Abstract
Background
Plasmodium vivax merozoites specifically invade reticulocytes. Until recently, two reticulocyte-binding proteins (Pvrbp1 and Pvrbp2) expressed at the apical pole of the P. vivax merozoite were considered to be involved in reticulocyte recognition. The genome sequence recently obtained for the Salvador I (Sal-I) strain of P. vivax revealed additional genes in this family, and in particular Pvrbp2a, Pvrbp2b (Pvrbp2 has been renamed as Pvrbp2c) and two pseudogenes Pvrbp2d and Pvrbp3. It had been previously found that Pvrbp2c is substantially more polymorphic than Pvrbp1. The primary goal of this study was to ascertain the level of polymorphism of these new genes.
Methodology/Principal Findings
The sequence of the Pvrbp2a, Pvrbp2b, Pvrbp2d and Pvrbp3 genes were obtained by amplification/cloning using DNA purified from four isolates collected from patients that acquired the infection in the four cardinal regions of Thailand (west, north, south and east). An additional seven isolates from western Thailand were analyzed for gene copy number variation. There were significant polymorphisms exhibited by these genes (compared to the reference Sal-I strain) with the ratio of mutations leading to a non-synonymous or synonymous amino acid change close to 3∶1 for Pvrbp2a and Pvrbp2b. Although the degree of polymorphism exhibited by these two genes was higher than that of Pvrbp1, it did not reach the exceptional diversity noted for Pvrbp2c. It was interesting to note that variations in the copy number of Pvrbp2a and Pvrbp2b occurred in some isolates.
Conclusions/Significance
The evolution of different members of the Pvrbp2 family and their relatively high degree of polymorphism suggests that the proteins encoded by these genes are important for parasite survival and are under immune selection. Our data also shows that there are highly conserved regions in rbp2a and rbp2b, which might provide suitable targets for future vaccine development against the blood stage of P. vivax.
Introduction
Plasmodium spp. merozoites invade the host red cell via a multistep invasion process [1]. Parasite species differ in the preference they exhibit with respect to the type of red blood cells they can invade, for example P. malariae is almost exclusively observed in normocytes, while P. vivax is confined to reticulocytes. Thus, one of the initial steps prior to invasion is the specific recognition of the red blood cell type. For P. vivax two proteins expressed at the apical pole of the merozoite have been identified and implicated in reticulocyte recognition/selection, and were named reticulocyte binding proteins (RBP) [2]. Genes related to those coding for the RBPs of P. vivax (Pvrbp) were found in Plasmodium species that infect humans [3], [4], [5], [6], simians [7], [8], [9] and rodents [10], [11], [12]. Given their crucial implication in the invasion process, PVRBP proteins are considered to be good vaccine candidates [1], [13], [14]. However, investigations aiming to define the functional domains of Pvrbp genes or their receptors on the reticulocyte were hampered by the fact that a practical invasion assay for P. vivax invasion have been developed only recently [15], and that the two genes are quite large in size (ca. 8 kb–9 kb). Thus, overlapping peptides were used to define the binding domains of Pvrbp1 [16], [17], [18]. The other investigation was confined to an assessment of Pvrbp1 and Pvrbp2 diversity in four isolates from different geographical regions [19], which revealed a remarkably high diversity in Pvrbp2 as compared to Pvrbp1 and its homolog in P. falciparum and provided an indication as to the sub-domains it might be worth to focus on in future studies.
When the complete genome of the Salvador I strain of P. vivax was obtained [20], it was noted that there were other Pvrbp genes present, which led to the reclassification of the Pvrbp family (Table 1). There were two partial genes, and 7 full-length genes of which two were pseudogenes (Pvrbp2d and Pvrbp3). The primary aim of this study was to ascertain whether the high sequence diversity observed for Pvrbp2c (the new name for the gene previously known as Pvrbp2) also characterises the newly uncovered members of the Pvrbp family.
