Abstract
In 2006, a severe foodborne EHEC outbreak occured in Norway. Seventeen cases were recorded and the HUS frequency was 60%. The causative strain, Esherichia coli O103:H25, is considered to be particularly virulent. Sequencing of the outbreak strain revealed resemblance to the 2011 German outbreak strain E. coli O104:H4, both in genome and Shiga toxin 2-encoding (Stx2) phage sequence. The nucleotide identity between the Stx2 phages from the Norwegian and German outbreak strains was 90%. During the 2006 outbreak, stx2-positive O103:H25 E. coli was isolated from two patients. All the other outbreak associated isolates, including all food isolates, were stx-negative, and carried a different phage replacing the Stx2 phage. This phage was of similar size to the Stx2 phage, but had a distinctive early phage region and no stx gene. The sequence of the early region of this phage was not retrieved from the bacterial host genome, and the origin of the phage is unknown. The contaminated food most likely contained a mixture of E. coli O103:H25 cells with either one of the phages.
Introduction
Enterohaemorrhagic Escherichia coli (EHEC) can cause serious disease in humans. Infection manifests itself as diarrhoea or haemorrhagic colitis. The life threatening haemolytic uraemic syndrome (HUS) is a potential sequelae. Previously, EHEC isolates belonging to serogroups O157, O26, O111, O145 and O103 were most frequently isolated in food borne outbreaks [1]. Recently, less common serotypes and pathotypes, have received more attention. This is illustrated by the enteroaggregative E. coli (EAEC) O104:H4 causing a large European outbreak in 2011 involving more than 4000 diseased patients, a 22% HUS incidence, and 50 fatalities [2], [3].
EHEC virulence is mainly attributed to the production of Shiga toxins (Stx), which are regarded as essential in EHEC disease. Shiga toxins are divided into two major families, Stx1 and Stx2. Toxins belonging to the Stx2 group are the most heterogenic and also include the most potent variants [4]. In E. coli, Stx are usually encoded by temperate, lamdoid bacteriophages whose genomes are mosaic in structure, and may integrate at several sites in the E. coli chromosome [5]–[7]. Upon certain stimuli of the bacterial cell, the phage enters a lytic cycle inducing production of Stx and phage particles. This culminates in bacterial cell lysis and release of toxin and infectious phages [8], [9]. Released Stx phages may infect new bacterial cells, playing an important role in the evolution of EHEC [10].
During the spring of 2006, Norway experienced a national disease outbreak caused by EHEC O103:H25. The outbreak was characterized by an extraordinary high frequency of HUS. Of seventeen recorded cases sixteen had diarrhoea, ten developed HUS and one case was fatal [11]. Stool cultures for E. coli O103:H25 were positive in 11 of the patients, but only two of the retrieved isolates were stx2-positive while the remaining nine were stx-negative [11]. The absence of stx genes in EHEC serotypes isolated from patients is not uncommon, and such strains are believed to be offspring of EHEC present at an earlier phase of the infection [12]. The more surprising finding in the Norwegian O103:H25 outbreak was that none of the food isolates had stx-genes [13]. Genotyping and MLVA revealed that the stx-negative isolates from food were indistinguishable or closely related to the two stx2-positive patient isolates [13], [14], and these results combined with epidemiologic investigation led to the conclusion that fermented sausage with the stx-negative isolates was the culprit of the outbreak [11], [13].
In this study, the genome of the outbreak strain was sequenced to elucidate its high virulence and examine potential relationship to other virulent EHEC strains. In addition, we aimed to characterize the Stx2 phage demonstrated in the stx2-positive isolates.
Results
Genome sequencing and phylogeny
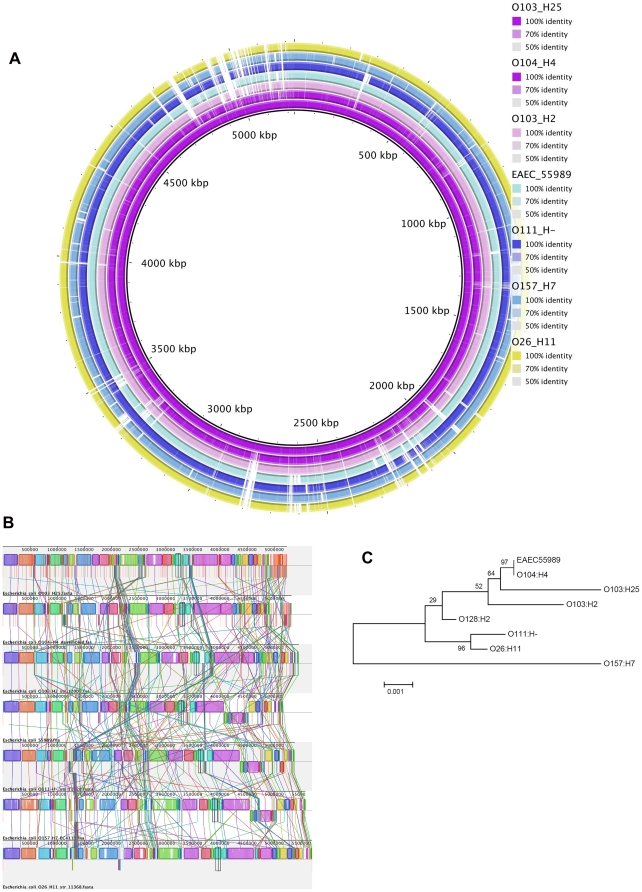
The sequence achieved through genome sequencing using 454 technology was assembled into 554 contigs and spanned altogether 5.2 Mb. Figure 1A shows a BRIG diagram comparing the EHEC O103:H25 NOS genome against the E. coli genomes of O104:H4 GOS1, O103:H2 str 12009, O104:H4 str 55989, O111:H- str 11128, O157:H7 and O26:H11 str 11368, revealing that EHEC O103:H25 NOS shows considerable resemblance to the German outbreak strain, EAEC O104:H4 GOS1.The Mauve alignment [15] shown in Figure 1B supports this finding. EHEC O103:H25 NOS was assigned to the sequence type 2523 (ST2523) complex (adk 77, fumC 7, gyrB 7, icd 151, mdh 65, purA 56, recA 7). A phylogenetic tree of allele sequences supports a close relationship between EHEC O103:H25 NOS and EAEC O104:H4 GOS (ST678 [16]) (Figure 1C).
Figure 1. Genome comparisons of EHEC O103:H25 NOS and related E. coli strains.
