Abstract
Background
Cold-water coral reef ecosystems are recognized as biodiversity hotspots in the deep sea, but insights into their associated bacterial communities are still limited. Deciphering principle patterns of bacterial community variation over multiple spatial scales may however prove critical for a better understanding of factors contributing to cold-water coral reef stability and functioning.
Methodology/Principal Findings
Bacterial community structure, as determined by Automated Ribosomal Intergenic Spacer Analysis (ARISA), was investigated with respect to (i) microbial habitat type and (ii) coral species and color, as well as the three spatial components (iii) geomorphologic reef zoning, (iv) reef boundary, and (v) reef location. Communities revealed fundamental differences between coral-generated (branch surface, mucus) and ambient microbial habitats (seawater, sediments). This habitat specificity appeared pivotal for determining bacterial community shifts over all other study levels investigated. Coral-derived surfaces showed species-specific patterns, differing significantly between Lophelia pertusa and Madrepora oculata, but not between L. pertusa color types. Within the reef center, no community distinction corresponded to geomorphologic reef zoning for both coral-generated and ambient microbial habitats. Beyond the reef center, however, bacterial communities varied considerably from local to regional scales, with marked shifts toward the reef periphery as well as between different in- and offshore reef sites, suggesting significant biogeographic imprinting but weak microbe-host specificity.
Conclusions/Significance
This study presents the first multi-scale survey of bacterial diversity in cold-water coral reefs, spanning a total of five observational levels including three spatial scales. It demonstrates that bacterial communities in cold-water coral reefs are structured by multiple factors acting at different spatial scales, which has fundamental implications for the monitoring of microbial diversity and function in those ecosystems.
Introduction
Cold-water coral (CWC) reef ecosystems are increasingly portrayed as biodiversity hotspots on continental margins, seamounts and mid-ocean ridges around the world [1]. They appear as speciose, abundant and widespread as their warm-water counterparts [2]–[5], and represent important species pools [6]–[8] and speciation centers [9] in the deep sea. Their potential to foster a high degree of local diversity and biomass is assumed to be rooted in the ecosystem engineering capacity of scleractinian corals [10], [11], such as the cosmopolitan key species Lophelia pertusa (L. 1758, Caryophylliidae) and Madrepora oculata (L. 1758, Oculinidae). By forming enormous dendritic skeletal frameworks, these corals provide complex three-dimensional living space for a plethora of mobile and sessile organisms [7], [8], [12]. They also alter flow regimes and sedimentation rates, thereby modifying the abiotic environment in time and space ([1] and references therein).
Often, structural complexity in CWC reefs is promoted by pronounced ecosystem heterogeneity. Unlike warm-water coral ecosystems that constitute relatively contiguous reef environments with clear wave and sun energy-related zoning [13], CWC ecosystems can consist of isolated colonies, small patch accumulations, large reefs, or giant carbonate mounds, and differ substantially with respect to their spatial configuration [14]–[19]. Often, individual clusters of coral frameworks form entire reef complexes which, depending on local seabed geology as well as community history (i.e. the combined effects of past community assembly, succession and interaction, including individual life trajectories and trade-offs), exhibit distinctive geomorphologic and taphonomic (i.e. seabed form- and fossilization-related) zoning, with marked transitions in sediment type, faunal composition and proliferation stage [7], [19]–[21].
Despite mounting evidence of warm-water coral reefs as structured landscapes of complex microbial communities [22]–[24], insights into the microbial diversity of CWC ecosystems are limited. Deciphering principle patterns of microbial community variation may, however, prove critical for a better understanding of factors contributing to CWC reef stability and functioning. Especially bacteria play important ecological roles for corals and entire reef systems by contributing substantially to biogeochemical processes, invertebrate life cycles, host metabolism, protection and adaptation, as well as to overall species diversity (e.g. [24] and references therein). Hence, their spatial and temporal dynamics are relevant features for the functioning of coral ecosystems.
So far, studies of CWC reef microbiology mainly focused on the identification of bacteria associated with scleractinians [25]–[32] or octocorals [33]–[37]. Community fingerprinting methods (e.g. Automated Ribosomal Intergenic Spacer Analysis, ARISA; Terminal Restriction Fragment Length Polymorphism, T-RFLP; Denaturing Gradient Gel Electrophoresis, DGGE) and 16 S rRNA gene sequencing were used to show that bacterial assemblages colonizing living scleractinians and octocorals differ from those of dead corals or from those of the ambient environment like seawater or sediments [25], [26], [28], [29], [32], [33]. Even distinct coral-generated microbial habitats, such as branch surface, mucus and tissue, were found to exhibit specific bacterial community signatures [26], [29], [32]. Spatial patterns of bacterial communities associated with different octocoral species appeared either highly distinct [37] or conserved between different reef sites (and across an environmental impact gradient [36]).
On L. pertusa, several bacterial sequences were shared across geographically separate regions, such as the Gulf of Mexico and the Trondheimsfjord in Norway [28], [30]. Strict host-specificity of L.pertusa-associated bacteria was so far not evidenced due to significant community variations between (i) sampling locations within the same geographic area or reef complex [26], [28], [29], [30], (ii) colonies of the same coral species [29], (iii) single polyps within the same coral colony [29], and (iv) differently colored types within the same coral species [28]. In fact, current evidence suggests that coral-bacteria associations considerably differ with both coral-derived microbial habitats and prevailing environmental conditions. However, due to the variations in spatial scale and methodology applied in aforementioned surveys, the relative importance of spatial and reef-organizational factors that determine bacterial biogeography across various scales is not evident.
The aim of the present study was to identify patterns of bacterial communities in CWC reefs (Fig. 1) using a multi-scale, hierarchical sampling approach spanning five study levels including three spatial scales (Fig. 2). Sources of bacterial community variation were assessed from local (intra-reef) to regional (inter-reef) scale by considering (i) microbial habitat type on and around coral colonies (coral branch surface, coral mucus, ambient seawater, proximal sediments), (ii) coral species (L. pertusa, M. oculata) and color phenotype (white, red), (iii) geomorphologic reef zoning (ridge crest, slope, depression), (iv) reef boundary (up-slope reef center, down-slope reef periphery), and (v) reef site (Røst Reef, Trænadjupet Reef, Tisler Reef, Langenuen Fjord) and proximity to shore (offshore, inshore).
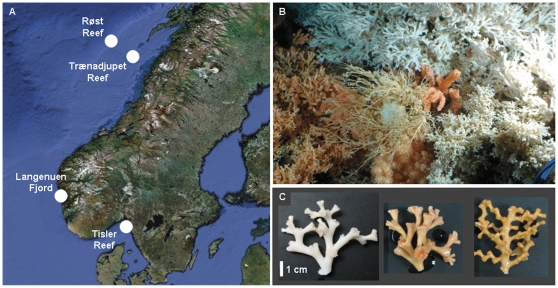
Figure 1. Reef sites and corals targeted in this study.
(A) Offshore and CWC ecosystems along the Norwegian continental margin. (B) Living colonies of Lophelia pertusa and Madrepora oculata in their natural environment at Røst, northern mid-Norwegian continental margin. (C) Fragments of freshly sampled white L. pertusa (left), red L. pertusa (middle), and red M. oculata (right).
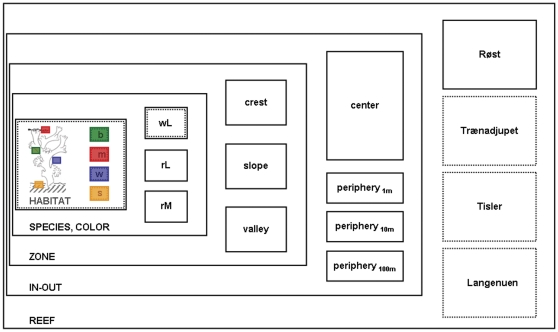
Figure 2. Multi-scale, hierarchical sampling design.