Table 1. Chromosomal location of the Plasmodium vivax Reticulocyte Binding Protein genes.
| Name | Strain | Chromosome |
| Pvrbp1a | Sal-I | 7 |
| Pvrbp1b | Sal-I | 7 |
| Pvrbp2a | Sal-I | 14 |
| Pvrbp2b | Sal-I | 8 |
| Pvrbp2c | Sal-I | 5 |
| Pvrbp2 (partial) | Sal-I | 5 |
| Pvrbp2d | Sal-I | 14 |
| Pvrbp2 (partial) | Sal-I | 14 |
| Pvrbp3 | Sal-I | 14 |
Materials and Methods
Ethics
This study was approved from Human Research Ethical Committee at Ministry of Public Health, Thailand (Reference no. 101/2550). This study utilized P. vivax infected blood samples collected prior to treatment and after informed and written consent from patients presenting in 2008–2009 at clinics from Tak, Mae Hong Sorn, Prachup Kririkhan or Chantaburi provinces, which are located in west, north, south and east Thailand, respectively.
Blood samples
Approximatley30 µl of blood were spotted onto filter paper (Protein saver, Whatman®) and kept in a dry and cool place until DNA extraction. Giemsa stained blood films were used to determine the species present and the parasitaemia. DNA from dried blood spots was extracted using QIAamp DNA Blood Mini Kit (QIAGEN, Germany) following the manufacturer's instructions, and the resulting DNA templates were stored at −20°C. Four samples, one from each region were selected for use in this study, after they were confirmed by PCR [21] to contain only P. vivax, and where parasitaemias were close to 400 parasites per microlitre of blood (about 1 parasites per 1000 RBC). Seven additional samples (numbered TK1 to TK7) were randomly selected from the west of Thailand (Tak province) for determination of the pvrbp2a and pvrbp2b copy number.
Reticulocyte Binding Protein (RBP) primer design and amplification
Reticulocyte Binding Protein (rbp genes) primers were designed based on the sequences obtained from the P. vivax Sal-I genome: Gene ID PVX_121920, PVX_094255, PVX_101585, and PVX_101495, which code for Pvrbp2a, Pvrbp2b, Pvrbp2d (pseudogene) and Pvrbp3 (pseudogene), respectively [22]. Given their large size (around eight kb), six overlapping 1.5 kb PCR fragments were amplified for each gene (eight fragments for the 8.7 kb Pvrbp3 gene). All primers and annealing temperature are shown in (Table S1). Two microlitres of the DNA template were used to initiate PCR amplification that was carried out in a total volume of 20 µl in the presence of 30 mM Tris-HCl, pH 8.3, 100 mM KCl, 2 mM MgCl2, 0.25 µM primers, 250 µM dNTPs and 0.02 U/µl of Ampli Taq Gold DNA Polymerase (Applied Biosystems). The cycling conditions were an initial denaturation 94°C for 5 minutes; followed by 35 cycles: annealing at a defined temperature (see Table S1) for 2 minutes, extension at 72°C for 2 minutes, denaturation 94°C for 1 minute; and a final extension at 72°C for 20 minutes. PCR products were analysed on an agarose gel stained with Gel red (Biotium, Inc., CA, USA) and visualized under UV light. All amplicons were immediately stored in −20°C until cloning process.
RBP cloning and sequence analysis
The PCR products from each gene were cloned into pCR4 Topo vector (Topo® TA cloning Kit, Invitrogen), and the resulting plasmids were sequenced using BigDye v3.1 Terminator (Applied Biosystems) and analyzed on an ABI 730XL (Applied Biosystems). Sequences were generated from both forward and reverse strands. Only single nucleotide polymorphisms (SNPs) that were observed in two or more fragments obtained from independent PCR amplifications were considered to be confirmed and unlikely to result as an artefact of the fidelity of the polymerase.
Copy number determination
The copy number of Pvrbp2a and of Pvrbp2b gene was estimated by a quantitative real-time SYBR Green PCR assay, as previously described [23]. Primers for these two genes were designed according to their conserved regions (Table S2). The qPCR method utilized a plasmid [vector pCR2.1® (Topo TA cloning kit, Invitrogen)] containing the reference P. vivax aldolase gene (GenBank Acc. No. AF247063) (single copy standard), and the gene of interest. Real-time PCR was run in the LightCycler® 480 (Roche Applied Science) using the following thermocycler program: 10 minutes of pre-incubation at 95°C before conducting 45 cycles of 10 seconds at 95°C, 5 seconds at 60°C and 10 seconds at 60°C. Subsequent to this, the melting curve was set for 0 second initiating temperature at 95°C, 15 seconds at 60°C, continuous at 72°C and cool down at 40°C 30 seconds. The reaction volume for each sample was 20 µl, and the reactions were placed in a 96-well plate (Applied Biosystems™) in the presence of 1× SYBRGreen buffer (Roche Applied Science), 2.5 mM MgCl2, 250 nM of each primer, and 2 µl of DNA. At the end of each reaction, Cycle threshold (Ct) was evaluated and melting curves were acquired and analyzed. The ΔΔCt calculation for the relative quantification of P. vivax gene target was used as follows ΔΔCt = Ct selected gene−Ct aldolase. The gene copy number in each isolate was calculated in N-fold changes by using equation as showing: N-fold copies in isolate = 2−ΔΔCt [24].