A. BRIG blast atlas of EHEC O103:H25 NOS compared to EAEC O104:H4 GOS, EHEC O103:H2 str 12009, EAEC O104:H4 str 55989, EHEC O111:H- str 11128, EHEC O157:H7 EDL933 and EHEC O26:H11 str 11368. The white regions represent absent genetic regions. B. Whole genome alignment of EHEC O103:H25 NOS, EAEC O104:H4 GOS1, EHEC O103:H2 str 12009, EAEC O104:H4 str 55989, EHEC O111:H- str 11128, EHEC O157:H7 EDL933 and EHEC O26:H11 str 11368 (top to bottom) genomes using Mauve [15]. Each chromosome has been laid out horizontally and homologous blocks in each genome are shown as identically colored regions linked across genomes. C. Phylogenetic analysis of concatenated MLST gene alleles (adk, fumC, gyrB, icd, mdh, purA, recA) of EHEC O103:H25 NOS (ST2523), EAEC O104:H4 GOS (ST678), EAEC O104:H4 str 55989 (ST678), EHEC O103:H2 str 12009 (ST17), EHEC O26:H11 str 11368 (ST21), EHEC O111:H- str 11128 (ST16), EHEC O157:H7 EDL933 (ST11) and EHEC O128:H2 str 3171/00 (ST25) obtained from GenBank.
Virulence features
All five LEE operons (LEE1 to LEE5) comprising the LEE core of 35 kb [17] were present in the EHEC O103:H25 NOS. The intimin gene, eae θ (theta), was present in LEE5. The sequence of the five LEE operons showed 97% and 86% identity to EHEC 0111:H- str 11128 and EHEC O103:H2 str 12009, respectively.
A complete OI122, including the genes sen (Shigella flexneri enterotoxin 2), pagC (required for bacterial survival within macrophages) and efa (EHEC factor for adherence – an adhesin) [18], was present in all of the Norwegian O103:H25 isolates listed in Table 1. The OI122 in these isolates exhibits the large version of efa/lifA gene, an orf of 9672 bp, which encodes a protein inhibiting lymphocyte activation and lymphokine production [19]. The genes encoding enterohemolysin (ehxCABD) were all present in the O103:H25 NOS genome, and ehxA was detected in all Norwegian strains listed in Table 1. Neither this subtilase cytotoxin genes subA-subB, nor the genes pic, aggA, aggR, aatA associated with enteroaggregative E. coli, was present in EHEC O103:H25 NOS or any of the other outbreak isolates included in this study.
Table 1. Bacterial isolates included in the study.
| Strain | Synonym | Year | Stx | Source | Origin |
| NVH-734a | NIPH-11060424 | 2006 | stx2 | Patient no 2 | Norway |
| NVH-847 | NIPH-11060708 | 2006 | stx2 | Patient no 9 | Norway |
| NVH-848 | NIPH-11060707 | 2006 | - | Patient no 9 | Norway |
| NVH-849 | NIPH-11060747 | 2006 | - | Patient no 10 | Norway |
| NVH-760 | 625/06 | 2006 | - | Fermented sausage, home of patient 7 | Norway |
| NVH-737 | 2006 | - | Fermented sausage | Norway | |
| NVH-763 | 2006 | - | Food | Norway | |
| NVH-661 | NIPH-10306923 | 2003 | stx2 | Patient | Norway |
| NVH-731 | NIPH-11051601 | 2005 | stx2 | Patient | Norway |
| cdc-08-201 | Not known | Maine, USA | |||
| cdc-08-202 | Not known | Virginia, USA |
All isolates are E. coli O103:H25.
Referred to as Norwegian outbreak strain (NOS).
Characteristics and virulence features of EHEC O103:H25 NOS compared to one of the first described EHEC isolates O157:H7 EDL933 and the related strains EAEC O104:H4 GOS [3], [16] and EHEC O103:H2 str 12009 are listed in Table 2.
Table 2. Characteristics of EHEC O103:H25 NOS, the related strains O104:H4 GOS and O103:H2 str 12009, and the EHEC reference strain O157:H7 EDL933.
| Characteristics | O157:H7 EDL933 | O103:H25NOS | O104:H4 GOS | O103:H2str 12009 |
| Outbreak | ||||
| Year of isolation | 1982 | 2006 | 2011 | 2001 |
| Country | USA | Norway | Germany | Japan |
| Pathotype | EHEC | EHEC | EAEC | EHEC |
| No of diseased | 47 | 16 | 4075 | 1 |
| No of HUS (%) | 0 | 10 (62.5%) | 908 (22%) | 0 |
| No of deaths | 1 | 50 | 0 | |
| Stx2 phage | ||||
| Insertion site | wrbA | wrbA | wrbA | argW |
| Stx2A, nucleotide position 867 | T | C | C | T |
| LEE | ||||
| LEE operons | five | five | none | five |
| Intimin type | gamma | theta | not present | epsilon |
| OI122 | ||||
| sen (Shet2) | yes | yes | no | yes |
| pagC | yes | yes | no | yes |
| efa1/lifA | 2.1 kb | 9.7 kb | not present | 9.7 kb |
| Accessory virulence | ||||
| set1A (shet1) | no | no | yes | no |
| Colicin | E2 | |||
| ehxA | yes | yes | not present | yes |
PFGE analysis
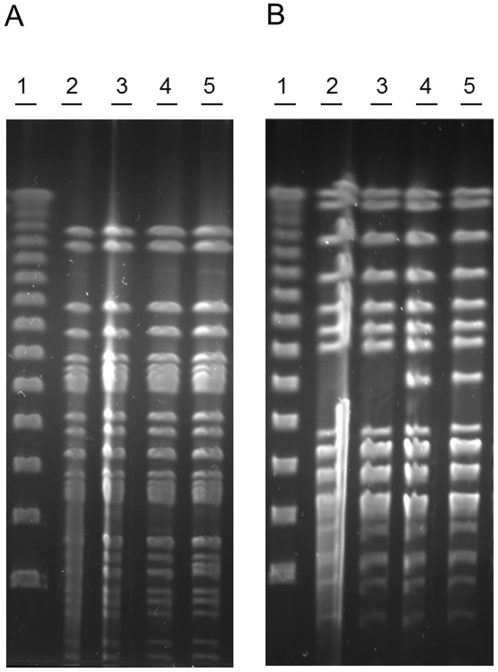
To distinguish between the stx2-positive and the stx-negative isolates the two stx2-positive isolates from patients and two stx-negative isolates, one from a patient and one from food (isolate NVH-848 and NVH-760, respectively), were analyzed by PFGE using the restriction enzyme AvrII. The PFGE patterns from XbaI digestion were indistinguishable as previously described by Sekse et al. [13] (Figure 2A), whereas the AvrII digestion exposed a difference in the PFGE pattern between the stx2-positive isolates and the stx-negative isolates (Figure 2B).
Figure 2. PFGE of outbreak associated isolates.
PFGE of E. coli O103:H25 NOS (lane 2), NVH-847 (lane 3), NVH-848 (lane 4), and NVH-760 (lane 5). Lambda ladder is used as marker (lane 1). Digestion with XbaI (A) showed indistinguishable PFGE patterns, while digestion with AvrII (B) exposed a difference between the stx2-positive and the stx-negative isolates.
