Abstract
Plague (Yersinia pestis infection) is a highly virulent rodent disease that persists in many natural ecosystems. The black rat (Rattus rattus) is the main host involved in the plague focus of the central highlands of Madagascar. Black rat populations from this area are highly resistant to plague, whereas those from areas in which the disease is absent (low altitude zones of Madagascar) are susceptible. Various lines of evidence suggest a role for the Major Histocompatibility Complex (MHC) in plague resistance. We therefore used the MHC region as a candidate for detecting signatures of plague-mediated selection in Malagasy black rats, by comparing population genetic structures for five MHC-linked microsatellites and neutral markers in two sampling designs. We first compared four pairs of populations, each pair including one population from the plague focus and one from the disease-free zone. Plague-mediated selection was expected to result in greater genetic differentiation between the two zones than expected under neutrality and this was observed for one MHC-class I-linked locus (D20Img2). For this marker as well as for four other MHC-linked loci, a geographic pattern of genetic structure was found at local scale within the plague focus. This pattern would be expected if plague selection pressures were spatially variable. Finally, another MHC-class I-linked locus (D20Rat21) showed evidences of balancing selection, but it seems more likely that this selection would be related to unknown pathogens more widely distributed in Madagascar than plague.
Introduction
Immune genes have been shown to be strongly affected by natural selection [1], [2]. In particular, the genes of the Major Histocompatibility Complex (MHC) have attracted the attention of evolutionary biologists, because of their extraordinary polymorphism and fundamental role in vertebrate immunity [3]. This genetic complex contains three gene subfamilies [4]. Class I genes are expressed by nucleated somatic cells and play an essential role in defense against intracellular pathogens, as their products present antigens derived from infected cells to cytolytic T-cells. Class II genes are expressed by antigen-presenting cells of the immune system and encode receptors presenting peptides mostly derived from extracellular pathogens to T helper cells. However, the functional difference between these two classes is not clear-cut, as both MHC class I and II may be involved in recognizing endogenous and exogenous antigens [5]. Finally, MHC class III includes a diverse array of structurally unrelated genes, including several involved in innate immunity (e.g., proinflammatory cytokines, complement components).
Pathogens are thought to be one of the main selective forces acting on the MHC genes of their hosts [3], [6]. Pathogen-mediated balancing selection is generally thought to account for the tremendous diversity observed at MHC loci, but the underlying mechanisms remain debated [6], [7]. Three main hypotheses have been proposed: heterozygote advantage, rare-allele advantage and spatio-temporal variation of selective pressures [7]. Pathogen selection pressure may also drive a decrease in genetic diversity at MHC markers through directional selection, with the spread of an advantageous allele (positive selection), or selection against disadvantageous alleles (negative or purifying selection) [8]. Directional selection may occur if a single pathogen strain or species is thought to exert a major selective pressure on the host species considered (see for example [8]). Various studies have shown that natural selection acts on MHC genes and have investigated its underlying mechanisms by comparing the population genetic structure of MHC genes with that of neutral loci (see for example [9], [10]; reviewed in [6]). Some of these studies have suggested that variability in pathogen exposure acts as a selective pressure on MHC loci [11], [12], but most did not include descriptions of the geographic distribution of pathogen prevalence (but see [13]).
Two main technical approaches are used to study MHC variation. First, gene sequences can be analyzed directly (using classical, or next-generation sequencing methods [14], [15] or by using a sequence library, e.g., by SSCP _Single Strand Conformation Polymorphism_ [16]). Second, MHC-linked microsatellites may be used, as linkage disequilibrium may stamp loci close to MHC genes with similar signatures of selection [17]. This latter approach has the advantages of allowing the analysis of several MHC loci at a moderate cost. It also allows the use of comparable markers (e.g., microsatellites) for the inference of population genetic structure and detection of signatures of selection [7]. Gene-linked microsatellites have already been shown to be useful for detecting natural selection acting on MHC genes (see for example [17]–[19]) or other candidate genes [20], [21]. The reliability of this indirect approach has been assessed by the observed correlation between the allelic variation of microsatellites and neighboring genes [22] and by the reported association between candidate gene-linked microsatellite diversity and parasite load [23].
Plague (Yersinia pestis infection) is among the most virulent of known pathogens for humans and susceptible rodents, resulting in high mortality rates in natural populations [24], [25]. Plague epidemics may therefore exert strong selective pressure on host immune genes. Resistance phenotypes may evolve rapidly in reservoir populations, as suggested by the association between the occurrence of plague and plague resistance reported for Onychomys leucogaster in North America [26] and Mastomys natalensis in South Africa [27]. This would also be the case for the black rat (Rattus rattus), the main reservoir of plague in Madagascar [28].
Plague was first reported in Madagascar in 1898. Areas of the central highlands of this country, at elevations above 800 meters have represented a plague focus since the 1920s, with hundreds of human cases reported each year and circulation within rodent populations. By contrast, there is no evidence of plague circulation in rural areas of the lowland zone of Madagascar [28], [29]. The black rat is usually considered to be highly susceptible to plague [24]. However, the R. rattus populations from the Malagasy plague focus (central highlands) are much more resistant (LD50 1000 times higher) than black rat populations from plague-free zones (low altitude regions) [30], [31]. This resistance is transmitted to laboratory-born descendants [30], suggesting a genetic basis.
MHC genes may be involved in the genetics of plague resistance. Indeed, a recent quantitative genetics study showed the presence of a resistance locus (which remains to be precisely identified) within the MHC region in the laboratory mouse [32]. In addition, various laboratory studies have demonstrated the potential importance of MHC products for the immune response to plague. Indeed, Y. pestis epitopes have been shown to be recognized by MHC class II receptors of the laboratory mouse [33], as expected for this mostly extracellular pathogen [34]. However, MHC class I molecules may also be involved, as human cytolytic T cells are stimulated during Y. pestis infection [35]. Finally, the Tnf-α (Tumor necrosis factor) gene, an MHC class III gene, encodes the proinflammatory cytokine TNF-α, which has been shown to protect against Y. pestis experimental infection in laboratory mice ([36], [37], see also [38]).
