2 or I.5, with a more marked effect in the M145 strain. Two genes related to amino acid metabolism, a cysteine desulfurase and a threonine synthase (SCO2146 and SCO4293) are included in this type.
Regulation of arginine biosynthesis and transport genes as measured by transcriptomics
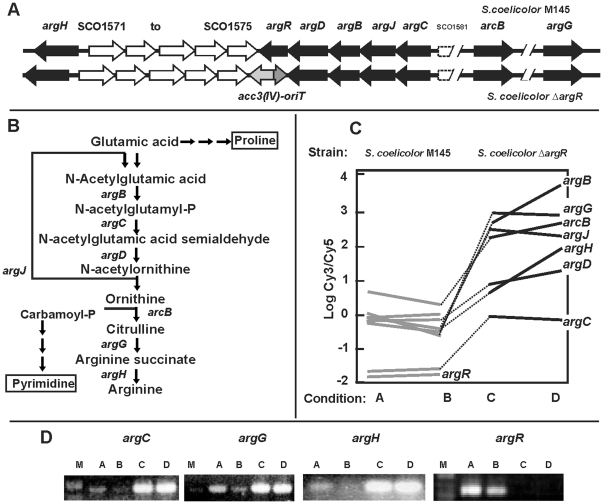
Arginine biosynthesis genes in S. coelicolor are organized in the argCJBDR cluster (SCO1580–1576) and in three separate genes: argH (SCO1570), arcB (SCO5976, located in the arc cluster) and argG (SCO7036) (Fig. 4). Genes involved in the formation of carbamoyl-phosphate (carAB) and required to form citrulline from ornithine will be considered in the next section. Our transcriptional data of the S. coelicolor parental strain (containing ArgR) did not show significant arginine-dependent repression for most arginine biosynthetic genes (argCJD, argG and arcB are in the arginine-not-affected type I.1), in contrast to E. coli [14]. On the other hand, arginine supplementation in the ΔargR mutant cultures caused a slight upregulation of some arg genes as argH (type I.3) and argB (type I.5) with a 2.4 and 2.1-fold increase, respectively. The main transcriptional differences were the higher transcription in the ΔargR mutant (from 1.6-fold in argD to seven fold in argB or arcB) in relation to the wild-type strain. A putative gene, SCO1581, divergent to argC, is not differentially expressed in any condition.
Figure 4. Arginine biosynthesis genes.
(A) Cluster of genes for arginine biosynthesis (black arrows) in S. coelicolor M145 (above) and the S. coelicolor ΔargR mutant (below). SCO1581 is indicated with broken lane since it might not be a real gene. (B) Arginine biosynthesis pathway showing its relation to proline and to pyrimidine biosynthesis via carbamoyl-phosphate incorporation. (C) Expression of the arginine biosynthesis genes in A, B, C and D conditions as indicated in Fig. 3. (D) RT-PCR amplification of the argC, argG, argH and argR genes in the same conditions and strains as above. M, Markers.
The SCO5258–5260 genes, homologous to Pseudomonas aeruginosa aotJQMOP encoding proteins for arginine/ornithine uptake, were similarly transcribed in all conditions. However, transcription of the SCO5776–5777 genes, annotated as a putative ATPase and binding proteins for glutamate uptake (an arginine precursor) were negatively modulated by arginine and activated in the ΔargR mutant.
Arginine catabolism
S. coelicolor M145 grew well on plates containing ornithine, arginine, urea or citrulline as sole carbon or nitrogen source. No significant differences were found in the growth of the S. coelicolor ΔargR mutant, with only a slightly lower growth on citrulline.
Of the multiple catabolic pathways for arginine catabolism found in other bacteria, only two catabolic genes, arcA and arcA2 (SCO0613 and SCO5975), both for arginine deiminase, have been identified in S. coelicolor genome. They have a high similarity to Pseudomonas or E. coli arcA genes but they were not differentially expressed in our experimental conditions. All the genes of the ADC pathway of Pseudomonas, forming succinate from arginine, have homologous genes scattered in S. coelicolor genome (SCO7311, SCO5527, SCO6414, SCO5655, SCO4913, SCO5676 and SCO7035) with e-value scores of 10−80 to 10−101. These genes were not affected by arginine or by the ArgR regulator, with the exception of gabT (SCO5676), which encodes the enzyme that catabolizes 4-aminobutyrate to form succinate semialdehyde. Expression of this type IV gene was twice as high in cultures supplemented with arginine than in unsupplemented cultures of both the M145 and the ΔargR strain.
The AST pathway is encoded by the aruCFGDBE genes in P. aeruginosa [4]. In S. coelicolor, an aruC homologue, gabT (SCO5676, discussed above) was stimulated by arginine and SCO1865 (subtype I.1), encoding an aminotransferase, was overexpressed in the ΔargR mutant. Homologues of aruF or aruG are not present in S. coelicolor and those genes encoding proteins similar to AruD did not show differential expression under any conditions.
Genes encoding standard arginases (EC.3.5.3.1) have not been found in the S. coelicolor genome. Nevertheless, arginase activity is possibly associated with the carbamoyl-phosphate transferase activity of ArcB, as occurs in S. clavuligerus [22]. Expression of arcB (and the putative arginase activity) was strongly repressed in the presence of ArgR, as indicated above. No effect due to arginine or to the lack of ArgR was observed in the urea-utilizing genes (ureDGFCBA, SCO1231–1236, and ureABC, SCO5525–5526).
Nucleotides and nucleic acid-biosynthesis-related genes
Pyrimidine biosynthesis is related to arginine biosynthesis through the utilization of carbamoyl-phosphate (CP). CP is formed by the CP synthase, encoded by pyrA and pyrAa (Figs. 4 and 5) and condensed to ornithine by the ornithine carbamoyltransferase (arcB) to yield citrulline, and to aspartate to form carbamoyl-aspartate, by the aspartate carbamoyl transferase (pyrB). Both arcB (I.1 subtype) and pyrB genes were upregulated in the ΔargR mutant.
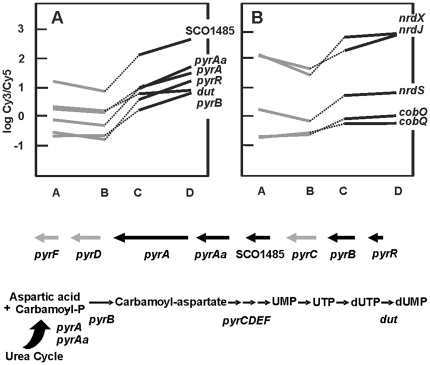
Figure 5. Differential expression of nucleotides biosynthesis genes.
(A) Expression of genes related to pyrimidine biosynthesis. A scheme of the gene cluster and biosynthesis pathway is shown below. Genes marked as black arrows were differentially expressed. (B) Expression of genes related to deoxyribonucleotides and cobalamin biosynthesis. A, B, C and D correspond to the conditions indicated in Fig. 3. Notice that nrdX is not a real gene but a transcribed regulatory intergenic region.
In the pyrimidine pathway, carbamoyl-aspartate is sequentially converted in UMP through the action of enzymes encoded by the pyrB, pyrC, pyrD, pyrE and pyrF genes [23], clustered in S. coelicolor, with exception of pyrE (Fig. 5A). Several genes in this cluster showed a significantly higher expression in the ΔargR mutant than in the parental strain (1.6–2.2-fold; contrast 1). Addition of arginine slightly reduced their transcription in the M145 strain but it increased in the ΔargR mutant (2.7–4-fold; contrast 4). Two sets of these genes were especially affected: pyrR–pyrB and SCO1485–pyrA–pyrAa showing profiles of type I.5, except for SCO1485 which was of type I.4. No significant differential expression of the pyrE gene, located outside of the cluster, was detected.
Expression of other genes directly or indirectly, involved in nucleotide metabolism was higher in the argR mutant. They included SCO2015 (encoding a putative secreted nucleotidase), SCO5662 (adenosine deaminase), SCO3060 (purK, phosphoribosylaminoimidazole carboxylase ATPase), SCO5743 (thyX, thymidylate synthase), and SCO5868 (dut, deoxyuridine 5′-triphosphate nucleotidohydrolase).