Stx2 phage
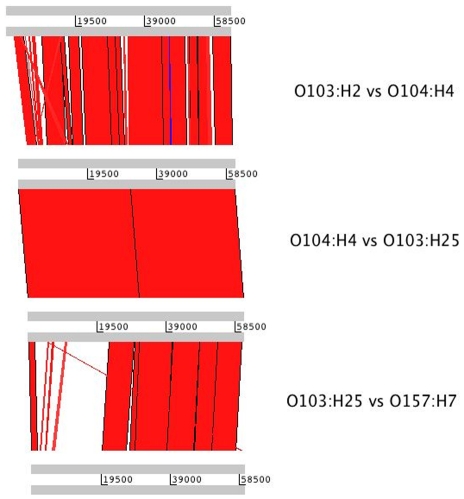
To retrieve the genome sequence of the Stx2 phage, contigs from the EHEC O103:H25 NOS genome sequence containing Stx2 phage DNA were assembled, followed by gap closure PCRs and sequencing. The Stx2 phage genome is 60595 bp in length with a G+C content of 50%. A schematic view of the Stx2 phage is shown in Figure 3. The entire phage sequence was blasted against the sequence of the Stx2 phages of EAEC O104:H4 str C227-11, EHEC O103:H2 str 12009 and EHEC O157:H7 EDL933, and an ACT comparison is shown in Figure 4. The nucleotide identity to the Stx2 phages of EAEC O104:H4 str C227-11 [2] and EHEC O103:H2 str 12009 [7] is 90 and 88%, respectively, and covers the entire sequence, while the similarity to the reference EHEC O157:H7 EDL933 phage is limited to the 35 kb late region (Figure 4).
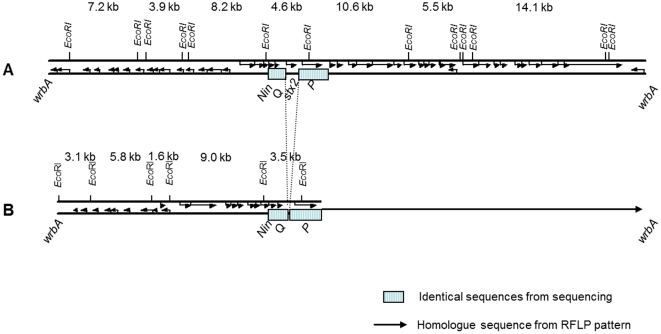
Figure 3. Comparison of Stx2 phage and stx-negative phage.
Comparison of Stx2 phage and stx-negative phage from EHEC O103:H25 NOS (A) and E. coli O103:H25 NVH-848 (B), respectively. The Stx2 phage is sequenced, while the illustration of the stx-negative phage is based on sequence (23 kb) and RFLP pattern (illustrated by arrow).
Figure 4. Stx2 phage from EHEC O103:H25 NOS compared to Stx2 phages from EAEC O104:H4, EHEC O103:H2 and EHEC O157:H7.
ACT visualization between stx2-positive phages from EHEC O103:H25 NOS, EAEC O104:H4 str C227-11, EHEC O103:H2 str 12009 and reference Stx2 phage 933W from EHEC O157:H7 EDL933. The phage genomes are compared using BLAST and the red regions represent hits. The white regions indicate absent genetic regions, which is especially noticeable in the comparison between the O103:H2 phage and the O157:H7 phage.
Stx2-related phage
In plaque hybridization assays, all plaques produced by EHEC O103:H25 NOS were stx2 positive, while all plaques produced by E. coli O103:H25 NVH-848 were stx2 negative. DNA from these latter phages (hereafter referred to as stx-negative phage) was isolated via plaque assay. RFLP patterns showed that phages from five stx-negative isolates were identical, but the restriction pattern differed from that of the phages of the two stx2-positive patient isolates (Figure 5). The integration site for the stx-negative phage was wrbA, which is the same site as the Stx2 phage [26].
Figure 5. RFLP of phage from stx2-positive and stx-negative isolates.
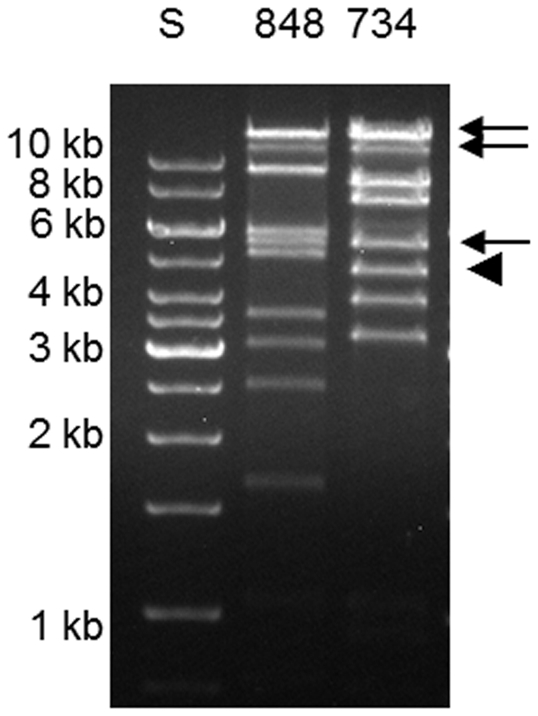
EcoRI restriction fragment length polymorphism analysis of Stx2 phage and stx-negative phage in EHEC O103:H25 NOS and E. coli O103:H25 NVH-848, respectively. The arrow indicates the EcoRI fragments that are indistinguishable between the strains, arrowhead indicates the EcoRI fragment of E. coli O103:H25 NOS where the stx2 gene is located.
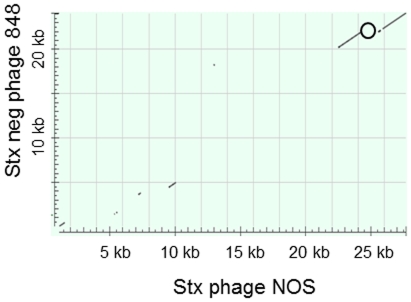
Hybridization located stx2 to a 4.5 kb EcoRI fragment not present in the stx-negative phage RFLP (Figure 5). Still, RFLP analysis revealed three bands of about 14, 11 and 5.5 kb that were common between the stx2-positive and -negative phages (Figure 5). The estimated sizes of the three bands correspond to the three large EcoRI fragments comprising the late region of the sequenced Stx2 phage NOS (Figure 3), indicating that the late region of the two phages are homologue sequences.
To sequence the early region of the stx-negative phage, two fragments of 3.2 and 1.6 kb (Figure 5) were cloned and sequenced. Primer walking was used to amplify PCR products from the stx-negative phage DNA to complete the sequence. The 23 kb sequence of the early region of the stx-negative phage is not related to the Stx2 phage from O103:H25 NOS and a schematic view of the two phages is shown in Figure 3. The sequence of the stx-negative phage revealed an AvrII restriction site not present in the Stx2 phage genome. The early phage regions of the two phages were blasted against each other and a Dot matrix view is shown in Figure 6.
Figure 6. Early region of Stx2 phage compared to early region of stx-negative phage.
Dot matrix view of 24 kb early region of the Stx2 phage from EHEC O103:H25 NOS and the 23 kb early region of the stx-negative phage from E. coli O103:H25 isolate NVH-848. The position of the stx2 gene in EHEC O103:H25 NOS is marked (o). Regions of similarity are based upon the BLAST results. Alignments are shown in the plot as lines. Plus strand and protein matches are slanted from the bottom left to the upper right corner, minus strand matches are slanted from the upper left to the lower right. The number of lines shown in the plot is the same as the number of alignments found by BLAST.