Results and Discussion
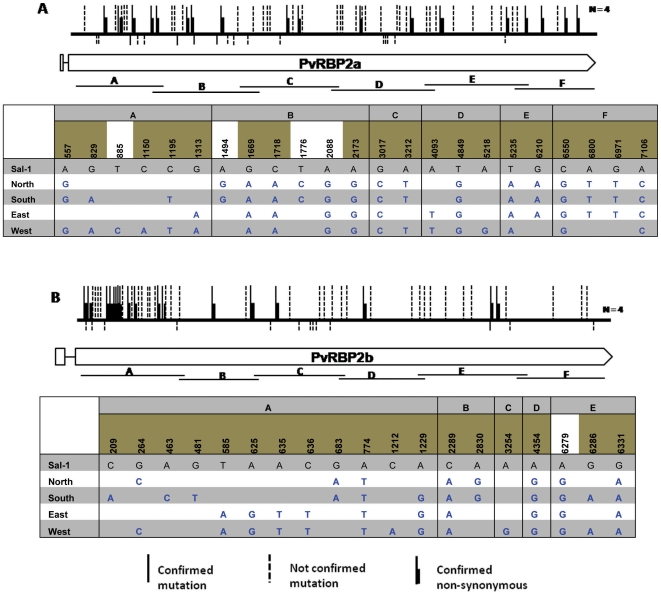
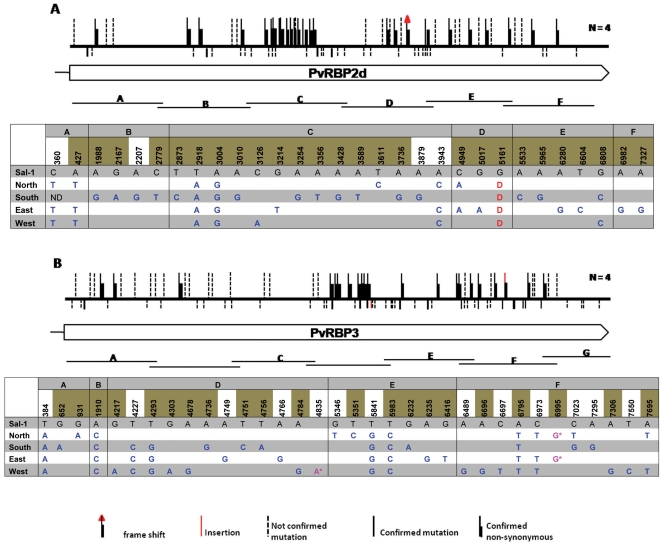
The sequences of the four recently discovered members of the Pvrbp genes (Pvrbp2a, Pvrbp2b, Pvrbp2d, and Pvrbp3) (Table 1) were obtained from 4 P. vivax isolates collected from distinct regions of Thailand (Accession numbers JN122379–JN122425 & JN172857–JN172908; totalling 130.5 kb of sequences). In all four P. vivax isolates from Thailand the Pvrbp2d, Pvrbp3 were confirmed to be pseudogenes, as was found for those in Sal-I. The sequences obtained for all the genes were then aligned with those of the P. vivax Sal-I strain genes. The overall results of this analysis are presented in Table 2, and schematically in Fig. 1 & 2. The sequences of the genes from the Thai P. vivax isolates differed significantly from those of the Sal-I strain with about 8 to 13 SNPs per 1000 bp. This level of diversity is closer to that observed for Pvrbp1 (3.3 SNP per 1000 bp) than to that for Pvrbp2c (76.4 SNPs per 1000 bp) (Table 2). It should however, be pointed out that more than half of the SNP's detected were observed only once, and it is possible that some were artefacts of the amplification reaction. Confirmation by sequencing of all the gene fragments from two independent PCR amplifications could not be carried out for logistic regions: this would have represented an extra 130 kb of sequences to obtain). If only the confirmed SNP'a are taken into consideration, then the level of diversity (3 to 4 SNPs per 1000 bp) nears that found previously for Pvrbp1 [19] (Table 2). The ratio of non-synonymous (NS) to synonymous (S) mutations for Pvrbp2a and Pvrbp2b was 3.0 and 3.1, respectively, which was comparable to that noted for Pvrbp2c (2.4 and 3.9) and lower than that recorded (4.5) for Pvrbp1 [19] (Table 2). The NS/S ratios for the two pseudogenes Pvrbp2d and Pvrbp3, was 1.2 and 1.4, respectively, as would be expected for non-functional genes. The extent of diversity observed was more restricted between the genes of the four isolates from Thailand, as compared to that noted when the Sal-I sequences were considered. It should also be noted that for all the mutations observed for any particular residue, the nucleotide substituted was invariably the same in the genes from the four isolates (except for one synonymous SNP at position 7023 of the Pvrbp3 pseudogene). There was some degree of clustering of the non-synonymous mutations in Pvrbp2b, suggesting that different domains might be under differential selective pressure. However, a similar pattern of clustering was observed for the two pseudogenes.