Two sets of independent ribonucleotide reductases (RNRs) in Streptomyces are encoded by the nrdABS (SCO5226–5224) and nrdRJ clusters (SCO5804–5805). The first cluster encodes the oxygen-dependent RNR and is regulated by a B12-dependent riboswitch located upstream of nrdA [24]. The microarray probes for the riboswitch region (associated to the fake gene nrdX because of the original genome annotation), the nrdS (encoding an AraC-like regulator) and the nrdJ (encoding the class II RNR) genes, indicated a 30–40% expression decrease in arginine supplemented cultures of the wild-type strain, and a higher expression in the ΔargR-mutant (up to 1.5-fold), especially in the presence of arginine (Fig. 5B). Two genes, cobQ and cobO (SCO1848 and SCO1851) for B12 biosynthesis had the type I.1 profile.
Effect of arginine on the nitrogen metabolic network
The nitrogen utilization network in Streptomyces uses the glutamine synthetase, encoded by glnA, as a key enzyme and is strictly regulated by the OmpR-like regulators GlnR and GlnRII [17].
Our data showed that the transcription of several nitrogen metabolism genes was controlled by ArgR. The glnK gene (SCO5584), encoding a PII-like regulator, was downregulated in the ΔargR mutant (II.2 type). The same regulation was found for the glnK flanking genes, amtB and glnD, encoding respectively an ammonium tranporter and the PII-modifying adenylyltransferase. Similarly, glnII (SCO2210) that encodes the eukaryotic-like glutamine synthetase GlnII, and glnA (SCO2198) that encodes the prokaryotic type I glutamine synthetase, showed profiles of type II.1 and II.4. Therefore, all these genes appeared to require ArgR for optimal expression. No differences in transcription were found for the regulators glnRII and glnR (SCO2213 and SCO4159, respectively). Thus, it seems that the positive regulation of ArgR on nitrogen metabolism is not mediated by GlnR or GlnRII.
Effect on morphology, differentiation and secondary metabolism genes
Arginine addition or deletion of argR resulted in a drastic reduction of actinorhodin and undecylprodigiosin, or even in complete lack of production of the latter pigment (Fig. 1). Indeed, two genes of the act pathway, actII-orf2 (SCO5083) encoding a putative actinorhodin transporter [25] (profile II.3) and the ketoacylreductase ActIII (SCO5086; II.4 profile) were affected. Probably due to the early time of culture used to extract RNA, no differences were found in expression of the undecylprodigiosin-encoding genes. The effect on production of this antibiotic (Fig. 1) was not related to the supply of the proline precursor, since the three genes of the proline pathway (proABC, SCO2585, SCO2587 and SCO3337) were equally expressed in both strains or culture conditions.
Two regulatory genes, essential for antibiotic formation, afsS (SCO4425) encoding a sigma-like protein and scbR (SCO6265) for a butyrolactone-receptor protein [26]–[27] showed II.5 and II.1 profiles, respectively, with a strong decrease in expression in the ΔargR mutant. The repression in afsS may also contribute to the drastic reduction of both pigmented antibiotics.
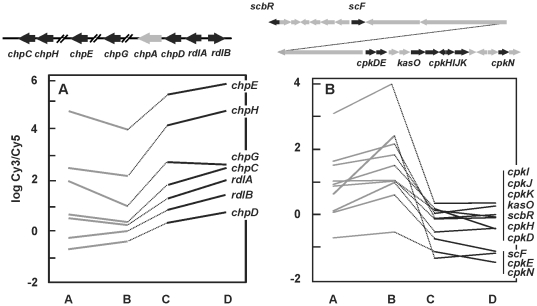
A very strong effect due to the lack of ArgR was observed on the biosynthesis of rodlins and chaplins, required to form aerial mycelium and spores [28], and on the genes encoding the yellow polyketide pigment CPK [29] (Fig. 6). Genes for chaplins are located in five different sites in the genome being chpA and chpD clustered with rdlA and rdlB for rodlins (SCO2716–2719). Although expression of chpF and chpA was not affected, the other genes showed profiles of the I.4 type (chpG and chpE, SCO2699 and SCO1800, respectively) and I.5 type (chpH, rdlA and chpC, SCO1675, SCO2718 and SCO1674, respectively).
Figure 6. Differential expression of genes related to morphology and secondary metabolism.
(A) Transcriptomic of genes for rodlins and chaplins. The cluster chp–rdl is shown above with differentially expressed genes in black. (B) Transcriptomic of the genes for CPK biosynthesis. The cpk cluster is shown above. Genes with differential expression are indicated in black. Included are the genes for scbR, for a butyrolactone receptor protein, and scF, for a putative secreted FAD-binding protein. A, B, C and D correspond to the conditions indicated in Fig. 3.
Most genes of the cpk cluster were downregulated in the ΔargR mutant. In the parental strain, higher transcription of some genes, especially cpkE, was detected in arginine-supplemented cultures. The profile of these genes was of II.1 type (kasO, cpkJ, cpkN, SCO6280, SCO6283 and SCO6288, respectively) or II.4 type (cpkDE, cpkHI, cpkK, SCO7276–7277, SCO6281–6282 and SCO6284, respectively) (Fig. 6B). The observed effect was probably due to a cascade effect mediated by the Streptomyces type of regulators encoded by kasO and cpkN, which were downregulated in the ΔargR mutant, but it may also reflect a general effect of ArgR on the glutamate supply, because the CPK antibiotic production is stimulated by glutamate [29]. Two genes located adjacent to the cpk cluster, scbR (already mentioned) and scF for a putative secreted FAD-binding protein, were also affected by the lack of ArgR (Fig. 6B).
Proteomic studies
Comparative analysis of the proteomes of S. coelicolor wild-type and ΔargR mutant grown with and without arginine supplementation revealed no significant differences, confirming the low effect of arginine detected in the transcriptomic studies. The largest proteome differences were found in the comparison of the wild-type and the ΔargR mutant. Twenty-six differentially represented proteins were identified, several of which were consistent with the transcriptomic results. They are listed in Table 1 (see Fig. 7 and Dataset S2 for details).
Table 1. Proteins differentially represented in S. coelicolor and S. coelicolor ΔargR.
| spot IN(1) | SCO n°(2) | gene name | annotated function(3) | p-value | up/down in ΔargR |
| 6149 | SCO0379 | katA | catalase | 0.000 | down |
| 6577 | SCO1570(a) | argH | argininosuccinate lyase | 0.005 | up |
| 6655 | SCO1580(a) | argC | N-acetyl-gamma-glutamyl-phosphate reductase | 0.000 | up |
| 6550 | SCO1814 | inhA | enoyl-(acyl carrier protein) reductase | 0.007 | Down |
| 6445 | SCO1935 | tktA1 | transketolase | 0.033 | Down |
| 6497 | SCO1945 | tpiA | triosephosphate isomerase | 0.015 | Down |
| 6233 | SCO1946 | pgk | phosphoglycerate kinase | 0.008 | Down |
| 6193 | SCO1947 | gap1 | glyceraldehyde-3-phosphate dehydrogenase | 0.029 | Down |
| 6666 | SCO1965 | export associated protein | 0.015 | Down | |
| 6494 | SCO1998 | rpsA | 30S ribosomal protein S1 | 0.009 | Up |
| 6143 | SCO2198(a) | glnA | glutamine synthetase I | 0.000 | Down |
| 6647 | SCO2368 | hypothetical protein | 0.007 | Down | |
| 6685 | SCO2633 | sodF | superoxide dismutase | 0.011 | Up |
| 6366 | SCO3649 | fba | fructose-bisphosphate aldolase | 0.012 | Down |
| 6264 | SCO4770 | guaB | inosine-5′-monophosphate dehydrogenase | 0.032 | Down |
| 6269 | SCO4771 | inositol-5′-monophosphate dehydrogenase | 0.040 | Down | |
| 6400 | SCO4809 | sucD | succinyl-CoA synthetase subunit alpha | 0.045 | Down |
| 6125 | SCO4814 | purH | formyltransferase/IMP cyclohydrolase | 0.030 | Down |
| 6167 | SCO4837(b) | glyA | serine hydroxymethyltransferase | 0.010 | Down |
| 6678 | SCO4856(a) | sdhA | succinate dehydrogenase flavoprotein subunit | 0.000 | Up |
| 6257 | SCO4958 | metB | cystathionine gamma-synthase | 0.027 | Down |
| 6340 | SCO6282(a) | cpkI | enoyl-(acyl carrier protein) reductase | 0.017 | Down |
| 6513 | SCO7036(a) | argG | argininosuccinate synthase | 0.000 | Up |
| 6771 | SCO7510 | cypH | peptidyl-prolyl cis-trans isomerase | 0.009 | Down |
| 6317 | SCO7511 | gap2 | glyceraldehyde 3-phosphate dehydrogenase | 0.026 | Down |
spot identification number automatically generated from ImageMaster software.