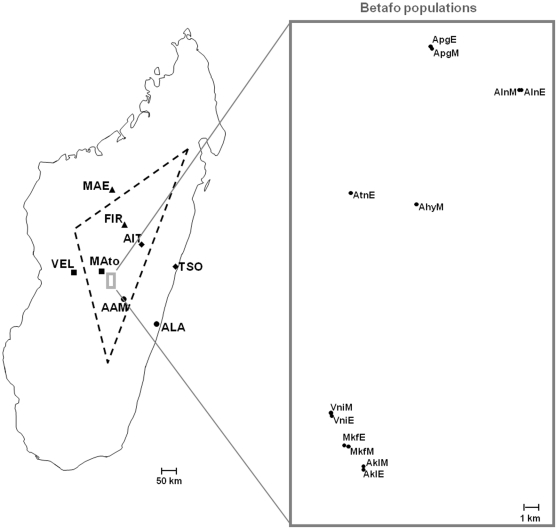
This study uses the MHC gene family as candidate for plague resistance in wild populations of Malagasy R. rattus. In addition to shedding light on the theoretical question of the way in which selection shapes MHC variation in wild rodents, this research is a step towards a better understanding of plague circulation in the central highlands of Madagascar, which remains one of the most important plague foci in the world [24], [39]. To this purpose, we used a population genetics approach based on the comparison of MHC-linked and neutral microsatellites in two contrasting population designs. The first dataset focuses on four pairs of populations, each consisting of one population from within and another from outside the plague focus of Madagascar (Figure 1 and Table 1). In this so-called “Madagascar” dataset, plague-driven divergent selection within these pairs of populations may imply higher levels of genetic differentiation at MHC markers than at neutral markers [3]. However, the presence of plague is not the only selective environmental factor characterizing the Malagasy plague focus. Elevation and the ecological conditions typical of regions more than 800 m above sea level may also have an effect. We thus also used a metapopulation sampling within a restricted area of the plague focus in which human cases of plague are reported annually (Figure 1 and Table 1). As plague is likely to exert a uniform selection pressure on the R. rattus populations of this area, the level of genetic differentiation at MHC markers found to be under divergent selection in the Madagascar dataset would be expected to be lower than that for neutral markers in the so-called “Betafo” dataset [40]. Alternatively, spatial variation in plague selection pressure may result in more marked geographic patterns of genetic differentiation at these markers than at neutral markers [3].
Figure 1. Location of the sampled sites.
For the Madagascar dataset, each population pair, consisting of one population from the plague focus and one from the plague-free zone, is identified by a different symbol. The principal Malagasy plague focus is outlined by a black dotted line and the region of Betafo is outlined in grey.
Table 1. List of the populations analyzed in this study.
| MADAGASCAR DATASET | |||||
| Population name | Distance(km) | Zone | Sampling year | Sample size | HE (13 presumed neutral markers) ± S.E. |
| AAM | 140 | C.H. | 2000 | 28 | 0.714±0.138 |
| ALA | L.A. | 1999 | 27 | 0.750±0.125 | |
| AIT | 139 | C.H. | 2006 | 22 | 0.717±0.118 |
| TSO | L.A. | 2007 | 22 | 0.723±0.144 | |
| FIR | 128 | C.H. | 1999 | 28 | 0.695±0.165 |
| MAE | L.A. | 1999 | 21 | 0.706±0.185 | |
| MAto | 103 | C.H. | 1996 | 28 | 0.681±0.170 |
| VEL | L.A. | 2000 | 22 | 0.697±0.128 | |
Sample sizes and genetic diversity (expected heterozygosity: HE) are shown. C.H.: central highlands; L.A.: lowland areas.
Results
Studied loci and within-population analyses
Five MHC-linked microsatellite markers designed on the basis of genome sequence of R. norvegicus were analyzed, each corresponding to one of the three MHC classes [4] (Table 2). These five MHC-linked loci comprised two (Msat-Tnf and D20Img2) present as single copies (maximum of two alleles per individual), and three duplicated loci: RT1N1 and D20Rat41 were present as two copies and D20Rat21 was present as three copies at least (the locus D20Rat21 was also found to be duplicated in R. norvegicus [41], but no such information was found for RT1N1 and D20Rat41). A set of 13 previously described microsatellites (see methods) was used as a control for non-selective factors and is referred to as “presumed neutral microsatellites”.
Table 2. Characteristics of the five MHC-linked microsatellites loci analyzed in this study.
| Class | Locus | Primer sequences | Closest gene | Repeat | Total number of alleles per locus | Number of alleles per ind. |
| (primer reference or design method) | (distance) | (size range) | ||||
| I | D20Rat21 | 5′-CTGTGCTATGGCAGGAGATT-3′ | RT1-M6 (500 bp) | TG and AG | 42 | 2–6 |
| 5′-GCCATCTTCAGCACTACAGG-3′ | (289–359) | |||||
| (Design on R. norvegicus genome) | ||||||
| RT1N1 | 5′-TCTCGTGGAATAGGCAGA-3′ | RT1N1 (1500 bp) | AG | 16 | 1–4 | |
| 5′-TGGCTGTCCTAGAACTCACT-3′ | (302–375) | |||||
| (Design on R. rattus sequences) | ||||||
| D20Img2 | 5′-CTGAGCTCCCTAGGACCTACAT-3′ | Ddr1 (42000 bp) | CA | 7 | 1–2 | |
| 5′-TCTCTTGTGTCAGGCTAATTAC-3′ | (277–333) | |||||
| [60] | ||||||
| III | Msat-Tnf | 5′-ACATAGGCATGGTGTCTCTG-3′ | Tnf (300 bp) | CA | 16 | 1–2 |
| 5′-CAGGATTCTGTGGCAATCTG-3′ | (147–180) | |||||
| (Design on R. rattus sequences) | ||||||
| II | D20Rat41 | 5′-AGTYCTCTTCTGGYCTCCAT -3′ | RT1-Bb (4 400 bp) | TG | 42 | 1–4 |
| 5′- TGGGACGATGTGTCATATCC -3′ | (162–220) | |||||
| (Design on R. rattus sequences) |
The five loci are ranked as on the chromosome, with D20Rat21 the closest to the telomere.
The neighboring genes indicated are based on the R. norvegicus genome sequence.
Linkage disequilibrium (LD) and Hardy-Weinberg (HW) tests were performed on non duplicated MHC-linked and presumed neutral microsatellites, for the both datasets. LD tests gave significant results after correction for multiple testing in 15 of 2087 cases. These significant tests accounted for only 0.7% of the tests carried out and concerned various locus pairs and populations. The 15 non duplicated loci were thus considered independent. HW tests indicated significant heterozygote deficiencies for some loci (Rr67, D11R56, D11M5, Msat-Tnf) that can be explained by null alleles following Micro-checker. Mean null alleles frequencies estimated using FreeNA over the 20 populations were however lower than 0.05 for these loci (table S1), having therefore little chance to bias genetic differentiation estimates [42], and thus outlier detection. None of the HW tests for heterozygote excess were significant for the two non duplicated MHC-linked loci: Msat-Tnf and D20Img2.
Detecting signatures of selection for the Madagascar dataset
For the Madagascar dataset, two model-based approaches were used to search for selection signatures (Fdist2 [43] and DetSel [44], [45]) in the two non duplicated MHC-linked and the 13 presumed neutral microsatellites. For MHC-linked loci, D20Img2 alone had a higher FST than expected under neutrality in some population pairs, in both Fdist2 (AIT/TSO: p = 0.003; marginally significant probability for FIR/MAE: p = 0.052) and DetSel (AIT/TSO: p = 0.018; AAM/ALA: p = 0.02) (Table 3 and figures S1 and S3). Msat-Tnf was not detected as outlier in any comparison (Table 3 and figures S1 and S2). However, a few presumed neutral loci were found to have significantly higher FST in some population pairs (see details in Table 3).