Table 2. Genetic diversity within the Plasmodium vivax Reticulocyte binding protein genes.
| Gene | n | Size | SNP (n) | Reference | |
| NS | S | ||||
| Pvrbp1 | 4 | 8499 | 25 | 3 | Rayner et al. [19] |
| Pvrbp 2c | 4 | 8454 | 489 | 151 | Rayner et al. [19] |
| Pvrbp2a | 4 | 7464 | 55 | 18 | This study |
| Pvrbp2b | 4 | 7959 | 51 | 13 | This study |
| Pvrbp2d | 4 | 8495 | 34 | 28 | This study |
| Pvrbp3 | 4 | 8702 | 67 | 48 | This study |
Note: n = number of sequenced isolates; Size = size of the gene analyzed with the non-coding regions excluded; SNP (n) = number of single nucleotide polymorphisms; NS = non-synonymous substitutions; S = synonymous substitutions.
Figure 1. Schematic representation of the Pvrbp2a (A) and Pvrbp2b (B) genes.
In each panel, the location of the fragments cloned is indicated below the gene model. Above the gene model, synonymous mutations are indicated by vertical bars below the horizontal bar that represents the gene, whereas non-synonymous mutations are place above this horizontal bar. SNPs that have been confirmed from two independent PCR amplifications are shown as solid lines, whereas those observed only once are represented by dotted lines. The nature and location of confirmed SNPs are also provided as tables, with the non-synonymous mutations highlighted in olive green.
Figure 2. Schematic representation of the Pvrbp2d (A) and Pvrbp3 (B) genes.
In each panel, the location of the fragments cloned is indicated below the gene model. Above the gene model, synonymous mutations are indicated by vertical bars below the horizontal bar that represents the gene, whereas non-synonymous mutations are place above this horizontal bar. SNPs that have been confirmed from two independent PCR amplifications are shown as solid lines, whereas those observed only once are represented by dotted lines. The nature and location of the confirmed SNPs are also provided in the tables, with the non-synonymous mutations highlighted in olive green. As these two genes are pseudogenes, the consequence of the SNP, synonymous or non-synonymous, has been predicted assuming that a continuous reading frame.
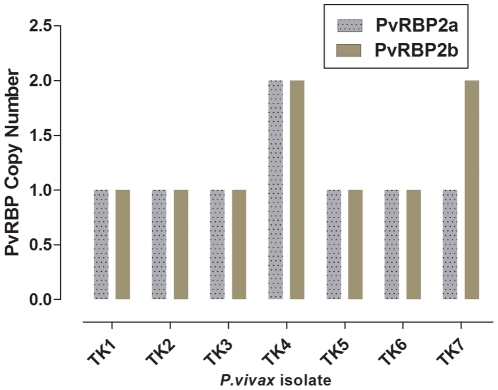
It had been previously noted that some P. falciparum lines harbour multiple copies of one of the Pvrbp homologues, Pfrh1 [25]. Variation in the copy number of Pvrbp2a and Pvrbp2b were sought in seven isolates collected from west Thailand (Tak province), and this was found in two of these P. vivax isolates: there were two copies of Pvrbp2b in one, and there were two copies of both Pvrbp2a and Pvrbp2b in the other (Fig. 3). The functional significance, if any, of these copy number variations is not known at present. This would also require a survey of a larger number of P. vivax isolates, and ultimately a means to ascertain if there is a link between the number of Pvrbp copy number and a particular parasite phenotype. Nonetheless, one could speculate that the pseudogenes (Pvrbp2d and Pvrbp3) might have been derived from genes present as multiple copies. It should be confirmed that Pvrbp2d and Pvrbp3 are indeed true pseudogenes. For example, evidence for the protein encoded by one of the homologues of the Pvrbp genes in P. falciparum (Pfrh3) that was considered to be a pseudogene, was detected by proteomic analysis in sporozoites extracts [26].