Among the 459 differentially transcribed genes, the products of seven genes were detected in the Proteomics experiments. Six genes showed the same transcriptomic and proteomic profile (a) and only one gene gave opposite results (b).
from ScoDB.
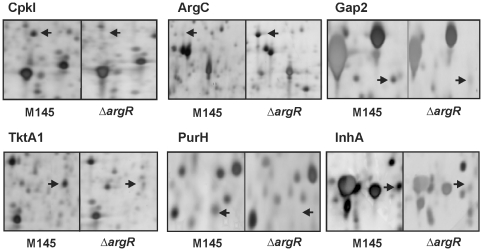
Figure 7. Proteomic Analysis.
Detailed view of 2D-SDS-PAGE gels showing differences on the proteomes of S. coelicolor M145 (left panels) and S. coelicolor ΔargR (right panels). Arrows indicate the protein spots differentially represented corresponding from left to right in the upper panels to CpkI (SCO6282), ArgC (SCO1580), and Gap2 (SCO7511), and in the lower panels to TktA1 (SCO1935), PurH (SCO4814) and InhA (SCO1814).
Arginine biosynthesis
In agreement with the transcriptomic results, several enzymes belonging to the arginine biosynthetic pathway were identified as protein spots only in the proteome of the mutant strain ΔargR. These included the N-acetyl-γ-glutamyl-phosphate reductase ArgC (SCO1580, spot IN 6655) and the last two enzymes in the pathway, argininosuccinate lyase ArgH (SCO1570, spot IN 6577), and argininosuccinate synthase ArgG (SCO7036, spot IN 6513).
Nitrogen metabolism proteins
Another transcriptomic and proteomic matching result was for glutamine synthetase I, GlnA (SCO2198, spot IN 6143), which was under-represented in the mutant proteome, confirming that glnA expression required ArgR as transcriptional activator. As glutamine is the amino donor for carbamoyl-phosphate synthesis, ArgR may influence the urea cycle and the pyrimidine biosynthesis by controlling the glutamine levels.
Pentose phosphate and glycolysis enzymes
Several proteins related to these pathways are under-represented in the S. coelicolor ΔargR proteome as compared with the wild-type. These the proteins are the transketolase TktA1 (encoded by SCO1935, spot IN 6445), that interconverts D-fructose-6-phosphate and D-xylulose-5-phosphate in the pentose phosphate pathway, which finally results in formation of D-glyceraldehyde-3-phosphate; the triose phosphate isomerase TpiA (SCO1945, spot IN 6497), which interconverts glyceraldehyde-3-phosphate and hydroxyacetone; the phosphoglycerate kinase Pgk (SCO1946, spot IN 6233) and a glyceraldehyde-3-phosphate dehydrogenase isoenzyme Gap1 (SCO1947, spot IN 6193). Also, the deoxyribose-phosphate aldolase encoded by SCO4914 (spot IN 6381), the fructose biphosphate aldolase Fba (SCO3649, spot IN 6366), and the second glyceraldehyde-3-phosphate dehydrogenase Gap2 (SCO7511, spot IN 6317), were under-represented in the ΔargR mutant when compared to the wild-type strain. These proteomic results revealed that glycolytic and pentose phosphate pathways were more active in the wild-type strain.
Proteins related to succinate metabolism
Two of the enzymes involved in arginine biosynthesis, ArgH and ArgG, have succinate either as substrate or reaction product. Surprisingly several proteins using succinate were under-represented in the mutant proteome. This is the case of MetB (SCO4958, spot IN 6257) a cystathionine γ-synthase that uses O-succinyl/acetyl homoserine as substrate and SucD (SCO4809, spot IN 6400) the α subunit of succinyl-CoA synthetase. Although no significant changes in the expression of these genes were observed, other genes related to succinate metabolism showed a differential expression in transcriptomic experiments. That is the case for gabT (forming succinate), the adenylosuccinate-synthetase-encoding gene purA (SCO3629, profile II.2) and for dhsA (SCO4856), dhsB (SCO4855) and SCO0922, three out of the 10 genes encoding succinate dehydrogenases which showed I.3 profile.
Purine metabolism
In the proteome of the wild-type strain, three protein spots involved in purine metabolism were detected, which were under-represented in the mutant strain. Proteins with inosine 5′-monophosphate (IMP) dehydrogenase (SCO4770) and 5-inositol-5-monophosphate dehydrogenase (SCO4771) were identified in spots IN 6264 and IN 6269, respectively. These activities carry out the conversion of IMP to xanthosine 5′-phosphate. The same profile was shown by the PurH protein, (spot IN 6125), encoded by SCO4814 involved in the synthesis of 1-(5′-phosphoribosyl)-5-formamide-4-imidazole carboxamide which connects purines and histidine pathways.
Other proteins
Three polyketide and fatty acid biosynthesis enzymes were also under-represented in the mutant proteome. The 3-oxoacyl-ACP reductase CpkI (encoded by SCO6282, spot IN 6648), required for the production of the yellow-pigmented CPK associated metabolite [29] which was consistent with the transcriptomic results. The enoyl-ACP-reductase FabI/InhA (SCO1814, spot IN 6550) was also under-represented.
Post-transcritional regulation, protein turnover, mRNA stability or translation efficiency might account for the low correlation in the transcriptomic and proteomic results in some cases [30].
Validation experiments: RT-PCR and heterologous genes expression
In addition to proteomic studies, validation of the transcriptomic results was performed for nearly 40 representative genes by either (i) RT-PCR, (ii) coupling promoters to the Vibrio harveyi promoter-less luxAB genes to measure luciferase activity and (iii) EMSA using a recombinant Strep-ArgR fusion protein (Table 2).
Table 2. Validation assays. Summary of EMSA, promoter probe assays or RT-PCR.
| EMSA | Promoter-luxAB fusion | RT-PCR | ||||||||||
| Promoter region | Probe chromosome coordinates | Ri (bits) of possible ARG boxes | Strep-ArgR binding(1) | Gene | Ratio of contrast(2) | Gene assayed | Results (3) | |||||
| 1 | 2 | 3 | 4 | |||||||||
| SCO0800–SCO0801 | 847420–847787 | 4,1 | ND | – | – | |||||||
| SCO1086 | 1146430–1146605 | 3,4 | 16,5 | + | – | – | ||||||
| SCO1220–SCO1221 | 1291917–1292299 | 5,2 | ND | – | – | |||||||
| SCO1236 (ureA) | 1310577–1310873 | 7,5 | + | – | – | |||||||
| SCO1483 (pyrA) | 1587383–1587719 | 1,8 | + | – | pyrA | ↑ΔargR | ||||||
| SCO1487 (pyrB) | 1591391–1591624 | 7,1 | + | pyrB | Low(4) | Low(4) | Low(4) | Low(4) | – | |||
| SCO1488 (pyrR) | 1592025–1592381 | 7,7 | 5,1 | + | pyrR | 2.3(5) | 2.3 | 1.6(5) | 1.6(5) | pyrR | ↑ΔargR | |
| SCO1570 (argH) | 1681863–1682008 | 17,3 | 7,7 | + | argH | 2.2 | 0.3 | 2.2(5) | 13.9(5) | argH | ↑ΔargR | |
| SCO1580 (argC)-SCO1581 | 1691340–1691628 | 12,1 | 9,4 | 6,1 | + | – | argC | ↑ΔargR | ||||
| SCO2014 (pyk1)-SCO2015 | 2157777–2157998 | 8,2 | + | pyk1 | Low(4) | Low(4) | Low(4) | Low(4) | – | |||
| SCO2054 (hisD)-SCO2055 | 2202564–2202769 | 5,7 | 4,4 | + | hisD | NS | 2.0 | NS | NS | – | ||
| SCO2210 (glnII)(6) | 2373630–2374004 | 5,3 | ND | – | glnII | ↓ΔargR | ||||||
| SCO2231 (malE)-SCO2232 (malR) | 2400346–2400648 | 9,3 | 8,6 | + | – | – | ||||||
| SCO2686 | 2930701–2931003 | 12,8 | + | – | – | |||||||
| SCO3034 (whiB) | 3321020–3321291 | 6,0 | + | whiB | NS | 0.5(5) | NS | 1.7(5) | whiB | ↑ΔargR | ||
| SCO3067–SCO3068 (sig15) | 3360383–3360707 | 5,2 | + | – | – | |||||||
| SCO3943 (rstP) | 4339152–4339343 | 9,2 | + | – | – | |||||||
| SCO3978–SCO3979 | 4381777–4382142 | 10,1 | + | – | – | |||||||
| SCO4158(7) | 4381777–4382142 | 7,4 | ND | – | – | |||||||
| SCO4293 | 4708341–4708657 | 9,1 | + | – | – | |||||||
| leuA-SCO2529 | 2727205–2727535 | 10,4 | + | leuA | NS | NS | NS | 1.3 | – | |||
| SCO5226 (nrdA) | 5688147–5688631 | 4,6 | 6,0 | + | nrdA | NS | NS | 1.6 | 1.4(5) | – | ||
| SC5583 (amtB) | 6085631–6086027 | 8,3 | + | amtB | NS | 1.5 | 0.01 | 0.01(5) | – | |||
| SCO5864 | 6421240–6421614 | 7,0 | + | – | – | |||||||
| SCO5976 (arcB) | 6550204–6550395 | 10,9 | 9,5 | + | arcB | 3.6(5) | 2.0 | 2.3 | 4.1(5) | – | ||
| SCO7036 (argG) | 7824665–7824928 | 8,1 | 16,7 | + | – | argG | ↑ΔargR | |||||
| SCO7302–SCO7303 | 8110855–8111154 | 7,4 | 4,6 | ND | – | – | ||||||
| SCO7314 | 8120320–8120720 | 7,5 | + | – | – | |||||||
| – | SCO1485(8) | ↑ΔargR | ||||||||||
| – | SCO1674 (chpC)(9) | ↑ΔargR | ||||||||||
| – | SCO1800 (chpE)(9) | ↑ΔargR | ||||||||||
| – | SCO2198 (glnA)(9) | ↓ΔargR | ||||||||||
| – | SCO2718 (rdlA)(9) | ↑ΔargR | ||||||||||
| – | SCO5584 (glnK)(10) | ↓ΔargR | ||||||||||
| – | SCO6265 (scbR)(11) | ↓ΔargR | ||||||||||
| – | SCO6282 (cpkI)(9) | ↓ΔargR | ||||||||||
| – | SCO6283 (cpkJ)(9) | ↓ΔargR | ||||||||||
ND, not detected.