Nested frames indicate the different levels of observation, increasing in scale from inside (left) to outside (right). Boxes within frames symbolize each lower level as integral part of respective next higher level. At the main study site, Røst, sampling was implemented on all levels (continuous line); at all other sites, it was performed only on the lowest and highest level, respectively (dotted line). The following levels and scales of observation were considered: 1) HABITAT (µm–cm): coral branch surface (“b”), coral mucus (“m”), ambient seawater (“w”), proximal sediments (“s”); 2) SPECIES (cm–m) – white Lophelia pertusa (“wL”), red Lophelia pertusa (“rL”), red Madrepora oculata (“rM”); 3) ZONE (1 m–10 m): ridge top with coral terraces (“crest”), ridge slope with single coral colonies on rubble (“slope”), ridge depression with single colonies on clay (“valley”); 4) IN-OUT (1 m–10 m–100 m): reef center (“reef-in”), reef periphery in distances of 1, 10, 100 m away from the apparent reef margin (“reef-out”); 5) REEF (km): Røst Reef (“Røst”), Trænadjupet Reef (“Trænadjupet”), Tisler Reef (“Tisler”), Langenuen Fjord (“Langenuen”).
For this purpose, bacterial community DNA derived from two constructional corals L. pertusa and M. oculata, as well as from associated seawater and surface sediments, was collected from four CWC reef ecosystems on the Norwegian continental margin (Fig. 1). The community fingerprinting approach ARISA then allowed for a time- and cost-effective analysis of the large, heterogeneous sample set. Despite the lack of information on OTU identity, ARISA was chosen for its proven ability to provide robust insights about bacterial community dynamics at different spatial (and temporal) scales (e.g. [38], [39]). Ultimately, the different but not mutually exclusive sources of bacterial community variation were disentangled by quantifying their respective effects with multivariate statistics.
Results
Variation in bacterial OTU number and occurrence
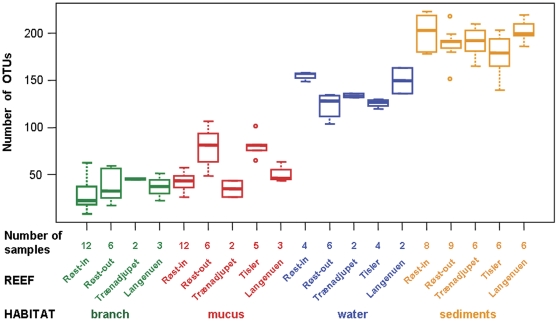
From a pool of 440 different operational taxonomic units (OTUs) occurring in the whole data set (104 samples), between 9–223 OTUs were obtained per sample. OTU number was strongly related to microbial habitat type (KW, P<0.001), and to the reef site (P = 0.05; Fig. 3). The most pronounced difference occurred between coral-generated surfaces and the ambient environment, with branch (34±15 OTUs) and mucus (58±22 OTUs) featuring 30–80% lower mean OTU numbers than water (135±16 OTUs) and sediments (192±19 OTUs). At Røst, mean OTU numbers showed a slight increase (P = 0.0398; Fig. 3) between reef center (Røst-in, 86±11 OTUs) and reef periphery (Røst-out, 116±62 OTUs), which was mainly related to branch and mucus variability. When studying local trends at Røst-in and Røst-out separately, however, neither geomorphologic reef zoning (P = 0.098), nor gradual distance away from the apparent reef margin (P = 0.956) resulted in any significant variation of OTU numbers. These were also not significantly related to coral species (L. pertusa and M. oculata with 36±13 and 35±17 OTUs, respectively; P>0.05) or to coral color (white and red colonies with 34±14 and 38±14 OTUs, respectively; P>0.05).
Figure 3. Number of ARISA-derived OTUs in distinct microbial habitats at each reef site.
Top, middle, and bottom lines of the boxes represent the 25th, 50th (median), and 75th percentiles, respectively, while the end of the whiskers represent the 5th and 95th percentiles, respectively; box height and symmetry around the median indicate the degree of dispersion and skewness in the data, respectively; outliers above and below the whiskers denote extreme values.
OTU partitioning among the four different microbial habitats showed that, from a total of 380–390 different OTUs detected in coral- water-, and sediment samples at Røst-in, only very few were exclusively present on L. pertusa and M. oculata surfaces of a given color phenotype (branch: <1–2%, mucus: 1%; Fig. S2A). Even with all coral samples from Røst-in combined, branch and mucus each contained only about 2% unique OTUs, from a total of 396 different OTUs (Fig. S2B). By contrast, the surrounding water and proximal sediments at Røst-in contained 4–5% and 19–28% unique OTUs, respectively, and therefore more of the microbial habitat-specific bacterial signatures (Fig. S2B). These patterns were confirmed for all study sites, with only minor variations (data not shown). OTU partitioning with the whole data set, i.e. with altogether 440 different OTUs from Røst, Trænadjupet, Tisler, and Langenuen combined (Fig. S2B), revealed clearly lower fractions of water- and sediment-specific OTUs (<1% and 5%, respectively), while those of branch- and mucus-specific OTUs remained virtually unchanged (branch: 2–3%, mucus: 1–2%). Concomitantly, the fraction of shared OTUs increased considerably from local (Røst-in: 4–55 and 9%, Fig. S2A) to regional scale (all sites: 27%; Fig. S2B), indicating a decrease in habitat-specificity for water- and sediment-associated OTUs, due to their partial presence in branch and mucus samples from other sites.
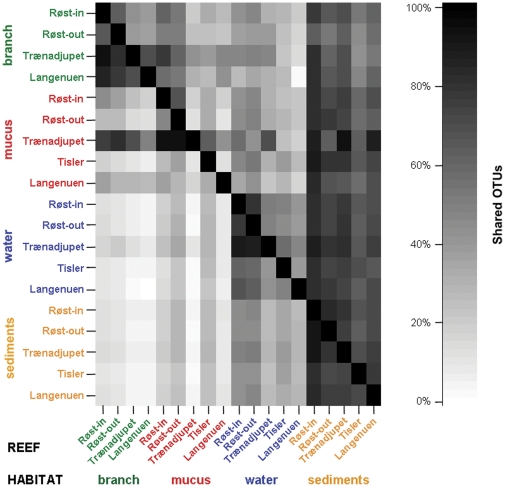
The detailed analyses of OTU overlap between samples confirmed those habitat-specific trends and distinguished particularly coral-derived surfaces from the surrounding environment (Fig. 4). Differences in OTU overlap clearly reflected variations in OTU number, with OTU-poor habitats (branch, mucus) sharing a much higher percentage of their OTU pool with OTU-rich habitats (water, sediments) than reciprocally. Branch and mucus shared at least half of their OTUs with sediments (50% and 73%, respectively), and a comparatively lower fraction with water (25% and 36%, respectively). Conversely, only 9–10% and 17–18% of all water and sediment OTU were found among branch and mucus OTUs, respectively. The number of OTUs shared solely between both coral-associated habitats amounted to a third of their respective OTU content (33–34%), whereas the water shared a much higher fraction of its OTU pool with the sediments (74%) than vice versa (34%). Between different reef sites (within each habitat separately; Fig. 4), the mean number of OTUs overlapping between any two reefs was the lowest in mucus (mean: 37%) and also the most variable (range: 11–94%). In contrast, reef-specific OTU fractions were usually the highest in sediments (76%), with relatively high and constant OTU overlap among reefs (63–90%). Overall, no difference between offshore (Røst, Trænadjupet) and inshore (Tisler, Langenuen) reef sites was detected (Fig. 4).
Figure 4. Pairwise comparison of OTU overlap between microbial habitats at each reef site.
Samples were grouped according to microbial habitat type and reef site. In this asymmetrical representation, rows correspond to the reference group and columns to the group being compared. It provides an overview of potential directional dynamics between different microbial habitat types, with the respective fraction (%) of shared OTUs indicated by different degrees of shading.