Contigs from the EHEC O103:H25 NOS genome did not contain the sequence of the stx-negative phage, and also PCR run on total DNA from EHEC O103:H25 NOS with primers specific for the early region of the stx-negative phage were negative or gave products of incorrect size.
Cloning of phage DNA
An attempt to isolate and clone phage DNA directly from EHEC O103:H25 NOS resulted in the discovery of another phage in this strain. This phage is 45 kb, has a G+C content of 47% and the sequence is 53% identical to bacteriophage ΦV10 which is a temperate phage that specifically infects E. coli serogroup O157:H7 [20]. The insertion site of the phage is within the guaA gene, and the phage does not infect E. coli DH5α.
Colicin and plasmids
Colicin from EHEC O103:H25 NOS in culture supernatants was phenotypically observed as clear zones in an E. coli DH5α cell lawn. In silico analysis of the sequenced genome demonstrated the presence of a colicin E2 gene and its associated immunity and lysis genes on a 6744 bp contig. A gap-closure PCR confirmed the contig to be a complete plasmid. Hybridization with a colE2 pcr template identified the presence of the colicin E2 carrying plasmid in outbreak-associated O103:H25 isolates from both patients and food, including the isolates from 2003 and 2005, while non-outbreak O103:H25 isolates associated with Norwegian sheep did not harbour the plasmid. The entire colicin plasmid was 97% identical to pO111_4 from E. coli O111:H- str 11128 [7].
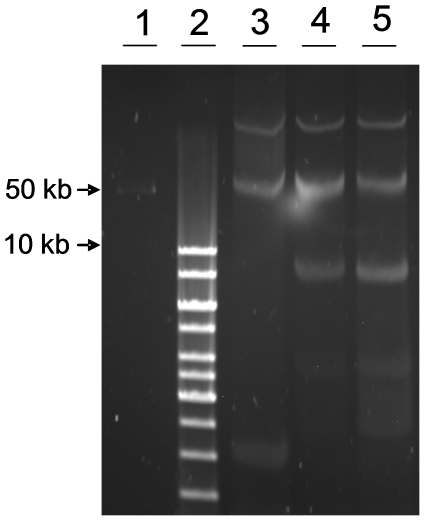
EHEC O103:H25 NOS and E. coli O103:H25 NVH-848 share identical plasmid profiles and harbour one large plasmid of approximately the same size as pO157 of E. coli O157:H7 EDL 933 (Figure 7).
Figure 7. Plasmid profiles of E. coli O103:H25.
Comparison of large plasmids in EHEC O103:H25 NOS (lane 4) and E. coli O103:H25 NVH-848 (lane 5) with EHEC O157:H7 EDL 933 (lane 3). GeneRuler 1 kb ladder (lane 2) and Lambda ladder (lane 1) were used as molecular size marker.
Discussion
Serotype O103:H25 is a rare cause of EHEC disease, and has not previously been associated with outbreaks [11], only with sporadic disease cases [21], [22]. A few E. coli O103:H25 strains have been characterized and found to carry stx1, but they have not been associated with severe disease [18]. The Norwegian 2006 outbreak caused by EHEC O103:H25 had a 60% HUS frequency, and the strain is considered to be particularly virulent [11].
We have shown that the genome of EHEC O103:H25 NOS resemble the EAEC O104:H4 GOS and EHEC O103:H2 str 12009. This is supported in a pan genomic study of 61 sequenced E. coli genomes, showing that the E. coli O103 Oslo (O103:H25 NOS) clusters closely together with the EHEC O103:H2 str 12009 [23], and a more recent study shows the similarities between the EHEC O103:H2 str 12009 and the EAEC O104:H4 GOS genomes [2]. EAEC O104:H4 GOS caused diarrhoea in approximately 4000 individuals, 22% of which developed HUS, and the strain is notably more virulent than most EHEC [2]. The E. coli O104:H4 outbreak began in Germany in May 2011, but was later identified in other European countries [2], [24]. Due to phenotypic and genotypic characteristics, the German O104:H4 outbreak strain is not classified as an EHEC, but rather as a Shiga toxin producing enteroaggregative E. coli (EAEC) [2], [3], [25]. Despite the lack of genes characteristic of EAEC in EHEC O103:H25 NOS, and thus differing in both pathotype and serotype, the genomes of the Norwegian and the German outbreak strains are highly similar, as illustrated in Figures 1A and 1B. The close relationship between the two strains is supported by MLST analysis (Figure 1C). Also the Stx2 phages in these two strains show a striking homology with a DNA sequence identity of 90% (Figure 4) [2]. The identity includes a 1 bp silent nucleotide mutation in the stx2A gene [3], [26] which is rare in other stx2 genes. This indicates a common origin for the two phages. The finding of closely related Stx2 phages and genomes in two outbreak strains of different serotypes and pathotypes, but with a high HUS incidence in common, is remarkable and will be investigated further.
Other strains with related Stx2 phages include EHEC O103:H2 str 12009 from a sporadic case of diarrhoea in Japan in 2001, and E. coli O111:H- str 11128. Similar Stx2 phages to the Norwegian outbreak strain are thus present in E. coli strains of serogroups O103, O104 and O111, and the phage seems to be rather promiscuous in nature. The similarity between the Stx2 phage of EHEC O103:H25 NOS and the reference Stx2 phage 933W from O157:H7 EDL933 is high in the 38 kb late region of the phages where the stx genes are located (95%), however, the early regions of these phages differ in composition (Figure 4). In EHEC O103:H25 NOS the Stx2 phage is inserted into wrbA, a previously described integration site of Stx2 phages, e.g. in EHEC O157:H7 EDL933 and Sakai strains [27], [28]. The wrbA had not been observed as an integration site in serogroup O103 prior to the Norwegian outbreak [26]. The closely related Stx2 phage in O104:H4 GOS is also inserted in wrbA, while the Stx2 phage in EHEC O103:H2 str 12009 is located within the argW gene.
The only observed feature distinguishing between the Norwegian outbreak strain and stx-negative isolates from the 2006 outbreak is the presence of either the stx2-positive or the stx-negative phage, respectively. These two phages are related and share parts of their sequences and insertion site. While the 30 kb late regions are similar in the two phages, the early regions are completely different. Interestingly, the shift between the similar and dissimilar parts is abrupt and in proximity of the stx genes (Figure 3). As Stx2 phages are mosaic by nature and rearrangements are not uncommon [6], the stx-negative phage could have developed from the Stx2 phage by acquiring its distinctive sequence from the chromosome of the E. coli O103:H25 host. However, the distinctive stx-negative phage sequence has not been identified in the EHEC O103:H25 NOS genome neither by in silico analysis nor by PCR. This indicates that this phage, or at least part of it, has another origin. The distinctive sequence of the stx-negative phage shows some similarities to phage related sequences in a BLAST search, but it seems to have a rather unique construction.