Figure 3. The copy number of Pvrbp2a, Pvrbp2b genes in seven P. vivax isolates (Numbered TK1 to TK7) collected in western Thailand (Tak province).
Given that the sequence data presented was derived from four isolates only, it is felt that any conclusions that can be drawn from the pattern of mutations are at best speculative. For example, it would be interesting to investigate whether a higher degree of genetic diversity would be observed for P. vivax from the west of Thailand as a result of the relatively higher incidence of this parasites as compared to the other areas of Thailand (Malaria Cases Report (Report 506): Annual report for the fiscal year 2006–2007 by provinces, Department of Communicable Disease Control, Ministry of Public Health, Bangkok 2008). Similarly, it is not known whether the nature and extent of the diversity observed is specific to the local parasite populations: P. vivax circulating in Thailand as compared to those circulating in geographically isolated populations of P. vivax, such as; Papua New Guinea, Afghanistan, Madagascar or South America. Until such a time when cloning and sequencing of high numbers of large genes becomes economically justifiable, one can envisage studies that target particular sub-domains of the Pvrbp genes.
Conclusion and Future Studies
To our knowledge, this is the first investigation of the genetic diversity of the recently uncovered full-length members of the Pvrbp family [20]. There is still very little known about the specific role of the corresponding proteins. Nonetheless, the high number of non-synonymous mutations in rbp2a and rbp2b suggest that they are likely to be under active selection and occurrence of P. vivax strains in which one or both are present as multiple copies is consistent with an important role in the survival of the parasite. A recently validated invasion inhibition assay employing P. vivax field isolates [15] makes it possible to envisage investigations of the functional role of the various PvRBP proteins or domains thereof, and ultimately to establish if any can serve as a useful target for a future vaccine against P. vivax.
Supporting Information
Sequence and annealing temperature of the primers used to amplify the Pvrbp genes.
(DOCX)
Primers set for Pvrbp genes copy number determination.
(DOCX)
Acknowledgments
We wish to thank staff and patients of the Thai MOH clinics from Tak, Mae Hong Sorn, Prachup Kririkhan and Chantaburi provinces.
Footnotes
Competing Interests: GS, ACG, BR and LR are PLoS ONE Editorial Board members. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The authors have no funding or support to report.
References
- 1.Galinski MR, Barnwell JW. Plasmodium vivax: Merozoites, invasion of reticulocytes and considerations for malaria vaccine development. Parasitol Today. 1996;12:20–29. doi: 10.1016/0169-4758(96)80641-7. [DOI] [PubMed] [Google Scholar]
- 2.Galinski M, Medina C, Ingravallo P, Barnwell J. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- 3.Kaneko O, Mu J, Tsuboi T, Su X, Torii M. Gene structure and expression of a Plasmodium falciparum 220-kDa protein homologous to the Plasmodium vivax reticulocyte binding proteins. Mol Biochem Parasitol. 2002;121:275–278. doi: 10.1016/s0166-6851(02)00042-7. [DOI] [PubMed] [Google Scholar]
- 4.Rayner JC, Galinski MR, Ingravallo P, Barnwell JW. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc Natl Acad Sci U S A. 2000;97:9648–9653. doi: 10.1073/pnas.160469097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor HM, Triglia T, Thompson J, Sajid M, Fowler R, et al. Plasmodium falciparum homologue of the genes for Plasmodium vivax and Plasmodium yoelii adhesive proteins, which is transcribed but not translated. Infect Immun. 2001;69:3635–3645. doi: 10.1128/IAI.69.6.3635-3645.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Triglia T, Thompson J, Caruana SR, Delorenzi M, Speed T, et al. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infect Immun. 2001;69:1084–1092. doi: 10.1128/IAI.69.2.1084-1092.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer EV, Semenya AA, Okenu DM, Dluzewski AR, Bannister LH, et al. The reticulocyte binding-like proteins of P. knowlesi locate to the micronemes of merozoites and define two new members of this invasion ligand family. Mol Biochem Parasitol. 