Table 3. Results of population differentiation tests for the Madagascar dataset analyses.
| Population pair | |||||
| AAM-ALA | AIT-TSO | FIR-MAE | MATo-VEL | ||
| Fdist2 analysis | Msat-Tnf | 0.441 | 0.226 | 0.272 | 0.457 |
| D20Img2 | 0.167 | 0.003 | 0.052 | 0.207 | |
| Supposed neutral loci revealing significant (p-value) | Rr067 (0.017) D11R56 (0.006) | D10R20 (0.045) | Rr067 (0.026) D7R13 (0.023) | Rr014 (0.005) Rr054 (0.011) | |
| DetSel analysis | Msat-Tnf | 0.61 | 0.41 | 0.84 | 0.49 |
| D20Img2 | 0.024 | 0.018 | 0.102 | 0.159 | |
| Supposed neutral loci revealing significant (p-value) | D11R56 (0.003) | Rr093 (0.003) | D10R20 (0.049) Rr114 (0.039) | D7R13 (0.001) | |
The p-values associated with each of the two tests (Fdist2 [43] and DetSel [44], [45] analyses) for each of the four population pairs are reported for the two MHC-linked loci: Msat-Tnf and D20Img2. The supposedly neutral microsatellites for which a significant result was obtained in the analyses are also indicated, together with their p-values (in the case of Fdist2 analysis, all the presumed neutral loci revealing significant had FST higher than expected).
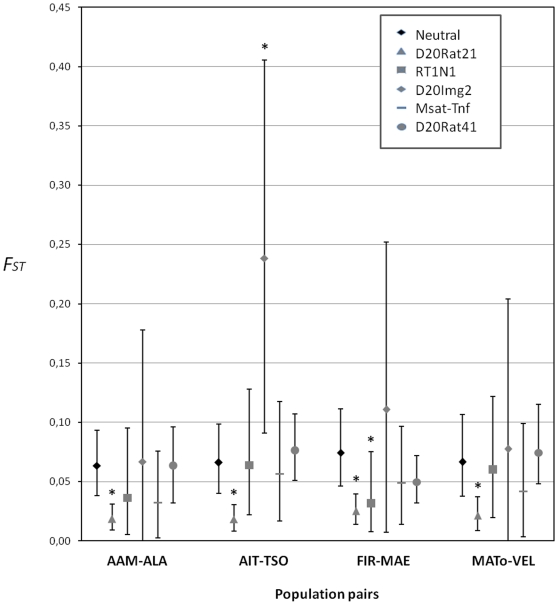
The pairwise FST comparisons carried out between MHC-linked microsatellites (either duplicated or not) and neutral loci (global FST) within each population pair of the Madagascar dataset are presented in Figure 2. Higher FST was obtained for and the locus D20Img2 in the population pair AIT-TSO (p-value obtained through bootstrapping over individuals: p = 0.007), in accordance with the results obtained with the simulation-based programs (see above). The duplicated loci D20Rat21 had significantly lower FST values than neutral markers in the four population pairs (p = 0.001 for AAM-ALA, p = 0 for AIT-TSO, p = 0.001 for FIR-MAE and p = 0.005 for MATo-VEL). Finally, the duplicated locus RT1N1 had low genetic differentiation estimate only in the population pair FIR-MAE (p = 0.033).
Figure 2. Comparison of pairwise population differentiation levels (FST) between the five MHC-linked microsatellites and the neutral loci for the Madagascar's dataset.
The points correspond to the mean FST obtained over 1000 bootstrap and the error bars to the 95% confidence intervals (determined .from the 25th and 975th value in the ranked list of 1000 bootstrap FST values). The neutral values are estimates from the global dataset of 13 presumed-neutral microsatellites. * indicate the MHC-linked markers significantly differing from neutral microsatellites for FST estimates.
Detecting signatures of selection for the Betafo dataset
For the Betafo dataset, the two non-duplicated MHC-linked microsatellites had overall FST values within the envelope expected under neutrality, by Beaumont's method [43] (Fdist2 program, data not shown) and were therefore not identified as being subject to natural selection (p = 0.260 for Msat-Tnf and p = 0.338 for D20Img2). However, two presumed neutral microsatellite loci were found to be subject to purifying selection: Rr017 (p = 0.037) and Rr054 (p = 0.040). Significant isolation by distance was observed for neutral markers (p = 0.005). Partial Mantel tests with neutral microsatellite variation kept constant showed a positive effect of geographic distance on D20Rat41 (r2 = 12.9; p = 0.003), RT1N1 (r2 = 16.8; p = 0.001), D20Img2 (r2 = 10.4; p = 0.006) and Msat-Tnf (r2 = 11.4; p = 0.005) (Table 4). By contrast, the duplicated locus D20Rat21 presented no significant evidence of isolation by distance (Table 4) and its mean FST value (0.014±0.008) indicated weaker genetic differentiation than for neutral loci (95% CI = (0.05; 0.07)).
Table 4. Partial Mantel tests carried out on the Betafo dataset for each of the five MHC-linked microsatellites.
| Locus | Model | Partial Mantel | |
| R2 (%) | P | ||
| D20Rat21 | Global model | 5.13 | 0.1906 |
| Geography | 0 | 0.3978 | |
| Neutral | 2.43 | 0.155 | |
| Geography*Neutral | 0.93 | ||
| RT1N1 | Global model | 17.05 | 0.0029 |
| Geography | 16.77 | 0.001 | |
| Neutral | 0 | 0.8457 | |
| Geography*Neutral | 0.25 | ||
| D20Img2 | Global model | 12.18 | 0.0162 |
| Geography | 10.38 | 0.0063 | |
| Neutral | 3.78 | 0.1134 | |
| Geography*Neutral | −1.99 | ||
| Msat-Tnf | Global model | 14.14 | 0.008 |
| Geography | 11.45 | 0.0049 | |
| Neutral | 0.91 | 0.4627 | |
| Geography*Neutral | 1.76 | ||
| D20Rat41 | Global model | 22.12 | 0.0008 |
| Geography | 12.87 | 0.0034 | |
| Neutral | 5.15 | 0.0674 | |
| Geography*Neutral | 4.06 | ||
Discussion
In this study, we developed five MHC-linked microsatellites from the three classes of the MHC gene family in R. rattus. No linkage disequilibrium was found between the two non duplicated MHC-linked loci, Msat-Tnf and D20Img2 (this analysis was not possible for the duplicated loci), which are about 500 kb apart within the R. norvegicus genome. The MHC is characterized by islands of strong linkage disequilibrium, interspersed with recombination hotspots. Many factors, such as the selective and demographic history of the populations, may have a major effect on recombination patterns [46]. The absence of significant linkage disequilibrium between MHC loci was not surprising, as the islands of strong linkage disequilibrium have been estimated to cover about 50 kb in the human MHC class II region [47], and recombination hotspots appear to be largely conserved between mammalian species [48]. Our results show that different selective pressures affected the MHC-linked microsatellites studied, as frequently reported in previous studies, even for markers from the same MHC class (see for example [10], [11], [49], [50]).