Differences in community structure
Bacterial community structure strongly differed between microbial habitat types, with the greatest difference between coral-associated surfaces and ambient environment (Fig. 5, Fig. S3) as confirmed by PERMANOVA (Table 1). At Røst-in, as well as for all study sites combined, distinct community patterns were associated with branch, mucus, water or sediments. For the other reef sites (Table 1), the microbial habitat-specific separation was also similarly pronounced (PERMANOVA R2 = 0.75–0.80). In addition, bacterial community structure associated with each of the four reef systems clearly differed from each other, especially between offshore (Røst, Trænadjupet) and inshore (Tisler, Langenuen) sites (Table 1). These regional, inter-reef differences appeared particularly pronounced for mucus communities (Fig. 5, Fig. S4), but also for water and sediment communities.
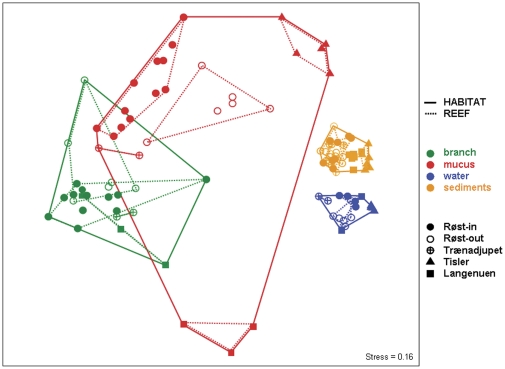
Figure 5. NMDS ordination of all ARISA community profiles.
For each sample, consensus signals of PCR triplicates were used. Objects represent consensus signals for all PCR triplicates per sample and share a more similar community structure when plotting closer to each other (Bray-Curtis distance). A posteriori groupings specify microbial habitat type and reef site. The low stress value indicates appropriate representation of the original Bray-Curtis dissimilarity matrix into a 2-dimensional space.
Table 1. PERMANOVA of regional, inter-reef bacterial variation at all study sites as related to reef site (REEF) and microbial habitat type (HABITAT).
| Test factora | Test group | R2 b | F-ratio | Pc |
| HABITAT | all samples | 0.467 | 17.222 | 0.001 ***d |
| HABITAT | Røst | 0.777 | 20.947 | 0.001 ***d |
| HABITAT | Trænadjupet | 0.794 | 10.273 | 0.001 ***d |
| HABITAT | Tisler | 0.801 | 24.135 | 0.001 ***d |
| HABITAT | Langenuen | 0.748 | 9.888 | 0.001 ***d |
| REEF | all samples | 0.124 | 2.795 | 0.001 ***d |
| REEF | branch | 0.393 | 2.262 | 0.032 * |
| REEF | mucus | 0.754 | 11.210 | 0.001 ***d |
| REEF | water | 0.835 | 13.476 | 0.001 ***d |
| REEF | sediments | 0.504 | 7.452 | 0.001 ***d |
Source of variation.
Amount of explained variation.
Significance level, assessed by 999 random permutations (*** P≤0.001, ** P≤0.01, * P≤0.05).
Significance level below Bonferroni correction threshold.
At Røst, significant intra-reef differences were detected between the up-slope reef center (Røst-in) and the down-slope reef periphery (Røst-out) (Table 2). Similarly to the aforementioned regional patterns, this local pattern was mainly evidenced in mucus, water, marginally in sediments, but not in branch (Fig. S4). While those intra-reef differences in the Røst area indicated some degree of separation, the pronounced geomorphologic (including vertical) zoning at the reef center itself (Røst-in) revealed only a very weak trend among bacterial communities (Table 3, Fig. 3, Fig. S4). Communities did not change significantly over distances of 1, 10, and 100 m in the reef periphery (Røst-out; P>0.05).
Table 2. PERMANOVA of local, intra-reef bacterial variation at Røst as related to reef boundary, i.e. in-/out-reef location (IN-OUT).
| Test factora | Test group | R2 b | F-ratio | Pc |
| IN-OUT | all Røst samples | 0.048 | 3.100 | 0.006 ** |
| IN-OUT | branch | 0.095 | 1.684 | 0.073 |
| IN-OUT | mucus | 0.319 | 7.478 | 0.001 ***d |
| IN-OUT | water | 0.300 | 3.433 | 0.01 * |
| IN-OUT | sediments | 0.165 | 2.967 | 0.003 ** |
Source of variation.
Amount of explained variation.
Significance level, assessed by 999 random permutations (*** P≤0.001, ** P≤0.01, * P≤0.05).
Significance level below Bonferroni correction threshold.
Table 3. PERMANOVA of local, intra-reef bacterial variation at Røst-in as related to geomorphologic reef zoning (ZONE), coral species (SPECIES), coral color phenotype (COLOR), and microbial habitat type (HABITAT).
| Test factora | Test group | R2 b | F-ratio | Pc |
| ZONE | all Røst-in samples | |||
| ZONE | branch | 0.143 | 0.752 | 0.752 |
| ZONE | mucus | 0.146 | 0.767 | 0.803 |
| ZONE | water | 0.399 | 1.326 | 0.260 |
| ZONE | sediments | 0.209 | 1.589 | 0.134 |
| SPECIESf | all Røst-in samples | 0.416 | 11.742 | 0.001 ***d |
| SPECIESf | branch | 0.236 | 3.086 | 0.001 ***d |
| SPECIESf | mucus | 0.163 | 1.942 | 0.043 * |
| COLORf | all Røst-in samples | 0.024 | 0.398 | 0.868 |
| COLORf | branch | 0.011 | 0.878 | 0.607 |
| COLORf | mucus | 0.082 | 0.631 | 0.794 |
| HABITATf | all Røst-in samples | 0.587 | 23.411 | 0.001 ***d |
Source of variation.
Amount of explained variation.
Significance level, assessed by 999 random permutations (*** P≤0.001, ** P≤0.01, * P≤0.05).
Significance level below Bonferroni correction threshold.
Hierarchical sampling design, with each factor nested within the next higher one.
L. pertusa and M. oculata harbored significantly different bacterial assemblages (Table 3) despite some overlap (Fig. S4), and exhibited slight differences in their response to local reef complexity (Table S2A, C, B): While M. oculata-associated seemed to reflect changes in in-/out-reef location and geomorphologic reef zoning, L. pertusa-associated bacteria appeared more stable over space. In contrast, bacterial community variation related to coral color was, albeit overall significant, never supported, neither on branch nor in mucus (Table 3). All described results were also generally confirmed by ANOSIM and cluster analysis (data not shown).
Discussion
Microbial habitat type
Bacterial communities associated with the CWCs L. pertusa and M. oculata substantially differed according to the type of microbial habitat sampled. Coral branch surface, coral mucus, ambient seawater and proximal sediments each featured a specific community structure that significantly varied both in OTU composition and relative abundance. This habitat specificity seemed valid at all study sites, and confirmed earlier findings based on samples from one reef location [32]. Other studies have already reported evidence for bacterial habitat specificity in CWC reefs from the North-East Atlantic [26], [28], [29], the Central Mediterranean [25] and the Gulf of Alaska [33], or warm-water coral reefs (e.g. [40], [41]). The most pronounced differences concerned the distinction between bacterial communities associated with coral-generated surfaces and the ambient environment, in both OTU number and composition.
Not surprisingly, the difference in bacterial OTU number between the OTU-poor coral-associated and OTU-rich ambient microbial habitats strongly determined the overall degree of bacterial community partitioning and overlap: Irrespective of reef site and local zoning, branch and mucus exhibited notably few specific OTUs, as most of their respective OTU pool was shared with water and sediments. As expected, sediments generally exhibited the highest OTU abundance and number of specific OTUs, which was also previously observed in other ARISA-based studies on warm-water coral reefs (e.g. [38]).