Values are ratios of specific luminescences at 32 h of culture. Values are shown when t-test p-value <0.05 (4 biological replicates for each condition). NS, no significant differences. 1, 2, 3 and 4 as in Figure 2.
Relative up or down expression of the gene in mutant strain.
Luminescence measures too low (<1 arbitrary unit).
Microarray results (when uncorrected p-value <0.05 for the contrast, if not, no comparison was done) showed the same sign of regulation.
The probe does not correspond to the promoter region but to the upstream gene SCO2209 which contains the predicted ARG box.
The predicted ARG box is located in the 25 nucleotides of the 3′end of SCO4159 (glnR).
Predicted polycistronic transcript from SCO1488 (pyrR).
Putative ARG boxes not found (Ri≥3, 800-nt region centered at the start codon).
Predicted polycistronic transcript from SCO5583 (amtB).
Predicted ARG box overlapping the start codon, Ri 7.7 bits.
Genes for arginine biosynthesis (argC, argR, argH and argG), secondary metabolism (cpkJ and cpkI) and morphology (whiB, scbR, rdlA, chpE and chpC), controlling nitrogen metabolism (glnA, glnK and glnII), and for pyrimidine biosynthesis (pyrA, pyrR and SCO1485), were tested. In all cases, the amplification pattern observed was concordant with the microarray experiment (Figs. 4D and 8A).
Figure 8. Validation experiments by RT-PCR and EMSA.
(A) RT-PCR amplification of mRNA corresponding to the rdlA, cpkJ, (upper panels), SCO1485, scbR (medium panels), glnA and glnII (lower panels) genes. A, B, C and D correspond to the conditions indicated in Fig. 3. M, Marker. (B) EMSA of 6-FAM labelled argH (199 bp) and arcB (251 bp) promoters using Strep-ArgR protein. P indicates the free probe and B the binding reaction.
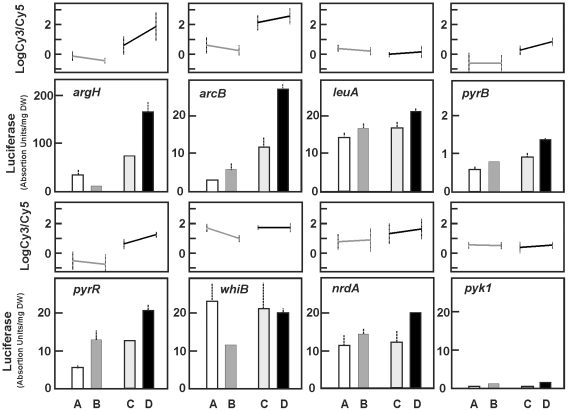
Promoters of differentially expressed genes were fused to the promoter-less luxAB genes; these constructions were introduced into S. coelicolor M145 and S. coelicolor ΔargR and cultured in MG medium with or without arginine supplementation. The promoters tested were those belonging to argH and arcB for arginine biosynthesis, pyrR, pyrB and nrdA (SCO5226) related to nucleotide biosynthesis, leuA for amino acids biosynthesis, whiB for morphological differentiation, and SCO1086, a strongly ArgR affected gene, of unknown function. The promoter of the pyk1 gene, which is constitutively expressed in our culture conditions, was used as a negative control. In general, the luminescence values in S. coelicolor ΔargR exconjugants correlated well with the transcriptomic experiment, although S. coelicolor M145 exconjugants in the presence of arginine gave a slightly higher activity than that expected from the microarray expression data (Fig. 9).
Figure 9. Validation experiments by heterologous gene expression.
Luciferase activity of luxAB-fused promoters transformed in the adequate S. coelicolor strain and grown in the A, B, C and D conditions indicated in Fig. 3. Activity corresponds to the promoter of argH, arcB, leuA, pyrB (upper panels) pyrR, whiB, nrdA and pyk1 (lower panels). On top of each panel is shown the expression profile of the corresponding gene in the transcriptomic studies. The luciferase activity values correspond to 32-h cultures. Vertical bars show standard deviation of two biological replicates measured twice.
Identification of ArgR binding sites
With the aim of finding putative ArgR binding sites that correlated with the differential expression results, we made a matrix (model 1) from the alignment of the 16 experimentally tested S. clavuligerus ARG boxes upstream of argC, argG, argH and arcB [6] and their complementary sequences. The S. coelicolor 5′ regions were scanned by means of the Patser algorithm and the RSA tools server. A neutral separator of two positions was inserted between a duplicated matrix to take into account the tandem arrangement of the ARG boxes. Using model 1 the only sequences with Ri values (individual information content) higher than 12.2 were those of the above ARG boxes, and a new one in the promoter region of SCO1086. This gene encodes a protein with a transglutaminase-like motif (PF01841) that might be a protease according to Makarova et al. [31] and is repressed by ArgR (1.5 profile) as predicted by Castro-Melchor et al. [32].
Although no more ArgR binding sites were detected, more ArgR binding sites in S. coelicolor genome could have been expected. They might include low-conserved ARG boxes, boxes with different length of separation, or single (half-length) ARG boxes, as occurs in Thermotoga or B. subtilis [33]–[34]. A single putative ARG box of Ri 10.4 was found upstream of the leuA (SCO2528) gene. This region was found to be clearly retarded by ArgR. The 22 ARG boxes of argC, argG, argH, arcB, SCO1086 and leuA were integrated in a new weight matrix. In the new ARG box Streptomyces model (model 2, Fig. 10C), the binding site was formed by an imperfect 20 nt palindromic sequence. This matrix allowed us to locate 1583 ARG box candidates with a Ri>5. Manual selection of them was performed, paying attention to those located in the neighbouring of differentially transcribed genes.
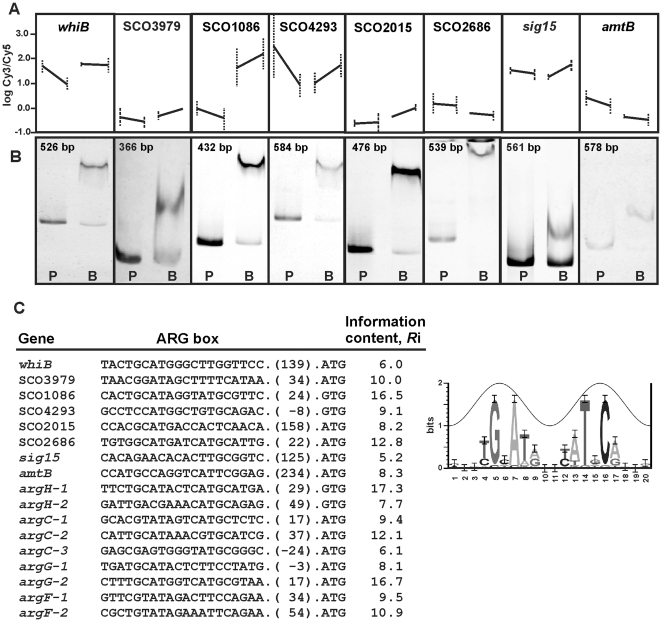
Figure 10. Functional analysis of ARG boxes by EMSA.