As parasite-mediated selection is thought to be the driving force behind MHC evolution, pathogens (rather than other non pathogenic selective pressures) may account for the selection signatures of MHC-linked markers [6]. In Madagascar, plague is the only known highly virulent pathogen capable of differentiating between the R. rattus populations of the highlands and lowland areas [51]. We therefore have good reason to think that the selection signatures detected for the MHC-linked markers in this study will enable us to identify valuable candidate genes for plague resistance.
Firm evidence was obtained for natural selection acting on the D20Img2 locus, which presented levels of differentiation higher than expected under neutrality in some population pairs of the Madagascar dataset, whether in analyses based on two simulation-based programs (Fdist2 or DetSel) or using an empirical test. A few presumed neutral loci also displayed significant levels of differentiation (Table 3), but D20Img2 was more frequently detected as an outlier. This pattern may be interpreted as a signal of directional selection between the plague focus and the plague-free zone, as expected in conditions of plague-mediated selective pressure. We therefore expected D20Img2 to display a lower level of differentiation than neutral markers within the Betafo dataset, in which every population was assumed to be subject to plague selection. Instead, we showed an effect of geographic location on both loci, after controlling for demographic and stochastic factors. This geographic pattern within the plague focus may reflect spatial heterogeneity in plague selective pressures between R. rattus populations and the occurrence of spatially localized epizootics and epidemics. This would be consistent with the spatial clustering of human plague cases reported in the Betafo region, and the high variability of plague seroprevalence (between 0% and 44% [52]) in the rat populations studied. Spatial heterogeneity in plague selection pressure may also account for the detection of selection signatures in only some of the population pairs of the Madagascar dataset: those at sites within the plague focus at which human plague cases had been recorded in the last ten years (AIT in 2003 and AAM in 1998; database of the Malagasy Health Ministry). Alternatively, the genetic diversity of plague bacteria may imply geographic variation in the type of selective pressure. Indeed, a recent study was able to detect genetic diversity in plague bacterial strains isolated from patients in Madagascar [53]. Our four central highlands populations in the Madagascar dataset are located in regions where isolated strains were different; and even at the small spatial scale of the region investigated for the Betafo dataset, at least two plague genotypes were found. These different strains would however consist in different selective pressure only if they not only vary for neutral loci but also in their infection phenotype, but the latter was not investigated to date.
For the other four MHC-linked markers, the evidence for a selection signature were either weak or selection was less clearly related to plague. Indeed, an effect of geographic location, after controlling for demographic and stochastic factors, was also found for D20Rat41, Msat-Tnf and RT1N1. However, as no selection signature was obtained for these markers in the Madagascar dataset, the geographic pattern observed may not be specifically plague-mediated. Instead, it may be accounted for by the spatial variation of other selective pressures within the studied area of the plague focus, probably mediated by unidentified pathogens. The lack of a clear selective signal on these MHC markers does not however imply that there is no plague-mediated selection acting on the related MHC regions. Indeed, the main drawback of the MHC-linked microsatellite approach lies in the low reliability of negative results as an absence of selection signal may only be due to a low linkage disequilibrium between the gene under selection and the studied microsatellite. Finally, the low FST estimates found in the duplicated locus D20Rat21 for both the Madagascar and Betafo datasets probably result from balancing selection [40]. The low genetic differentiation between plague focus and plague-free zone suggests similar selective pressures in these two regions. Consequently, balancing selection acting on D20Rat21 could most likely be exerted by undetermined pathogens homogeneously distributed in Madagascar.
The intensity of plague-mediated natural selection observed in this study did not depend on the distance between microsatellite markers and identified MHC candidate genes. The best a priori candidate among the five markers investigated was Msat-Tnf, because this marker was the closest to an identified gene (230 bp from Tnf in the R. norvegicus genome) and various studies have shown that the cytokine TNF-α plays an important role in host defense against plague (see introduction). However, this marker showed no clear evidence of plague-mediated selection. By contrast, the D20Img2 locus, for which stronger evidence of plague-mediated selection was obtained with both datasets, was located further from any known MHC gene in the R. norvegicus genome. The closest MHC gene to D20Img2 was Ddr1 (epithelial Discoidin domain receptor 1, 42 kb from D20Img2), which has no known function in defense against pathogens. The Ier3 (Immediate early response 3, located about 77 kb from D20Img2) gene, which encodes a protein involved in T-cell proliferation (according to the Ensembl database) and apoptosis [54], is the most probable best candidate for pathogen-mediated natural selection in the vicinity of D20Img2.
In terms of the selection mechanisms acting on MHC-linked loci, we found no evidence of heterozygote advantage, as non duplicated loci (D20Img2 and Msat-Tnf) were at Hardy-Weinberg equilibrium. As in other studies (reviewed in [6]), it remains possible that the selection of heterozygotes is not strong enough to be detected within a single generation [3]. Spatial variation in selective pressure was observed for all MHC-linked loci except D20Rat21, through geographic effects in the Betafo dataset and the high FST for D20Img2 found within the population pairs in the Madagascar dataset. Plague-specific seroprevalence levels differed markedly within the plague focus region studied (see above), and a fortiori between the plague focus and the plague-free zone. Unfortunately, comparing allelic frequencies between seroprevalence categories within each population was not possible here because the sample size was too small given the number of alleles. However, this type of analysis would improve characterization of the spatial variability of plague-mediated selective pressure. Consequently, as in other studies reporting an effect of geographic location on the spatial genetic structure of MHC genes [11], [12], [55] or higher differentiation than for neutral loci [9], [11], our study highlights the need to characterize the variability of pathogen exposure more accurately, for evaluation of the contribution of pathogen-mediated fluctuating selection to these patterns.
Materials and Methods
Rat sampling
Field sampling was carried out systematically by joint teams comprising staff from the IRD (Institut de Recherche pour le Développement), from the IPM (Institut Pasteur de Madagascar) and from the Malagasy Ministry of Health (Communicable Disease Control Department). Each trapping campaign was validated by the national, regional and local health authorities. Rats were caught alive in wire-mesh and Sherman traps placed within houses and outdoors, according to a previously described protocol [56]. Outside houses, traps were set with the agreement of the village head, and always on the edge of cultivated fields, so as not to cause any damage in crops. Within houses, traps were set after the approval of the owner or tenant of the house. The only sampled rodents were introduced rodents (house mouse, black rat and Norway rat), which are classified as harmful and with no protected status in Madagascar. No permission was therefore required for their capture.