Albeit strongly reduced in overall bacterial OTU number and specificity, bacterial communities on coral surfaces were characterized by high inter-sample, intra-habitat differences, clearly exceeding those of OTU-rich water and sediment communities. This may result from both stochastic events during community assembly, such as the random attachment of environmental bacteria on coral surfaces [42], and deterministic processes, such as the selection of few opportunists through the coral host (e.g. [43]), or antagonistic interactions between bacterial types [44]. Furthermore, this high variation in coral-associated assemblages may reflect local, inter-colony differences in host status, such as genetic identity [45], physiological condition [46], or developmental state ([47] and references therein). In their study of M. oculata-associated microbes, Hansson et al. [29] also reported significant inter-colony differences, which may even be further enhanced by intra-colony differences between single polyps [48], [49]. In addition to passive controls, bacterial colonization may also be actively regulated by the coral host (e.g. [50]) in adaptation to changing environmental conditions [51].
Reef zoning, boundary and location
In general, bacterial communities mapped within the Røst reef center (Røst-in) revealed surprisingly similar patterns, despite the pronounced geomorphologic reef zoning (Table 3). This was unexpected, because seabed features often strongly affect and reflect local environmental dynamics of e.g. current regime, sediment deposition and diagenesis as well as organic matter quality, transport and remineralization within only few tens of meters (e.g. [18], [19]). Hence, the clearly distinguishable reef features present at Røst-in were assumed to significantly contribute to the structuring of bacterial assemblages, particularly so in water and sediments, but also in coral-derived microbial habitats.
The observed local similarity of bacterial community structure across reef-internal zones was not maintained beyond the reef center (Røst-in), due to significant community changes towards the reef periphery (Røst-out; Table 2). Only the branch communities remained similar, thereby marking an intriguing partition between the two coral-generated microbial habitats, branch and mucus. The scale-independent similarity of communities in branch versus mucus may be attributed to the circumstance that branch surface samples also included traces of coenosarc tissue, which may not only contain internal bacterial cells [31] but also exhibit external biofilm formation [43]. Those tissue- and biofilm-associated cells could be buffered more from exogenous change than mucus-inhabiting assemblages due to their embedding in the respective intra- and/or extra-cellular matrices, while the latter are more exposed to water column processes and thereby more prone to mirroring local, meso-scale spatial and environmental shifts. Within Røst, such shifts may occur as a function of the marked zone transition from the reef center to the periphery, with the latter representing an interface (“ecotone”; sensu [52]) between the structurally complex reef ecosystem and the more uniform, level bottom down-sloping into the abyssal plain [7]. Consequently, the underlying spatial and environmental changes that select for certain community structures may not necessarily follow linear distance relationships, but rather be subject to a whole interplay of locally different, ecosystem-specific factors.
The presence of spatial and environmental imprinting became even more evident by the finding of significant bacterial community differences between the four reef sites (Table 1). Remarkably, observed patterns not only reflected local site specificity (i.e. characteristic assemblages at different reefs), but also regional specificity (i.e. marked separation between both offshore versus each of the inshore reefs; Fig. S4). Although all reef sites share specific geological and hydrological features that are pivotal for local coral recruitment and proliferation (e.g. [1] and references therein), Røst, Trænadjupet, Tisler and Langenuen substantially differ in their geo- and hydrographical setting and structure. This seemed clearly reflected in the pronounced site specificity of water- and sediment-inhabiting bacteria. But also coral-generated microbial habitats exhibited reef-specific bacterial differences, more so in mucus than in branch samples, which basically confirmed the aforementioned differences from the local (meso-) to the regional (large-) scale. Those differences may involve reef-specific variations in mucus composition [53] as governed by environmental controls [54], or geographic fluctuations in coral reproduction strategy and genetic variability [45] as well as local coral food supply and quality [55], [56].
Coral species and color
L. pertusa and M. oculata exhibited significantly different bacterial community structures, largely due to differences in OTU relative abundances. This corroborates evidence presented by Hansson et al. [29] who found DGGE signals from both species to group separately in NMDS and cluster analyses (yet, with >50% similarity). Like in our study, those corals originated from the same sampling location, where they occurred right next to each other, hence, a mere spatial separation of bacterial assemblages may not explain this pattern. Also, comparisons of 16S rRNA gene sequences from different studies [28]–[30] suggested divergence between L. pertusa and M. oculata-associated bacterial communities. As both coral species differ with respect to tissue-contained acid concentrations [57] as well as to the carbohydrate composition of their mucus [53], the involvement of host-related traits in structuring bacterial assemblages seems likely. Species-specific differences in mucus composition are also known from warm-water corals [58] and even held responsible for a close attuning of bacterial communities to host metabolism [59]. Host-related traits may also explain why L. pertusa and M. oculata-associated communities slightly varied in their response to spatial heterogeneity (Table S2A, C, B), indicating possible coral-specific differences in host-microbe interactions.
In contrast to the marked species-related patterns, no bacterial community differences were significantly linked to L. pertusa color type. This was due to the fact that none out of the 387–390 L. pertusa-associated OTUs were exclusively attributable to either white or red specimens. In contrast, Neulinger et al. [28] who previously studied bacterial associates of white and red L. pertusa from the Tautra Reef by 16S rRNA-based T-RFLP and sequence analysis, observed color-specific associations of distinct 16S rRNA gene phylotypes, but could not resolve community differences among color phenotypes by fingerprinting. In principle, ARISA offers more resolution than T-RFLP to detect OTU changes [60] as well as intra-genomic heterogeneities within closely related gene clusters [61]. In addition, the two studies differ with respect to sample origin and processing: While the Tautra corals were completely homogenized, introducing considerable amounts of tissue and carbonate skeleton into the analysis, the Røst corals were distinctly sampled for branch surface plaques and mucus exudates, targeting mainly interfacial communities on the coral surface. Hence, the consistent absence of significant phenotypic community change in both these fingerprinting studies may indeed indicate that bacterial associates of white and red L. pertusa are largely indiscriminative, at least by those techniques.
Noticeably, the finding of low numbers of shared bacterial types combined with significant community differences related to host taxonomy, are often interpreted as signs of host specificity, implying the selection of few beneficial associates as part of commensalistic or mutualistic relationships (e.g. [62] and references therein). Yet, host-microbe associations may only be called “specific” if the respective bacterial signatures are maintained over time and space, which is so far unresolved for bacteria associated with CWCs due to the lack of studies across spatial and temporal scales. Here, habitat-specific community patterns were conserved over all sites, including differences between low and high numbers of unique OTUs in coral-derived versus ambient microbial habitats, respectively. The divergence between bacterial communities associated with L. pertusa and M. oculata also appeared consistent, at least within a highly heterogeneous reef site such as Røst. Further, 3 out of 8 OTUs unique to L. pertusa branch at Røst-in also occurred in the branch samples at all other sites (i.e. Røst-out, Trænadjupet, Tisler, Langenuen), potentially indicating specialized bacterial associates. By contrast, such property was not identified for any of the mucus-contained OTUs, suggesting variable colonization of the mucus matrix by locally occurring communities. Most of the coral-associated OTUs were also found in the proximal sediments, suggesting a potential source for coral-associated bacteria. This was corroborated by the change of coral-associated bacterial communities from local (small- and meso-) to regional (large-) scale, resulting in biogeographic patterns comparable to those of ambient bacterial communities. Host specificity of bacterial types and communities should therefore be further investigated by colonization experiments with artificial surfaces.
Bacterial biodiversity hotspots?
By providing a high degree of structural complexity and habitat heterogeneity, CWC reef ecosystems locally promote faunal diversity in the deep sea [7], [12]. The potential of CWC reefs to also represent biodiversity hotspots for bacterial communities seems, however, rather questionable, given the low bacterial OTU number (as proxy for bacterial richness) and the limited OTU specificity (as indicator of conserved bacterial signatures) associated with coral-generated surfaces detected in this study. Nevertheless, CWCs contributed 5% (OTUs found exclusively on corals) to 44% (OTUs shared between corals and water/sediments) of all bacterial OTUs detected at a single proliferating reef center (Røst-in). Furthermore, coral-generated surfaces, such as branch and mucus, were characterized by high bacterial community variation. Although high variability does not automatically translate into high local diversity, these small-scale differences may, combined with the meso- and large-scale community changes occurring within and between reef sites, increase bacterial community variation (beta diversity) in CWC ecosystems and contribute to specific source-sink dynamics in bacterial dispersal. In terms of bacterial types, however, it may rather be the reef sediments, water column or filtering invertebrates such as sponges, that contribute more to the overall bacterial diversity in CWC reefs than surfaces generated by L. pertusa or M. oculata. Those microbial habitats can feature high levels of organic matter and nutrients ([1] and references therein; [63]) and are usually characterized by diverse bacterial communities (e.g. [64], [65]).