(A) Transcriptomic expression of the tested genes in the A, B, C and D conditions indicated in Fig. 3. (B) EMSA of PCR-amplified DNA fragments containing putative ARG boxes located upstream of the indicated genes. In all cases, P indicates the free probe and B binding reaction using ArgR (0.8 µM protein). The size of the probes is indicated in the panels. (C) Sequence of the putative ARG box present upstream of the indicated gene and Ri value determined using the ARG box model represented on the right. The numbers in parenthesis indicate the nucleotides to the translation start codon. The argH, argC, argG and arcB ARG boxes are included for comparison.
Nearly thirty DNA fragments containing putative ARG boxes were used in binding assays in the presence of Strep-ArgR in EMSA experiments. They correspond to upstream regions of: (i) genes related to amino acids biosynthesis (other than arginine) such as SCO4293 for a putative threonine synthase, leuA or the bidirectional promoter region hisD–SCO2055; (ii) genes for nucleotide biosynthesis or regulation as nrdA, SCO2015 or rstP; (iii) genes related to nitrogen or carbon metabolism: glnII, amtB, glnR (internal), SCO1086, ureA (SCO1236) or the bidirectional promoter region pyk1–SCO2015; (iv) genes for transcriptional regulators: whiB, malE–malR, SCO2686, SCO7302–7303, SCO1220–1221, SCO3978–3979, SCO0800–0801, or for sigma or anti-sigma factors: SCO3067–sig15, SCO7314; (v) genes encoding hypothetical conserved proteins (SCO5864); and (vi) genes related to arginine (argH, arcB; argG, argC and SCO1086) or pyrimidine (pyrR, pyrB and pyrA) biosynthesis as positive controls. The tested ARG boxes have Ri values that range from 5 (intergenic SCO3067–sig15 region) to 17 (upstream of argH). In most of the analyzed regions the ARG box (20 nucleotides) is unique, but in some cases two ARG boxes, separated from 1 to 208 nucleotides, can be identified. The two ARG boxes upstream of argH, arcB, argG and argC are adyacent.
Twenty-four fragments were bound by ArgR. In general a good correlation with the gene expression in transcriptomic studies was found (Figs. 8, 10 and Table 2). Only occasionally, probe shift was unclear or not present (i.e. for glnII, glnR, SCO7302–7303, SCO0800–0801 and SCO1220–1221). The occasional lack of relation between ARG box and differential expression in the microarray experiments will be discussed below.
Discussion
ArgR is known as the regulator of the arginine biosynthesis pathway and, in most bacteria, the argR gene is clustered with arginine biosynthesis or transport genes [7], [35].
The transcriptomic and proteomic studies shown in this work indicated that in addition to arginine biosynthesis ArgR controls directly or indirectly 452 genes (contrasts 1 and 4, Fig. 2) involved in different aspects of nitrogen metabolism, purine and pyrimidine biosynthesis, cell morphology, and antibiotic production. More interesting is the effect of ArgR on the expression of genes encoding general regulators (GntR-, AbaA- and TetR-like regulators), sigma factors (BldN, SigR, AfsS), or two component regulatory systems, which could be mediated by a cascade regulatory mechanism. Binding assays validated the ARG boxes detected in Figure 10 and in most of the promoters shown in Table 2.
In E. coli arginine acts as co-repressor binding ArgR and controlling arginine biosynthesis. However, in Streptomyces the effect of arginine as co-repressor is weak and high levels of arginine (25 mM) were required to exert effect on the expression of only 27 genes (contrasts 2 and 3, Fig. 2).
Addition of arginine to S. coelicolor cultures reduced the production of actinorhodin and undecylprodigiosin by 92 and 99%, respectively, even though none of the pigmented antibiotic derives directly from arginine. Even more interesting was the observation that the ΔargR mutant showed a strong reduction (80%) in undecylprodigiosin production in relation to the wild-type strain and lacked detectable actinorhodin production. Therefore, ArgR is a regulator required for production of these secondary metabolites.
This drastic effect of ArgR on the production of both antibiotics might be mediated by the transcriptional response of the global positive regulator afsS. In addition, the effect on actinorhodin production could be correlated with the lower expression of actIII and actII-orf2 in the ΔargR mutant. The transporter protein encoded by actII-orf2 is essential for actinorhodin secretion [25] because accumulation of intracellular antibiotic feedback represses the biosynthetic genes. Identification of additional control mechanisms related to antibiotic production will require further experiments using RNA samples taken during the production phase, because the samples for this omics analyses were taken at an earlier time (32 h) than that of intense antibiotic production.
The ArgR protein acts as a transcriptional regulator after binding to the promoters of the arg regulon genes. The binding sites are composed of imperfect palindromes, known as ARG boxes [36]–[38]. Both the argR genes and the ARG boxes are well conserved among different bacteria [33], [39]. In E. coli the binding sites are composed of two ARG boxes of 18 bp that, with only one known exception, overlap the promoter. The separation between boxes is 3 bp except the 2 bp of separation of the argR operator [40], [41]. ArgR functions in E. coli as a repressor on arginine biosynthesis and transport genes [14], [42] and on the glutamate synthase operon [43], but as activator of the ast operon [44]. The B. subtilis AhrC protein – the ArgR orthologue –, which was found to bind S. coelicolor argC promoter [45], is also a repressor of the arginine biosynthesis genes [46], [47] and an activator of the arginine catabolism genes in Bacillus [48]; similar regulatory behavior is found for ArgR in P. aeruginosa and Salmonella [35], [49]. In B. subtilis argC gene two operators have been found. The operator with the highest affinity for AhrC is formed by two ARG boxes, separated by 11 bp. The second operator, within argC coding region, has a single ARG box. A model for the holo-AhrC-argC complex proposes that the AhrC hexamer interacts with the three ARG boxes [34]. Other operators formed by a single ARG box are found in the Bacillus rocA and rocD catabolic genes and show a lower affinity for AhrC [48].
We identified by DNase I footprinting the ArgR binding site that controls the argCJBDR operon of S. clavuligerus. This operator is formed by two 18 nucleotides ARG boxes separated by 2 bp (6). This arrangement is conserved in ArgR putative binding sites of other Streptomyces promoter and in the upstream sequences of S. coelicolor genes (argC, argG, arcB and argH) involved in arginine biosynthesis.
To fit with the experimental transcriptomic and gel-shift data presented in this work a new ArgR binding site model has been designed (Fig. 10C). This sequence is formed by a 20 nucleotide-single ARG box and permitted the location of new ARG boxes with different Ri values. The presence of two tandem ARG boxes in the arginine operon genes suggests that these genes are more tightly controlled than others containing a single ARG box. In some of the DNA fragments tested by EMSA two separated ARG boxes were predicted (Table 2). In the intergenic malE–malR region the two putative boxes with Ri values of 9.3 and 8.6 are separated 83 nucleotides. The upstream region of nrdA and the intergenic SCO7302–SCO7303 or SCO2055–hisD regions also presented two putative sites with lower Ri. Additional studies will be necessary to demonstrate whether or not ArgR binds both or only one of these ARG boxes.
In most of the cases shown in Fig. 10, the gel shift correlated with a statistically significant differential expression of the gene whose promoter was tested, indicating that the model 2 is functional, although additional EMSA should be done with increasing amounts of protein to determine the different ArgR affinity to the operators. However, this was not always the case, as occurred with the amtB, SCO3067–sig15 or SCO2686 probes. This apparent lack of correlation in a few cases might be explained by the specific experimental conditions used in this study. The differential transcription observed in the microarray reflects the sampling of a 32-h culture grown in MG medium; however, in other media or culture times, these genes giving only in vitro binding might also show in vivo significant differential expression.
Gel-shift assays confirmed that the ArgR-dependent luciferase activity of these promoters results from the direct control of ArgR on these regions. As shown in Fig. 10 and Table 2, ArgR is a transcriptional regulator of pyrimidines (pyrR, pyrA, pyrB), nucleotides (i.e. SCO2015, nrdA), several amino acids (SCO4293), nitrogen metabolism (amtB, ureA) and several regulators (whiB, rstP, SCO2686 and SCO3979). ArgR also bound the intergenic region SCO3067–SCO3068 (for anti-anti-sigma and sigma factors) and the upstream region of SCO7314 encoding a sigma factor suggesting that many of the observed transcriptional effects are likely to be due to a cascade mechanism promoted by ArgR.