2009;165:111–121. doi: 10.1016/j.molbiopara.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okenu DM, Meyer EV, Puckett TC, Rosas-Acosta G, Barnwell JW, et al. The reticulocyte binding proteins of Plasmodium cynomolgi: a model system for studies of P. vivax. Mol Biochem Parasitol. 2005;143:116–120. doi: 10.1016/j.molbiopara.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Rayner JC, Huber CS, Galinski MR, Barnwell JW. Rapid evolution of an erythrocyte invasion gene family: the Plasmodium reichenowi Reticulocyte Binding Like (RBL) genes. Mol Biochem Parasitol. 2004;133:287–296. doi: 10.1016/j.molbiopara.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Galinski MR, Xu M, Barnwell JW. Plasmodium vivax reticulocyte binding protein-2 (PvRBP-2) shares structural features with PvRBP-1 and the Plasmodium yoelii 235 kDa rhoptry protein family. Mol Biochem Parasitol. 2000;108:257–262. doi: 10.1016/s0166-6851(00)00219-x. [DOI] [PubMed] [Google Scholar]
- 11.Gruner AC, Snounou G, Fuller K, Jarra W, Renia L, et al. The Py235 proteins: glimpses into the versatility of a malaria multigene family. Microbes Infect. 2004;6:864–873. doi: 10.1016/j.micinf.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Keen JK, Sinha KA, Brown KN, Holder AA. A gene coding for a high-molecular mass rhoptry protein of Plasmodium yoelii. Mol Biochem Parasitol. 1994;65:171–177. doi: 10.1016/0166-6851(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 13.Galinski MR, Barnwell JW. Plasmodium vivax: who cares? Malar J. 2008;7(Suppl 1):S9. doi: 10.1186/1475-2875-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polley SD, McRobert L, Sutherland CJ. Vaccination for vivax malaria: targeting the invaders. Trends Parasitol. 2004;20:99–102. doi: 10.1016/j.pt.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Russell B, Suwanarusk R, Borlon C, Costa FT, Chu CS, et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood. 2011;118:e74–81. doi: 10.1182/blood-2011-04-348748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantor EM, Lombo TB, Cepeda A, Espinosa AM, Barrero CA, et al. Plasmodium vivax: functional analysis of a highly conserved PvRBP-1 protein region. Mol Biochem Parasitol. 2001;117:229–234. doi: 10.1016/s0166-6851(01)00355-3. [DOI] [PubMed] [Google Scholar]
- 17.Rojas Caraballo J, Delgado G, Rodriguez R, Patarroyo MA. The antigenicity of a Plasmodium vivax reticulocyte binding protein-1 (PvRBP1) recombinant fragment in humans and its immunogenicity and protection studies in Aotus monkeys. Vaccine. 2007;25:3713–3721. doi: 10.1016/j.vaccine.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 18.Urquiza M, Patarroyo MA, Mari V, Ocampo M, Suarez J, et al. Identification and polymorphism of Plasmodium vivax RBP-1 peptides which bind specifically to reticulocytes. Peptides. 2002;23:2265–2277. doi: 10.1016/s0196-9781(02)00267-x. [DOI] [PubMed] [Google Scholar]
- 19.Rayner JC, Tran TM, Corredor V, Huber CS, Barnwell JW, et al. Dramatic difference in diversity between Plasmodium falciparum and Plasmodium vivax reticulocyte binding-like genes. Am J Trop Med Hyg. 2005;72:666–674. [PubMed] [Google Scholar]
- 20.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol. 2003;97:131–137. doi: 10.1179/000349803125002977. [DOI] [PubMed] [Google Scholar]
- 22.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suwanarusk R, Russell B, Chavchich M, Chalfein F, Kenangalem E, et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira ID, Rosario VE, Cravo PV. Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malar J. 2006;5:1. doi: 10.1186/1475-2875-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triglia T, Duraisingh MT, Good RT, Cowman AF. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol Microbiol. 2005;55:162–174. doi: 10.1111/j.1365-2958.2004.04388.x. [DOI] [PubMed] [Google Scholar]
- 26.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence and annealing temperature of the primers used to amplify the Pvrbp genes.
(DOCX)
Primers set for Pvrbp genes copy number determination.
(DOCX)