Given that ARISA is based on the discrimination of ITS length among bacterial types, the technique cannot be used to precisely delineate the taxonomic levels at which the observed community changes operate [61]. Yet, based on our evaluation of community shifts at multiple levels of observation, future investigations may involve sequencing of ribosomal genes on representative samples to explore finer taxa-environment relationships. As reported by previous studies (e.g. [61], [66], [67]), bacterial community patterns derived from ARISA and sequencing (both Sanger and high-throughput) may, after all, be highly comparable and lead to very similar ecological conclusions.
Conclusion
This study presents the first multi-scale survey of bacterial communities associated with L. pertusa and M. oculata in different CWC reefs, spanning a total of five study levels: (i) Microbial habitat type and (ii) coral species and color, as well as the spatial components (iii) geomorphologic reef zoning, (iv) reef boundary, and (v) reef location. Our findings revealed fundamental differences in bacterial specificity and community structure between distinct coral-generated (branch surface, mucus) and ambient microbial habitats (seawater, sediments), which appeared pivotal for determining bacterial variation over all observational levels investigated. Especially the high community variability associated with coral-derived surfaces represented a consistent feature of all four CWC reef sites under study. In addition, bacterial communities changed markedly from local (small- and meso-) to regional (large-) scale, suggesting significant biogeographic imprinting of seawater-, sediment- and even coral-associated communities, but weak microbe-host specificity. Overall, the relative effects of the different test parameters did not reflect any linear or even hierarchical relationship of bacterial community organization, but the deterministic effect of microbial habitat type and the strong effect of reef location seemed to play dominant roles in structuring bacteria in CWC reefs. The bacterial communities were structured across different spatial scales, from local within-reef habitats to regional across-reef systems, which may reflect the combined effects of local community history (e.g., community assembly and interactions) and environmental filtering. As exploring bacterial diversity in CWC reefs is but one of the first steps to a better understanding of coral-microbe relationships in complex deep-sea environments, further studies need to address how changes in bacterial (and general microbial) diversity affect the dynamics and functioning of CWC reef ecosystems – especially in the context of global environmental changes and the protection of CWC reefs as biodiversity hotspots.
Materials and Methods
Study sites
The four Norwegian CWC reef ecosystems sampled (Fig. 1A) comprised two offshore sites on the northern mid-Norwegian continental shelf (Røst, Trænadjupet), as well as two inshore sites located in the Norwegian Skagerrak (Tisler) and on the Norwegian South-West coast (Langenuen). A more detailed description, including sampling times, sampling coordinates, water depths and sample type (i.e. coral, seawater and sediment) yields, is provided as Supporting Information (Text S1; Fig. S1, Table S1).
Hierarchical sampling design
Sampling was performed hierarchically, encompassing five levels of observation, including three spatial scales (Fig. 2). The first study level (HABITAT) comprised four potentially distinct types of microbial habitats associated with and surrounding a scleractinian coral colony in its reef environment: Coral branch surface, coral mucus, ambient seawater, and proximal sediments. The second level (COLOR, SPECIES) featured specific coral species (L. pertusa, M. oculata; Fig. 1B) and coral color types (white and red individuals of L. pertusa). Geomorphologic reef zoning, as prevailing at Røst, determined the third level (ZONE), including the terrace-covered ridge crest, the rubble- and sponge-dominated ridge slope, and the clay-bearing, sparsely populated inter-ridge depression in the reef center. At the fourth level (IN-OUT), the up-slope reef center (Røst-in) was compared with the down-slope reef periphery (Røst-out) in distances of 1 m, 10 m and 100 m away from the reef margin. The fifth level (REEF) allowed a comparison of the offshore Røst site with the nearby Trænadjupet site and the two inshore sites, Tisler and Langenuen. Arranged in a nested layout, each of these study levels was considered as integral part of the respective next higher level, along an increasing gradient of complexity ranging from level one (HABITAT) to level five (REEF).
At Røst, the main study site, where sampling focused on intra-reef differences (HABITAT, SPECIES/COLOR, ZONE, IN-OUT; Fig. 2), the collection of corals (L. pertusa, M. oculata), seawater and surface sediments was performed during two manned submersible dives down-slope across the reef (Fig. S1, Table S1). The first dive (ship station: PS 70/17-1) traversed two of the uppermost ridges in the reef center, while the second dive (ship station: PS 70/31-1) extended further down-slope to the reef periphery at a distance of approx. 2.5 km. At the other study sites (inter-reef differences: REEF; Fig. 2), sampling involved the collection of L. pertusa, seawater and sediments at random locations within the respective main reef area (Table S1).
In-situ sample collection
Specimens of living CWCs were sampled by manned submersible (Røst, Trænadjupet) or video-assisted remotely operated vehicle (Tisler, Langenuen; Table S1). After visual assessment of each target colony in situ, one healthy looking fragment was picked from the colony's living outer rind using the manipulator arm, and placed into a separate compartment of the sampling reservoir. Onboard, each specimen was inspected for epigrowth, impurities or degeneration, before selecting an intact fragment (5–15 cm in length) for sub-sampling of coral-associated microbial habitats. Fragments needed for branch surface and mucus sampling were maintained in flow-through tanks with in-situ water at a temperature of 10–11°C for ≤30 min until subsequent processing. Seawater was sampled with 2l-Niskin bottles attached to the submersible (Røst, Trænadjupet), or mounted on a conductivity-temperature-depth rosette sampler (Langenuen) or video-assisted steel cable (Tisler; Table S1). Immediately after retrieval, 2l-water samples were kept at 4°C, filtered in 500 ml aliquots onto sterile polycarbonate membranes (0.22 µm pore size, Millipore, Billerica, MA), and stored at −20°C until further treatment. Surface sediments (approximately 0–5 cm sediment depth) were collected by custom sampling scoops operated via the submersible and vehicle manipulator arm (Røst, Trænadjupet, Tisler) or by Van-Veen grab (Langenuen; Table S1). Upon retrieval, sediment samples were immediately transferred into sterile 50-ml vials and stored at −20°C until further processing. At Røst, sediment sampling was not possible on the ridge crests owing to the density of the prevailing coral framework cover.
Coral sub-sampling procedures
After gentle rinsing with sterile-filtered (Whatman, Maidstone, UK) local seawater, branch surfaces of living corals were sampled by scraping an area of up to 5 cm2 per fragment with sterile scalpel blades, yielding a mixture of surface plaques, coenosarc tissue, and calcareous particles. Scraping was carried out on the primary, and partly secondary, branches of each fragment, avoiding fragile outer branches as well as polyp calices. All material accumulated per fragment was directly transferred into a DNA extraction tube (see below). Freshly produced coral mucus was sampled by gently rinsing living coral fragments with sterile-filtered seawater and inducing mucus exudation through 2–5 min air exposure. After discarding exudate released during the first minute, subsequent production of up to 0.5 ml per fragment was collected directly from polyp surfaces by using sterile syringes. Resulting mucus-seawater mixture was concentrated onto sterile polycarbonate filters (Whatman), and frozen at −20°C until DNA extraction.
DNA extraction
Total community DNA was extracted and purified with the Ultra Clean Soil DNA Kit (MoBio, Carlsbad, CA, USA) following the manufacturer's instructions for maximum yield, with slight modifications. Branch samples (scrapings from up to 5 cm2 surface per fragment) and sediment samples (3×1 g per scoop) were directly transferred into extraction tubes, mucus samples (up to 0.5 ml per fragment) and water samples (2–4×500 ml per Niskin bottle) on respective filter membranes. DNA yields were quantified by NanoDrop spectrophotometry (NanoDrop, Wilmington, DE). For a complete overview of sample units and replicates subjected to DNA extraction and subsequent community analyses, see Table S1.