Regarding nitrogen control, putative ARG boxes were observed upstream of amtB (Ri 8.3), upstream of glnII (into the SCO2209 coding region, Ri 5.3) and in the 3′ end of glnR (Ri 7.4). However, only the one located upstream of amtB was controlled by ArgR, as indicated by the luciferase assay and EMSA. Therefore, ArgR did not regulate glnR and the down-expression of glnII observed in the deleted mutant was not directly due to ArgR. Interestingly the amtB operator also binds GlnR but this regulatory protein and ArgR have different binding sites [50].
Methods
Culture conditions
Growth and manipulation of Streptomyces strains were carried out according to standard procedures [51]. Spore suspensions were obtained in TBO medium: 20 g/L tomato paste, 20 g/L oat flakes, and 20 g/L agar, pH 6.5. S. coelicolor cultures were grown at 30°C, 300 rpm (2.5 cm orbit), in starch and glutamate defined MG medium [16] that contained 2.5 mM potassium phosphate and 25 mM arginine when indicated. Baffled flasks (500 mL), that contained 100 mL of MG medium, were inoculated with 106 spores/mL. Dry weight was determined in 2-mL culture samples that were washed twice with MilliQ water and dried at 65°C for 4 days. Arginine consumption was followed using the method of Hess et al. [52]. Cultures of plasmid-bearing cells were supplemented with ampicillin (50 µg/mL), chloramphenicol (25 µg/mL), kanamycin (25 µg/mL) or apramycin (50 µg/mL), as appropriate. E. coli DH5α was used as the general cloning host.
Cultures of 32 h were selected for the expression analysis due to the intense growth and good expression of amino acid biosynthesis pathways at this culture time. The experimental conditions chosen were as follows: (A) S. coelicolor M145 grown in MG medium, (B) S. coelicolor M145 grown in MG medium supplemented with 25 mM arginine, (C) S. coelicolor ΔargR grown in MG medium and (D) S. coelicolor ΔargR grown in MG medium supplemented with 25 mM arginine.
Construction of a mutant of S. coelicolor with argR gene deletion
An argR-deleted mutant was constructed by PCR targeting [53] using oligonucleotides Coe-argR1 and Coe-argR2 on plasmid pTC123-aphII, a pTC182-derived plasmid [54] that contained the neo gene and a 5.7-kb SphI DNA insert carrying S. coelicolor argBDR genes as well as upstream and downstream sequences. After conjugation, four apramycin-resistant, kanamycin-sensitive S. coelicolor recombinants were characterized by Southern hybridization. We used as probes a SalI–PauI DNA fragment containing argR and the flanking sequences, and the apramycin resistance acc(3)IV gene. The hybridization pattern obtained confirmed the argR replacement deletion in the four identical clones that were named S. coelicolor ΔargR.
Nucleic acid isolation and purification
Samples (2 mL) from 32-h cultures of S. coelicolor M145 and S. coelicolor ΔargR grown in MG medium, with or without arginine, were stabilized with RNA Protect Bacteria Reagent (Qiagen). For RNA isolation, mycelia were treated with lysozyme (30 mg/mL); the lysates were extracted with phenol and then transferred to RNeasy Midi Spin Columns (Qiagen), according to the manufacturer's instructions. RNA preparations were incubated with Turbo DNase (Ambion) to eliminate chromosomal DNA contamination. Sample quantification was done with a NanoDrop ND-1000 UV-Vis Spectrophotometer. Total genomic DNA (gDNA) was isolated from a stationary-phase culture following the Kirby mix procedure [51].
PCR and semi-quantitative RT-PCR analysis
Oligonucleotide primers used in this study are shown in Table S2. All PCRs were performed in a TGradient (Biometra) thermocycler using Platinum Pfx DNA Polymerase (Invitrogen). The dNTP mix contained a higher proportion of G-C (35% each) than A-T nucleotides (15% each), to improve the amplification of high G+C DNA content. PCR products subcloned into pBluescript II SK(+) were sequenced to check the amplification fidelity. Gene expression analysis by RT-PCR was done with the SuperScript One-Step System (Invitrogen) using 150 ng total RNA as a template. For semi-quantitative analysis, samples were taken at three-cycle intervals between cycles 24 and 33, to compare non-saturated PCR product formation. Negative controls were carried out with each set of primers and Platinum Taq DNA polymerase (Invitrogen) to verify the absence of contaminating DNA in the RNA preparations.
Purification of Strep-tagged ArgR
To purify the ArgR protein, plasmid pET-Strep-argR was constructed. The argR gene was amplified by PCR using oligonucleotides ArgR17/18 to place the Strep tag upstream of argR. The amplified product was subcloned in NdeI/HindIII digested plasmid pET-24a(+) (Novagen, Merck). The E. coli BL21(DE3)pLysS (Invitrogen) transformants carrying pET-Strep-argR were grown at 37°C to OD 0.6, induced with 1 mM IPTG, and the growth was continued at 20°C. After 18–20 h, the cells were harvested by centrifugation at 2640 rcf and kept at −80°C. Cells were broken with a Misonix XL-2000 sonifier, centrifuged at 16 100 rcf, and the supernatant was applied to a 1-mL StrepTrap HP column (GE Healthcare) and purified in an Akta Prime FPLC Protein Purification System (GE Healthcare) following the manufacturer's instructions.
Electrophoresis mobility shift assay (EMSA)
The promoters cloned in pBluescript SK+ were sequenced and amplified by PCR using specific or universal 6-FAM labelled oligonucleotides (Table S2). The amplification products were used for EMSA as follows: the reaction contained 5 µL buffer (10 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 2.5 mM CaCl2, 250 mM KCl, 0.5 mM DTT, 10 mM L-arginine, pH 7.4), poly-(dIdC) 1,3 µg/mL, 6-FAM-labelled probe 2 nM, glycerol 10% and Strep-ArgR protein 0.8 µM in a total volume of 15 µL. The reaction was maintained for 30 min at 30°C, and then the DNA was separated in a 5% acrylamide gel using 0.5× TBE as developing buffer at 50 V. The bands were visualized in an Ettan DIGE imager (GE Healthcare). In all cases, competition and specificity experiments were done with increasing amounts of unlabelled specific probe and with BSA. The argH promoter-probe was used to test the effect of L-arginine. ArgR affinity was higher in the presence of L-arginine, so it was mantained in the binding reaction mixture.
Luciferase assay
For luciferase reporter analysis, promoter regions were amplified with primers containing NdeI and BamHI restriction sites (Table S2) to clone the promoters into the ATG codon of the luxA gene in pLUXAR-neo [55]. Cultures of S. coelicolor exconjugants harbouring the promoter–probe constructs were carried out in MG medium. Samples at 32, 47, 55 and 79 h were taken, spun down, kept frozen and processed simultaneously. Riboflavine was added to the cell suspension to improve the sensitivity of the luciferase assays, which were measured in a Luminoskan luminometer (Labsystems) [18], [55]. At least two different cultures from the same strain were analyzed and measured by duplicate.
Labelling and microarray hybridizations
S. coelicolor microarrays (SCo3 design) were obtained from the Functional Genomics Laboratory, Surrey University (UK). They contained duplicated probes (50-mer) for 7728 chromosomal genes (out of 7825). The experimental design used S. coelicolor M145 gDNA as a common reference. RNA was extracted from two nutritional states, MG and MG with 25 mM arginine, and from two strains, S. coelicolor M145 and S. coelicolor ΔargR. Four biological replicates were made for each condition. The Pronto! Universal Microarray Hybridization kit (Corning) was used for prehybridization of the slides. Labelling reactions were performed according to the recommendations described in http://www.surrey.ac.uk/SBMS/Fgenomics. Total RNA was labelled with Cy3-dCTP (Amersham) using random primers and Superscript II reverse transcriptase (Invitrogen). gDNA was labelled with Cy5-dCTP (Amersham) from random primers extended with the Klenow fragment of DNA polymerase (Roche). The final products were purified with MinElute columns (Qiagen) and labelling efficiencies were quantified spectrophotometrically. Cy3-cDNA (100 pmol) and Cy5-labeled gDNA (20 pmol) were mixed, vacuum dried and resuspended in 40 µL Pronto! Long Oligo/cDNA Hybridization Solution (Corning), to be applied on the microarray surface. Hybridizations were carried out at 42°C and extended to 72 h to improve the quality of the results [56]. Washing, scanning with an Agilent DNA Microarray Scanner G2565BA, and image quantification were carried out as indicated previously [16].