The 2006 EHEC O103:H25 outbreak is remarkable because all food isolates were stx-negative, and only two of 11 isolates from patients were stx2-postive [11], [13]. The lack of stx genes in EHEC serotypes isolated from patients is not uncommon and has been reported in both O157 and non-O157 isolates [12], [29]. In studies where the mechanism of stx gene loss has been investigated, it has been shown that the stx-negative strains lack the entire Stx-encoding bacteriophage. This has been demonstrated by the presence or reappearance of an intact integration site for the Stx phage and an altered PFGE pattern [29]–[32]. Such stx-negative isolates from patients are believed to be progenies of an EHEC that lost the stx genes during the course of illness, and might be referred to as EHEC-LST (lost Shiga toxin) [12]. The EHEC-LST model is supported by the finding that EHEC are difficult to isolate from patients late in illness [33], and the theory is further confirmed by Mellmann and Karch who demonstrated the presence of stx-negative strains of O26:H11/NM and sorbitol fermenting (SF) O157:NM subsequent to stx2-positive isogenic isolates in the same patients [12], [34], [35].
In contrast to the EHEC-LST phenomenon, we find that the integration site of the Stx2 phage is occupied in the stx-negative isolates by a partly related phage but without stx genes. The stx-negative isolates thus differ from the stx2-positive isolates not only by the lack of the Stx2 phage but also by the presence of this stx-negative phage. The similar size and the lack of an XbaI restriction site in both the Stx2 phage and the stx-negative phage explain the finding of identical XbaI digested PFGE profiles of the stx2-positive and stx-negative isolates shown by Sekse et al. [13]. Digestion with AvrII, however, revealed a difference in PFGE pattern, and sequencing of the two phage genomes identified an AvrII restriction site in the Stx2 phage which is not present in the stx-negative phage and that most likely explains the observed difference.
Only two stx2-positive patient isolates were retrieved in 2006, and as no stx2-positive isolates were retrieved from food, it could be speculated that the stx-negative E. coli acquired the Stx2 phage in the patients' gut. However, the Stx2 phages from the two patient isolates have identical RFLP pattern and a rare silent nucleotide mutation in stxA, which both are found in two EHEC O103:H25 isolates from sporadic cases in Norway in 2003 and 2005 [26]. This strongly indicates that the Stx2 phages in these four isolates are epidemiologically linked and that the stx2-positive isolates from 2006 originates from the same source.
The peculiar circumstance is that both stx2-positive and stx-negative O103:H25 E. coli cells must have been present in the contaminated fermented sausage in 2006. Which of the two variants is the ancestor is difficult to predict, but there is reason to believe that the stx2-positive clone preceded the stx-negative clone because the related Stx2 phage was identified in the same E. coli serotype three years prior to the 2006 outbreak. On the other hand, stx-negative isolates could have been overlooked in earlier cases, and the origin and history of this clone is difficult to evaluate. The hypothesis that bacterial cells with stx-negative or stx2-positive phage were both present in the food is strengthened by the finding of Sekse et al. [26]. They were not able to isolate infective phage particles from food samples in the 2006 outbreak, but stx2 was detected in food samples by PCR. Although the food most likely contained a mixture of stx2-positive and stx-negative bacterial cells, the two clones may not have been present in equal numbers. As only the stx-negative clone is isolated from food the proportion of stx2-positive E. coli was probably considerably lower. In the diseased patients, this imbalance might have been reversed during the course of illness. All investigated non-stx2 isolates from patients have been shown to be the same stx-negative clone as is isolated from food, and not offspring from the stx2-positive clone in form of EHEC-LST.
Several factors may contribute to the virulence of EHEC including factors within the individual (age, microbiota, number of Gb3 receptors), the bacterial host (intimin type), the toxin itself (subtype, synergy, Stx toxin levels), and other virulence factors (e.g. subtilase cytotoxin) [4], [18], [36]–[38]. The pathogenicity island locus of enterocyte effacement (LEE) is associated with virulence, and encodes the intimin gene (eae), the translocated intimin receptor (Tir) and a type III secretion system (TTSS). These are all involved in the intimate attachment of EHEC to enterocytes [39]. All five LEE operons are present in EHEC O103:H25 NOS, however, LEE is not part of the EAEC O104:H4 GOS genome. Another genomic island present in the 2006 outbreak strain is the putative pathogenicity island OI122 with the genes sen, pagC and efa1 which have been strongly correlated with virulence and disease severity [18], [40]. The complete efa1/lifA gene is 9672 kb and is a bifunctional protein for adherence and inhibition of lymphocyte activation [19]. The distribution of this large toxin is limited to less than 30 strains of heterologous serogroups (BLAST search), only one of the sequenced O157:H7 strains and two O157:NM strains exhibit it [41], [42]. EAEC O104:H4 GOS does not exhibit any version of efa/lifA, while O103:H2 str 12009 exhibits the large version of efa/lifA. Enterohemolysin (Ehx) is regarded a virulence factor of EHEC and the genes are located on large plasmids like pO157 [43]–[45]. One large plasmid is detected in O103:H25 NOS, in contrast to O104:H4 GOS which carries two large plasmids [25].
The colicin production of E. coli O103:H25 may have provided an advantage for the bacterial cells in the harsh competition of the gut, making the colonization more efficient. In addition, the colicin E2 is a DNAse colicin which has been found to increase the in vitro production of Stx in EHEC cells exposed to it [46]. It is however unlikely that colicin E2 can affect the production of Stx when the genes coexist in the same cell, but it has been shown that intestinal E. coli cells can act as chaperones and contribute to the production of Stx [47] and the colicin could possibly play a role here. Similar colicin plasmids are found in EHEC O26:H11 and EHEC O111:H- [7], and in a study by Karama et al. [48], 38.4% of E. coli O103:H2 strains were found to have a colicin producing phenotype.
The phi-like phage isolated from the wild-type EHEC O103:H25 NOS has an unknown function in the E. coli O103:H25 host. However, the number of the phi-like phage particles released from EHEC O103:H25 NOS after induction with Mitomycin C is estimated to be approximately ten times the number of Stx2 phage particles released (data not shown), and hence this abundant phi-like phage is a practical challenge as it complicates the isolation of the Stx2 phage and stx-negative phage directly from wild types.
Conclusion
Both the Stx2 phage and the bacterial genome from EHEC O103:H25 NOS are related to the Shiga toxin producing EAEC O104:H4 that caused a large European E. coli outbreak in 2011. Two patient isolates from the Norwegian O103:H25 outbreak carry an Stx2 phage, while other outbreak associated isolates carry a related phage in the same insertion site. The two variants, E. coli O103:H25 with the Stx2 phage or the stx-negative phage, have probably both been present in the contaminated food which caused the Norwegian outbreak.
Materials and Methods
Bacterial isolates
The E. coli O103:H25 isolates included in the study are presented in Table 1. NVH-734 is the outbreak reference strain and is also referred to as the Norwegian outbreak strain (NOS). E. coli DH5α was used as recipient strain in the plaque assay. E. coli strains were cultured in Luria-Bertani (LB) broth or on LB agar plates (LB containing 1% agar).