Community fingerprinting and multivariate analyses
Universal bacterial ARISA [68] based on 3 PCR replicates per sample, and subsequent binning into OTUs were carried out as described previously [69]. Based on a threshold of ≥0.09% in relative fluorescence intensity (individual peak areas divided by the total peak area of the respective sample) and 50 in fluorescence units, only ARISA fragments in the size range of 100–1000 bp were subjected to the binning procedure with a window size of 2 bp. Total numbers of ARISA-based OTUs (i.e. the number of bacterial types contained in each ARISA sample; used as relative proxy for richness), were assessed for mean difference by applying the non-parametric, omnibus Kruskal-Wallis test (KW), followed by pairwise Wilcoxon-Mann-Whitney tests (WMW). Multivariate patterns in community structure were analyzed based on Bray-Curtis dissimilarity matrices which were visually inspected by Non-metric MultiDimensional Scaling (NMDS), cluster analysis, and heat-mapping. Differences in community overlap between a posteriori defined sample categories were tested by Analysis of Similarity (ANOSIM) and corrected for multiple tests according to the Bonferroni criterion. Sources of bacterial community differences were further assessed by Permutational Multivariate Analysis of Variance (PERMANOVA; [70]) at three levels: (i) regional, inter-reef differences between the four different reef sites (REEF, HABITAT; Fig. 2) (ii) local, intra-reef differences in the whole Røst area (IN-OUT; Fig. 2), and (iii) local, intra-reef differences (ZONE, SPECIES, COLOR, HABITAT; Fig. 2) at the reef center of Røst. Numerical analyses were implemented in PAST v2.0 (Palaeontological Statistics) and in R v.2.9 (The R Project for Statistical Computing) using the standard and vegan packages, as well as custom scripts.
Supporting Information
More detailed description of the study sites.
(DOC)
Geographical and topographical setting of sampling events at Røst. (A) Røst bathymetry, including dive transects at Røst-in (reef center) and Røst-out (reef periphery; map: courtesy of V. Unnithan, JUB), (B) Røst transversal scheme (not to scale) indicating topographical reef structure, geomorphological reef zoning and single sampling stations (reef center: val = valley, slo = slope, cre = crest; reef periphery: 1/10/100 m = 1/10/100 m beyond the apparent reef margin).
(TIF)
Partitioning of bacterial OTUs between distinct coral-associated and ambient microbial habitats. Numbers indicate the amount of OTUs unique to each microbial habitat, or common to any two or all microbial habitats: (A) Bacterial OTUs associated with samples of white L. pertusa (left), red L. pertusa (middle) or M. oculata (right) and their ambient environment at Røst-in, (B) Bacterial OTUs associated with samples of all coral species/colors and their ambient environment at Røst-in (left) or at all sites combined (right).
(TIF)
Pairwise Bray-Curtis dissimilarity relationships between all community profiles. Samples are grouped according to microbial habitat type, coral species and color, geomorphologic reef zoning, reef boundary (incl. distances away from the apparent reef margin), and reef site. Cell position corresponds to the symmetrical pairing of single sample groups. Cell shading indicates the magnitude of dissimilarity between sample pairs.
(TIF)
NMDS ordinations of ARISA community profiles per microbial habitat. For each microbial habitat type, differences in bacterial community structure are plotted as related to reef site, reef boundary, geomorphologic reef zoning, coral species and color. Objects represent consensus signals for all PCR triplicates per sample and share a more similar community structure when plotting closer to each other (Bray-Curtis distance). Stress values indicate the goodness-of-fit of the 2-dimensional representation compared to the original multi-dimensional matrix.
(TIF)
Overview on sampling events of living coral specimens, seawater and sediments at the different study sites. wL = white L. pertusa, rL = red L. pertusa, rM = red M. oculata. aSample lost during processing. The full station list of ARKXXII/1a is available via the PANGAEA database at: http://www.pangaea.de/ddi?retr=events/HERMES/ARK-XXII_1a.retr&conf=events/CruiseReportHTML.conf&title=StationlistofcruiseARK-XXII/1a&format=html.
(DOC)
PERMANOVA of coral-associated bacterial variation done at Røst. A) Analyses considering A) reef boundary, i.e. in/out-reef location (IN-OUT) and B) geomorphologic reef zoning (ZONE) within Røst-in. aSource of variation. bAmount of explained variation. cSignificance level, assessed by 999 random permutations (*** P≤0.001, ** P≤0.01, * P≤0.05). dSignificance level below Bonferroni correction threshold.
(DOC)
Acknowledgments
Our sincere thanks go to the JAGO Team (IfM Geomar) as well as the crew and scientific shipboard parties of RV Polarstern (ARK XXII/1a), RV G.O. Sars, RV Lophelia, and RV Hans Brattstrøm for excellent support during in-situ sampling and onboard sample processing. We also greatly acknowledge Hans Tore Rapp and Christopher Schander (UiB), as well as Tomas Lundälv, Lisbeth Jonsson, and Anne Larsson (SLCMS) for fruitful ‘joint ventures’, helpful discussions, and logistical support. We further thank Laura Wehrmann and Verena Witt (MPI-MM) for assistance in the field and in the lab. Special thanks to Stefanie Grünke (MPI-MM) for valuable comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the German Research Foundation (DFG) (grant Wi 2677/3-1) to CW within the EuroDIVERSITY project MiCROSYSTEMS, by the German Research Foundation to FH, and by the EU 6th FP HERMES and the EU 7th FP HERMIONE (grant agreement n° 226354) to AB. Further support was received from the Max Planck Society and the Helmholtz Association. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Roberts M, Wheeler A, Freiwald A, Cairns S. Cold-water corals: The biology and geology of deep-sea coral habitats. New York: Cambridge University Press; 2009. [Google Scholar]
- 2.Jensen A, Frederiksen R. The fauna associated with bank-forming deepwater coral Lophelia pertusa (Scleractinia) on the Faroe shelf. Sarsia. 1992;77:53–69. [Google Scholar]
- 3.Freiwald A, Fosså JH, Grehan A, Koslow T, Roberts JM. Cold-water coral reefs. Cambridge: UNEP-WCMC; 2004. [Google Scholar]
- 4.Henry LA, Roberts JM. Biodiversity and ecological composition of macrobenthos on cold-water coral mounds and adjacent off-mound habitat in the bathyal Porcupine Seabight, NE Atlantic. Deep Sea Res I. 2007;54:654–672. [Google Scholar]
- 5.Hovland M. Deep-water coral reefs: Unique biodiversity hotspots. Berlin: Springer; 2008. [Google Scholar]
- 6.Roberts JM. Reefs of the deep: The biology and geology of cold-water coral ecosystems. Science. 2006;312:543–547. doi: 10.1126/science.1119861. [DOI] [PubMed] [Google Scholar]
- 7.Buhl-Mortensen L, Vanreusel A, Gooday AJ, Levin LA, Priede IG, et al. Biological structures as source of habitat heterogeneity and biodiversity on deep-ocean margins. Mar Ecol. 2010;31:21–50. [Google Scholar]
- 8.Henry LA, Davies AJ, Roberts JM. Beta diversity of cold-water coral reef communities off western Scotland. Coral Reefs. 2010;29:427–436. [Google Scholar]
- 9.Lindner A, Cairns SD, Cunningham CW. From offshore to onshore: Multiple origins of shallow-water corals from deep-sea ancestors. PLoS ONE. 2008;3:e2429. doi: 10.1371/journal.pone.0002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones CG, Lawton JH, Shachak M. Organisms as ecosystem engineers. Oikos. 1994;69:373–386. [Google Scholar]