Identification of differentially transcribed genes and transcription profile classification
Microarray data were normalized and analysed with the Bioconductor package limma [57], [58]. Spot quality weights were estimated as indicated in Text S1. Local and global normalizations were both used [59]. First, weighted medians of log2 Cy3/Cy5 intensities were calculated for print-tip correction, and afterwards, global Loess was applied [57]. The normalized log2 of Cy3/Cy5 intensities is referred to in this work as the Mg value, which is proportional to the abundance of transcripts for a particular gene [60]. The information from within-array spot duplicates [61] and empirical array weights [62] were taken into account in the linear models [58]. The transcription results of the four experimental conditions were compared using five contrasts. For each contrast, p-values and Mc values (log measure of the differential transcription) were calculated. False-discovery rate (FDR) correction for multiple testing was applied. For each contrast or comparison between two experimental conditions, a result was considered as statistically significant if the FDR-corrected p-value was <0.05. A total of 459 genes showed statistically significant results in at least one contrast.
To classify the transcription profiles observed for this set of genes, we used the Hr values, which summarized the results of the hypothesis tests. For each contrast and each gene, the Hri value of each gene was calculated as follows: (i) Hri = 0 indicated that the contrast result was not significant using uncorrected p-values (α = 0.05); and (ii) if the uncorrected p-value was <0.05, then Hri = 1 (indicating upregulation) if the respective Mc value was positive, or Hri = −1, for a negative Mc value (downregulation). The set of 459 genes yielded a total of 52 observed combinations of their Hr values. Visual inspection of the transcription profiles allowed us to group a subset of 365 genes, which showed the profiles with more likely biological meaning, into 5 main types (I–V) and 13 subtypes as shown in Fig. 3 (see also Dataset S1 and Text S1).
Bioinformatic analysis of ArgR binding sites
ArgR binding sites composed of two palindrome sequences, known as ARG boxes, have been identified previously in S. clavuligerus [6], [7]. ARG boxes were easily identified upstream of the arginine biosynthesis genes argH, argG, argC and arcB of S. coelicolor. The sequences of ARG boxes and their complementaries –due to the symmetry of the palindromic site– were used to create information theory models by the Delila programs makebk, encode, rseq, dalvec, ri and makelogo [63], [64]. To find new operator sites, the promoter regions (−300, +100 nt) of the S. coelicolor chromosome were scanned by means of the Patser algorithm and the RSA tools server [65].
Proteomic
The mycelia from 20-mL culture samples of S. coelicolor M145 and S. coelicolor ΔargR grown for 32 h in MG medium were harvested by centrifugation (10 min at 2640 rcf), immediately frozen in liquid nitrogen, and stored at −80°C until use. Proteomic analysis and Mass Spectrometry identification were done as described by Santamarta et al. [66]. Positive identifications were based on the MOWSE score algorithm. A MOWSE score of 52 or higher was significant at the 5% level or better, and proteins typically gave scores well above 70. The analysis was performed in the Proteomic Service of the National Center for Biotechnology (Madrid, Spain).
Supporting Information
Selected differentially expressed I and II profile genes.
(DOC)
Primers used within this manuscript.
(DOC)
Estimation of spot weights for microarray data analysis.
(DOC)
Hypothesis-testing results for profile classification.
(XLS)
Peptide mass fingerprints.
(PDF)
Acknowledgments
We appreciate the technical collaboration of Dr. Carlos Barreiro, Dr. Fernando Santos and Nuria Nárdiz.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Grants from the Spanish Comisión Interministerial de Ciencia y Tecnología GEN2003-20245, BIO2009-09820, and by the European Project LSHM-CT-2004-005224. AB received a fellowship from the Ministry of Science and Innovation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Maas WK. The arginine repressor of Escherichia coli. Microbiol Rev. 1994;58:631–640. doi: 10.1128/mr.58.4.631-640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu CD, Yang Z, Li W. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol. 2004;186:3855–3861. doi: 10.1128/JB.186.12.3855-3861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.North AK, Smith MC, Baumberg S. Nucleotide sequence of a Bacillus subtilis arginine regulatory gene and homology of its product to the Escherichia coli arginine repressor. Gene. 1989;80:29–38. doi: 10.1016/0378-1119(89)90247-3. [DOI] [PubMed] [Google Scholar]
- 4.Larsen R, van Hijum SA, Martinussen J, Kuipers OP, Kok J. Transcriptome analysis of the Lactococcus lactis ArgR and AhrC regulons. Appl Environ Microbiol. 2008;74:4768–4771. doi: 10.1128/AEM.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SY, Park JM, Lee JH, Chang ST, Park JS, et al. Interaction of transcriptional repressor ArgR with transcriptional regulator FarR at the argB promoter region in Corynebacterium glutamicum. Appl Environ Microbiol. 2011;77:711–718. doi: 10.1128/AEM.01610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez-García A, Ludovice M, Martín JF, Liras P. Arginine boxes and the argR gene in Streptomyces clavuligerus: evidence for a clear regulation of the arginine pathway. Mol Microbiol. 1997;25:219–228. doi: 10.1046/j.1365-2958.1997.4511815.x. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez-García A, de la Fuente A, Pérez-Redondo R, Martín JF, Liras P. Characterization and expression of the arginine biosynthesis gene cluster of Streptomyces clavuligerus. . J Mol Microbiol Biotechnol. 2000;2:543–550. [PubMed] [Google Scholar]
- 8.Walker JB. Pathway of the guanidinated inositol moieties of streptomycin and bluensomycin. Methods in Enzymol. 1975;43:429–470. doi: 10.1016/0076-6879(75)43097-x. [DOI] [PubMed] [Google Scholar]
- 9.Hornemann U, Eggert H. Utilization of the intact carbamoyl group of L- [NH2CO-13C, 15N] citrulline in mitomycin biosynthesis by Streptomyces verticillatus. J Antibiot. 1975;28:841–843. doi: 10.7164/antibiotics.28.841. [DOI] [PubMed] [Google Scholar]
- 10.Martinkus KJ, Tann C, Gould SJ. The biosynthesis of the streptolidine moiety in streptothricin F. Tetrahedron. 1983;39:3493–3505. [Google Scholar]
- 11.Romero J, Liras P, Martin JF. Utilization of ornithine and arginine as specific precursors of clavulanic acid. Appl Env Microbiol. 1986;52:892–897. doi: 10.1128/aem.52.4.892-897.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas MG, Burkart MD, Walsh CT. Conversion of L-proline to pyrrolyl-2-carboxyl-S-PCP during undecyl-prodigiosin and pyoluteorin biosynthesis. Chem Biol. 2002;9:171–184. doi: 10.1016/s1074-5521(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Lu CD. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J Bacteriol. 2007;189:3945–3953. doi: 10.1128/JB.00261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldara M, Charlier D, Cunin R. The arginine regulon of Escherichia coli: whole-system transcriptome analysis discovers new genes and provides an integrated view of arginine regulation. Microbiology. 2006;152:3343–3354. doi: 10.1099/mic.0.29088-0. [DOI] [PubMed] [Google Scholar]
- 15.Hara H, Ohnishi Y, Horinouchi S. DNA microarray analysis of global gene regulation by A-factor in Streptomyces griseus. . Microbiology. 2007;155:2197–2210. doi: 10.1099/mic.0.027862-0. [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez-García A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martín JF. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics. 2007;7:2410–2429. doi: 10.1002/pmic.200600883. [DOI] [PubMed] [Google Scholar]
- 17.Reuther J, Wohlleben W. Nitrogen metabolism in Streptomyces coelicolor: transcriptional and post-transcriptional regulation. J Mol Microb Biotechnol. 2007;12:139–146. doi: 10.1159/000096469. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-García A, Sola-Landa A, Apel K, Santos-Beneit F, Martín JF. Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 2009;37:3230–3242. doi: 10.1093/nar/gkp162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos-Beneit F, Rodríguez-García A, Sola-Landa A, Martín JF. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol Microbiol. 2009;72:53–68. doi: 10.1111/j.1365-2958.2009.06624.x. [DOI] [PubMed] [Google Scholar]