Whole genome sequencing, alignments and Multilocus sequence typing (MLST)
Isolate NVH-734 (EHEC O103:H25 NOS) was sequenced using 454 technology (454 Life Sciences, Branford, Connecticut, USA). Initial genome analysis revealed similarity to the E.coli O103:H2 str 12009 (accession: AP010958.1), and this strain was used as template for the alignment of contigs from the EHEC O103:H25 Norwegian Outbreak Strain (NOS) using Mauve [15]. Subsequently, genome alignment of EHEC O103:H25 NOS and the EAEC O104:H4 str German Outbreak Strain (GOS)1 (distributed on 208 contigs, accessions: AFWO01000001.1-AFWO01000208.1) was also carried out with Mauve, using the progressive alignment option. Whole genome BLAST-comparison was performed using the BRIG software package [49] on the following genomes: EHEC O103:H25 NOS, EAEC O104:H4 GOS1, EHEC O103:H2 str 12009, EAEC O104:H4 str 55989, EHEC O111:H- str 11128, EHEC O157:H7 EDL933 and EHEC O26:H11 str 11368. MLST was performed according to Wirth et al. [50], using the seven housekeeping genes adk, fumC, gyrB, icd, mdh, purA, and recA. A maximum likelihood test using PhyML [51] was carried out to assess the best nucleotide substitution matrix in R (http://www.R-project.org/) with the package ‘ape’ [52]. Based on this, Tamura-Nei with invariant sites was shown to be the best model, and subsequently MEGA 5 [53] was used to generate a maximum likelihood tree, which was bootstrapped 500 times. Sequence type (ST) of EHEC O103:H25 NOS was obtained from MLST Databases at the ERI, University College Cork (http://mlst.ucc.ie). The O103:H25 NOS, O104:H4 str C227-11 (originating from the German outbreak), O103:H2 str 12009 and O157:H7 EDL933 phages were BLASTed against each other and compared using the ACT tool [54].
PCR and sequencing
Primers for gap-closure PCR for completing the sequence of the Stx2 phage, and for detection of genes in other isolates than EHEC O103:H25 NOS were designed on the basis of the genome sequence. All PCRs were carried out in an Eppendorf Mastercycler gradient (Eppendorf AG, Hamburg, Germany). DyNAzyme II DNA polymerase (supplied with 10× buffer) and dNTP Mix from Finnzymes (Vantaa, Finland) were used as instructed by the manufacturer. The standard program was as follows: 95°C for 1 min, 30 cycles of 95°C for 1 min, 52°C for 1 min and 72°C for 1 min, and finally 72°C for 5 min. Sequencing of PCR products was performed by Source BioScience geneservice (United Kingdom), and DNA sequences were analyzed using Vector NTI Advance 11 (Invitrogen, Carlsbad, USA) and BLAST. Primers used for PCR detection of other virulence genes are listed in Table 3.
Table 3. Primers used in the study.
| Primer | Sequence (5′-3′) | Reference | Probe |
| colE2 | F ATGAGCGGTGGCGATGGACGC | This study | Hybridisation of plasmid profiles |
| R GCCCGGCCATTTGCCACATTCT | |||
| pagC | F ATGAGTGGTTCAAGACTGG | [18] | |
| R CCAACTCCAACAGTAAATCC | |||
| sen | F GGATGGAACCATACCTGG | [18] | |
| R CGCAATCAATTGCTAATGC | |||
| efa1 | F CTCCCAGAGATAATTTTGAGG | [18] | |
| R CAACTGTATGCGAATAGTACTC | |||
| efa2 | F CTGTCAGACGATGACATTGG | [18] | |
| R GAAGGATGGGCATTGTGTC | |||
| stx2 | F GCGTTTTGACCATCTTCGT R ACAGGAGCAGTTTCAGACAG | [57] | Hybridisation of RFLP and PFGE |
Plaque assay
E. coli O103:H25 LB broth cultures were incubated at 37°C with shaking at 200 rpm to an OD600 of 0.3–0.5 (∼2 h). To induce phage production Mitomycin C (Sigma-Aldrich, St. Louis, MO, USA) was added to a final concentration of 0.5 µg/ml and incubation was continued overnight in the dark. The cultures were centrifuged (2000×g, 10 min) to remove bacterial cells and debris, and the supernatants (phage lysates) were sterile filtered (0.22 µm; Minisart, Sartorius Stedim Biotech). As the wildtype strains also produced colicin (see below) that lysed the DH5α cell lawn, lysates were treated with 100 µg/µl trypsin (Sigma) at 37°C for 60 minutes prior to use to destroy colicin. The E. coli DH5α recipient was grown in LB broth to OD 0.4–0.6 at 37°C and shaking at 200 rpm. For determination of infectious phage particles, 100 µl of tenfold dilutions of phage lysates were added 900 µl E. coli DH5α. CaCl2 was added to a final concentration of 10 mM. The phage-recipient mixture was incubated for 30 min at 37°C before 2.5 ml molten soft agar (0.7% LB broth) was added and the mixture poured onto LB agar plates with CaCl2 (10 mM). The plates were incubated at 37°C overnight, plaques were counted by visual examination and phage titres were calculated.
Isolation and RFLP analysis of phage DNA
To ensure that DNA was isolated from only the Stx2- or Stx-related phage, DNA extraction was performed via the plaque assay. This was necessary because an abundant phi like phage is also present in the wild type strains, but this phage is not able to infect E. coli DH5α. Phage isolation was done from seven strains from the 2006 outbreak, in which five were stx-negative strains from either food or patients, and two were stx2-positive isolates from patients. The isolates are listed in Table 1. Briefly, 0.1 ml from an overnight starter culture of E. coli DH5α was transferred to 10 ml LB- broth and incubated at 37°C with shaking until bacterial growth reached mid log phase. Approximately 50 plaque were picked (from the plaque assay described above), dissolved in 50 µl MQ, and added to 0.5 ml of the E. coli DH5α culture together with 12.5 µl 1 M CaCl2 to facilitate bacteriophage infection. The culture was incubated at 37°C for two hours before 9.5 ml of LB-broth was added and then incubated at 37°C overnight. The overnight culture was centrifuged at 2000×g for 10 minutes and the supernatant containing phage particles was sterile filtered (0.22 µm, Minisart, Sartorius Stedim Biotech, Aubagne, France), precipitated with 0.18×PEG 8000/NaCl at 4°C for 2 hours, and centrifuged at 10 000×g for 1 hours. The pellet was dissolved in 0.5 ml TE buffer with proteinase K (Sigma-Aldrich, 50 mg/ml) and SDS (final concentration of 0.5%) and incubated for one hour at 56°C. The DNA was extracted using phenol/chloroform/isoamylalcohol (25∶24∶1), and the DNA was precipitated using equal amounts of isopropanol. Phage DNA was used as template in restriction fragment length polymorphism (RFLP) analysis and in PCR reactions. Phage DNA was digested with restriction enzymes EcoRI (New England BioLabs, Hertfordshire, England) and the restriction fragments separated by 1% agarose gels. A probe for detection of stx2A in RFLP hybridization was made using primers listed in Table 3. To complete the phage genomes of the stx2-positive and stx-negative phages, phage DNA from O103:H25 NOS and NVH-848, respectively, were used as templates in PCR reactions.