Animals were euthanized by cervical dislocation once trapped (as previously recommended [57]). Madagascar has no ethics committee that oversees animal experimentation, and the IRD has no ethics board to review animal experimentation protocols. However, the ANR-SEST (Agence Nationale pour la Recherche, Santé-Environnement et Santé-Travail) program on plague diffusion, which partially funded this project, has been approved by the managing director of the IRD. Additionally, regional approval was obtained from the regional head of veterinary service (Hérault, France), for the rodent sampling, euthanasia and tissue harvesting (approval no. B 34-169-1) carried out during this study. An autopsy was carried out and a piece of ear or tail was stored in 95% ethanol for genetic analyses.
The Madagascar dataset consisted of four pairs of populations, each consisting of one population from the plague focus and one from the plague-free zone, separated by a distance of 103 to 140 km (Figure 1, Table 1). These populations were sampled between 1996 and 2007, but each population pair was sampled in less time (maximum 4 years, Table 1) so that temporal variation is unlikely to drastically affect our results. The Betafo dataset consisted of 12 populations from seven villages sampled between 2006 and 2007 within an area of 150 km2 close to the town of Betafo, within the plague focus. In this area, the most recent human plague cases were reported in 2004. At least 14 rats were trapped at each study site (see Table 1 for sample sizes).
MHC-linked microsatellite loci
The MHC-linked microsatellites used in this study on R. rattus were inferred from R. norvegicus genome. Indeed, the two species only diverged 2.9My ago [58], and the gene syntheny was shown to be extremely high between both species [59]. Consequently, it appears safe to assume that locations of the studied microsatellites are the same in R. rattus as in R. norvegicus. We obtained specific primers for five MHC-linked microsatellites (Table 2). Previously published primers, designed for use in R. norvegicus, were used for the D20Img2 locus [60]. We used the R. norvegicus genome sequence to design primers for the D20Rat21 locus. Finally, the primers used for the three other loci (D20Rat41, RT1.N1 and Msat-Tnf) were designed into two steps: amplification of the R. rattus sequences with R. norvegicus primers, followed by sequencing and the design of primers more specific to R. rattus, based on the sequences obtained. The distance between each MHC-linked microsatellite and the closest known MHC gene was between 300 and 42,000 bp in R. norvegicus (Table 2).
Molecular biology methods
Genomic DNA was extracted from rat tissues with the DNeasy® Tissue Kit (Qiagen), according to the manufacturer's instructions. Two final elutions, each in 50 µl of AE buffer were carried out.
We genotyped 13 presumed neutral microsatellites as previously described [56], [61]. All these loci are polymorphic (5–20 alleles per population) dinucleotide microsatellites.
The MHC-linked microsatellites were amplified in two multiplex PCRs: one for Msat-Tnf and RT1N1 (PCR1) and the other for D20Rat41, D20Rat21 and D20Img2 (PCR2). Both PCRs were performed in a final volume of 10 µl, containing 0.2 µl of each primer, 5 µl of multiplex premix (Qiagen) and DNA (2 µl for PCR1, and 1 µl for PCR2). The two PCRs were performed according to the same program: initial denaturation (15 min at 95°C), followed by 35 cycles of denaturation (30 s at 94°C), hybridization (90 s at 60°C) and elongation (60 s at 72°C), and a final extension (30 min at 72°C). All PCR products for MHC-linked microsatellites were pooled together in a single run, and were detected with an automated ABI Prism 3130XL sequencer (Applied Biosystems). Electrophoretic gels were read with GeneMapper 3.7 (Applied Biosystems). Stuttering was relatively low in duplicated loci, and profiles were mostly clear. Each individual with an ambiguous profile was however re-amplified once by simple PCR (to avoid primer competition), and considered as a null genotype when ambiguity remains (less than 4% of the genotyped individuals).
Data analysis
For non-duplicated MHC-linked and presumed neutral loci, we checked for Hardy-Weinberg equilibrium and genotypic linkage disequilibrum between pairs of loci with Genepop v. 4 [62]. In both cases, we corrected for multiple testing by the false discovery rate (FDR) approach, with Q-value software (available from http://genomics.princeton.edu/storeylab/qvalue/); the tuning parameter λ was fixed at 0 [63]. As some loci exhibited heterozygote deficiencies, we used Micro-checker 2.2.3 [64] to evaluate whether it could be explained by the occurrence of null alleles. We then used the software Freena (INRA Montpellier website, available www.montpellier.inra.fr/URLB, accessed 2012 February 7) [42] to verify that mean null allele frequencies were low at these loci, as high null alleles frequencies can impact genetic differentiation estimations [42].
Under the hypothesis of overdominance (or heterozygote advantage, see introduction), an excess of heterozygotes would be expected at MHC loci. We thus tested for Hardy-Weinberg disequilibrium (H1: heterozygote excess) within each studied population for the two non duplicated MHC-linked loci using Genepop program. The FDR approach was also used to correct for multiple testing.
For the “Madagascar” dataset, we expected the level of genetic differentiation to be higher for MHC-linked microsatellites than for neutral loci within population pairs. We evaluated the deviation from neutrality for all non-duplicated MHC-linked and presumed neutral microsatellites, using two approaches for the detection of selection, based on the FST values for each of the four population pairs. We first used the method of Beaumont & Nichols [43], as implemented in Fdist2 (available at http://www.rubic.rdg.ac.uk/~mab/software.html). This approach uses an island model and simulates the distribution of FST conditioned on heterozygosity, under the null hypothesis of drift and migration only. We carried out 50,000 simulations of two demes, assuming a stepwise mutation model. We then used the Vitalis et al. [44] method, implemented in DetSel software [45]. This method performs coalescent simulations under a divergence model in which an ancestral population splits into two isolated populations. The simulations were performed with a stepwise mutation model and a mean mutation rate μ = 0.0005. We used several different values for ancestral population size (N e = 1,000, 10,000, 100,000), and the time since divergence was set at 1,200 generations (because R. rattus probably colonized the central highlands about 700 years ago [65]). For each pairwise analysis, we performed a total of 1,000,000 simulations, with each set of nuisance parameter values representing a third of the total simulations. We applied a minimum allele frequency of 0.01 to the observed and simulated data.
We then used a method similar to the one described in Neff & Fraser [66] to empirically compare the pairwise FST value for each MHC-linked (duplicated and non-duplicated) locus with the global F ST for the neutral loci within the four population pairs. For each of the five MHC-linked markers, as well as for the global neutral dataset (13 loci), 1000 datasets were obtained by bootstrapping over individuals. For all the datasets, pairwise F ST were computed within each population pair from allelic frequencies (see also [12], [55]) using the package Arlecore from Arlequin v 3.5 software [67]. Note that allelic frequencies cannot be directly estimated for the duplicated microsatellites (data available only consist in allele presence/absence, genotypes may be unknown). Allelic frequencies were thus inferred from the number of individuals carrying a given allele divided by the total number of alleles observed in a population (see also [12], [55]). The proportion of bootstrap replicates in which the F ST of MHC-linked markers were higher or lower than neutral F ST was used as p-value for the null hypothesis that one of MHC-linked markers had higher or lower F ST than the neutral markers (see also [66]).