- 21.Ludovice M, Martín JF, Carrachas P, Liras P. Characterization of the Streptomyces clavuligerus argC gene encoding N-acetylglutamyl-phosphate reductase: expression in Streptomyces lividans and effect on clavulanic acid production. J Bacteriol. 1992;174:4606–4613. doi: 10.1128/jb.174.14.4606-4613.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De la Fuente JL, Martín JF, Liras P. New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers Streptomyces clavuligerus and Nocardia lactamdurans. Biochem J. 1996;320:173–179. [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borovok I, Gorovitz B, Schreiber R, Aharonowitz Y, Cohen G. Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J Bacteriol. 2006;188:2512–2520. doi: 10.1128/JB.188.7.2512-2520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernández-Moreno MA, Caballero JL, Hopwood DA, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. . Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 26.Floriano B, Bibb M. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- 27.Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol. 2001;41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- 28.Claessen D, Stokroos I, Deelstra HJ, Penninga NA, Bormann C, et al. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplains. MolMicrobiol. 2004;53:433–443. doi: 10.1111/j.1365-2958.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- 29.Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology. 2010;156:2343–2353. doi: 10.1099/mic.0.038281-0. [DOI] [PubMed] [Google Scholar]
- 30.Jayapal KP, Philp RJ, Kok YJ, Yap MG, Sherman DH, et al. Uncovering genes with divergent mRNA-protein dynamics in Streptomyces coelicolor. PLoS One. 2008;3(5):e2097. doi: 10.1371/journal.pone.0002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova KS, Aravind L, Koonin EV. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 1999;8:1714–1719. doi: 10.1110/ps.8.8.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro-Melchor M, Charaniya S, Karypis G, Takano E, Hu WS. Genome-wide inference of regulatory networks in Streptomyces coelicolor. . BMC Genomics. 2010;11:578. doi: 10.1186/1471-2164-11-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlier D. Arginine regulation in Thermotoga neapolitana and Thermotoga maritime. . Biochem Soc Trans. 2004;32:310–313. doi: 10.1042/bst0320310. [DOI] [PubMed] [Google Scholar]
- 34.Garnett JA, Marincs F, Baumberg S, Stockley PG, Phillips SEV. Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J Mol Biol. 2008;379:284–298. doi: 10.1016/j.jmb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Park S-M, Lu C-D, Abdelal AT. Cloning and characterization of argR, a gene that participate in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J Bacteriol. 1997;179:5300–5308. doi: 10.1128/jb.179.17.5300-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunin R, Eckhardt T, Piette J, Boyen A, Piérard A, et al. Molecular basis for modulated regulation of gene expression in the arginine regulon of Escherichia coli K-12. Nucleic Acids Res. 1983;11:5007–5019. doi: 10.1093/nar/11.15.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SY, Kim YH, Min J. The effect of ArgR-DNA binding affinity on ornithine production in Corynebacterium glutamicum. . Curr Microbiol. 2009;59:483–488. doi: 10.1007/s00284-009-9467-y. [DOI] [PubMed] [Google Scholar]
- 38.Lee SY, Shin HS, Park JS, Kim YH, Min J. Proline reduces the binding of transcriptional regulator ArgR to upstream of argB in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2010;86:235–242. doi: 10.1007/s00253-009-2264-5. [DOI] [PubMed] [Google Scholar]
- 39.Makarova KS, Mironov AA, Gelfand MS. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2001;2(4):1–8. doi: 10.1186/gb-2001-2-4-research0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlier D, Roovers M, Vliet FV, Boyen A, Cunin R, et al. Arginine regulon of Escherichia coli K-12 A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J Mol Biol. 1992;226:367–386. doi: 10.1016/0022-2836(92)90953-h. [DOI] [PubMed] [Google Scholar]
- 41.Tian G, Lim D, Carey J, Maas WK. Binding of the arginine repressor of Escherichia coli K12 to its operator sites. J Mol Biol. 1992;226:387–397. doi: 10.1016/0022-2836(92)90954-i. [DOI] [PubMed] [Google Scholar]
- 42.Caldara M, Le Minh PN, Bostoen S, Massant J, Charlier D. ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. Journal of Molecular Biology. 2007;373:251–267. doi: 10.1016/j.jmb.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 43.Paul L, Mishra P, Blumenthal R, Matthews R. Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, Crp, and ArgR. BMC Microbiol. 2007;7:2–19. doi: 10.1186/1471-2180-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiupatkis AK, Reitzer L. ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J Bacteriol. 2002;184:2940–2950. doi: 10.1128/JB.184.11.2940-2950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soutar A, Baumberg S. Implication of a repression system, homologous to those of other bacteria, in the control of arginine biosynthesis genes in Streptomyces coelicolor. . Mol Gen Genet. 1996;251:245–251. doi: 10.1007/BF02172924. [DOI] [PubMed] [Google Scholar]
- 46.Smith MC, Czaplewski L, North AK, Baumberg S, Stockley PG. Sequences required for regulation of arginine biosynthesis promoters are conserved between Bacillus subtilis and Escherichia coli. Mol Microbiol. 1989;3:23–28. doi: 10.1111/j.1365-2958.1989.tb00099.x. [DOI] [PubMed] [Google Scholar]
- 47.Czaplewski LG, North AK, Smith MCM, Baumberg S. Purification and characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. . Mol Microbiol. 1992;6:267–275. doi: 10.1111/j.1365-2958.1992.tb02008.x. [DOI] [PubMed] [Google Scholar]
- 48.Miller CM, Baumberg S, Stockley PG. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. MolMicrobiol. 1997;26:37–48. doi: 10.1046/j.1365-2958.1997.5441907.x. [DOI] [PubMed] [Google Scholar]
- 49.Lu CD, Abdelal AT. Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J Bacteriol. 1999;181:1934–1938. doi: 10.1128/jb.181.6.1934-1938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martín JF, Sola-Landa A, Santos-Beneit F, Fernández-Martínez LT, Prieto C, et al. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb Biotechnol. 2011;4:165–174. doi: 10.1111/j.1751-7915.2010.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. Norwich: John Innes Foundation; 2000. 613 [Google Scholar]
- 52.Hess J, Kito E, Martin RP, van Pilsum JF. Determination of creatine, creatinine, arginine, guanidinoacetic acid, guanidine, and methylguanidine in biological fluids. J Biol Chem. 1956;222:225–235. [PubMed] [Google Scholar]
- 53.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuente A, Cisneros E, Talavera A. Restriction end-converting vectors with tandem repeated multiple cloning sites. Gene. 1994;139:83–86. doi: 10.1016/0378-1119(94)90527-4. [DOI] [PubMed] [Google Scholar]
- 55.Santos-Beneit F, Rodríguez-García A, Franco-Domínguez E, Martín JF. Phosphate-dependent regulation of the low- and high-affinity transport systems in the model actinomycete Streptomyces coelicolor. Microbiology. 2008;154:2356–2370. doi: 10.1099/mic.0.2008/019539-0. [DOI] [PubMed] [Google Scholar]
- 56.Sartor M, Schwanekamp J, Halbleib D, Mohamed I, Karyala S, et al. Microarray results improve significantly as hybridization approaches equilibrium. Biotechniques. 2004;36:790–796. doi: 10.2144/04365ST02. [DOI] [PubMed] [Google Scholar]
- 57.Smyth GK, Speed TP. Normalization of cDNA microarray data. Methods 2003. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 58.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(Iss 1):Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 59.Wu W, Xing EP, Myers C, Mian IS, Bissell MJ. Evaluation of normalization methods for cDNA microarray data by k-NN classification. BMC Bioinformatics. 2005;6:191–212. doi: 10.1186/1471-2105-6-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehra S, Lian W, Jayapal KP, Charaniya SP, Sherman DH, et al. A framework to analyze multiple time series data: A case study with Streptomyces coelicolor. J Ind Microbiol Biotechnol. 2006;33:159–172. doi: 10.1007/s10295-005-0034-7. [DOI] [PubMed] [Google Scholar]
- 61.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 62.Ritchie M, Diyagama D, Neilson J, van Laar R, Dobrovic A, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 2006;7:261–277. doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider TD. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- 64.Schneider TD. Information content of individual genetic sequences. J Theor Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 65.Van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santamarta I, López-García MT, Kurt A, Nárdiz N, Álvarez-Álvarez R, et al. Characterization of DNA-binding sequences for CcaR in the cephamycin-clavulanic acid supercluster of Streptomyces clavuligerus. Mol Microbiol. 2011;81(4):968–981. doi: 10.1111/j.1365-2958.2011.07743.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selected differentially expressed I and II profile genes.
(DOC)
Primers used within this manuscript.
(DOC)
Estimation of spot weights for microarray data analysis.
(DOC)
Hypothesis-testing results for profile classification.
(XLS)
Peptide mass fingerprints.
(PDF)