Colicin production
Colicin production was detected by observation of a lytic effect of sterile filtered supernatant from overnight culture of EHEC O103:H25 NOS on E. coli DH5 cell lawns. The identification of a colicin E2 encoding gene was performed by in silico analysis of the genome sequence, and gap closure PCR was performed to confirm a plasmid configuration of the contig. Primers colE2F and colE2R (Table 3) and standard PCR conditions were used to generate a probe for plasmid hybridization.
Pulsed-Field Gel Electrophoresis (PFGE) and plasmid isolation
The E. coli isolates associated with the outbreak were analyzed by PFGE as described previously [13] using the restriction enzymes XbaI and AvrII (New England BioLabs). DNA for detection of colicin E2 encoding plasmids were isolated by Qiagen plasmid purification kit (Qiagen, Hilden, Germany), and separated by electrophoresis in 2% agarose gels. Large plasmids were isolated by the method of Kado and Liu [55] with modifications. Bacteria were grown in 5 ml LB broth (Oxoid) over night at 37°C, 250 rpm of which 1.5 ml were centrifuged. The pellet was suspended in 200 ml cold (4°C) TAE-buffer (40 mM Tris-acetate, 2 mM EDTA, pH 7.6) and 400 µl 50 mM Tris/3% SDS, pH 12.55 were added prior to incubation at 55°C for 60 min. Plasmid DNA was extracted twice with phenol-chloroform (1∶1, vol/vol) and 25 µl were applied directly to 0.7% agarose gel. Plasmids were separated by electrophoresis at 120 V for 3 h at 4°C.
Southern blotting and hybridization
Plasmid DNA and digested phage DNA were transferred to nylon membranes (Hybond-N, Amersham International plc, Amersham, United Kingdom) by Southern blotting [56]. For detection of the colicin E2 encoding gene and phage genes probes were labelled with digoxigenin (DIG) and hybridized with a DIG DNA labelling and detection kit (Roche Diagnostics, Basel, Switzerland) according to the instructions by the manufacturer.
Cloning of phage DNA
Phage DNA isolated directly from EHEC O103:H25 NOS culture supernatants was digested with EcoRI and cloned in pUC18, and the clones were subsequently sequenced.
GenBank accession numbers
The genome sequence of EHEC O103:H25 NOS has been deposited at DDBJ/EMBL/GenBank under the accession no AGSG00000000. The version described in this paper is the first version, AGSG01000000. The 61 kb Stx2 phage genome has accession no JQ011318 and the 24 kb early region of the stx-negative phage has accession no JQ011316. The 45 kb phi-like phage has accession no JQ011317.
Acknowledgments
We thank Evangeline Sowers at the Centers for Disease Control and Prevention (USA) for providing control isolates.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by The Research Council of Norway (project 178274/I10). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gyles CL. Shiga toxin-producing Escherichia coli: an overview. J Anim Sci. 2007;85:E45–E62. doi: 10.2527/jas.2006-508. jas.2006-508 [pii];10.2527/jas.2006-508 [doi] [DOI] [PubMed] [Google Scholar]
- 2.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. 10.1056/NEJMoa1106920 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. S1473-3099(11)70165-7 [pii];10.1016/S1473-3099(11)70165-7 [doi] [DOI] [PubMed] [Google Scholar]
- 4.Fuller CA, Pellino CA, Flagler MJ, Strasser JE, Weiss AA. Shiga toxin subtypes display dramatic differences in potency. Infect Immun. 2011;79:1329–1337. doi: 10.1128/IAI.01182-10. IAI.01182-10 [pii];10.1128/IAI.01182-10 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fogg PC, Gossage SM, Smith DL, Saunders JR, McCarthy AJ, et al. Identification of multiple integration sites for Stx-phage Phi24B in the Escherichia coli genome, description of a novel integrase and evidence for a functional anti-repressor. Microbiology. 2007;153:4098–4110. doi: 10.1099/mic.0.2007/011205-0. [DOI] [PubMed] [Google Scholar]
- 6.Johansen BK, Wasteson Y, Granum PE, Brynestad S. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology. 2001;147:1929–1936. doi: 10.1099/00221287-147-7-1929. [DOI] [PubMed] [Google Scholar]
- 7.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamage SD, Patton AK, Hanson JF, Weiss AA. Diversity and host range of Shiga toxin-encoding phage. Infect Immun. 2004;72:7131–7139. doi: 10.1128/IAI.72.12.7131-7139.2004. 72/12/7131 [pii];10.1128/IAI.72.12.7131-7139.2004 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, et al. lambda repressor and cro-components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 10.Allison HE. Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2007;2:165–174. doi: 10.2217/17460913.2.2.165. 10.2217/17460913.2.2.165 [doi] [DOI] [PubMed] [Google Scholar]
- 11.Schimmer B, Nygard K, Eriksen HM, Lassen J, Lindstedt BA, et al. Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect Dis. 2008;8:41. doi: 10.1186/1471-2334-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellmann A, Bielaszewska M, Zimmerhackl LB, Prager R, Harmsen D, et al. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin Infect Dis. 2005;41:785–792. doi: 10.1086/432722. CID36325 [pii];10.1086/432722 [doi] [DOI] [PubMed] [Google Scholar]
- 13.Sekse C, O'Sullivan K, Granum PE, Rørvik LM, Wasteson Y, et al. An outbreak of Escherichia coli O103:H25 - Bacteriological investigations and genotyping of isolates from food. Int J Food Microbiol. 2009;133:259–264. doi: 10.1016/j.ijfoodmicro.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 14.Lindstedt BA, Brandal LT, Aas L, Vardund T, Kapperud G. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J Microbiol Methods. 2007;69:197–205. doi: 10.1016/j.mimet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. 10.1101/gr.2289704 [doi];14/7/1394 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzuszkiewicz E, Thurmer A, Schuldes J, Leimbach A, Liesegang H, et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Arch Microbiol. 2011 doi: 10.1007/s00203-011-0725-6. 10.1007/s00203-011-0725-6 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, et al. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol. 2003;41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klapproth JM, Scaletsky IC, McNamara BP, Lai LC, Malstrom C, et al. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect Immun. 2000;68:2148–2155. doi: 10.1128/iai.68.4.2148-2155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry LL, SanMiguel P, Minocha U, Terekhov AI, Shroyer ML, et al. Sequence analysis of Escherichia coli O157:H7 bacteriophage PhiV10 and identification of a phage-encoded immunity protein that modifies the O157 antigen. FEMS Microbiol Lett. 2009;292:182–186. doi: 10.1111/j.1574-6968.2009.01511.x. [DOI] [PubMed] [Google Scholar]