For the “Betafo” dataset, we expected the level of genetic differentiation to be lower for MHC-linked microsatellites than for presumed neutral loci, provided that the selection pressure exerted by plague was uniform over the whole area [40]. We tested this hypothesis for non duplicated microsatellites, with Fdist2 (with the same parameters as for the analysis of the Madagascar dataset, see above). We did not use DetSel on the “Betafo” dataset: it does not conform to the model of a single ancestral population diverging into two isolated populations with no migration between them, as it corresponds to a small scale metapopulation sampling design.
A significant isolation by distance (IBD) pattern was found for presumed neutral microsatellites within the “Betafo” dataset (as already reported [52]). For each MHC-linked locus, we performed partial Mantel tests [68], which evaluated the correlative link between the pairwise FST at the MHC locus and geographic distance while keeping constant differentiation at neutral microsatellites (excluding those identified by FDist2 as corresponding to selection signature in the “Betafo” dataset). Under the hypothesis of strong selection pressures acting on MHC loci, we would expect a lower isolation by distance pattern than under neutrality for these loci. Pairwise FST values were estimated for each locus with Genepop v. 4 for non duplicated loci, or from allelic frequencies for duplicated loci. Partial Mantel tests were performed with Fstat v. 2.9.3.2 [69], with 20,000 permutations.
Supporting Information
Selection signatures detected with Beaumont & Nichols's approach (Fdist2 program [43] ) applied to the Madagascar dataset and the 15 loci: Msat-Tnf, D20Img2 and the 13 apparently neutral microsatellites. The analysis was performed for the four population pairs, each consisting of one population from the plague focus and one population form the plague-free zone. Genetic differentiation between the two populations (FST) is plotted against heterozygosity. Median and 95% confidence interval are indicated with solid lines, whereas the 99% confidence interval is indicated with a dotted line.
(PDF)
Results of the test of natural selection for Msat-Tnf, carried out with the method of Vitalis et al. (DetSel program [44], [45]) for the four population pairs studied for the Madagascar dataset. Genetic differentiation for the first population (F1) is plotted against genetic differentiation for the second population (F2).
(PDF)
Results of the test of natural selection for D20Img2, carried out with the method of Vitalis et al. (DetSel program [44], [45]) for the four population pairs studied for the Madagascar dataset. Genetic differentiation for the first population (F1) is plotted against genetic differentiation for the second population (F2).
(PDF)
Null allele frequencies estimates obtained using FreeNA software [42] for each of the 15 non duplicated microsatellite loci in the 20 populations.
(PDF)
Acknowledgments
We are grateful to the staff of the Plague Laboratory of the Institut Pasteur de Madagascar for their excellent assistance during the fieldwork, to Sylvain Piry for creating the datasets and formatting the results of the boostrap analysis and to Renaud Vitalis for assistance with statistical analyses carried out with DetSel. We warmly thank Nathalie Charbonnel as well as anonymous reviewers for helpful comments that significantly improved the manuscript. The data used in this work were generated through use of the molecular genetic analysis technical facilities of IFR119 “Montpellier Environnement Biodiversité”.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the Institut de Recherche pour le Développement, the Institut Pasteur de Madagascar and an Agence Nationale pour la Recherche, Santé-Environnement et Santé-Travail program on plague diffusion. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jiggins FM, Kim KW. A screen for immunity genes evolving under positive selection in Drosophila. Journal of Evolutionary Biology. 2007;20:965–970. doi: 10.1111/j.1420-9101.2007.01305.x. [DOI] [PubMed] [Google Scholar]
- 2.Vallender EJ, Lahn BT. Positive selection on the human genome. Human Molecular Genetics. 2004;13:R245–R254. doi: 10.1093/hmg/ddh253. [DOI] [PubMed] [Google Scholar]
- 3.Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years?. Journal of Evolutionary Biology. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 4.Hurt P, Walter L, Sudbrak R, Klages S, Muller I, et al. The genomic sequence and comparative analysis of the rat major histocompatibility complex. Genome Research. 2004;14:631–639. doi: 10.1101/gr.1987704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nature Reviews Immunology. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piertney SB, Oliver MK. The evolutionary ecology of the Major Histocompatibility Complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- 7.Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proceedings of the Royal Society of London B. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teacher AGF, Garner TWJ, Nichols RA. Evidence for directional selection at a novel major Histocompatibilty Class I marker in wild common frogs (Rana temporaria) exposed to a viral pathogen (Ranavirus). PLoS One. 2009;4(2):e4616. doi: 10.1371/journal.pone.0004616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilar A, Garza A. A comparison of variability and population structure for major histocompatibility complex and microsatellite loci in California coastal steelhead (Oncorhynchus mykiss Walbaum). Molecular Ecology. 2006;15:923–937. doi: 10.1111/j.1365-294X.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- 10.Evans ML, Neff BD, Heath DD. MHC genetic structure and divergence across populations of Chinook salmon (Oncorhynchus tshawytscha). Heredity. 2010;104:449–459. doi: 10.1038/hdy.2009.121. [DOI] [PubMed] [Google Scholar]
- 11.Bryja J, Charbonnel N, Berthier K, Galan M, Cosson JF. Density-related changes in selection pattern for major histocompatibility complex genes in fluctuating populations of voles. Molecular Ecology. 2007;16:5084–5097. doi: 10.1111/j.1365-294X.2007.03584.x. [DOI] [PubMed] [Google Scholar]
- 12.Loiseau C, Richard M, Garnier S, Chastel O, Julliard R, et al. Diversifying selection on MHC class I in the house sparrow (Passer domesticus). Molecular Ecology. 2009;18:1331–1340. doi: 10.1111/j.1365-294X.2009.04105.x. [DOI] [PubMed] [Google Scholar]
- 13.Dionne M, Miller KM, Dodson JJ, Caron F, Bernatchez L. Clinal variation in MHC diversity with temperature: Evidence for the role of host-pathogen interaction on local adaptation in Atlantic salmon. Evolution. 2007;61:2154–2164. doi: 10.1111/j.1558-5646.2007.00178.x. [DOI] [PubMed] [Google Scholar]
- 14.Babik W, Taberlet P, Jan Ejsmond M, Radwan J. New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Molecular Ecology Research. 2009;9:713–719. doi: 10.1111/j.1755-0998.2009.02622.x. [DOI] [PubMed] [Google Scholar]
- 15.Galan M, Guivier E, Caraux G, Charbonnel N, Cosson J-F. A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genomics. 2010;11:296. doi: 10.1186/1471-2164-11-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryja J, Galan M, Charbonnel N, Cosson J-F. Analysis of major histocompatibility complex class II gene in water voles using capillary electrophoresis-single stranded conformation polymorphism. Molecular Ecology Notes. 2005;5:173–176. [Google Scholar]
- 17.Santucci F, Ibrahim KM, Bruzzone A, Hewit GM. Selection on MHC-linked microsatellite loci in sheep populations. Heredity. 2007;99:340–348. doi: 10.1038/sj.hdy.6801006. [DOI] [PubMed] [Google Scholar]
- 18.Charbonnel N, Pemberton J. A long-term genetic survey of an ungulate population reveals balancing selection acting on MHC through spatial and temporal fluctuations in selection. Heredity. 2005;95:377–388. doi: 10.1038/sj.hdy.6800735. [DOI] [PubMed] [Google Scholar]