- 11.Berke SK. Functional groups of ecosystem engineers: A proposed classification with comments on current issues. Integrat Comparat Biol. 2010;50:147–157. doi: 10.1093/icb/icq077. [DOI] [PubMed] [Google Scholar]
- 12.Bongiorni L, Mea M, Gambi C, Pusceddu A, Taviani M, et al. Deep-water scleractinian corals promote higher biodiversity in deep-sea meiofaunal assemblages along continental margins. Biol Conserv. 2010;143:1687–1700. [Google Scholar]
- 13.Spalding M, Ravilious C, Green P. UNEP-WCMC world atlas of coral reefs. Berkeley: University of California Press; 2001. [Google Scholar]
- 14.Wilson JB. ‘Patch’ development of the deep-water coral Lophelia pertusa (L.) on Rockall Bank. J Mar Biol Assoc UK. 1979;59:165–177. [Google Scholar]
- 15.Freiwald A, Henrich R, Pätzold J. Anatomy of a deep-water coral reef mound from Stjernsund, West-Finnmark, northern Norway. In: James NP, Clarke JAD, editors. Cool-water carbonates. SEPM Spec Publ. 1997;56:141–161. [Google Scholar]
- 16.Rogers AD. The biology of Lophelia pertusa (Linnaeus 1758) and other deep-water reef-forming corals and impacts from human activities. Intl Rev Hydrobiol. 1999;84:315–406. [Google Scholar]
- 17.Mortensen PB, Hovland MT, Fosså JH, Furevik DM. Distribution, abundance and size of Lophelia pertusa coral reefs in mid-Norway in relation to seabed characteristics. J Mar Biol Assoc UK. 2001;81:581–597. [Google Scholar]
- 18.Freiwald A. Reef-forming cold-water corals. In: Wefer G, Billett D, Hebbeln D, Jørgensen BB, Schlüter M, van Weering T, editors. Ocean margin systems. Berlin: Springer; 2002. pp. 365–385. [Google Scholar]
- 19.Freiwald A, Hühnerbach V, Lindberg B, Wilson B, Campbell J. The Sula Reef complex, Norwegian shelf. Facies. 2002;47:179–200. [Google Scholar]
- 20.Mortensen PB, Hovland M, Brattegard T, Farestveit R. Deep water bioherms of the scleractinian coral Lophelia pertusa (L.) at 64°N on the Norwegian Shelf: Structure and associated megafauna. Sarsia. 1995;80:145–158. [Google Scholar]
- 21.Freiwald A, Wilson JB. Taphonomy of modern, deep, cold-temperate water coral reefs. Historical Biol. 1998;13:37–52. [Google Scholar]
- 22.Knowlton N, Rohwer F. Multispecies microbial mutualisms on coral reefs: The host as a habitat. Am Nat. 2003;162:S51–S62. doi: 10.1086/378684. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg E, Kellogg CA, Rohwer F. Coral microbiology. Oceanogr. 2008;20:146–154. [Google Scholar]
- 24.Ainsworth TD, Thurber RV, Gates RD. The future of coral reefs: A microbial perspective. Trends Ecol Evol. 2010;25:233–240. doi: 10.1016/j.tree.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Yakimov MM, Cappello S, Crisafi E, Tursi A, Savini A, et al. Phylogenetic survey of metabolically active microbial communities associated with the deep-sea coral Lophelia pertusa from the Apulian plateau, Central Mediterranean Sea. Deep Sea Res I. 2006;53:62–75. [Google Scholar]
- 26.Großkurth AK. Analysis of bacterial community composition on the cold-water coral Lophelia pertusa and antibacterial effects of coral extracts. Diploma thesis. Oldenburg: Carl von Ossietzky University; 2007. [Google Scholar]
- 27.Försterra G, Häussermann V. Unusual symbiotic relationships between microendolithic phototrophic organisms and azooxanthellate cold-water corals from Chilean fjords. Mar Ecol Prog Ser. 2008;370:121–125. [Google Scholar]
- 28.Neulinger SC, Järnegren J, Ludvigsen M, Lochte K, Dullo WC. Phenotype-specific bacterial communities in the cold-water coral Lophelia pertusa (Scleractinia) and their implications for the host's nutrition, health, and distribution. Appl Environ Microbiol. 2008;74:7272–7285. doi: 10.1128/AEM.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansson L, Agis M, Maier C, Weinbauer G. Community composition of bacteria associated with cold-water coral Madrepora oculata: within and between colony variability. Mar Ecol Prog Ser. 2009;397:89–102. [Google Scholar]
- 30.Kellogg CA, Lisle JT, Galkiewicz J. Culture-independent characterization of bacterial communities associated with the cold-water coral Lophelia pertusa in the Northeastern Gulf of Mexico. Appl Environ Microbiol. 2009;75:2294–2303. doi: 10.1128/AEM.02357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neulinger SC, Gärtner A, Järnegren J, Ludvigsen M, Lochte K, et al. Tissue-associated “Candidatus Mycoplasma corallicola” and filamentous bacteria on the cold-water coral Lophelia pertusa (Scleractinia). Appl Environ Microbiol. 2009;75:1437–1444. doi: 10.1128/AEM.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schöttner S, Hoffmann F, Wild C, Rapp HT, Boetius A, et al. Inter- and intra- habitat bacterial diversity associated with cold-water corals. ISME J. 2009;3:756–759. doi: 10.1038/ismej.2009.15. [DOI] [PubMed] [Google Scholar]
- 33.Penn K, Wu D, Eisen JA, Ward NL. Characterization of bacterial communities associated with deep-sea corals on Gulf of Alaska seamounts. Appl Environ Microbiol. 2006;72:1680–1683. doi: 10.1128/AEM.72.2.1680-1683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brück TB, Brück WM, Santiago-Vázquez LZ, McCarthy PJ, Kerr RG. Diversity of the bacterial communities associated with the azooxanthellate deep water octocorals Leptogorgia minima, Iciligorgia schrammi, and Swiftia exertia. Mar Biotechnol. 2007;9:561–576. doi: 10.1007/s10126-007-9009-1. [DOI] [PubMed] [Google Scholar]
- 35.Hall-Spencer JM, Pike J, Munn CB. Diseases affect cold-water corals too: Eunicella verrucosa (Cnidaria: Gorgonacea) necrosis in SW England. Dis Aquat Org. 2007;76:87–97. doi: 10.3354/dao076087. [DOI] [PubMed] [Google Scholar]
- 36.Webster N, Bourne D. Bacterial community structure associated with the Antarctic soft coral, Alcyonium antarcticum. FEMS Microbiol Ecol. 2007;59:81–94. doi: 10.1111/j.1574-6941.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 37.Gray MA, Stone RP, McLaughlin MR, Kellogg CA. Microbial consortia of gorgonian corals from the Aleutian Islands. FEMS Microbiol Ecol. 2011;76:109–120. doi: 10.1111/j.1574-6941.2010.01033.x. [DOI] [PubMed] [Google Scholar]
- 38.Hewson I, Fuhrman JA. Spatial and vertical biogeography of coral reef sediment bacteria and diazotroph communities. Mar Ecol Prog Ser. 2006;306:79–86. [Google Scholar]
- 39.Böer SI, Hedtkamp SIC, van Beusekom JJE, Fuhrman JA, Boetius A, et al. Time and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 2009;3:780–791. doi: 10.1038/ismej.2009.29. [DOI] [PubMed] [Google Scholar]
- 40.Bourne DG, Munn CB. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol. 2005;7:1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- 41.Koren O, Rosenberg E. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl Environ Microbiol. 2006;72:5254–5259. doi: 10.1128/AEM.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser. 2006;322:1–14. [Google Scholar]
- 43.Sweet MJ, Coquer A, Bythell JC. Development of bacterial biofilms in comparison to surface-assoiated microbes of hard corals. PLoS ONE. 2011;6:e21195. doi: 10.1371/journal.pone.0021195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rypien KL, Ward JR, Azam F. Antagonistic interactions among coral-associated bacteria. Environ Microbiol. 2010;12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- 45.Le Goff-Vitry MC, Pybus OG, Rogers AD. Genetic structure of the deep-sea coral Lophelia pertusa in the northeast Atlantic revealed by microsatellites and internal transcribed spacer sequences. Mol Ecol. 2004;13:537–349. doi: 10.1046/j.1365-294x.2004.2079.x. [DOI] [PubMed] [Google Scholar]
- 46.Dodds LA, Roberts JM, Taylor AC, Marubini F. Metabolic tolerance of the cold-water coral Lophelia pertusa to temperature and dissolved oxygen change. J Exp Mar Biol Ecol. 2007;349:205–214. [Google Scholar]