- 21.Mackenzie AM, Lebel P, Orrbine E, Rowe PC, Hyde L, et al. Sensitivities and specificities of premier E. coli O157 and premier EHEC enzyme immunoassays for diagnosis of infection with verotxin (Shiga-like toxin)-producing Escherichia coli. J Clin Microbiol. 1998;36:1608–1611. doi: 10.1128/jcm.36.6.1608-1611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivas M, Miliwebsky E, Chinen I, Roldán CD, Balbi L, et al. Characterization and epidemiologic subtyping of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome and diarrhea cases in Argentina. Foodborne Pathog Dis. 2006;3:88–96. doi: 10.1089/fpd.2006.3.88. 10.1089/fpd.2006.3.88 [doi] [DOI] [PubMed] [Google Scholar]
- 23.Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. 10.1007/s00248-010-9717-3 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu CJ, Hsueh PR, Ko WC. A new health threat in Europe: Shiga toxin-producing Escherichia coli O104:H4 infections. J Microbiol Immunol Infect. 2011 doi: 10.1016/j.jmii.2011.07.001. S1684–1182(11)00152-6 [pii];10.1016/j.jmii.2011.07.001 [doi] [DOI] [PubMed] [Google Scholar]
- 25.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. 10.1371/journal.pone.0022751 [doi];PONE-D-11-11826 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekse C, Muniesa M, Wasteson Y. Conserved Stx2 phages from Escherichia coli O103:H25 isolated from patients suffering from hemolytic uremic syndrome. Foodborne Pathog Dis. 2008;5:801–810. doi: 10.1089/fpd.2008.0130. [DOI] [PubMed] [Google Scholar]
- 27.Makino K, Yokoyama K, Kubota Y, Yutsudo CH, Kimura S, et al. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet Syst. 1999;74:227–239. doi: 10.1266/ggs.74.227. [DOI] [PubMed] [Google Scholar]
- 28.Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. 10.1038/35054089 [doi] [DOI] [PubMed] [Google Scholar]
- 29.Bielaszewska M, Prager R, Köck R, Mellmann A, Zhang W, et al. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl Environ Microbiol. 2007;73:3144–3150. doi: 10.1128/AEM.02937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bielaszewska M, Prager R, Zhang W, Friedrich AW, Mellmann A, et al. Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol. 2006;72:1900–1909. doi: 10.1128/AEM.72.3.1900-1909.2006. 72/3/1900 [pii];10.1128/AEM.72.3.1900-1909.2006 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng P, Dey M, Abe A, Takeda T. Isogenic strain of Escherichia coli O157:H7 that has lost both Shiga toxin 1 and 2 genes. Clin Diagn Lab Immunol. 2001;8:711–717. doi: 10.1128/CDLI.8.4.711-717.2001. 10.1128/CDLI.8.4.711-717.2001 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murase T, Yamai S, Watanabe H. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr Microbiol. 1999;38:48–50. doi: 10.1007/pl00006771. [DOI] [PubMed] [Google Scholar]
- 33.Tarr PI, Neill MA, Clausen CR, Watkins SL, Christie DL, et al. Escherichia coli O157:H7 and the hemolytic uremic syndrome: importance of early cultures in establishing the etiology. J Infect Dis. 1990;162:553–556. doi: 10.1093/infdis/162.2.553. [DOI] [PubMed] [Google Scholar]
- 34.Karch H, Mellmann A, Bielaszewska M. Epidemiology and pathogenesis of enterohaemorrhagic Escherichia coli. Berl Munch Tierarztl Wochenschr. 2009;122:417–424. [PubMed] [Google Scholar]
- 35.Mellmann A, Lu S, Karch H, Xu JG, Harmsen D, et al. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl Environ Microbiol. 2008;74:67–72. doi: 10.1128/AEM.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Sablet T, Bertin Y, Vareille M, Girardeau JP, Garrivier A, et al. Differential expression of stx2 variants in Shiga toxin-producing Escherichia coli belonging to seropathotypes A and C. Microbiology. 2008;154:176–186. doi: 10.1099/mic.0.2007/009704-0. 154/1/176 [pii];10.1099/mic.0.2007/009704-0 [doi] [DOI] [PubMed] [Google Scholar]
- 37.de Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, et al. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2009;77:783–790. doi: 10.1128/IAI.01048-08. IAI.01048-08 [pii];10.1128/IAI.01048-08 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paton AW, Paton JC. Escherichia coli subtilase cytotoxin. Toxins (Basel) 2010;2:215–228. doi: 10.3390/toxins2020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt MA. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol. 2010;12:1544–1552. doi: 10.1111/j.1462-5822.2010.01518.x. CMI1518 [pii];10.1111/j.1462-5822.2010.01518.x [doi] [DOI] [PubMed] [Google Scholar]
- 40.Wickham ME, Lupp C, Mascarenhas M, Vázquez A, Coombes BK, et al. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J Infect Dis. 2006;194:819–827. doi: 10.1086/506620. JID36198 [pii];10.1086/506620 [doi] [DOI] [PubMed] [Google Scholar]
- 41.Bielaszewska M, Köck R, Friedrich AW, von Eiff C, Zimmerhackl LB, et al. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One. 2007;2:e1024. doi: 10.1371/journal.pone.0001024. 10.1371/journal.pone.0001024 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedrich AW, Zhang W, Bielaszewska M, Mellmann A, Köck R, et al. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin Infect Dis. 2007;45:39–45. doi: 10.1086/518573. CID50097 [pii];10.1086/518573 [doi] [DOI] [PubMed] [Google Scholar]
- 43.Caprioli A, Morabito S, Brugere H, Oswald E. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. 10.1051/vetres:2005002 [doi];v4055 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt H, Kernbach C, Karch H. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142(Pt 4):907–914. doi: 10.1099/00221287-142-4-907. [DOI] [PubMed] [Google Scholar]
- 46.Toshima H, Yoshimura A, Arikawa K, Hidaka A, Ogasawara J, et al. Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7 by DNase colicins. Appl Environ Microbiol. 2007;73:7582–7588. doi: 10.1128/AEM.01326-07. AEM.01326-07 [pii];10.1128/AEM.01326-07 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gamage SD, Strasser JE, Chalk CL, Weiss AA. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect Immun. 2003;71:3107–3115. doi: 10.1128/IAI.71.6.3107-3115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karama M, Johnson RP, Holtslander R, Gyles CL. Phenotypic and genotypic characterization of verotoxin-producing Escherichia coli O103:H2 isolates from cattle and humans. J Clin Microbiol. 2008;46:3569–3575. doi: 10.1128/JCM.01095-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. 1471-2164-12-402 [pii];10.1186/1471-2164-12-402 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirth T, Falush D, Lan R, Colles F, Mensa P, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. MMI5172 [pii];10.1111/j.1365-2958.2006.05172.x [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. 10.1007/978-1-59745-251-9_6 [doi] [DOI] [PubMed] [Google Scholar]
- 52.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 53.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. msr121 [pii];10.1093/molbev/msr121 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carver T, Berriman M, Tivey A, Patel C, Bohme U, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. btn529 [pii];10.1093/bioinformatics/btn529 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Muniesa M, Jofre J. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl Environ Microbiol. 1998;64:2443–2448. doi: 10.1128/aem.64.7.2443-2448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]