- 19.Hansen MM, Skaala O, Jensen LF, Bekkevold D, Mensberg KLD. Gene flow, effective population size and selection at major histocompatibility complex genes: brown trout in the Hardanger Fjord, Norway. Molecular Ecology. 2007;16:1413–1425. doi: 10.1111/j.1365-294X.2007.03255.x. [DOI] [PubMed] [Google Scholar]
- 20.Jensen LF, Hansen MM, Mensberg KL, Loeschcke V. Spatially and temporally fluctuating selection at non-MHC immune genes: evidence from TAP polymorphism in populations of brown trout (Salmo trutta, L.). Heredity. 2008;100:79–91. doi: 10.1038/sj.hdy.6801067. [DOI] [PubMed] [Google Scholar]
- 21.Oetjen K, Ferber S, Dankert I, Reusch TBH. New evidence for habitat-specific selection in Wadden Sea Zostera marina populations revealed by genome scanning using SNP and microsatellite markers. Marine Biology. 2010;157:81–89. [Google Scholar]
- 22.Doxiadis GGM, de Groot N, Claas FHJ, Doxiadis IIN, van Rood JJ, et al. A highly divergent microsatellite facilitating fast and accurate DRB haplotyping in humans and rhesus macaques. Proceedings National Academy of Science USA. 2007;104:8907–8912. doi: 10.1073/pnas.0702964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luikart G, Pilgrim K, Visty J, Ezenwa VO, Schwartz MK. Candidate gene microsatellite variation is associated with parasitism in wild bighorn sheep. Biology Letter. 2008;4:228–231. doi: 10.1098/rsbl.2007.0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis DT, Gage KL, Gratz N, Poland J, Tikhomirov E. 1999. Plague manual: epidemiology, distribution, surveillance and control: Geneva.
- 25.Stenseth NC, Atshabar BB, Begon M, Belmain SR, Bertherat E, et al. Plague: past, present and future. PLoS Medicine. 2008;5(1):e3. doi: 10.1371/journal.pmed.0050003. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas RE, Barnes AM, Quan TJ, Beard ML, Carter LG, et al. Susceptibility to Yersinia pestis in the northern grasshopper mouse (Onychomys leucogaster). Journal of Wildlife Disease. 1988;24:327–333. doi: 10.7589/0090-3558-24.2.327. [DOI] [PubMed] [Google Scholar]
- 27.Shepherd AJ, Leman PA, Hummitzsch DE. Experimental plague infection in South-African wild rodents. Journal of Hygiene. 1986;96:171–183. doi: 10.1017/s0022172400065943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brygoo ER. Epidémiologie de la peste à Madagascar. Archives de l'Institut Pasteur de Madagascar. 1966;35:9–147. [Google Scholar]
- 29.Migliani R, Chanteau S, Rahalison L, Ratsitorahina M, Boutin JP, et al. Epidemiological trends for human plague in Madagascar during the second half of the 20th century: a survey of 20 900 notified cases. Tropical Medicine and International Health. 2006;11:1228–1237. doi: 10.1111/j.1365-3156.2006.01677.x. [DOI] [PubMed] [Google Scholar]
- 30.Rahalison L, Ranjalahy M, Duplantier J-M, Duchemin J-B, Ravelosaona J, et al. Susceptibility to plague of the rodents in Antananarivo, Madagascar The genus Yersinia. 2003. Kluwer Academic/Plenum Publishers: New York. [DOI] [PubMed]
- 31.Tollenaere C, Rahalison L, Ranjalahy M, Duplantier JM, Rahelinirina S, et al. Susceptibility to Yersinia pestis infection in wild Rattus rattus, reservoir of plague in Madagascar. EcoHealth. 2010;7:242–247. doi: 10.1007/s10393-010-0312-3. [DOI] [PubMed] [Google Scholar]
- 32.Turner JK, McAllister MM, Xu JL, Tapping RI. The resistance of BALB/cJ mice to yersinia pestis maps to the Major Histocompatibility Complex of chromosome 17. Infection and Immunity. 2008;76:4092–4099. doi: 10.1128/IAI.00488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shim HK, Musson JA, Harper HM, McNeill HV, Walker N, et al. Mechanisms of major histocompatibility complex class II-restricted processing and presentation of the V antigen of Yersinia pestis. Immunology. 2006;119:385–392. doi: 10.1111/j.1365-2567.2006.02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pujol C, Bliska JB. Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clinical Immunology. 2005;114:216–226. doi: 10.1016/j.clim.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Saikh KU, Kissner TL, Dyas B, Tropea JE, Waugh DS, et al. Human cytolytic T cell recognition of Yersinia pestis virulence proteins that target innate immune responses. Journal of Infectious Diseases. 2006;194:1753–1760. doi: 10.1086/509507. [DOI] [PubMed] [Google Scholar]
- 36.Nakajima R, Brubaker RR. Association between virulence of Yersinia pestis and suppression of gamma interferon and tumor necrosis factor. Infection and Immunity. 1993;61:23–31. doi: 10.1128/iai.61.1.23-31.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parent MA, Wilhelm LB, Kummer LW, Szaba FM, Mullarky IK, et al. Gamma interferon, tumor necrosis factor alpha, and nitric oxide synthase 2, key elements of cellular immunity, perform critical protective functions during humoral defense against lethal pulmonary Yersinia pestis infection. Infection and Immunity. 2006;74:3381–3386. doi: 10.1128/IAI.00185-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert ND, Nilles ML, Bradley DS. Characterization of immune responses to Yersinia pestis infection: resistant versus susceptible mice. Journal of Immunology. 2009;182:129.29. [Google Scholar]
- 39.World Health Organization. Human plague: review of regional morbidity and mortality, 2004–2009. Weekly Epidemiological Record. 2010;85:40–45. [PubMed] [Google Scholar]
- 40.Schierup MH, Vekemans X, Charlesworth D. The effect of subdivision on variation at multi-allelic loci under balancing selection. Genetical Research. 2000;76:51–62. doi: 10.1017/s0016672300004535. [DOI] [PubMed] [Google Scholar]
- 41.Lambracht-Washington D, Moore YF, Wonigeit K, Fisher Lindahl K. Structure and expression of MHC class Ib genes of the central M region in rat and mouse: M4, M5 and M6. Immunogenetics. 2008;60:131–145. doi: 10.1007/s00251-008-0282-6. [DOI] [PubMed] [Google Scholar]
- 42.Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution. 2007;24(3):621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- 43.Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London B. 1996;263:1619–1626. [Google Scholar]
- 44.Vitalis R, Dawson K, Boursot P. Interpretation of variation across marker loci as evidence of selection. Genetics. 2001;158:1811–1823. doi: 10.1093/genetics/158.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vitalis R. Pompanon F, Bonin A, editors. DetSel: a R-package to detect marker loci responding to selection. Population Genomics: Methods and Protocols, Molecular Biology series. 2011. Humana Press, USA. [DOI] [PubMed]
- 46.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nature Review Genetics. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 47.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genetics. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 48.Guryev V, Smits BMG, van de Belt J, Verheul M, Hubner N, et al. Haplotype block structure is conserved across mammals. PLoS Genetics. 2006;2:1111–1118. doi: 10.1371/journal.pgen.0020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cizkova D, de Bellocq J, Baird SJE, Pialek J, Bryja J. Genetic structure and contrasting selection pattern at two major histocompatibility complex genes in wild house mouse populations. Heredity. 2011;106(5):727–740. doi: 10.1038/hdy.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babik W, Pabijan M, Radwan J. Contrasting patterns of variation in MHC loci in the Alpine newt. Molecular Ecology. 2008;17(10):2339–2355. doi: 10.1111/j.1365-294X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 51.Duplantier JM, Duchemin JB. Human diseases and introduced small mammals. 2003. pp. 158–161. In: The natural history of Madagascar, (Goodman SM & Benstead JP, eds.) University of Chicago Press, Chicago.