- 47.Brown BE, Bythell JC. Perspectives on mucus secretion in reef corals. Mar Ecol Prog Ser. 2005;296:291–395. [Google Scholar]
- 48.Brooke S, Young CM. In-situ measurement of survival and growth of Lophelia pertusa in the northern Gulf of Mexico. Mar Ecol Prog Ser. 2009;397:153–161. [Google Scholar]
- 49.Maier C, Hegemann J, Weinbauer MG, Gattuso JP. Calicification of the cold-water coral Lophelia pertusa under ambient and reduced pH. Biogeosci. 2009;6:1671–1680. [Google Scholar]
- 50.Rohwer F, Kelley S. Culture independent analyses of coral associated microbes. In: Rosenberg E, Loya Y, editors. Coral health and disease. Berlin: Springer; 2004. pp. 265–278. [Google Scholar]
- 51.Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 52.Costello MJ. Distinguishing marine habitat classification concepts for ecological data management. Mar Ecol Progr Ser. 2009;397:253–268. [Google Scholar]
- 53.Wild C, Naumann M, Niggl W, Haas A. Carbohydrate composition of mucus released by scleractinian warm- and cold-water reef corals. Aquat Biol. 2010;10:131–138. [Google Scholar]
- 54.Drollet JH, Teai T, Faucon M, Martin PMV. Field study of compensatory changes in UV-absorbing compounds in the mucus of the solitary coral Fungia repanda (Scleractinia: Fungiidae) in relation to solar UV-radiation, seawater temperature and other coincident physico-chemical parameters. Mar Freshw Res. 1997;48:329–333. [Google Scholar]
- 55.Kiriakoulakis K, Fisher L, Freiwald A, Grehan A, Roberts JM, et al. Lipids and nitrogen isotopes of two deep-water corals from the North-East Atlantic: Initial results and implications for their nutrition. In: Freiwald A, Roberts JM, editors. Cold-water corals and ecosystems. Berlin: Springer; 2005. pp. 715–729. [Google Scholar]
- 56.Dodds LA, Black KD, Orr H, Roberts JM. Lipid biomarkers reveal geographical differences in food supply to the cold-water coral Lophelia pertusa (Scleractinia). Mar Ecol Prog Ser. 2009;397:113–124. [Google Scholar]
- 57.Mancini I, Guerriero A, Guella G, Bakken T, Zimbrowius H, et al. Novel 10-hydroxydocosapolyenoic acids from deep-water scleractinian corals. Helv Chim Acta. 1999;82:677–684. [Google Scholar]
- 58.Meikle P, Richards GN, Yellowlees D. Structural investigation on the mucus from six species of coral. Mar Biol. 1988;99:187–193. [Google Scholar]
- 59.Ducklow HW, Mitchell L. Bacterial populations and adaptations in the mucus layers on living corals. Limnol Oceanogr. 1979;24:715–725. [Google Scholar]
- 60.Danovaro R, Luna GM, Dell'Anno A, Pietrangeli B. Comparison of two fingerprinting techniques, terminal restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis, for determination of bacterial diversity in aquatic environments. Appl Environ Microbiol. 2006;72:5982–5989. doi: 10.1128/AEM.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown M, Fuhrman JA. Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat Microb Ecol. 2005;41:15–23. [Google Scholar]
- 62.Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nature Rev Microbiol. 2008;6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 63.Hentschel U, Usher KM, Taylor MW. Marine sponges as microbial fermenters. FEMS Microbiol Ecol. 2006;55:167–177. doi: 10.1111/j.1574-6941.2005.00046.x. [DOI] [PubMed] [Google Scholar]
- 64.Jensen S, Neufeld JD, Birkeland DK, Hovland M, Murrell JC. Insight into the microbial community structure of a Norwegian deep-water coral reef environment. Deep Sea Res I. 2008;55:1554–1563. [Google Scholar]
- 65.Lee OO, Wang Y, Yang JK, Lafi FF, Al-Suweilem K, et al. Pyrosequencing reveals highly diverse and species-specific communities in sponges from the Red Sea. ISME J. 2011;5:650–664. doi: 10.1038/ismej.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schöttner S, Pfitzner B, Grünke S, Rasheed M, Wild C, Ramette A. Drivers of bacterial diversity dynamics in permeable carbonate and silicate coral reef sands from the Red Sea. Environ Microbiol. 2011;13:1815–1826. doi: 10.1111/j.1462-2920.2011.02494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bienhold C, Boetius A, Ramette A. The energy-diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J. 2011 doi: 10.1038/ismej.2011.140. doi: 10.1038/ismej.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fisher MM, Triplett EW. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater communities. Appl Environ Microbiol. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramette A. Quantitative community fingerprinting methods for estimating the abundance of operational taxonomic units in natural microbial communities. Appl Environ Microbiol. 2009;75:2495–2505. doi: 10.1128/AEM.02409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
More detailed description of the study sites.
(DOC)
Geographical and topographical setting of sampling events at Røst. (A) Røst bathymetry, including dive transects at Røst-in (reef center) and Røst-out (reef periphery; map: courtesy of V. Unnithan, JUB), (B) Røst transversal scheme (not to scale) indicating topographical reef structure, geomorphological reef zoning and single sampling stations (reef center: val = valley, slo = slope, cre = crest; reef periphery: 1/10/100 m = 1/10/100 m beyond the apparent reef margin).
(TIF)
Partitioning of bacterial OTUs between distinct coral-associated and ambient microbial habitats. Numbers indicate the amount of OTUs unique to each microbial habitat, or common to any two or all microbial habitats: (A) Bacterial OTUs associated with samples of white L. pertusa (left), red L. pertusa (middle) or M. oculata (right) and their ambient environment at Røst-in, (B) Bacterial OTUs associated with samples of all coral species/colors and their ambient environment at Røst-in (left) or at all sites combined (right).
(TIF)
Pairwise Bray-Curtis dissimilarity relationships between all community profiles. Samples are grouped according to microbial habitat type, coral species and color, geomorphologic reef zoning, reef boundary (incl. distances away from the apparent reef margin), and reef site. Cell position corresponds to the symmetrical pairing of single sample groups. Cell shading indicates the magnitude of dissimilarity between sample pairs.
(TIF)
NMDS ordinations of ARISA community profiles per microbial habitat. For each microbial habitat type, differences in bacterial community structure are plotted as related to reef site, reef boundary, geomorphologic reef zoning, coral species and color. Objects represent consensus signals for all PCR triplicates per sample and share a more similar community structure when plotting closer to each other (Bray-Curtis distance). Stress values indicate the goodness-of-fit of the 2-dimensional representation compared to the original multi-dimensional matrix.
(TIF)
Overview on sampling events of living coral specimens, seawater and sediments at the different study sites. wL = white L. pertusa, rL = red L. pertusa, rM = red M. oculata. aSample lost during processing. The full station list of ARKXXII/1a is available via the PANGAEA database at: http://www.pangaea.de/ddi?retr=events/HERMES/ARK-XXII_1a.retr&conf=events/CruiseReportHTML.conf&title=StationlistofcruiseARK-XXII/1a&format=html.
(DOC)
PERMANOVA of coral-associated bacterial variation done at Røst. A) Analyses considering A) reef boundary, i.e. in/out-reef location (IN-OUT) and B) geomorphologic reef zoning (ZONE) within Røst-in. aSource of variation. bAmount of explained variation. cSignificance level, assessed by 999 random permutations (*** P≤0.001, ** P≤0.01, * P≤0.05). dSignificance level below Bonferroni correction threshold.
(DOC)