- 52.Rahelinirina S. Le risque pesteux dans les foyers ruraux du Moyen-Ouest malgache: déplacements et structuration des populations de rats noirs de l'échelle de l'habitat à celle du paysage. 2009. PhD Thesis, Antananarivo, Antananarivo.
- 53.Vogler AJ, Chan F, Wagner DM, Roumagnac P, Lee J, et al. Phylogeography and molecular epidemiology of Yersinia pestis in Madagascar. Plos Neglected Tropical Diseases, 2011;5(9):e1319. doi: 10.1371/journal.pntd.0001319. doi: 10.1371/journal.pntd.0001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ao L, Liu JY, Gao LH, Liu SX, Yang MS, et al. Differential expression of genes associated with cell proliferation and apoptosis induced by okadaic acid during the transformation process of BALB/c 3T3 cells. Toxicology in Vitro. 2008;22:116–127. doi: 10.1016/j.tiv.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 55.Ekblom R, Saether SA, Jacobsson P, Fiske P, Sahlman T, et al. Spatial pattern of MHC class II variation in the great snipe (Gallinago media). Molecular Ecology. 2007;16:1439–1451. doi: 10.1111/j.1365-294X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- 56.Gilabert A, Loiseau A, Duplantier JM, Rahelinirina S, Rahalison L, et al. Genetic structure of black rat populations in a rural plague focus in Madagascar. Canadian Journal of Zoology. 2007;85:965–972. [Google Scholar]
- 57.Mills JN, Yates TL, Childs JE, Parmenter RR, Ksiazek TG, et al. Guidelines for working with rodents potentially infected with hantavirus. Journal of Mammalogy. 1995;76:716–722. [Google Scholar]
- 58.Robins JH, McLenachan PA, Philips MJ, Craig L, Ross HA, et al. Dating of divergences within the Rattus genus phylogeny using whole mitochondrial genomes. Molecular Phylogenetics and Evolution. 2008;49(2):460–466. doi: 10.1016/j.ympev.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Cavagna P, Stone G, Stanyon R. Black rat (Rattus rattus) genomic variability characterized by chromosome painting. Mammalian Genome. 2002;13(3):157–163. doi: 10.1007/BF02684021. [DOI] [PubMed] [Google Scholar]
- 60.Ioannidu S, Walter L, Dressel R, Gunther E. Physical map and expression profile of genes of the telomeric class I gene region of the rat MHC. Journal of Immunology. 2001;166:3957–3965. doi: 10.4049/jimmunol.166.6.3957. [DOI] [PubMed] [Google Scholar]
- 61.Loiseau A, Rahelinirina S, Rahalison L, Konecny A, Duplantier JM, et al. Isolation and characterization of microsatellites in Rattus rattus. Molecular Ecology Resources. 2008;8:916–918. doi: 10.1111/j.1755-0998.2008.02115.x. [DOI] [PubMed] [Google Scholar]
- 62.Rousset F. GENEPOP ′ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Ressources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 63.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 64.Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- 65.Tollenaere C, Brouat C, Duplantier JM, Rahalison L, Rahelinirina S, et al. Phylogeography of the introduced species Rattus rattus in the western Indian Ocean, with special emphasis on the colonization history of Madagascar. Journal of Biogeography. 2010;37:398–410. [Google Scholar]
- 66.Neff BD, Fraser BA. A program to compare genetic differentiation statistics across loci using resampling of individuals and loci. Molecular Ecology Resources. 2010;10:546–550. doi: 10.1111/j.1755-0998.2009.02786.x. [DOI] [PubMed] [Google Scholar]
- 67.Excoffier L, Lischer HLE. Arlequin suite ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 68.Smouse PE, Long JC, Sokal RR. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Systematics and Zoology. 1986;35:627–632. [Google Scholar]
- 69.Goudet J. FSTAT: A Program to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9.3). 2001. Population Genetics Laboratory Lausanne. Available: http://www2.unil.ch/popgen/softwares/fstat.htm. Accessed 2012 February 7.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selection signatures detected with Beaumont & Nichols's approach (Fdist2 program [43] ) applied to the Madagascar dataset and the 15 loci: Msat-Tnf, D20Img2 and the 13 apparently neutral microsatellites. The analysis was performed for the four population pairs, each consisting of one population from the plague focus and one population form the plague-free zone. Genetic differentiation between the two populations (FST) is plotted against heterozygosity. Median and 95% confidence interval are indicated with solid lines, whereas the 99% confidence interval is indicated with a dotted line.
(PDF)
Results of the test of natural selection for Msat-Tnf, carried out with the method of Vitalis et al. (DetSel program [44], [45]) for the four population pairs studied for the Madagascar dataset. Genetic differentiation for the first population (F1) is plotted against genetic differentiation for the second population (F2).
(PDF)
Results of the test of natural selection for D20Img2, carried out with the method of Vitalis et al. (DetSel program [44], [45]) for the four population pairs studied for the Madagascar dataset. Genetic differentiation for the first population (F1) is plotted against genetic differentiation for the second population (F2).
(PDF)
Null allele frequencies estimates obtained using FreeNA software [42] for each of the 15 non duplicated microsatellite loci in the 20 populations.
(PDF)